Abstract
The study of germ cell-specific gene regulation in vitro is challenging. Here we report that the promoter of the oocyte-specific gene, Gdf9, is active in a population of cultured murine embryonic stem cells (ES) which have a phenotype resembling ooocytes. The promoter region of the murine Gdf9 coupled to enhanced green fluorescent protein (eGFP) was stably transfected into XX mouse ES cells. eGFP was expressed only in oocytes of chimeric mice generated from the transfected XX ES cells. The transfected ES cells were examined when cultured on feeder layers or as embryoid bodies. Large eGFP-positive cells, surrounded by a structure resembling a zona pellucida appeared transiently in cultures of the ES cells on feeder layers. Surprisingly, they were detectable on days 1 and 2 of culture but virtually absent on day 3. Addition of leukemia inhibitory factor (LIF) to the media significantly increased the number of eGFP-positive cels resembling oocytes. Quantitative real-time PCR demonstrated a parallel increase in Gdf9 and Zp3 mRNA with changes in the abundance of eGFP-positive cells. In embryoid body cultures, eGFP-positive cells appeared transiently and then re-appeared in regional clusters after 30−45 days of culture. These findings demonstrate that a population of cultured murine ES cells contain the transcriptional machinery to drive expression of an oocyte-specific gene, and that those cells phenotypically resemble oocytes.
Keywords: Embryonic Stem cells, Gdf9, oocyte, leukemia inhibitory factor
1. Introduction
Embryonic stem (ES) cells are derived from the inner cell mass of blastocyst stage embryos and possess both pluripotency and the capacity for self-renewal. When ES cells are cultured in vitro, they have the ability to differentiate into a variety of cell types dependent upon the culture conditions employed [1, 2]. Primordial germ cells (PGCs) give rise to the mammalian gametes, oocytes and sperm, and are separated from somatic cell lineages early on in embryonic development. This process, called specification is one of the most crucial steps in mammalian development [3]. However, the precise mechanisms by which specification occurs remains unclear, especially in humans, where research limitations make similar studies done in animals difficult. While the ability of ES cells to give rise to germ cells in vitro was once thought to be implausible, recently several studies have demonstrated the ability of ES cells to give rise not only to PGCs, but also to oocyte and sperm-like cells in vitro [4-10]. In vivo, several growth factors are responsible for the differentiation of PGCs into oogonia after the arrival at the genital ridge. These include kit ligand (KL), bone morphogenic proteins (BMPs), stem cell factor (SCF), fibroblast growth factor (βFGF) and members of the transforming growth factor β (TGFβ) superfamily [11]. Several studies have established conditions by which ES cells generate germ cells in vitro through the use of PGC and germ cell enrichment techniques [4-10]. These in vitro systems of germ cell generation have helped to establish the mechanisms by which germ cells are specified. Hübner et al previously reported that mouse embryonic stem cells could develop into mature oocytes as well as form follicle-like structures when allowed to differentiate in culture [8, 12]. A modified Oct 4/GFP reporter construct containing the germ cell-specific enhancer was utilized to track ES cell differentiation into a germ cell lineage. This suggested that ES cells contain the transcriptional machinery to drive expression of germ cell genes, which raises the possibility of using cultured ES cells to explore function of germ cell-specific gene regulation.
Here we report the that the promoter of the oocyte-specific gene, growth differentiation factor-9 (Gdf9), drives expression of enhanced green fluorescent protein (eGFP) in murine XX ES cells that have phenotypic features of oocyte-like cells. This opens up the possibility of in vitro analysis of promoter function of germ cell-specific genes.
2. Methods
The following were kindly provided: JM21 XX ES cells by Dr. John MacLaughlin of the University of Pennsylvania School of Veterinary Medicine (New Bolton, PA); PGK 12.1 XX ES cells by Neil Brockdorff of MRC Clinical Sciences Centre (London, UK); and Gdf9/eGFP pE42 plasmid by Dr. Martin Matzuk of Baylor College of Medicine (Houston, TX); Plasmid 375 (Sf1 promoter and gene fragments) by Dr. Keith Parker of University of Texas Southwestern (Dallas, TX). The following were purchased: Proteinase K, DNA Rapid Ligation kit and FUGENE® from Roche Diagnostics (Indianapolis, IN); ESGRO® LIF from Chemicon (Temecula, CA); ES cell grade fetal bovine serum (FBS) from HyClone (Logan, UT); β-mercaptoethanol, DMSO and Hoechst 33342 stain from Sigma-Aldrich (St. Louis, MO); PuReTaq® Ready-to-Go PCR beads, rainbow protein marker and Hybond C+ nitrocellulose from Amersham Biosciences (Piscataway, NJ); Restriction enzymes, MMLV reverse transcriptase, DNAse, dNTPs and random primers from Promega (Madison, WI); All QPCR materials, GAPDH VIC probe and primers, SYBR® green and Taqman® from Applied Biosystems (Foster City, CA); Activin and TGFβ growth factors from Calbiochem (La Jolla, CA); pDsRED Express-1 vector from BD Biosciences (Mountain View, CA); Recombinant brain-derived neurotropic factor (BDNF), G418 antibiotic, Trizol® and all culture media and supplements from Invitrogen Life Technologies (Carlsbad, CA).
Sf1/DsRED plasmid construction
A 674 base pair fragment of the mouse Sf1 promoter from −585 to +84 was isolated from the 375 plasmid [13] by PCR using the following primers: Forward: 5’-AATTAAGCTTCACCCTTAGCCCAGCAGTCCTGGCAC–3’, Reverse: 5’- AATTGAATTCAGGTAGGGCAGTGGCTAGCGGGCTCT–3’, digested with EcoRI and Hind III and cloned into pDsRED Express-1. Positive clones were obtained through restriction digestion screening followed by sequencing analysis. In vitro expression of Sf1/DsRED was confirmed by transfecting MA10 steroidogenic cells with the Sf1/DsRED purified plasmid. Cells were analyzed for the expression of the plasmid using a Nikon TS100 Eclipse fluorescent microscope. DsRED fluorescence was seen only in MA10 cells transfected with Sf1/DsRED and not in cells transfected with DsRED alone (data not shown). Purified Gdf9/eGFP [14] and Sf1/DsRED plasmid DNA for use in ES cells was obtained through cesium chloride purification.
Embryonic stem cell expansion
XX ES cells were cultured on γ-irradiated mouse embryonic fibroblasts (MEF) [15] on 0.1% gelatin coated tissue culture plates in Dulbecco's modified Eagle's medium (DMEM) supplemented with 15% FBS, 1% glutamine, 1% non-essential amino acids, 1 mM β-mercaptoethanol, and LIF (1000 IU/ml). Plates were incubated at 37°C with 5% CO2. Cells were grown in 6 well dishes for 2−3 days and the media was changed every day. On day 2 or 3, cells were washed with phosphate buffered saline (PBS), trypsinized with 0.125% trypsin-EDTA (2.5% trypsin in Versene (Invitrogen)) and split onto two 60 mm dishes 2 hours after refreshing the media. Cells were split again 2−3 days later onto two 10 cm dishes and allowed to grow in the presence of LIF for 3 days. Cells were frozen (6 vials/plate) in 0.5 ml freezing media (90% FBS, 10% DMSO) 2 hours after refreshing the media for use in future experiments.
