Three sets of novel 4-deoxy-L-arabinose analogs were synthesized and evaluated as potential inhibitors of the bacterial resistance mechanism in which lipid A, on the outer membrane, is modified with 4-amino-4-deoxy-L-arabinose (L-Ara4N). One compound diminished the transfer of L-Ara4N onto lipid A. These results suggest that small molecules might be designed that would effect the same reversal of bacterial resistance observed in genetic knockouts.
Polymyxin is a currently prescribed antibiotic whose use is predominately topical as a result of its systemic toxicity. With the advent of devises that include as ‘topical’ the aerosol route of administration, polymyxin is now widely used for chronic airway infections, notably in cystic fibrosis patients for whom this is a significant cause of morbidity and mortality.1 Polymyxin, like all cationic antimicrobial peptides (CAPs), binds to negative charges on phosphates of the lipopolysaccharide component lipid A that makes up the outer leaflet of the Gram-negative bacterial outer membrane. Lipid A binding by CAPs results in critical disorganization of the outer membrane.2 Bacteria can resist the action of polymyxin by modification of lipid A phosphates, which decreases surface negative charge and presumably reduces polymyxin binding to the altered electrostatic topography of the remodeled membrane. 3 4 5
The pathway that results in aminoarabinose incorporation into lipid A has been elucidated in some detail. Five enzymes catalyze seven reactions that produce an undecaprenylated aminosugar at the cytoplasmic surface of the inner membrane. This glycolipid is then translocated to the periplasmic side of the inner membrane where ArnT/PmrK catalyzes the displacement of the prenyl group by the 4’- and/or 1’- phosphate(s) of lipid A.6
If the mechanisms of lipid A modification could be disabled, CAP-resistant bacteria might be returned to a susceptible state. An agent capable of this restoration could also promote the activity of host antimicrobial peptides in controlling infection, and indeed, mutants defective for the addition of aminoarabinose are attenuated for virulence in mouse models of infection. A resensitizing agent could also expand the repertoire of utility for polymyxin and enable the durable therapeutic utility of the newer CAPs under investigation.7 While the ultimate goal of the project is to develop small molecule inhibitors of the L-Ara4N-lipidA that could exogenously reproduce the effects of genetic knockouts, we have initially focused on the synthesis of compounds with inhibitory activity in vitroWe now report on the synthesis and evaluation of a series of 4-modified arabinose analogs.
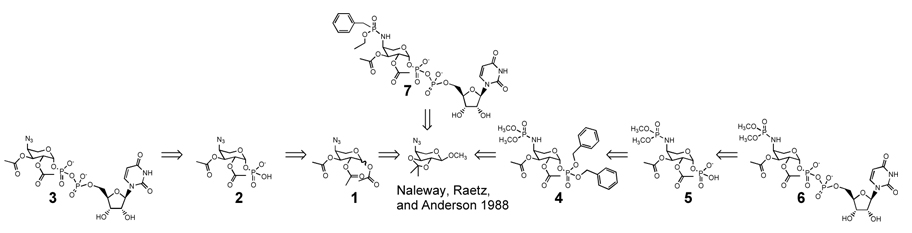
The 4-amino function of aminoarabinose is formylated for several of the cytoplasmic biosynthetic steps. N-formylation drives the otherwise-unfavorable transamination reaction (ArnB/PmrH) forward,8 and the formyl group is subsequently removed by ArnD.9 We constructed analogs (Figure 1) that might mimic the N-formylation transition state or serve as surrogates for the formylated amine. Three isosteres for the N-C(OH)-N framework were selected, phosphonamidate, phosphoramidate, and azide. For the azide and phosphoramidate, several substituents at the anomeric carbon were explored: O- phosphoryl, O-acetyl, O-benzyl, and, the most substrate-mimetic, O-uridyldiphosphoryl. Although the initial biological characterization was to be in cell-free enzyme preparations, we designed in some structural features that anticipated our longer term plans for compounds having good stability in the human host and the ability to penetrate bacterial outer membranes. Since five of the L-Ara4N biosynthetic enzymes, both the soluble and membrane bound, lie beyond the inner membrane, while the lipid A modifying enzyme, ArnT/PmrK, is on the inner membrane, access to these enzymes was expected to be a significant determinant of our compounds’ ultimate capabilities against intact bacteria. With this in mind, the 2- and 3-hydroxyls were acetylated, and the phorphoramidates/phosphonamidates maintained as esters. We anticipated that these esters would encounter nonselective esterases and be hydrolyzed in vivo. The selection of the compounds benefited from an economy in design, since many candidates were also intermediates en route to the more complex structures. The 4-azido-2,3-isopropylidene-1-methyl acetal10 served as the single common precursor.
