Abstract
Humans are exposed to arsenicals through many routes with the most common being in drinking water. Exposure to arsenic has been associated with an increase in the incidence of cancer of the skin, lung and bladder. Although the relationship between exposure and carcinogenesis is well documented, the mechanisms by which arsenic participates in tumorigenesis are not fully elucidated. We evaluated the potential epigenetic component of arsenical action by assessing the histone acetylation state of 13 000 human gene promoters in a cell line model of arsenical-mediated malignant transformation. We show changes in histone H3 acetylation occur during arsenical-induced malignant transformation that are linked to the expression state of the associated gene. DNA hypermethylation was detected in hypoacetylated promoters in the select cases analyzed. These epigenetic changes occurred frequently in the same promoters whether the selection was performed with arsenite [As(III)] or with monomethylarsonous acid, suggesting that these promoters were targeted in a non-random fashion, and probably occur in regions important in arsenical-induced malignant transformation. Taken together, these data suggest that arsenicals may participate in tumorigenesis by altering the epigenetic terrain of select genes.
Introduction
Humans are exposed to many forms of environmental arsenic with the inorganic arsenicals, arsenate [As(V)] and arsenite [As(III)], being the most abundant forms. Human exposure to inorganic arsenic occurs most commonly through contaminated drinking water. Once ingested, inorganic arsenic is enzymatically biotransformed to a number of metabolites including both trivalent and pentavalent methylated species (1,2). The methylated trivalent arsenicals are highly toxic forms of arsenic and have been detected in human urine with monomethylarsonous acid [MMA(III)] being of particular interest (3–5). MMA(III) is a more potent toxicant than arsenite, with its cytotoxic potency being 20 times that of the parent compound (6,7). Epidemiological studies have indicated that exposure to arsenicals leads to a multitude of health problems including cancers of the lung, skin and bladder (8–11). A recent report detailed a 50 years epidemiological study that examined the incidence of bladder cancer relative to arsenic exposure in Chilean populations. Results suggested that a significant increase in bladder cancer mortality, peaking with a rate ratio of 6.1:13.8, was linked to higher levels of arsenic in the drinking water (10).
Although arsenic is classified as a human carcinogen, the mechanisms by which arsenicals lead to the formation of neoplasms remain unclear and are probably multifactorial. Previous studies have shown that arsenicals can alter growth signaling pathways (12,13). Arsenicals are classified as weak mutagens by most classical mutagenesis assays; however, they can indirectly damage DNA via the generation of reactive oxygen species and inhibition of DNA replication and repair enzymes (14–20). These mechanisms clearly play a role in arsenical-mediated effects on cells; however, the long-term effects of arsenicals on cellular phenotype may be mediated through additional mechanisms as well.
Cells may adapt to long-term arsenical exposure through epigenetic mechanisms of gene control. Molecular toxicological studies on the heavy metals nickel and arsenic support this possibility. Using various cell types, it has been demonstrated that exposure to nickel can result in an altered chromatin state, specifically a decrease in histone acetylation (21–23). In addition, it has been shown, both in vitro and in vivo, that arsenic exposure results in the loss of genomic DNA methylation in rat hepatocytes (24,25). This relationship between the DNA methylation status of select promoters and arsenic exposure is also observed in human bladder cancer specimens (26). These studies shed light upon gene–environment interactions that involve the epigenetic control of carcinogenesis. Currently, however, little is known about the histone modification profile that results from chronic, low-level exposure to As(III) and MMA(III).
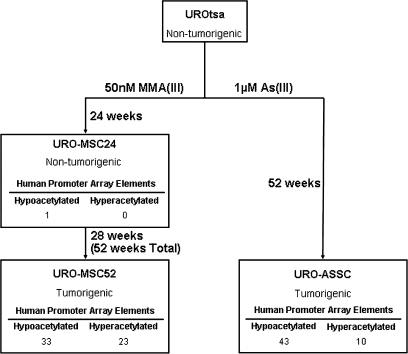
In light of recent research suggesting that environmental agents can perturb the epigenetic terrain, we initiated studies to address the role of arsenicals on the epigenome during malignant transformation. We used an immortalized, non-tumorigenic cell line model of human urothelial cells, UROtsa, and malignantly transformed variants of UROtsa that emerged from long-term exposure to arsenicals (27,28). These cells were malignantly transformed through chronic exposure to either 1 μM As(III) or 50 nM MMA(III), resulting in two independent cell line derivatives termed URO-ASSC and URO-MSC, respectively (7,29). Initial studies showed chronic treatment of UROtsa cells with As(III) resulted in the generation of cells showing cancer phenotypes including hyperproliferation, anchorage-independent growth and tumor formation after heterotransplantation into nude mice (29). Similar findings were independently obtained by Bredfeldt et al. after exposing UROtsa cells to 50 nM MMA(III). After 24 weeks of treatment (URO-MSC24), cells showed anchorage-independent growth and hyperproliferation but did not form tumors when injected into severe combined immunodeficiency mice. After 52 weeks of MMA(III) treatment, these cells (URO-MSC52) acquired the ability to form tumors in SCID mice in addition to exhibiting anchorage-independent growth (7). These UROtsa cell lines provide an unique model in which to study the epigenomic events that occur during arsenical-mediated tumorigenesis.
To study the epigenetic effects that occur during As(III)- and MMA(III)-induced malignant transformation, we employed an epigenomic scanning approach using a human promoter microarray platform. In this study, we show that the transformation of UROtsa cells with both As(III) and MMA(III) is associated with changes in histone H3 acetylation patterns. Additionally, the promoters that lost this permissive histone modification could be associated with an increase in DNA methylation in these regions and a repression of gene expression. Conversely, promoters that showed an increase in histone acetylation also revealed an increase in gene expression. Thus, these multilayered epigenetic changes probably play a functional role since the expression of the associated genes is linked to the epigenetic landscape present in their promoter regions. Taken together, these data provide a genome-wide view of the epigenetic changes associated with chronic arsenical exposure and provide new gene targets that participate in arsenical-induced tumorigenesis.
Materials and methods
Cell culture
UROtsa and URO-ASSC cells were kindly provided by the laboratory of Donald and Mary Ann Sens. URO-MSC24 and URO-MSC52 cell lines were provided by the laboratory of A.J.G. All cells were cultured in Dulbecco’s modified Eagle’s medium (Cellgro, Herndon, VA) supplemented with 5% vol/vol fetal bovine serum (Omega Scientific, Tarzana, CA) and 1% vol/vol penicillin–streptomycin (Cellgro) and maintained in 150 cm2 culture flasks (Greiner Bio-One, Monroe, NC) at 37°C with 5% CO2 as described by Bredfeldt et al. (7).