Embryonic stem cell electroporation
Fifty μg of Gdf9/eGFP and Sf1/DsRED DNA were linearized with either EcoRV or Hind III respectively, purified by phenol-chloroform extraction and ethanol precipitation and resuspended in sterile HEPES buffered saline (HBS). ES cells were split 1:6 as usual 2 days prior to electroporation and cells were fed everyday. In addition, media was changed 2 hours prior to electroporation. DNA in a 3:1 molar ratio of Gdf9/eGFP: Sf1/DsRED was delivered into cells via Gene Pulser (BioRad, Hercules, CA). Positive clones were selected for using G418 for 8 days. On day 8, 100 colonies were isolated, seeded in duplicate in 24 well plates and allowed to grow for 2−4 days. One well of each duplicate was processed for DNA isolation, the other for cell freezing. DNA was isolated by lysing cells in a DNA lysis solution (150mM NaCl, 20mM Tris (pH 7.5), 5 mM EDTA, 0.5% SDS, proteinase K (0.25 mg/ml)) followed by isopropanol /ethanol precipitation. Positive clones were identified using PCR with the following primers: Sf1/DsRED Forward: 5’-TGAAGAAGTTTCTGAGAGCCCG–3’, Sf1/DsRED Reverse: 5’-ACAGGATGTCCCAGGCGAAGGG–3’, Gdf9/eGFP Forward: 5’–TAACCACCTGGACGTGGGAGCT–3’, Gdf9/eGFP Reverse: 5’ –ACGCCGTAGGTCAGGGTGGTCA–3’. Specific positive clones were chosen randomly for expansion and manipulation.
ES cell differentiation/Visualization
ES cells were thawed and plated on 0.1% gelatin coated 60 mm plates on a MEF monolayer. These cells were allowed to grow for 2−4 days until approximately 80% confluent and in large defined colonies. For embryoid body formation, the cells were transferred to a 10 cm bacterial culture dish and incubated for 5 days to allow for formation of the embryoid bodies. Media was changed on the fifth day and the embryoid bodies were split 1:2 back onto tissue culture dishes without MEFs in either the presence or absence of LIF, EBs were analyzed the next day (d1) for oocyte formation. Media was changed every 5 days. For ES cell differentiation studies, the cells were split 1:4 on either: MEF monolayer in LIF media; no MEF monolayer, LIF media; or no MEF monolayer, no LIF in media. Cells were then visualized starting on d1 of differentiation (24 h after split) using a Nikon TS100 Eclipse fluorescent microscope with a FITC and TrITC filter. Green cells were counted in a 7-cm2 region of the dish. Cells were either allowed to differentiate further, changing the media every 4−5 days or were collected at various timepoints after the 1:4 split for RNA or protein isolation. Supernatant fractions were obtained by collecting the media after loosening any unattached or poorly attached cells by manual agitation, centrifuging for 1 min at 5000 rpm and resuspending the pellet in either 1 ml of Trizol® (RNA) or in 200 μl 4X SDS sample buffer (0.3125 M Tris pH 6.8, 10% SDS, 50% glycerol, 0.005% bromophenol blue, 25% β-mercaptoethanol) (protein). Plate fractions were obtained by scraping cells into 1 ml Trizol® or 4X SDS sample buffer added directly to the dish. Hoescht staining was done by adding Hoescht 33342 stain (10 μM) directly to the dish and incubating for 15 min at 37°C. Cells were visualized on a Nikon Eclipse TE300 Inverted fluorescent microscope and images captured and analyzed using MetaMorph® Software (Molecular Devices, Downingtown, PA).
Blastocyst injection/In vivo analysis
A Gdf9/eGFP transfected ES cell clone were seeded onto two 0.1% gelatin coated 60 mm plates on a MEF feeder layer in the presence of LIF and allowed to grow overnight. Cells were trypsinized 2 hours after a media change and plated back onto a 60 mm dish for 1 hour to remove the MEF feeders. The cells were washed extensively and resuspended in 1 ml of ES cell media for use in the blastocyst injection. Blastocyst injections were performed by the University of Pennsylvania's transgenic core. 26 embryos were injected and transplanted into 2 host C57BL/6 females resulting in 2 founder females with approximately 40% chimerism. These females were mated with C57BL/6 black males 3 different times producing 3 litters with no chimerics. The founder females were then sacrificed, their ovaries harvested and the oocyte-cumulus cell complexes isolated using a 30g needle to puncture the follicles. Oocyte-cumulus cell complexes were then visualized using a Nikon T5100 Eclipse fluorescent microscope with a FITC filter.
Quantitative Polymerase Chain Reaction (QPCR)
RNA from both supernatant and plate fractions was isolated and resuspended in 20 μl sterile H2O. 5 μg of RNA was reverse transcribed after DNAse treatment and the DNA was diluted 1:10 for use in the QPCR analysis. QPCR was done in a 20 μl final volume in 386 well plates in a 7600T Sequence Detection System (Applied Biosystems). Results were analyzed using SDS 2.2 software (Applied Biosystems) and statistical analysis was done using Graph Pad Prism (Graph Pad Software, Inc, San Diego, CA). The following primers were used for the amplification: Gdf9 Forward: 5’ – ATCGAGTGCAGTGTCCGTAGGT–3’, Gdf9 Reverse: 5’–TTCACTTGGTTTATGGCAACGA – 3’, Zp3 Forward: 5’ –CGGCCAGAGACTCTCCAGTT-3’, Zp3 Reverse: 5’ –ATGTAGAGCGTATTTCTGGAGCTGTT–3’, Gapdh VIC probe primers were obtained by Applied Biosystems.
Immunoblot Analysis
Samples collected in 4X sample buffer were boiled 5 min and 30 ml of each sample was analyzed on a 4−15% Tris-HCl gradient denaturing gel. Protein was transferred to Hybond C+ Nitrocellulose and incubated with ZP3 IE-10 Antibody [16] (1:50) which was derived from a hybridoma cell line (ATCC, Manassas, VA) and purified IgG was obtained by the University of Pennsylvania Cell Center cell culture hybridoma core. The blot was then incubated with anti-mouse HRP IgG (1:500) (Cell Signaling Technologies, Danvers, MA) and visualized using enhanced chemiluminescence (Pierce Chemical, Boston, MA).
3. Results
Gdf9/eGFP is expressed in oocytes of chimeric mice generated from stably transfected murine XX ES cells
Gdf9 is an oocyte-specific gene that is expressed in ovarian follicles beginning at the primary follicle stage, when the oocyte is surrounded by a single layer of cuboidal granulosa cells [17]. To establish the ability of this oocyte-specific gene promoter to drive expression of eGFP specifically in oocytes of ES cell-derived ovarian follicles, we transfected a plasmid [14] , in which eGFP is expressed under control of a 3.1 kilobase (kb) region of the Gdf9 promoter, into JM21 XX ES cells. This region of the Gdf9 promoter has been shown to limit its expression to the oocyte of primary and larger size follicles in transgenic mice [14]. The plasmid was transfected into the ES cells together with the Sf1/DsRED plasmid, engineered by restriction digest to contain an inactivate DsRED gene, for use solely as a neomycin selection plasmid. Positive clones were selected for using G418 and screened by PCR using primers specific to the Gdf9/eGFP transgene.