Figure 1.
The 4-azido analogs 1, 2, and 3 were synthesized as shown in Scheme I. Conversion of the isopropylidene methyl acetal directly to the triacetate was accomplished in a three-step sequence using Takeo’s procedure.11 Selective 1-O-deacetylation was achieved by treatment of the triacetate with a saturated solution of dimethylamine in acetonitrile at −20 °C.12 Installation of the phosphate via the two-step method13 gave the β-L-arabinose phosphate 2 in modest yield but good stereochemical purity.14 Reaction of the 1-phosphate, as its tri n-octylamine salt, with uridine monophosphoryl morpholidate15 in the presence of tetrazole, completed the synthesis of 3. For the synthesis of 4, 5, and 6 the azide was converted via a modified Staudinger reaction to the phosphoramidate (Scheme II).16 Without the synthetic incompatibility presented by the azide function, dibenzylphosphate could be directly introduced (8 to 10) and readily removed (4 to 5).
Scheme I.
a. (i) 1 M HCl/acetone, then NaHCO3, (ii) 1M HCl, 90 °C, (iii) Li2CO3, (iv)Ac2O, NaOAc, reflux; b. (CH3)2NH, CH3CN, −20 °C; c. (i) 2-chloro-4[H]-benzodioxaphosphorin-4-one, NEt3, THF-dioxane, 0 °C, (ii) H2O; d. (i) Dowex 50W × 8, H+ form, THF-dioxane, (ii) tBuOOH/decane, cat. I2, NEt3; e. (i) tri n-octylalmine/pyridine, (ii) UMP morpholidate, DCCM salt, tetrazole, pyridine
Scheme II.
a. (i) n-BuLi, THF, (ii) tetrabenzylpyrophosphate, THF; b. (CH3)3P, pyridine, CH2Cl2; c. (i) H2, Pd-C, CH3OH (ii) NEt3; d. UMP morpholidate, tetrazole, pyridine
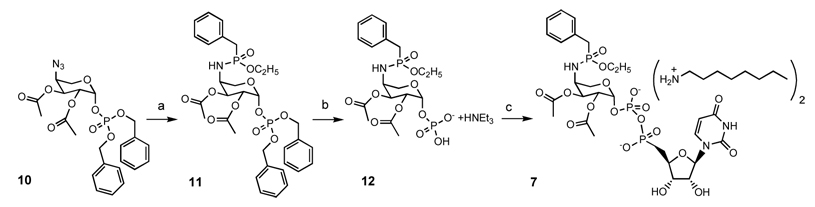
The efficiency of the Staudinger reaction that retains the N-P bond for the preparation of phosphoramidates encouraged us to develop a similar strategy for the synthesis of phosphonamidate 7. Intermediate 10 was reacted with ethyl benzyl H-phosphinate17, 18in the presence of 6 equivalents of BTSTFA, followed by thermal rearrangement to give phosphonamidate 11 (Scheme III)16, 19. Hydrogenolysis of the benzyl esters gave the free phosphate 12. Solubility mandated prior formation of the bis-n-octylammonium salt for the reaction with UMP morpholodate, giving 7 as the corresponding bis-n-octylammonium salt.
Scheme III.
a. (i) ethyl benzyl H-phosphinate, BTSTFA, pyridine (ii) 1:3 CH3OH-CHCl3 , Δ; b. (i) H2, Pd-C, CH3OH (ii) NEt3; c. (i) tri-n-octylamine, pyridine, (ii) UMP morpholidate, tetrazole, pyridine
Compounds 1–7 were evaluated in vitro in a single global assay for the decarboxylase and transformylase activity encoded in ArnA/PmrI as well as the trasnaminase activity of ArnB/PmrH as previously described.20, 21 These enzymes were unaffected at compound concentrations up to 100 µM. Crystal structures of ArnA and ArnB20–22 suggest that the 2- and 3- acetates, as well as excessive functionality at the 4-position, may be impeding rather than promoting activity against these two aminoarabinose biosynthetic enzymes.