Drug treatment of URO-ASSC cells
URO-ASSC cells were treated with 5-aza-2′-deoxycytidine (5-aza-dCyd), trichostatin A (TSA) or both of these drugs as described previously (30). For 5-aza-dCyd treatment, cells were treated with 10 μM 5-aza-dCyd (Sigma, St Louis, MO) for 96 total hours with media and 5-aza-dCyd replaced after 48 h. TSA treatments were performed using 300 nM TSA (Sigma) for 24 h. Combination treatment with both 5-aza-dCyd and TSA was performed following the procedure as described previously for 5-aza-dCyd treatment with TSA added for the final 24 h.
Chemicals
Sodium arsenite, phenylmethylsulphonyl fluoride, aprotinin and pepstatin A were obtained from Sigma–Aldrich (St Louis, MO). Diiodomethylarsine [MMA(III) iodide, CH3AsI2] was prepared by the Synthetic Chemistry Facility Core (Southwest Environmental Health Sciences Center, Tucson, AZ) using the method of Millar et al. (31).
Isolation of nucleic acids
Nucleic acids were isolated as described previously (32). All samples were quantified using absorbance at 260 nm using the NanoDrop 1000 Spectrophotometer (NanoDrop, Wilmington, DE).
Chromatin immunoprecipitation
Chromatin immunoprecipitations (ChIPs) were performed as described previously (33). Briefly, cells were treated with 1% formaldehyde for 10 min to cross-link DNA and protein. Cells were then scraped from culture plates in Hank’s balanced salt solution (Cellgro) containing the protease inhibitors phenylmethylsulphonyl fluoride, aprotinin and pepstatin A. Resulting DNA–protein complexes were sonicated and subjected to gel electrophoresis to ensure proper sonication. A portion of the product was removed for later analysis as input DNA. The remaining portion was precleared using protein A Sepharose GL-4B beads (GE Healthcare, Piscataway, NJ) and incubated overnight with an antibody directed toward a specific histone modification. Antibodies against acetylated histone H3 and trimethylated histone H3 lysine-27 were purchased from Upstate/Millipore (Billerica, MA). Antibodies against dimethylated histone H3 lysine-4 and dimethylated histone H3 lysine-9 were purchased from Abcam (Cambridge, MA). After incubation, the bound DNA was immunoprecipitated, washed and treated with 5 M NaCl to reverse DNA–protein cross-links, after which protein was digested with proteinase K (Fermentas, Glen Burnie, MD). Immunoprecipitated and input DNA samples were purified using the PCR Purification Kit (Qiagen, Valencia, CA) and were quantified using the Quant-IT Picogreen dsDNA detection kit (Invitrogen, Carlsbad, CA). Fluorescence was measured using the Biotek FLx800 microplate reader (Biotek,Winooski, VT).
Human promoter microarray
Primers for the human promoter microarray probes were obtained from the Whitehead Institute (Cambridge, MA) (34). Microarray probes were generated by adding 100 ng genomic DNA from normal, human mononuclear cells to 45 μl of PCR master mix (Eppendorf, Hamburg, Germany) with polymerase chain reaction (PCR) primers (20 pmoles each) added. PCRs were performed in a 96-well format (ABgene, Rochester, NY) using MJ thermal cyclers (MJ Research, Waltham, MA). After completion of PCR, a 3 μl aliquot of PCR product was analyzed by gel electrophoresis in a 96-well format using the Invitrogen 2% agarose E-Gel 96 system (Invitrogen). Remaining product was then purified using the QIAquick 96 PCR Purification Kit (Qiagen). After clean up, PCR products were quantified. DNA was then lyophilized and resuspended in 10 μl 3× sodium chloride-sodium citrate buffer for printing onto activated microarray slides (Corning, Lowell, MA) using the OmniGrid Robot (Gene Machines, San Carlos, CA).
Promoter microarray hybridization
Equal amounts of input and ChIP DNA (100 ng) were amplified using the BioPrime Array CGH Genomic Labeling Module (Invitrogen) according to the modified manufacturer’s protocol. Resulting amplified DNA was purified using the PCR Purification Kit (Qiagen) and quantified, after which equal amounts of input and ChIP DNA (1 μg) were subjected to another round of amplification using the BioPrime Array CGH Genomic Labeling Module (Invitrogen), this time incorporating cyanine-labeled deoxyuridine triphosphate (GE Healthcare). Input DNA was labeled with cyanine-5, whereas the immunoprecipitated DNA was labeled with cyanine-3. Labeled DNA was again purified using the PCR Purification Kit (Qiagen) and quantified to ensure proper amplification and incorporation of labeled deoxyuridine triphosphate using the microarray function of the NanoDrop 1000 Spectrophotometer (NanoDrop). Input and ChIP DNA were then combined, to which was added human COT-I DNA (Invitrogen) and yeast transfer RNA (Invitrogen), and the resulting mix was lyophilized to dryness. Dried target was then resuspended in Domino Oligo Hybridization Buffer (Gel Company, San Francisco, CA), denatured and applied to the human promoter microarray slide. Slides were incubated for 16 h, washed and scanned using an Axon GenePix 4000B microarray scanner (Axon, Sunnyvale, CA).
ChIPs coupled to real-time PCR
Equal amounts of input and immunoprecipitated DNA (1 ng) were added to IQ supermix (Bio-Rad, Hercules, CA), promoter-specific primers and fluorescent probes (Roche, Basel, Switzerland) and were analyzed using the ABI 7500 Real-Time Detection System (Applied Biosystems, Foster City, CA). Values were calculated using the delta Ct method normalizing to the respective input for each sample. Primers were designed using Primer3 in conjunction with ProbeFinder version 2.40 software (Roche Applied Science, Basel, Switzerland). Statistics were calculated using an unpaired t-test between each transformed cell line and UROtsa. Primer sequences are available upon request.