We generated transgenic mice through injection of a JM21 XX ES cell clone containing the Gdf9/eGFP transgene into blastocysts. The injected blastocysts were transferred into host females and generated two founder females with approximately 40% chimerism. While these mice did not produce agouti offspring after 3 matings, the ovaries of the founder female mice were evaluated and were found to exhibit eGFP expression specifically in the oocytes, demonstrating the ability of these transfected XX ES cells to contribute to the germline (Figure 1). eGFP expression was not seen in other organs such as brain, adrenals, liver or kidneys and was therefore specific to the oocytes of the developing gonad (data not shown).
Figure 1. In vivo expression of Gdf9/eGFP transgene.
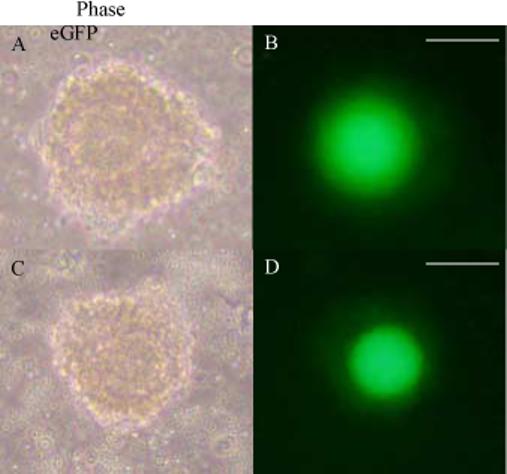
Oocyte-cumulus cell complexes expressed from the ovaries of a founder female generated by blastocyst injection of JM21 XX ES cells transfected with the Gdf9/eGFP transgene. Phase images (A, C) or eGFP fluorescent images (B, D) obtained using a Nikon TS100 Eclipse microscope at 20X magnification. Two different oocyte-cumulus cell complexes are represented. Scale bar = 75μm.
The Gdf9 promoter is active in a population of cultured stably transfected ES cells
When ES cells containing the Gdf9/eGFP that gave rise to appropriate oocyte eGFP expression in chimeric mice were allowed to differentiate on a MEF monolayer through the removal of LIF from the culture media [20], eGFP-positive cells were present by day 1 (d1). These eGFP-positive structures were large and mostly found floating in the culture medium (supernatant).
The presence of DNA in the eGFP-positive cells was confirmed by Hoechst 33342 staining (Figure 2). Upon further study, we found that many of the eGFP-positive cells contained structures that resembled polar bodies (Figure 3 A and B, arrow). In addition, many of the were surrounded by an extracellular membrane-like component consistent with a zona pellucida (Fig 3 A, red arrow). Hoechst staining revealed that some of the eGFP-positive oocyte-like cells seemed to be undergoing mitosis or neiosis. Figure 3C depicts a representative eGFP-positive cell in what appears to be the metaphase stage, where the chromosomes are condensed and lined up along a metaphase plate (arrow). Other eGFP-positive oocyte-like cells resembled embryos at the 2-cell and 4-cell stage of development (Figure 3 D and E) suggesting that, as seen in previous reports [5, 8], these ES cell-derived oocyte-like cells were capable of undergoing parthenogenesis. When the ES cells were allowed to differentiate further, we saw a substantial decrease in the number of eGFP-positive oocyte-like cells by d2, suggesting that most of the eGFP-positive cells had degenerated (Table I). However, at d15, a few of these eGFP-positive cells were seen embedded in large colonies growing on the plate. These cells were larger than the surrounding ES cells and many seemed to contain a structure resembling a polar body (Figure 4D). If these ES cells were allowed to differentiate even further, large clusters of eGFP-positive oocyte-like cells were found enriched in defined areas of the dish, increasing in number until around d45 (Figure 4J).
Figure 2. In vitro expression of Gdf9/eGFP in cells with an oocyte-like phenotype.
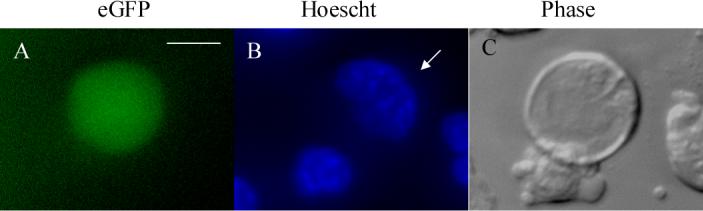
eGFP-positive cell at d1 of in vitro differentiation of JM21 XX ES cells. The eGFP-positive oocyte-like cell (A) was also stained with Hoechst 33342 to detect DNA (B, arrow). Phase image represented in C. Images obtained on Nikon TE300 Eclipse Inverted Fluorescent microscope at 40X magnification. Scale bar = 25μm
Figure 3. Expression of eGFP driven by the Gdf9 promoter in cultured ES cells.
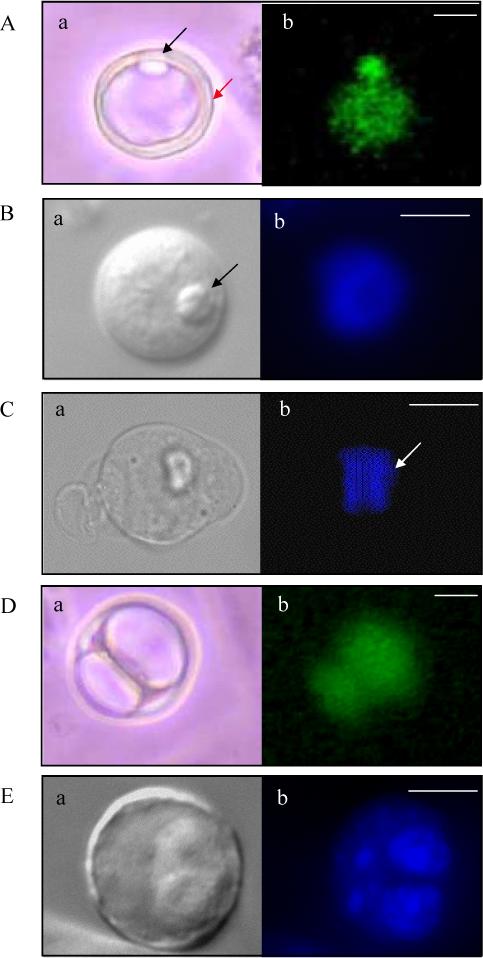
(A, B) eGFP-positive cells seen at d1 of in vitro differentiation of JM21 XX ES cells. Structures resembling polar bodies (black arrow) and a zona pellucida (red arrow) are seen. eGFP fluorescence represented in A, Hoechst DNA stain in B. (C) Hoechst stain of an oocyte-like cell at d1 of differentiation in metaphase stage of mitosis as determined by chromosomal alignment along a metaphase plate (white arrow). (D) eGFP positive zygote-like structure seen at d2 of in vitro differentiation and (E) Four-cell Hoechst stained cell seen at d2 of in vitro differentiation. (A-E, a): Phase images. Images in A and D = 20X magnification, Images in B, C and E = 40X magnification. Scale bars = 20μm (A, D), 25μm (B, C, E).