Considering that the 4-azido group might be a neutral surrogate for the 4-amino group, compounds 1, 2, 3, were selected for evaluation in an assay against PmrK/ArnT.6 Compound 7 was included as a structural (and presumed negative) control of the most substrate-like of the azide trio, compound 3. To monitor PmrK/ArnT activity, whole membranes from the Gram-negative bacterium S. typhimurium strain JSG43523were assayed for the addition of aminoarabinose to 32P labeled precursors of lipid A.6 Salmonella JSG435 overproduces the enzymatic machinery required for the modification of lipid A with aminoarabinose, including the membrane-bound glycosyl transferase PmrK/ArnT.6 As determined by densiometry24 of the 32P TLC (Figure 2), compound 1 effected a three-fold diminution of aminoarabinose addition to the phosphate groups of the lipid A precursor lipid IVA, demonstrating inhibition of PmrK/ArnT.
Figure 2.
Assay of PmrK/ArnT aminoarabinose transferase activity. Membranes from S. typhimurium strain JSG435 were assayed for the transfer of L-Ara4N from endogenous sources to the tetra-acylated lipid A precursor 4’-32P-lipid IVA. The protein concentration was 1 mg/ml and the concentration of the inhibitor when present was 1 mM. Assays were carried out as previously described6 for 0.5 h (Panel A) or for 6 h (Panel B) at 30 C using 5 µM 4’-32P-lipid IVA (20,000 cpm/nmol). The reaction products were separated by thin layer chromatography in the solvent chloroform, pyridine, 88% formic acid, water (50:50:16:5, v/v) and visualized by Phosphorimaging. Lipid IIA arises from the addition of L-Ara4N to the starting substrate lipid IVA. Lipids IIB and IVB arise from the PagP-dependent addition of palmitate to lipid IIA and lipid IVA, respectively. Reaction products modified with L-Ara4N are indicated by an asterisk.
Acting against the transferase PmrK/ArnT, azido compound 1 represents the first reported small molecule to interfere with Lipid A modification. With the synthetic methods developed, the structural biology of the enzymes well-characterized, and with compound 1 as a potential lead, additional analogs may now be prepared with increased activity against the lipid A modifying enzymes and against resistant bacteria.25
Supplementary Material
Acknowledgments
The authors would like to thank Stona Jackson and Kathleen Barry for technical assistance in the synthetic chemistry. This work was supported by an NIAID award for the Northwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases U54 AI057141 The generous support of the National Institutes of Health NIH (RO1-AI064184 to M. S. T), and AI060841-01 to M. C. S.) is also acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.Beringer P. The clinical use of colistin in patients with cystic fibrosis. Curr Opin Pulm Med. 2001;7(6):434–440. doi: 10.1097/00063198-200111000-00013. [DOI] [PubMed] [Google Scholar]
- 2.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3(3):238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 3.Peschel A. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 2002;10(4):179–186. doi: 10.1016/s0966-842x(02)02333-8. [DOI] [PubMed] [Google Scholar]
- 4.McPhee JB, Lewenza S, Hancock RE. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol. 2003;50(1):205–217. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- 5.Bader MW, Navarre WW, Shiau W, Nikaido H, Frye JG, McClelland M, Fang FC, Miller SI. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol Microbiol. 2003;50(1):219–230. doi: 10.1046/j.1365-2958.2003.03675.x. [DOI] [PubMed] [Google Scholar]
- 6.Trent MS, Ribeiro AA, Lin S, Cotter RJ, Raetz CR. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-L-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J Biol Chem. 2001;276(46):43122–43131. doi: 10.1074/jbc.M106961200. [DOI] [PubMed] [Google Scholar]
- 7.Baron C, Coombes B. Targeting bacterial secretion systems: benefits of disarmament in the microcosm. Infect Disord Drug Targets. 2007;7(1):19–27. doi: 10.2174/187152607780090685. [DOI] [PubMed] [Google Scholar]
- 8.Breazeale SD, Ribeiro AA, McClerren AL, Raetz CR. A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-Amino-4-deoxy-L-arabinose. Identification and function oF UDP-4-deoxy-4-formamido-L-arabinose. J Biol Chem. 2005;280(14):14154–14167. doi: 10.1074/jbc.M414265200. [DOI] [PubMed] [Google Scholar]