Real-time reverse transcription–PCR
Total RNA (250 ng) was converted to complementary DNA according to the manufacturer’s instructions (Applied Biosystems). Converted complementary DNA (10 ng) was added to IQ supermix (Bio-Rad), gene-specific primers and fluorescent probes (Roche) and subjected to real-time PCR analysis using Roche UniversalProbe technology (Roche) using the ABI 7500 Real-Time Detection System (Applied Biosystems). Results were calculated using the delta Ct method normalizing to β-actin expression for each sample. Primers were designed using Primer3 in conjunction with ProbeFinder version 2.40 software (Roche Applied Science). Statistics were calculated using an unpaired t-test between each transformed cell line and UROtsa. Primer sequences are available upon request.
Methylcytosine immunoprecipitation
RNase-treated genomic DNA (15 μg) was sonicated and analyzed using gel electrophoresis to ensure proper sonication. DNA was incubated overnight with an antibody to 5-methylcytosine (Aviva Systems Biology, San Diego, CA). Antibody–DNA complexes were then immunoprecipitated, washed and eluted using the QiaQuick Gel Extraction Kit (Qiagen). DNA was quantified using absorbance at 260 nm on the NanoDrop 1000 Spectrophotometer (NanoDrop). Real-time PCR for these samples was performed as described previously for histone H3 acetylation. Statistics were calculated using an unpaired t-test between each transformed cell line and UROtsa.
Sodium bisulfite sequencing
Sodium bisulfite sequencing was performed as described previously (35). Primers were designed using Methyl Primer Express v1.0 software (Applied Biosystems) to amplify a sequence located with the probe sequence located on the promoter microarray. For each cell line, 12 clones were sequenced with samples exhibiting <80% identity with the predicted sequence or <90% bisulfite conversion removed from analysis. Sequencing data were analyzed using the BiQ Analyzer (36). Primer sequences are available upon request.
Data analysis
All microarray data were processed in R programming environment (37). For normalization of all data, the Linear Models for Microarray Data (Limma) package was used (38). Differentially acetylated elements were identified using statistical approaches as described previously (39). To control for false discovery rate, a multiple testing correction was performed according to the methods described by Benjamini et al. (40).
Results
Histone H3 exhibits altered acetylation patterns in gene promoters
UROtsa cells are a non-tumorigenic cell line model of human urothelium. Independent exposures of UROtsa to As(III) and MMA(III) resulted in two distinct cell line models of arsenical-selected malignant transformation termed URO-ASSC and URO-MSC cell lines, respectively (Figure 1). We used these cell lines to examine whether there were consistent changes in the histone H3 acetylation state linked to arsenical-induced malignant transformation. In order to examine the effect of arsenical selection on histone modification state, we coupled ChIP to microarray hybridizations that probed 13 000 human gene promoters. A minimum of three independent experiments were analyzed using the promoter microarrays in order to confirm reproducibility and minimize the number of false positives. To account for false discovery rate, P-values were adjusted according to the methods described by Benjamini et al. (40).
Fig. 1.
Arsenical-induced malignant transformation results in histone H3 acetylation changes in gene promoters. UROtsa cells were exposed to As(III) (URO-ASSC) and MMA(III) (URO-MSC24, 52) with exposure time for each cell line described. Tumorigenicity classification is based on the ability of each cell line to form tumors when heterotransplanted into mice. ChIP experiments were coupled to human promoter microarrays in order to assess H3 acetylation state in UROtsa parent and the arsenical-exposed cell lines. Numbers shown represent the number of elements on the microarray showing significantly less (hypoacetylated) or significantly more (hyperacetylated) histone acetylation relative to the parental UROtsa cells (adjusted P-value < 0.05). Adjusted P-values were calculated according to the methods described by Benjamini et al. (40).
When compared with the UROtsa parental cell line, each transformed cell line showed changes in histone H3 acetylation in a select number of promoters (Figure 1). URO-MSC52 and URO-ASSC, the two cell lines able to form tumors when heterotransplanted into animals, displayed a similar number of promoters altered in a statistically significant fashion; 56 promoters in URO-MSC52 and 53 promoters in URO-ASSC (adjusted P < 0.05). When examining the promoter regions affected, it was striking that of the promoters affected in URO-MSC52 and URO-ASSC, 17 were altered in both cell lines. To determine if this number of changes could have co-occurred randomly, observed/expected ratios were calculated. If the changes were to have simply occurred as a result of random chance, one would expect that 0.26 elements from the microarray would be altered in both URO-ASSC and URO-MSC52 cell lines. The number of co-occurring changes observed (17) is 65 times higher than would have been expected by random chance, suggesting that the observed changes were arsenic specific.
The number of promoters altered in URO-MSC24 in relation to URO-MSC52 provides additional insights since it is an antecedent to URO-MSC52 and differs only by the duration of the chronic MMA exposure. In URO-MSC24, only one gene promoter showed a statistically significant change (decrease) in histone H3 acetylation, the promoter for ZSCAN12, a zinc finger protein from the Kruppel family of transcription factors (41). Significant changes in the ZSCAN12 gene promoter were also seen in URO-MSC52, as well as URO-ASSC cells. The data in Figure 2 further show that the gene promoters that show statistically significant changes that are common to both URO-MSC52 and URO-ASSC are already beginning to show these changes in URO-MSC24.
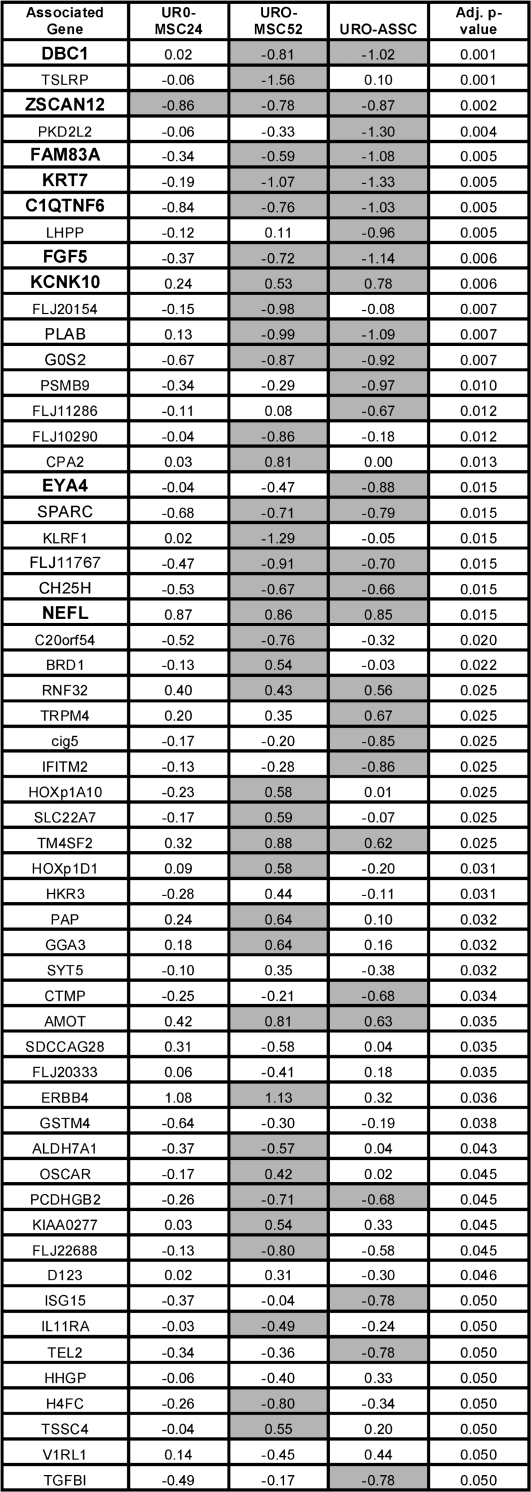
Fig. 2.