Table I.
Effects of different culture conditions on eGFP-positive cell number in JM21 XX ES cells
| Culture conditions | Day 1 | Day 2 |
|---|---|---|
| No LIF | 608 | 21 |
| Plus LIF | 1884 | 156 |
| No LIF/MEFs present | 41.3 | 2 |
| Embryoid bodies, no LIF | 3307 | 1634 |
| Embryoid bodies, plus | 9230 | 4337 |
| LIF |
Triplicate cultures were prepared for each treatment group and the average number of eGFP-positive cells/dish in each treatment group was determined by visual inspection. The results shown are representative of three separate experiments.
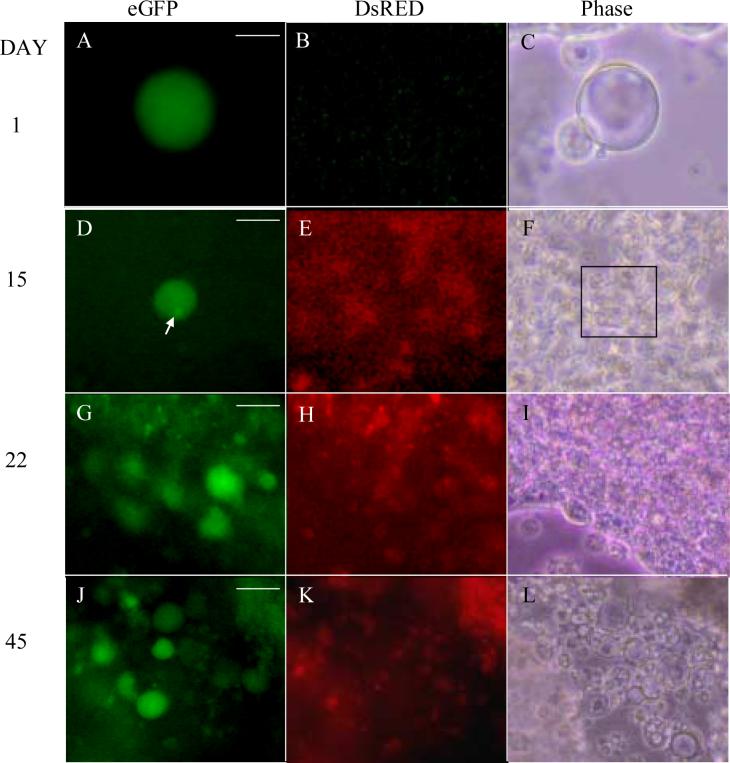
Figure 4. Gdf9 promoter and Sf1 promoter activity in cultured murine ES cells.
In vitro differentiation ofPGK12.1 XX ES cells transfected with the Gdf9/eGFP and Sf1/DsRED dual reporter system. Oocyte-like structures seen at d1 (A-C), d15 (D-F), d22 (G-I) and d45 (J-L) of culture. Polar body-like structure seen in D (arrow). DsRED expression is seen beginning at d15 (E) and continues through d22 (H) and d45 (K), although in no discernable structure. eGFP images (A, D, G, J); DsRED images (B, E, H, K) and Phase images (C, F, I, L) are shown. Magnification = 20X. Scale bars = 25μm (A), 50μm (B), 75μm (G, J).
As a control, we created two XX ES cell lines, one in JM21 XX ES cells, the other in PGK12.1 XX ES cells that were co-transfected the Gdf9/eGFP construct and an active DsRED gene driven by the Sf1 promoter. These two co-transfected cell lines behaved in a qualitatively similar fashion to the JM21 XX ES transfected with only the active Gdf9/eGF promoter construct. Using the dual promoter cell lines, DsRED-positive cells were seen in the culture dish by d15, though in no discernable structure or pattern about the eGFP-positive cells (Fig 4E). Moreover, DsRED and eGFP expression were never co-localized. By d22, more of the eGFP-positive oocyte-like structures were found embedded in the culture dish, many found in pockets of eGFP expression (Fig 4G). DsRED expression was again present, but not co-localized with eGFP (Figure 4H). These findings demonstrate selective activation of the Gdf9 and Sf1 promoters in specific cell populations.
Gdf9 promoter function in embryoid bodies
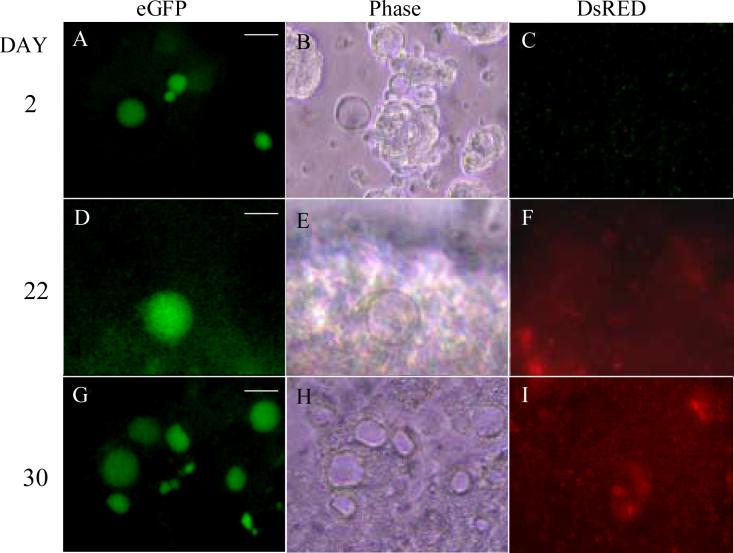
When the transfected ES cells were allowed to form embryoid bodies (EBs) in culture [4, 10], the appearance of eGFP-positive cells was again seen by d1. The eGFP-positive cells were seen clustered around the EBs and their structure was similar to that of the eGFP-positive cells that formed during in vitro differentiation of ES cells (Fig 5A). Most importantly, EB formation led to a substantial increase in the number of eGFP-positive cells formed by d1 of culture (Table I). In addition, degeneration of the GFP-expressing cells was slower in the EB culture with only a ∼50% decrease in eGFP-positive cell number by d2 (Table I), suggesting that embryoid body culture leads to an increase not only in eGFP-positive oocyte-like cell formation but also in apparent cell viability. The eGFP-positive cells were present in much lower numbers by d3 and were absent by d5. If the EBs remained in culture for up to 40 days, we observed a gradual increase in eGFP-positive cells beginning at around d22 (Figure 5D) and reaching its peak by d30 (Fig 5G). The eGFP-positive cells seen in the EB dish at d30 were found enriched in distinct clusters around the plate. These cells were found in close proximity to the EBs but not directly in or on them. They varied in size and were mostly found attached to the culture dish.