- 9.Williams GJ, Breazeale SD, Raetz CR, Naismith JH. Structure and function of both domains of ArnA, a dual function decarboxylase and a formyltransferase, involved in 4-amino-4-deoxy-L-arabinose biosynthesis. J Biol Chem. 2005;280(24):23000–23008. doi: 10.1074/jbc.M501534200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naleway JJ, Raetz CR, Anderson L. A convenient synthesis of 4-amino-4-deoxy-L-arabinose and its reduction product, 1,4-dideoxy-1,4-imino-L-arabinitol. Carbohydr Res. 1988;179:199–209. doi: 10.1016/0008-6215(88)84118-1. [DOI] [PubMed] [Google Scholar]
- 11.Takeo K, Murata Y, Kitamura S. A facile synthesis of 4-O-allyl-D-xylopyranose and its use in the preparation of xylo-oligosaccharides. Carbohydr Res. 1992;224:311–318. doi: 10.1016/0008-6215(92)84118-c. [DOI] [PubMed] [Google Scholar]
- 12.Upreti, Dipali MR, Vishwakarma Ram A. Synthesis of the Tetrascaccharide CapDomain of the Antigenic Lipphosphoglycan of Leishmania donovani Parasite. Tetrahedron. 2000;56:6577–6584. [Google Scholar]
- 13.Takaya K, Nagahori N, Kurogochi M, Furuike T, Miura N, Monde K, Lee YC, Nishimura S. Rational design, synthesis, and characterization of novel inhibitors for human beta1,4-galactosyltransferase. J Med Chem. 2005;48(19):6054–6065. doi: 10.1021/jm0504297. [DOI] [PubMed] [Google Scholar]
- 14.For the synthesis of compound 2 and analytical data please see the Supplementary Information.
- 15.Wittmann V, Wong CH. 1H-Tetrazole as Catalyst in Phosphomorpholidate Coupling Reactions: Efficient Synthesis of GDP-Fucose, GDP-Mannose, and UDP-Galactose. J Org Chem. 1997;62(7):2144–2147. doi: 10.1021/jo9620066. [DOI] [PubMed] [Google Scholar]
- 16.For the synthesis of compound 4 and analytical data please see the Supplementary Information.
- 17.Abrunhosa-Thomas I, Ribiere Patrice, Adcock Alicia C, Montchampt, Jean-Luc Direcct Monoalkylation of Alkyl Phosphinates to Access H-Phosphinic Acid Esters. Synthesis. 2006;2006(2):325–331. [Google Scholar]
- 18.Deprele SM, Jean-Luc A Novel and Convenient Preparation of Hypophosphite Esters. Journal of Organiometallic Chemistry. 2002;634–644:154–163. [Google Scholar]
- 19.For the synthesis of compound 11 and analytical data please see the Supplementary Information. Compound 11.
- 20.Gatzeva-Topalova PZ, May AP, Sousa MC. Crystal structure and mechanism of the Escherichia coli ArnA (PmrI) transformylase domain. An enzyme for lipid A modification with 4-amino-4-deoxy-L-arabinose and polymyxin resistance. Biochemistry. 2005;44(14):5328–5338. doi: 10.1021/bi047384g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gatzeva-Topalova PZ, May AP, Sousa MC. Structure and mechanism of ArnA: conformational change implies ordered dehydrogenase mechanism in key enzyme for polymyxin resistance. Structure. 2005;13(6):929–942. doi: 10.1016/j.str.2005.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gatzeva-Topalova PZ, May AP, Sousa MC. Crystal structure of Escherichia coli ArnA (PmrI) decarboxylase domain. A key enzyme for lipid A modification with 4-amino-4-deoxy-L-arabinose and polymyxin resistance. Biochemistry. 2004;43(42):13370–13379. doi: 10.1021/bi048551f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunn JS, Ryan SS, Van Velkinburgh JC, Ernst RK, Miller SI. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar typhimurium. Infect Immun. 2000;68(11):6139–6146. doi: 10.1128/iai.68.11.6139-6146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.TLC plates were exposed overnight to a PhosphorImager Screen and product formation detected using a Bio-Rad Molecular Imager PhosphorImager. Enzyme activity was calculated by determining the percentage of the substrate converted to product by densitometry using Quantity One Software.
- 25.A preliminary assay against whole bacteria showed that None of the compounds, at highest concentrations ranging from 90mM (compound 1) to 500 mM (compound 7), were able to restore susceptibility to the CAP polymyxin in Salmonella strain JSG435that constitutively synthesizes a lipid A modified with aminoarabinose. The lack of effect against intact bacteria could result from insufficient activity against the target, poor permeability/retention, metabolic inactivation, or a combination of these factors.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.