Arsenicals selectively alter histone H3 acetylation. Associated gene names of the significantly affected promoters are shown with promoters validated by real-time PCR shown in bold. Values represent log 2 ratios of each cell line derivative relative to UROtsa. Negative values are representative of hypoacetylated elements, whereas positive values represent hyperacetylated promoters. Promoters are sorted by adjusted P-value relative to differences between all three treated cell lines and UROtsa. All changes represented as significant for that particular cell line when compared with UROtsa (adjusted P-value < 0.05) are shaded. Adjusted P-values were calculated according to the methods described by Benjamini et al. (40). For a complete list of promoters exhibiting altered histone acetylation, refer to supplementary Figure 1 (available at Carcinogenesis Online).
Figure 2 shows the 57 statistically significant gene promoters sorted by adjusted P-value relative to differences between the three arsenical-exposed cell lines and the parental UROtsa and includes all 17 of the gene promoters common to both URO-MSC52 and URO-ASSC. The remaining 43 statistically significant genes (adjusted P < 0.05) are provided in supplementary Figure 1 (available at Carcinogenesis Online). The values are the log 2 ratios of histone acetylation change for that cell line compared with the UROtsa parent cell line; the shaded values indicate the gene promoters that show statistically significant changes in the given cell lines. Interestingly, the most significant gene promoter identified was for DBC1, which stands for ‘deleted in bladder cancer’, a gene with tumor suppressor function whose activity is frequently lost in bladder as well as other cancers (42,43). Taken together, these data indicate that arsenical-selected malignant transformation may result in non-random, gene-specific changes in histone H3 acetylation and that the genes associated with these promoters could be important participants in arsenical-mediated malignant transformation.
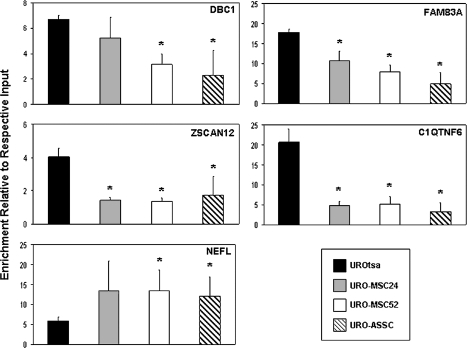
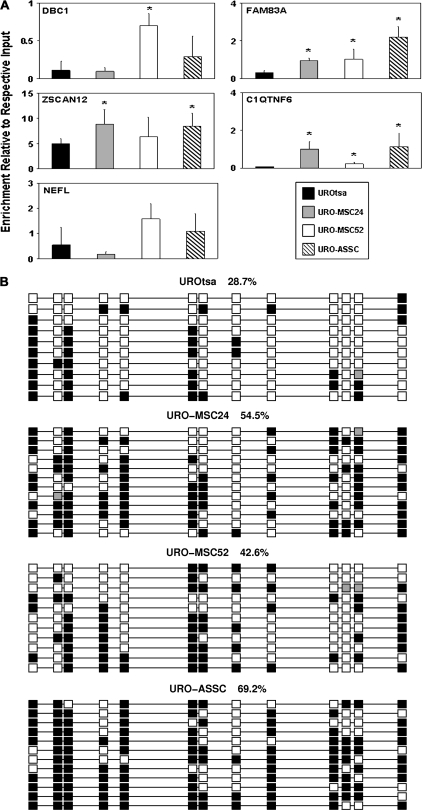
In order to confirm the promoter microarray results obtained, we validated a subset of these genes using the ChIP DNA coupled to real-time PCR. Five promoter regions that showed differential acetylation were selected for confirmatory real-time PCR analysis. Four of these regions (DBC1, FAM83A, ZSCAN12 and C1QTNF6) showed hypoacetylated histone H3 in the transformed cell lines relative to UROtsa, whereas one showed hyperacetylation (NEFL), roughly reflecting the ratio of hypoacetylated to hyperacetylated elements observed in the microarray analysis. PCR primers were designed to amplify a region located within the microarray probe sequence. Equal amounts of input DNA and acetyl-H3-immunoprecipitated DNA were analyzed using promoter-specific primers with the results displayed as the enrichment of each individual sample over its respective input DNA according to the delta Ct method (Figure 3; supplementary Figure 2 is available at Carcinogenesis Online). The enrichment of each promoter examined via real-time PCR correlated well with what was observed using the human promoter microarray, showing that the results obtained were reliable. The promoter region of GAPDH, an ubiquitously expressed housekeeping gene whose promoter is known to be enriched for acetylated histone H3, was used as a positive control for successful ChIP. All samples showed similar enrichment levels for GAPDH (supplementary Figure 3 is available at Carcinogenesis Online). Overall, the real-time PCR analysis of selected promoters confirmed the results obtained from the promoter microarray experiments.
Fig. 3.
Real-time PCR confirms the changes in histone H3 acetylation observed using the human promoter microarray. ChIP experiments were coupled to real-time PCR to confirm the histone H3 acetylation levels present with the promoter regions of selected genes in UROtsa, URO-MSC24, URO-MSC52 and URO-ASSC cells. Mean fold enrichment over the respective input DNA for three independent samples is represented by changes along the y-axis. Error bars are representative of the standard deviation of three independent experiments. Changes significantly (P < 0.05) different than UROtsa are indicated by *.