Figure 5. Activity of the Gdf9 promoter in embryoid body culture.
Transfected PGK2.1 ES cells on d2 after embryoid body formation (A-C) with many oocyte-like eGFP-positive cells seen around the embryoid bodies. eGFP-positive cells disappeared by d5 and did not return until d22 (D-F). Clusters of GFP-positive oocyte-like cells were seen by d30 (G-I) in distinct regions of the dish. GFP images (A, D, G); DsRED images (C, F, I) and phase images (B, E, H) are shown. Magnification = 20X. Scale bars = 50μm (A, G), 25μm (D).
Effects of LIF on Gdf9 promoter activity and gene expression
To examine conditions that would optimize the activity of the Gdf9 promoter in cultured ES cells, we tested the effects of growth factors thought to be important in female gamete formation in vivo on gene expression in culture. We first focused our attention on the media additive LIF. While LIF has been extensively studied as an important factor in stem cell self-renewal [21-24], several studies have reported the importance of LIF in the development of primordial and primary follicles of the ovary and have determined LIF is necessary for oocyte maturation [25-29]. PGK12.1 XX ES cells transfected with the Gdf9/eGFP were grown on a MEF monolayer in the presence of LIF for two days. On the second day, the cells were split 1:4 onto tissue culture plates without MEFs in either the presence or absence of LIF (d0). Cells were then analyzed by fluorescence microscopy 24h and 48h later. The eGFP-positive cells were counted and either RNA or protein was isolated for QPCR or immunoblot analysis.
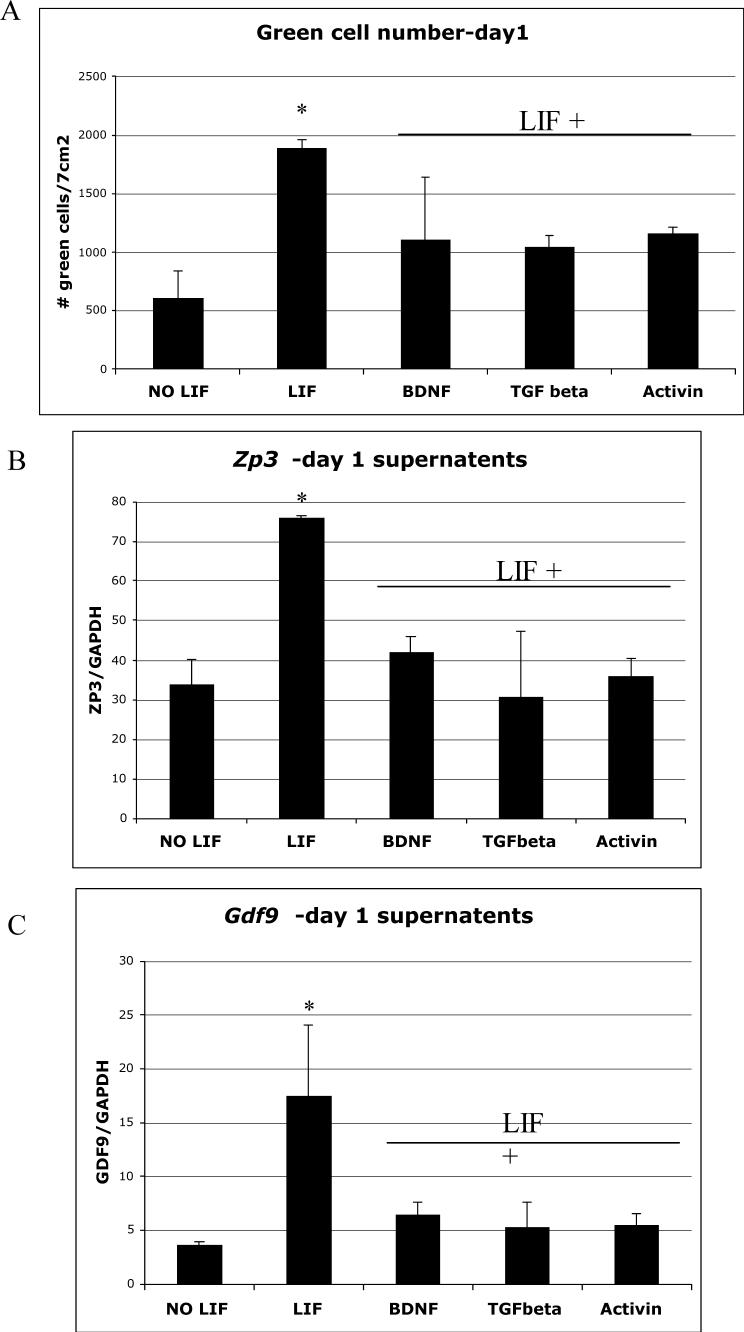
Removal of the MEF feeder layer and addition of LIF to the media led to a 68 % (+/−12.5%) increase in the number of eGFP-positive oocyte-like cells formed by d1 (Figure 6A). While LIF increased the number of eGFP-positive cells on d1, it was not sufficient to prevent the loss of these cells seen on d2 (Table I). While more eGFP-positive cells were seen on d2 in the LIF treated plates, the number seen still represented a ∼90% decrease in green cell number similar to results seen when no LIF was present. Since most of the eGFP-positive structures were found floating in the culture media, RNA from the cells in the supernatant was isolated and subjected to QPCR. QPCR analysis of the supernatant fractions for genes specifically expressed in oocytes, such as Gdf9 and the zona pellucida gene Zp3, revealed a 76% (+/−6%) and a 55.8% (+/−8.5%) increase in gene expression respectively (Figure 6B and C). The increase in oocyte-specific gene expression in response to LIF on d1 of differentiation correlated with the increase in eGFP-positive oocyte-like cell number seen on d1 when LIF was added to the media. In addition, immunoblot analysis of the supernatant fractions revealed an increase in ZP3 protein expression in response to LIF treatment (Figure 7).
Figure 6. Effect of LIF on Gdf9 promoter activity and gene expression.
(A) PGK2.1 XX ES cells transfected with the dual reporter system were cultured in media without LIF (NO LIF), media with LIF (LIF), or in media with LIF plus either BDNF (0.5 ng/ml), TGFβ (10 ng/ml) or Activin (25 ng/ml). On d1, eGFP-positive oocyte-like cells were counted visually in a 7cm2 region in the center of a 60 mm tissue culture dish and represented graphically here (n=3). (B, C) From the above treatments, RNA from supernatant fractions (collected by gentle agitation of the culture dish) was isolated; reverse transcribed and the DNA subjected to QPCR analysis using primers for Zp3 (B) and Gdf9 (C) with Gapdh as a control. Results are representative of 3 separate experiments.
Figure 7. Effect of LIF on ZP3 protein expression.
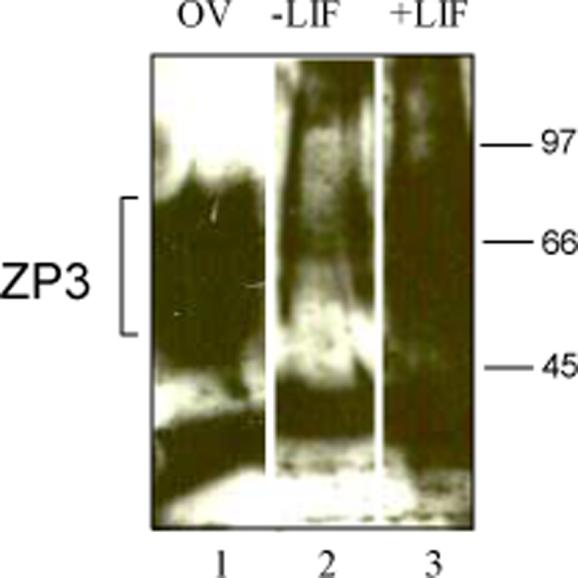
PGK21.1 XX ES cells transfected with the dual reporter system were cultured in media without LIF (Minus LIF, lane 2) or with LIF (Plus LIF, lane 3). On d1, protein was collected in 4X sample buffer from cells spun down from the supernatant fraction, subjected to SDS-PAGE gel electrophoresis in a 4−15% Tris-HCl gradient gel, transferred to Hybond C+ nitrocellulose and incubated with ZP3 (IE-10) antibody. Note the smear of the highly glycosylated ZP3 protein band. Ovarian extract was used as a positive control in lane 1.
We next examined the effects of specific growth factors such as brain-derived neurotropic factor (BDNF), transforming growth factor-β (TGFβ) and the TGFβ family member, activin on eGFP expression when added to the LIF treated plates on d0. BDNF is a member of the neurotrophin family of proteins that are widely expressed in the central nervous system and important for neuronal function [30]. Several studies have shown that BDNF plays a role in non-neuronal tissues, including in the ovary, where it was found to be essential for the development of early follicles and for oocyte maturation [30-32]. Members of the TGFβ superfamily have also been shown to play important roles in ovarian development starting with the specification of primordial germ cells by the bone morphogenetic proteins through to the recruitment of primordial follicles by anti-Mullerian hormone and, finally, their transformation into preantral and antral follicles in response to activin and TGFβ [33−35]. When BDNF, TGFβ or activin were added to the LIF treated plates, we saw a significant decrease in eGFP-positive oocyte-like cells (Figure 6A). Similarly, we found that BDNF, TGFβ and activin all cause an approximate 2-fold decrease in Zp3 gene expression (Figure 6B) and an approximate 3.5-fold decrease in Gdf9 gene expression (Figure 6C). These data suggest that while these factors have been shown to be important in oocyte maturation, they act to inhibit activity of the Gdf9 promoter. Taken together, these data suggest that LIF acts in a specific manner through a pathway that is inhibited by TGFβ, BDNF and activin, to increase the expression of eGFP driven by the Gdf9 promoter XX ES cells, as determined by an increase in green oocyte-like cells and an increase in Gdf9 and Zp3 gene expression.
4. Concluding Remarks
One of the most challenging aspects of the study of gene expression is the analysis of germ cell-specific genes in in vitro systems. Since ES cells and PGCs share the expression of many transcription factors and cell surface receptors, we tested the notion that cultured ES cells could be used to examine the function of an oocyte-specific gene promoter. Early PGCs can be distinguished from ES cells and epiblast cells by their differential expression of Oct-4 transgenes containing germ cell-specific regulatory elements [36-39]. This is possible because the expression of Oct-4 in ES cells, epiblast and PGCs relies on different promoter elements providing a means for identification of these different cell populations. Detection of postmigratory PGC markers, like mouse vasa homolog (mvh) have also been used to demonstrate the presence and subsequent isolation of germ cells in cultures of ES cells [5, 6, 8, 9]. In our initial studies, ES cells transfected with a modified Oct-4 promoter construct were employed to establish that ES cells have the propensity to differentiate into oocyte and follicle-like cells [8, 12].
Building on these findings, we transfected XX ES cells with transgenes that would express fluorescent reporters under the control of an unmodified oocyte-specific gene promoter. We found that XX ES cells transfected with the Gdf9/eGFP transgene expressed eGFP in cells that had a phenotype of oocyte-like cells. It was surprising that cells expressing eGFP appeared so rapidly in vitro. Most of these structures formed only 1 day after removal of the MEF monolayer. This rapid activation of the Gdf9 promoter suggests that within the initial population of undifferentiated ES cells there is a subset of cells that are already pre-programmed to undergo female germ line differentiation and is consistent with observations of genetic similarity between ES cells and early germ cells [40]. When these cells were injected into blastocysts and transplanted into host females, transgenic mice expressing eGFP specifically in the oocytes of the founder females were developed. This proves that these ES cells have the ability to develop into PGCs and ultimately migrate to the developing gonad and that the transfected ES cells contribute to the germline. In addition, these transgenic mice are a possible source of blastocysts from which new ES cell lines containing the Gdf9/eGFP transgene can be generated, eliminating some of the problems that arise when ES cells are passaged numerous times. Moreover, the Gdf9/eGFP transgenic mice derived from the transfected XX ES cells could be employed to examine the ability of stem cells from other locations (e.g., bone marrow) to produce female germ cells.
The oocyte-like cells expressing eGFP that appeared transiently in culture were not normal and rapidly degenerated. The presence of structures that resemble polar bodies, Hoechst staining patterns resembling metaphase plates, and the detection of zygote-like structures suggest that meiosis and parthenogenesis may have occurred in a number of the oocyte-like cells, which is consistent with our earlier findings [8]. However, the majority of the eGFP-positive cells appear to have degenerated, perhaps due to their inability to properly execute meiosis [7].
We singly and doubly transfected two separate XX ES cell lines with the Gdf9/eGFP and Sf1/DsRED fluorescent marker transgenes and obtained similar eGFP-positive oocyte-like cells by d1 of differentiation from both cell lines. This suggests that the rapid differentiation of ES cells into female germ cells is not limited to one particular ES cell line. When the doubly transfected XX ES cell line was allowed to differentiate in culture we observed the presence of oocyte-specific structures including polar bodies and zona pellucida. We also observed eGFP-positive cells in various stages of mitosis, leading to the development of 2-cell and 4-cell stage embryo-like structures. Previous studies have also reported the formation of blastocyst-like structures from ES cells in culture, presumably through parthenogenesis [5, 8]. While we observed DsRED expression, turned on at specific times during ES cell differentiation, it was not found in any discernable structure around the oocyte, and while the cells positive for eGFP were not positive for DsRED expression, no distinct cluster of DsRED positive cells was seen. The expression of DsRED was absent in the first week of differentiation and steadily increased throughout the 45 days of culture. The region of the Sf1 promoter used to drive DsRED expression was shown to be specific for gonadal cells and not present in other steroidogenic cells [13]. This suggests that while these ES cells were able to generate steroidogenic cells of the ovary, neither the monolayer nor EB culture was a sufficient environment for the recruitment of these cells into a follicle-like structure.