Gene expression correlates with the histone H3 acetylation pattern
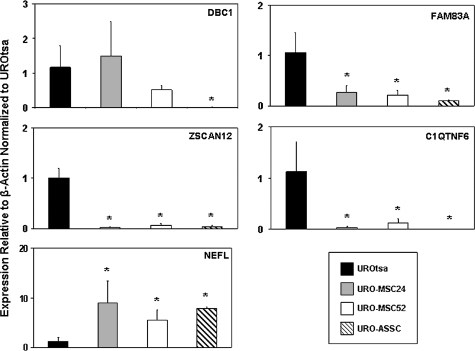
To determine if the observed changes in histone acetylation are linked to changes in gene expression, we analyzed these genes by quantitative real-time reverse transcription–PCR. The genes DBC1, FAM83A, ZSCAN12 and C1QTNF6 whose promoters showed a decrease in H3 acetylation also showed a corresponding decrease in associated gene expression (Figure 4). In addition, NEFL, a gene whose promoter region showed a significant increase in histone acetylation in the arsenical-selected cell lines, also showed an increase in gene expression in these malignantly transformed cells. Taken together, these data suggest that the changes observed in histone acetylation levels are probably closely linked to changes in gene expression in malignantly transformed cell lines, thereby potentially playing a functional role in a cancer-specific fashion.
Fig. 4.
Gene expression levels correlate with changes in histone acetylation. Expression levels were examined for genes that were verified to have differentially acetylated promoters. Quantitative real-time reverse transcription–PCR was performed on three independent samples for each cell line with the average expression value shown. Expression levels presented on the y-axis are relative to β-actin expression and normalized to UROtsa. Error bars are representative of the standard deviation. Changes significantly (P < 0.05) different than UROtsa are indicated by *.
Since histone acetylation state is an important component that affects gene transcription and is a highly dynamic modification that may be rapidly changed by acute arsenical exposure, we looked to see if 24 h exposure altered the gene expression of a subset of identified genes. To do so, UROtsa cells were exposed to either 50 nM MMA(III) or 1 μM As(III) for 24 h and analyzed for gene expression by real-time reverse transcription–PCR. After exposure, none of the arsenical target genes examined showed changes that approached those seen in the malignantly transformed cell lines, suggesting that these genes are not part of an early arsenical response (supplementary Figure 4 is available at Carcinogenesis Online).
Aberrant DNA methylation occurs in the differentially acetylated promoter regions
As histone deacetylation has been mechanistically linked to DNA hypermethylation, we wanted to determine if DNA methylation was increased in the same gene promoter regions that showed changes in histone H3 acetylation. We isolated genomic DNA and performed immunoprecipitation experiments with an antibody directed toward 5-methylcytosine methylated DNA immunoprecipitation (MeDIP) and coupled this with real-time PCR to assess relative DNA methylation levels. The same primers that were used to examine histone H3 acetylation were used to analyze relative methylation. The DNA methylation level observed in each of these hypoacetylated genes was inversely correlated with the histone acetylation levels at the same locus in the promoter regions of DBC1, FAM83A, ZSCAN12 and C1QTNF6 (Figure 5A). Although not all these changes reached a statistically significant level of P < 0.05 when using an unpaired t-test, the pattern of increased DNA methylation corresponding with decreased histone H3 acetylation was evident. Conversely, DNA hypomethylation could be associated histone hyperacetylation. DNA methylation levels in the hyperacetylated promoter region of NEFL, however, showed no significant change in DNA methylation between the malignantly transformed cell lines and UROtsa. The promoter region of GAPDH, a gene whose promoter is known to be unmethylated, was used as a negative control. All samples showed similar low levels of enrichment for GAPDH (supplementary Figure 3 is available at Carcinogenesis Online). Taken together, these data allow us to conclude that aberrant DNA methylation in selected promoter regions is associated with a decrease in H3 acetylation and decreased gene expression during arsenical-induced malignant transformation.
Fig. 5.
DNA methylation is altered in selected gene promoter regions. (A) Relative DNA methylation levels were measured for the same promoters that were found to be hypoacetylated. MeDIP was performed on three independent samples from each cell line and coupled to real-time PCR with the mean enrichment relative to respective input shown along the y-axis. Error bars are representative of the standard deviation. Changes significantly (P < 0.05) different than UROtsa are indicated by *. (B) Sodium bisulfite sequencing confirms MeDIP results. Sodium bisulfite sequencing was performed to examine the DNA methylation levels and pattern in the promoter region of ZSCAN12. Each column represents an individual CpG site, whereas each row is representative of an individual sequenced clone. Spacing is representative of the underlying DNA sequence with the methylation state at each CpG dinucleotide [methylated (filled squares); unmethylated (open squares) and poor sequence (gray squares)] shown. Numbers describe the methylation level for each cell line in the region shown. Clones were sorted for presentation.
To verify and extend the DNA methylation data obtained using MeDIP, we performed clonal sodium bisulfite sequencing. We chose to examine a region located within the promoter of ZSCAN12 (Figure 5B). Results show that the pattern of DNA methylation observed, UROtsa and URO-MSC52 having lower levels of DNA methylation than URO-MSC24 and URO-ASSC, is consistent with MeDIP PCR results. Taken together, these results suggest that the data obtained using MeDIP coupled to real-time PCR are reliable and the promoter region of ZSCAN12 is hypermethylated in the exposed cell lines.
Histone H3 methylation is altered in a subset of promoter regions examined
To gain a more comprehensive knowledge of the histone modification profile in each of the promoter regions examined, we performed ChIPs to examine the levels of the repressive histone modifications histone H3 lysine-27 trimethylation and histone H3 lysine-9 dimethylation as well as the permissive histone modification histone H3 lysine-4 dimethylation.
While there were statistically significant small changes in histone H3 lysine-27 trimethylation (supplementary Figure 5 is available at Carcinogenesis Online), histone H3 lysine-9 dimethylation (supplementary Figure 6 is available at Carcinogenesis Online) and histone H3 lysine-4 dimethylation (supplementary Figure 7 is available at Carcinogenesis Online) in a subset of promoter regions, the majority of the promoters examined did not exhibit altered levels of histone methylation. It is important to note that while many of these regions do not have significantly altered levels of histone H3 methylation, none of the significant changes measured was discordant with histone H3 acetylation. Although clearly not the case in all the promoter regions measured, examination of the levels of histone H3 histone H3 lysine-27 trimethylation, histone H3 lysine-9 dimethylation and histone H3 lysine-4 dimethylation suggests that, in addition to histone acetylation and DNA methylation, histone H3 methylation may be altered in select promoter regions, providing further evidence of epigenetic remodeling during arsenical-induced malignant transformation.
Pharmacologic reactivation of silenced genes confirms an epigenetic mechanism
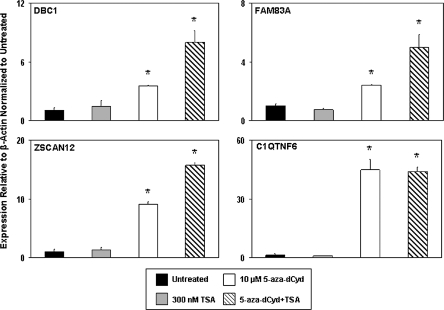
Epigenetic modifications are targets of cancer therapy as they are inherently reversible. To confirm that the differential epigenetic profile in the promoter regions of the downregulated genes DBC1, FAM83A, ZSCAN12 and C1QTNF6 was playing a functional role in controlling gene expression, we treated the malignantly transformed URO-ASSC cell line with the DNA methyltransferase inhibitor 5-aza-dCyd, the histone deacetylase inhibitor TSA or a combination treatment with both. Treatment with 10 μM 5-aza-dCyd was performed for 96 h, whereas treatment with 300 nM TSA was performed for 24 h. After the completion of treatment, gene expression levels were measured using real-time reverse transcription–PCR. Each experiment was performed in triplicate to ensure reproducibility.
Pharmacologic reactivation with the epigenetic modifying drugs suggests that the expression of each of the genes examined is mediated by epigenetic mechanisms in URO-ASSC cells (Figure 6). Treatment of these cells with TSA alone did not significantly change the expression of any of the hypoacetylated genes examined. Conversely, treatment with 5-aza-dCyd alone significantly increased the expression of each of these genes (P < 0.001). Dual treatment with 5-aza-dCyd and TSA resulted in an additive effect in terms of increasing gene expression. These data suggest that both of these epigenetic marks probably contribute and that epigenetic remodeling has a functional effect in controlling the expression level of associated genes.
Fig. 6.
Pharmacologic reactivation of gene expression using epigenetic inhibitors. URO-ASSC cells were treated with 10 μM 5-aza-dCyd, 300 nM TSA or both. Gene expression levels were measured using real-time reverse transcription–PCR with the values on the y-axis of each graph representing expression levels relative to β-actin and normalized to the untreated control. Error bars are representative of the standard deviation of three independent experiments. Changes significantly (P < 0.001) different than the untreated control are indicated by *.
Discussion
Arsenic exposure is associated with the etiology of various types of cancer including cancer of the bladder. The mechanisms by which arsenicals result in malignancy include the alteration of signaling pathways and indirect DNA damage, although they are only mildly mutagenic (12–19,24,25). We have demonstrated herein that malignant transformation of human urothelial cells by arsenicals is also associated with changes in histone acetylation and DNA methylation in gene promoter regions. Independent exposures to As(III) and MMA(III) resulted in a non-random perturbation of the epigenomic landscape and allowed us to define genes that may be important in tumorigenesis associated with arsenical exposure. Taken together, these data suggest that epigenetic remodeling is linked to malignant transformation associated with chronic, environmentally relevant exposure to As(III) and MMA(III).
Exposure to heavy metals, in particular nickel, has been shown to alter the histone code (21–23). Our data suggest that this phenomenon also occurs as a result of arsenical exposure. This is supported by a recent study suggesting that exposure to As(III) results in the alteration of multiple methylation marks on histone H3 in a global fashion (44). Microarray experiments showed numerous differentially acetylated promoter regions between the parental and malignantly transformed cell lines. Although many promoter regions were found to be differentially acetylated, it is probably that due to the sensitivity of the microarray and the stringency of our statistical criteria, we are potentially underestimating the number of changes in histone H3 acetylation in these cell lines since we were able to identify statistically significant changes in the URO-MSC24 cell lines utilizing real-time PCR that had not reached statistical significance by microarray.
Aberrant DNA methylation is a hallmark of nearly all types of cancer including bladder tumors and has been functionally linked to histone acetylation (45–48). We performed MeDIP coupled to real-time PCR to determine if relative DNA methylation levels were changed in the promoter regions of genes showing significant decreases in histone acetylation in the exposed cells. Previous studies in rodents demonstrated a loss in genomic DNA methylation in hepatocytes exposed to arsenic (24,25). The data presented here compliment these earlier studies as human tumors generally exhibit genomic hypomethylation while possessing focal regions of DNA hypermethylation, notably in gene promoter regions.
Importantly, the relative amount of DNA methylation increased with decreasing histone acetylation levels in the same promoters, providing additional evidence of the inverse relationship between these two epigenetic marks and further suggesting functional significance of these epigenetic changes. These data suggest the possibility of a mechanism of action of arsenicals whereby they induce yet unknown signaling pathways, resulting in aberrant DNA methylation in critical promoter regions. Methylated DNA can recruit histone deacetylase complexes through the protein MeCP2, resulting in a subsequent loss of histone acetylation in the promoter regions exhibiting increased DNA methylation (47,48). The result of such a mechanism, DNA hypermethylation with a concomitant loss of histone acetylation and corresponding gene expression, is the predominant pattern observed in this study.
This study has uncovered numerous genes that act as epigenetic targets of As(III) and MMA(III) during malignant transformation. These changes occurred in two independent models of arsenical-induced malignant transformation; thus, they may potentially play a role in this conversion, providing targets for future study and therapeutic design. Many of these genes are newly annotated and their cellular functions are not clear. Two genes that are particularly intriguing are DBC1 and ZSCAN12.
Loss of heterozygosity on chromosome 9q is the most common genetic aberration in transitional cell carcinoma, suggesting the presence of a tumor suppressor gene (42). Analysis of this region revealed the gene to be DBC1 or deleted in bladder cancer 1. DBC1 is downregulated by genetic and epigenetic mechanisms in multiple cancers including non-small-cell lung cancer and bladder cancer (42,43). DBC1 is transcriptionally silenced in some bladder tumor cell lines and this silencing can be reversed after treatment with 5-aza-dCyd, suggesting an epigenetic mechanism of repression (42). The mechanism of action of DBC1 may involve regulation of cellular proliferation as the reintroduction of this gene induces cell death in bladder cells (49). This possible mechanism is reinforced as URO-ASSC and URO-MSC cell lines exhibit an increased rate of proliferation compared with UROtsa (7,29). The detection of a gene frequently seen to be dysregulated in bladder tumor cells suggests the possibility that this study may not only be important in uncovering genes related to arsenical-induced malignant transformation but also those playing a role in the conversion of a bladder cell to a neoplastic phenotype.