Further examination of the culture conditions for these transfected XX ES led to several interesting observations. First, when the ES cells were allowed to form EBs in culture, we saw a substantial increase in the number of eGFP-positive oocyte-like cells formed. In addition, eGFP-positive cell viability was increased since less degeneration was seen by day 2 in the EB culture when compared to the ES cells differentiated in monolayer. Several other investigators have demonstrated the ability of EBs to give rise to germ cells (both male and female) in culture [4, 10, 41, 42]. This suggests that the three-dimensional structure of the EB may preserve greater intercellular communication between the cells, allowing for activation f the transcriptional machinery for oocyte-specific genes [43]. Second, when LIF is present in the media of the differentiating ES cells a significant increase in eGFP-positive oocyte-like cells was observed. LIF has been widely studied as a media additive important for the maintenance of ES cells in a self-renewing, undifferentiated state [21-24]. However, based on several reports demonstrating the importance of LIF not only in the survival and self-renewal of PGCs [25-27, 44], but also in the maturation of oocytes and ovarian follicles [26, 28, 31, 45], we decided to study its role in the formation of germ cells from ES cells in vitro. We found that LIF, when added to the media of ES cells differentiating in the absence of a MEF monolayer, significantly increased not only the number of eGFP-positive oocyte-like cells, but also the expression of oocyte specific genes in vitro.
Activin and TGFβ have been shown to be important factors in oocyte and follicle maturation through the activation of the Smad pathway in the cells of the ovary [33], while BDNF has also been shown to be important in the development of early follicles [30]. We observed, however, a decrease in LIF-stimulated eGFP expression when these growth factors were present in the culture media. While activin and TGFβ have been shown to inhibit the proliferation of mouse PGCs in culture, this data suggests they may also inhibit the formation of germ cells from PGCs. This may occur through the inhibition of a specific JAK/STAT signaling pathway activated by LIF binding to the GP130 receptor [22], possibly by the activation of Smad 5 [33].
In summary, our observations demonstrate the utility of cultured murine ES cells to study oocyte-specific gene expression in culture. This technology has revealed that eGFP-positive oocyte-like cells appear rapidly, and that LIF increases the number appearing in culture. Further exploitation of this methodology may help achieve better systems to examine oocyte-specific gene expression, and possibly the creation of competent female germ from ES cells in vitro.
Acknowledgments
We thank Dr. John MacLaughlin of the University of Pennsylvania School of Veterinary Medicine (New Bolton, PA) and Neil Brockdorff of MRC Clinical Sciences Centre (London, UK) for ES cells, Dr. Martin Matzuk of Baylor College of Medicine (Houston, TX) and Dr. Keith Parker of University of Texas Southwestern (Dallas, TX) for plasmids. This work was supported in part by HD-06274, a grant from the Health Research Formula Fund from the Pennsylvania Department of Health. This department specifically disclaims responsibility for any analyses, interpretations and conclusions, and in part by T32-HD-007305 and T32-HD-040135.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Terskikh AV, Bryant PJ, Schwartz PH. Mammalian stem cells. Pediatr Res. 2006;59:13R–20R. doi: 10.1203/01.pdr.0000205154.86517.2a. [DOI] [PubMed] [Google Scholar]
- 2.Naveiras O, Daley GQ. Stem cells and their niche: a matter of fate. Cell Mol Life Sci. 2006;63:760–766. doi: 10.1007/s00018-005-5469-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donovan PJ, De Miguel MP, Hirano MP, et al. Germ cell biology--from generation to generation. Int J Dev Biol. 2001;45:523–531. [PubMed] [Google Scholar]
- 4.Lacham-Kaplan O, Chy H, Trounson A. Testicular cell conditioned medium supports differentiation of embryonic stem cells into ovarian structures containing oocytes. Stem Cells. 2006;24:266–273. doi: 10.1634/stemcells.2005-0204. [DOI] [PubMed] [Google Scholar]
- 5.Dyce PW, Wen L, Li J. In vitro germline potential of stem cells derived from fetal porcine skin. Nat Cell Biol. 2006;8:384–390. doi: 10.1038/ncb1388. [DOI] [PubMed] [Google Scholar]
- 6.Clark AT, Bodnar MS, Fox M, et al. Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum Mol Genet. 2004;13:727–739. doi: 10.1093/hmg/ddh088. [DOI] [PubMed] [Google Scholar]
- 7.Novak I, Lightfoot DA, Wang H, et al. Mouse embryonic stem cells form follicle-like ovarian structures but do not progress through meiosis. Stem Cells. 2006 doi: 10.1634/stemcells.2005-0520. (in press) [DOI] [PubMed] [Google Scholar]
- 8.Hubner K, Fuhrmann G, Christenson LK, et al. Derivation of oocytes from mouse embryonic stem cells. Science. 2003;300:1251–1256. doi: 10.1126/science.1083452. [DOI] [PubMed] [Google Scholar]
- 9.Toyooka Y, Tsunekawa N, Akasu R, et al. Embryonic stem cells can form germ cells in vitro. Proc Natl Acad Sci U S A. 2003;100:11457–11462. doi: 10.1073/pnas.1932826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geijsen N, Horoschak M, Kim K, et al. Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature. 2004;427:148–154. doi: 10.1038/nature02247. [DOI] [PubMed] [Google Scholar]
- 11.Farini D, Scaldaferri ML, Iona S, et al. Growth factors sustain primordial germ cell survival, proliferation and entering into meiosis in the absence of somatic cells. Dev Biol. 2005;285:49–56. doi: 10.1016/j.ydbio.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 12.Kehler J, Hubner K, Garrett S, et al. Generating oocytes and sperm from embryonic stem cells. Semin Reprod Med. 2005;23:222–233. doi: 10.1055/s-2005-872450. [DOI] [PubMed] [Google Scholar]
- 13.Wilhelm D, Englert C. The Wilms tumor suppressor WT1 regulates early gonad development by activation of Sf1. Genes Dev. 2002;16:1839–1851. doi: 10.1101/gad.220102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan C, Elvin JA, Lin YN, et al. Regulation of growth differentiation factor 9 expression in oocytes in vivo: a key role of the E-box. Biol Reprod. 2006;74:999–1006. doi: 10.1095/biolreprod.105.050013. [DOI] [PubMed] [Google Scholar]
- 15.George EL, Hynes RO. Gene targeting and generation of mutant mice for studies of cell-extracellular matrix interactions. Methods Enzymol. 1994;245:386–420. doi: 10.1016/0076-6879(94)45021-8. [DOI] [PubMed] [Google Scholar]