ZSCAN12 is a member of the Kruppel family of zinc fingers proteins. While little is known about this particular gene, zinc fingers typically act as transcription factors, therefore it stands to reason that the downregulation of this gene by arsenicals could affect other genes, hence amplifying its effects. Additionally, the loss of histone acetylation and associated gene expression of ZSCAN12 is an early event in arsenic selection and may merit further evaluation as a biomarker of pre-cancerous lesions associated with arsenical exposure in the bladder.
In conclusion, this study suggests that the carcinogenic activity of arsenicals may be mediated by the disruption of normal epigenetic control at specific loci. The observed epigenetic changes are correlated with the expression of associated genes, thereby uncovering a collection of genes probably to play an important role in malignant transformation associated with arsenicals. To the best of our knowledge, the data presented herein is the first to show that epigenomic remodeling occurs at specific promoters during arsenical-induced malignant transformation in human cells, providing further understanding of the molecular etiology of arsenical-induced bladder carcinogenesis as well as describing critical genes that may play a role in this process.
Supplementary material
Supplementary Figures 1–7 can be found at http://carcin.oxfordjournals.org/
Funding
The National Institute of Environmental Health Sciences (ES06694), National Institutes of Health (CA23074), Bio-5 Institute to Genomics Shared Service; National Institute of Environmental Health Sciences (CA127989) to B.W.F. and T.J.J.; National Institute of Environmental Health Sciences Training Grant (ES007091) to K.E.
Supplementary Material
Acknowledgments
The authors would like to thank Dr Donald Sens and Dr Mary Ann Sens for kindly providing the UROtsa and URO-ASSC cell lines. In addition, the authors would like to thank Dr Ryan Wozniak, Dr Tiffany Bredfeldt and Jocelyn Tye for assistance on various aspects of this project. The authors thank Dr George Watts and the University of Arizona Genomics Shared Service at the Arizona Cancer Center for allowing the use of their equipment, facilities and expertise on the microarray studies.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- 5-aza-dCyd
5-aza-2′-deoxycytidine
- MeDIP
methylated DNA immunoprecipitation
- MMA
monomethylarsonous acid
- PCR
polymerase chain reaction
- TSA
trichostatin A
References
- 1.Aposhian HV. Enzymatic methylation of arsenic species and other new approaches to arsenic toxicity. Annu. Rev. Pharmacol. Toxicol. 1997;37:397–419. doi: 10.1146/annurev.pharmtox.37.1.397. [DOI] [PubMed] [Google Scholar]
- 2.Zakharyan RA, et al. Enzymatic methylation of arsenic compounds. VII. Monomethylarsonous acid (MMAIII) is the substrate for MMA methyltransferase of rabbit liver and human hepatocytes. Toxicol. Appl. Pharmacol. 1999;158:9–15. doi: 10.1006/taap.1999.8687. [DOI] [PubMed] [Google Scholar]
- 3.Styblo M, et al. Comparative toxicity of trivalent and pentavalent inorganic and methylated arsenicals in rat and human cells. Arch. Toxicol. 2000;74:289–299. doi: 10.1007/s002040000134. [DOI] [PubMed] [Google Scholar]
- 4.Aposhian HV, et al. Occurrence of monomethylarsonous acid in urine of humans exposed to inorganic arsenic. Chem. Res. Toxicol. 2000;13:693–697. doi: 10.1021/tx000114o. [DOI] [PubMed] [Google Scholar]
- 5.Le XC, et al. Speciation of key arsenic metabolic intermediates in human urine. Anal. Chem. 2000;72:5172–5177. doi: 10.1021/ac000527u. [DOI] [PubMed] [Google Scholar]
- 6.Styblo M, et al. The role of biomethylation in toxicity and carcinogenicity of arsenic: a research update. Environ. Health Perspect. 2002;110(suppl. 5):767–771. doi: 10.1289/ehp.110-1241242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bredfeldt TG, et al. Monomethylarsonous acid induces transformation of human bladder cells. Toxicol. Appl. Pharmacol. 2006;216:69–79. doi: 10.1016/j.taap.2006.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen CJ, et al. Arsenic and cancers. Lancet. 1988;1:414–415. doi: 10.1016/s0140-6736(88)91207-x. [DOI] [PubMed] [Google Scholar]
- 9.Chen SL, et al. Trace element concentration and arsenic speciation in the well water of a Taiwan area with endemic blackfoot disease. Biol. Trace Elem. Res. 1995;48:263–274. doi: 10.1007/BF02789408. [DOI] [PubMed] [Google Scholar]
- 10.Marshall G, et al. Fifty-year study of lung and bladder cancer mortality in Chile related to arsenic in drinking water. J. Natl Cancer Inst. 2007;99:920–928. doi: 10.1093/jnci/djm004. [DOI] [PubMed] [Google Scholar]
- 11.Hopenhayn-Rich C, et al. Bladder cancer mortality associated with arsenic in drinking water in Argentina. Epidemiology. 1996;7:117–124. doi: 10.1097/00001648-199603000-00003. [DOI] [PubMed] [Google Scholar]
- 12.Eblin KE, et al. Mitogenic signal transduction caused by monomethylarsonous acid in human bladder cells: role in arsenic-induced carcinogenesis. Toxicol Sci. 2007;95:321–330. doi: 10.1093/toxsci/kfl160. [DOI] [PubMed] [Google Scholar]
- 13.Simeonova PP, et al. Quantitative relationship between arsenic exposure and AP-1 activity in mouse urinary bladder epithelium. Toxicol Sci. 2001;60:279–284. doi: 10.1093/toxsci/60.2.279. [DOI] [PubMed] [Google Scholar]
- 14.Hei TK, et al. Mutagenicity of arsenic in mammalian cells: role of reactive oxygen species. Proc. Natl Acad. Sci. USA. 1998;95:8103–8107. doi: 10.1073/pnas.95.14.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynn S, et al. Arsenite retards DNA break rejoining by inhibiting DNA ligation. Mutagenesis. 1997;12:353–358. doi: 10.1093/mutage/12.5.353. [DOI] [PubMed] [Google Scholar]