- 16.East IJ, Gulyas BJ, Dean J. Monoclonal antibodies to the murine zona pellucida protein with sperm receptor activity: effects on fertilization and early development. Dev Biol. 1985;109:268–273. doi: 10.1016/0012-1606(85)90454-3. [DOI] [PubMed] [Google Scholar]
- 17.Su YQ, Wu X, O'Brien MJ, et al. Synergistic roles of BMP15 and GDF9 in the development and function of the oocyte-cumulus cell complex in mice: genetic evidence for an oocyte-granulosa cell regulatory loop. Dev Biol. 2004;276:64–73. doi: 10.1016/j.ydbio.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 18.Hanley NA, Ikeda Y, Luo X, et al. Steroidogenic factor 1 (SF-1) is essential for ovarian development and function. Mol Cell Endocrinol. 2000;163:27–32. doi: 10.1016/s0303-7207(99)00237-3. [DOI] [PubMed] [Google Scholar]
- 19.Parker KL, Ikeda Y, Luo X. The roles of steroidogenic factor-1 in reproductive function. Steroids. 1996;61:161–165. doi: 10.1016/0039-128x(96)00006-2. [DOI] [PubMed] [Google Scholar]
- 20.Lee CK, Piedrahita JA. Effects of growth factors and feeder cells on porcine primordial germ cells in vitro. Cloning. 2000;2:197–205. doi: 10.1089/152045500454753. [DOI] [PubMed] [Google Scholar]
- 21.Chambers I. The molecular basis of pluripotency in mouse embryonic stem cells. Cloning Stem Cells. 2004;6:386–391. doi: 10.1089/clo.2004.6.386. [DOI] [PubMed] [Google Scholar]
- 22.Metcalf D. The leukemia inhibitory factor (LIF). Int J Cell Cloning. 1991;9:95–108. doi: 10.1002/stem.5530090201. [DOI] [PubMed] [Google Scholar]
- 23.Metcalf D. The unsolved enigmas of leukemia inhibitory factor. Stem Cells. 2003;21:5–14. doi: 10.1634/stemcells.21-1-5. [DOI] [PubMed] [Google Scholar]
- 24.Niwa H. Molecular mechanism to maintain stem cell renewal of ES cells. Cell Struct Funct. 2001;26:137–148. doi: 10.1247/csf.26.137. [DOI] [PubMed] [Google Scholar]
- 25.Cheng L, Gearing DP, White LS, et al. Role of leukemia inhibitory factor and its receptor in mouse primordial germ cell growth. Development. 1994;120:3145–3153. doi: 10.1242/dev.120.11.3145. [DOI] [PubMed] [Google Scholar]
- 26.De Felici M, Klinger FG, Farini D, et al. Establishment of oocyte population in the fetal ovary: primordial germ cell proliferation and oocyte programmed cell death. Reprod Biomed Online. 2005;10:182–191. doi: 10.1016/s1472-6483(10)60939-x. [DOI] [PubMed] [Google Scholar]
- 27.Donovan PJ. Growth factor regulation of mouse primordial germ cell development. Curr Top Dev Biol. 1994;29:189–225. doi: 10.1016/s0070-2153(08)60551-7. [DOI] [PubMed] [Google Scholar]
- 28.Nilsson EE, Kezele P, Skinner MK. Leukemia inhibitory factor (LIF) promotes the primordial to primary follicle transition in rat ovaries. Mol Cell Endocrinol. 2002;188:65–73. doi: 10.1016/s0303-7207(01)00746-8. [DOI] [PubMed] [Google Scholar]
- 29.Nilsson EE, Skinner MK. Kit ligand and basic fibroblast growth factor interactions in the induction of ovarian primordial to primary follicle transition. Mol Cell Endocrinol. 2004;214:19–25. doi: 10.1016/j.mce.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Kawamura K, Kawamura N, Mulders SM, et al. Ovarian brain-derived neurotrophic factor (BDNF) promotes the development of oocytes into preimplantation embryos. Proc Natl Acad Sci U S A. 2005;102:9206–9211. doi: 10.1073/pnas.0502442102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paredes A, Romero C, Dissen GA, et al. TrkB receptors are required for follicular growth and oocyte survival in the mammalian ovary. Dev Biol. 2004;267:430–449. doi: 10.1016/j.ydbio.2003.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spears N, Molinek MD, Robinson LL, et al. The role of neurotrophin receptors in female germ-cell survival in mouse and human. Development. 2003;130:5481–5491. doi: 10.1242/dev.00707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drummond AE. TGFbeta signalling in the development of ovarian function. Cell Tissue Res. 2005;322:107–115. doi: 10.1007/s00441-005-1153-1. [DOI] [PubMed] [Google Scholar]
- 34.Elvin JA, Yan C, Matzuk MM. Oocyte-expressed TGF-beta superfamily members in female fertility. Mol Cell Endocrinol. 2000;159:1–5. doi: 10.1016/s0303-7207(99)00185-9. [DOI] [PubMed] [Google Scholar]
- 35.Richards AJ, Enders GC, Resnick JL. Activin and TGFbeta limit murine primordial germ cell proliferation. Dev Biol. 1999;207:470–475. doi: 10.1006/dbio.1998.9174. [DOI] [PubMed] [Google Scholar]
- 36.Noaksson K, Zoric N, Zeng X, et al. Monitoring differentiation of human embryonic stem cells using real-time PCR. Stem Cells. 2005;23:1460–1467. doi: 10.1634/stemcells.2005-0093. [DOI] [PubMed] [Google Scholar]
- 37.Ovitt CE, Scholer HR. The molecular biology of Oct-4 in the early mouse embryo. Mol Hum Reprod. 1998;4:1021–1031. doi: 10.1093/molehr/4.11.1021. [DOI] [PubMed] [Google Scholar]
- 38.Pesce M, Scholer HR. Oct-4: control of totipotency and germline determination. Mol Reprod Dev. 2000;55:452–457. doi: 10.1002/(SICI)1098-2795(200004)55:4<452::AID-MRD14>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 39.Yoshimizu T, Sugiyama N, De Felice M, et al. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev Growth Differ. 1999;41:675–684. doi: 10.1046/j.1440-169x.1999.00474.x. [DOI] [PubMed] [Google Scholar]
- 40.Zwaka TP, Thomson JA. A germ cell origin of embryonic stem cells? Development. 2005;132:227–233. doi: 10.1242/dev.01586. [DOI] [PubMed] [Google Scholar]
- 41.West JA, Daley GQ. In vitro gametogenesis from embryonic stem cells. Curr Opin Cell Biol. 2004;16:688–692. doi: 10.1016/j.ceb.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 42.Zhao X, Teng R, Asanuma K, et al. Differentiation of mouse embryonic stem cells into gonadotrope-like cells in vitro. J Soc Gynecol Investig. 2005;12:257–262. doi: 10.1016/j.jsgi.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 43.Tiedemann H, Asashima M, Grunz H, et al. Pluripotent cells (stem cells) and their determination and differentiation in early vertebrate embryogenesis. Dev Growth Differ. 2001;43:469–502. doi: 10.1046/j.1440-169x.2001.00599.x. [DOI] [PubMed] [Google Scholar]
- 44.De Felici M, Scaldaferri ML, Lobascio M, et al. Experimental approaches to the study of primordial germ cell lineage and proliferation. Hum Reprod Update. 2004;10:197–206. doi: 10.1093/humupd/dmh020. [DOI] [PubMed] [Google Scholar]
- 45.Skinner MK. Regulation of primordial follicle assembly and development. Hum Reprod Update. 2005;11:461–471. doi: 10.1093/humupd/dmi020. [DOI] [PubMed] [Google Scholar]