- 16.Schwerdtle T, et al. Induction of oxidative DNA damage by arsenite and its trivalent and pentavalent methylated metabolites in cultured human cells and isolated DNA. Carcinogenesis. 2003;24:967–974. doi: 10.1093/carcin/bgg018. [DOI] [PubMed] [Google Scholar]
- 17.Eblin KE, et al. Arsenite and monomethylarsonous acid generate oxidative stress response in human bladder cell culture. Toxicol. Appl. Pharmacol. 2006;217:7–14. doi: 10.1016/j.taap.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Jacobson-Kram D, et al. The reproductive effects assessment group’s report on the mutagenicity of inorganic arsenic. Environ. Mutagen. 1985;7:787–804. doi: 10.1002/em.2860070515. [DOI] [PubMed] [Google Scholar]
- 19.Wang TC, et al. Trivalent arsenicals induce lipid peroxidation, protein carbonylation, and oxidative DNA damage in human urothelial cells. Mutat. Res. 2007;615:75–86. doi: 10.1016/j.mrfmmm.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Smith AH, et al. Marked increase in bladder and lung cancer mortality in a region of Northern Chile due to arsenic in drinking water. Am. J. Epidemiol. 1998;147:660–669. doi: 10.1093/oxfordjournals.aje.a009507. [DOI] [PubMed] [Google Scholar]
- 21.Broday L, et al. Nickel compounds are novel inhibitors of histone H4 acetylation. Cancer Res. 2000;60:238–241. [PubMed] [Google Scholar]
- 22.Lee YW, et al. Carcinogenic nickel silences gene expression by chromatin condensation and DNA methylation: a new model for epigenetic carcinogens. Mol. Cell. Biol. 1995;15:2547–2557. doi: 10.1128/mcb.15.5.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ke Q, et al. Alterations of histone modifications and transgene silencing by nickel chloride. Carcinogenesis. 2006;27:1481–1488. doi: 10.1093/carcin/bgl004. [DOI] [PubMed] [Google Scholar]
- 24.Zhao CQ, et al. Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression. Proc. Natl Acad. Sci. USA. 1997;94:10907–10912. doi: 10.1073/pnas.94.20.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, et al. Chronic inorganic arsenic exposure induces hepatic global and individual gene hypomethylation: implications for arsenic hepatocarcinogenesis. Carcinogenesis. 2004;25:1779–1786. doi: 10.1093/carcin/bgh161. [DOI] [PubMed] [Google Scholar]
- 26.Marsit CJ, et al. Carcinogen exposure and gene promoter hypermethylation in bladder cancer. Carcinogenesis. 2006;27:112–116. doi: 10.1093/carcin/bgi172. [DOI] [PubMed] [Google Scholar]
- 27.Petzoldt JL, et al. Immortalisation of human urothelial cells. Urol. Res. 1995;23:377–380. doi: 10.1007/BF00698738. [DOI] [PubMed] [Google Scholar]
- 28.Rossi MR, et al. The immortalized UROtsa cell line as a potential cell culture model of human urothelium. Environ. Health Perspect. 2001;109:801–808. doi: 10.1289/ehp.01109801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sens DA, et al. Inorganic cadmium- and arsenite-induced malignant transformation of human bladder urothelial cells. Toxicol Sci. 2004;79:56–63. doi: 10.1093/toxsci/kfh086. [DOI] [PubMed] [Google Scholar]
- 30.Novak P, et al. Epigenetic inactivation of the HOXA gene cluster in breast cancer. Cancer Res. 2006;66:10664–10670. doi: 10.1158/0008-5472.CAN-06-2761. [DOI] [PubMed] [Google Scholar]
- 31.Millar I, et al. Methyldiiodoarsine. In: Rochow E.G., editor. Inorganic Synetheses. Vol. 6. New York: McGraw-Hill Book Company, Inc; 1960. pp. 113–115. [Google Scholar]
- 32.Oshiro MM, et al. Epigenetic silencing of DSC3 is a common event in human breast cancer. Breast Cancer Res. 2005;7:R669–R680. doi: 10.1186/bcr1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oshiro MM, et al. Mutant p53 and aberrant cytosine methylation cooperate to silence gene expression. Oncogene. 2003;22:3624–3634. doi: 10.1038/sj.onc.1206545. [DOI] [PubMed] [Google Scholar]
- 34.Odom DT, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clark SJ, et al. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bock C, et al. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–4068. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- 37.R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria: 2007. [Google Scholar]
- 38.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 39.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 40.Benjamini Y, et al. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 41.Lee PL, et al. Three genes encoding zinc finger proteins on human chromosome 6p21.3: members of a new subclass of the Kruppel gene family containing the conserved SCAN box domain. Genomics. 1997;43:191–201. doi: 10.1006/geno.1997.4806. [DOI] [PubMed] [Google Scholar]
- 42.Habuchi T, et al. Structure and methylation-based silencing of a gene (DBCCR1) within a candidate bladder cancer tumor suppressor region at 9q32-q33. Genomics. 1998;48:277–288. doi: 10.1006/geno.1997.5165. [DOI] [PubMed] [Google Scholar]
- 43.Izumi H, et al. Frequent silencing of DBC1 is by genetic or epigenetic mechanisms in non-small cell lung cancers. Hum. Mol. Genet. 2005;14:997–1007. doi: 10.1093/hmg/ddi092. [DOI] [PubMed] [Google Scholar]
- 44.Zhou X, et al. Arsenite alters global histone H3 methylation. Carcinogenesis. 2008 doi: 10.1093/carcin/bgn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neuhausen A, et al. DNA methylation alterations in urothelial carcinoma. Cancer Biol. Ther. 2006;5:993–1001. doi: 10.4161/cbt.5.8.2885. [DOI] [PubMed] [Google Scholar]
- 46.Salem C, et al. Progressive increases in de novo methylation of CpG islands in bladder cancer. Cancer Res. 2000;60:2473–2476. [PubMed] [Google Scholar]
- 47.Jones PL, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 48.Nan X, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 49.Wright KO, et al. DBCCR1 mediates death in cultured bladder tumor cells. Oncogene. 2004;23:82–90. doi: 10.1038/sj.onc.1206642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.