Abstract
Transcriptional factor 4 (TCF4), encoding a basic helix-loop-helix transcriptional factor, has recently been demonstrated as a causative gene for Pitt-Hopkins syndrome, a neurodevelopmental disease. Examination of gastric cancers using the restriction landmark genomic scanning technique revealed methylation at a NotI enzyme site in TCF4 intron 8 and further identified CpG dinucleotide hypermethylation in TCF4 exon 1, strongly associated with gene silencing in gastric cancer cell lines. Treatment with 5-aza-2′-deoxycytidine and/or trichostatin A restored TCF4 expression in TCF4-silenced gastric cancer cell lines. Real-time reverse transcription–polymerase chain reaction analysis of 77 paired primary gastric tumor samples revealed that 38% of analyzed tumors had a >2-fold decrease in TCF4 expression compared with adjacent normal-appearing tissue, and the decrease significantly correlated with increased CpG methylation in TCF4 exon 1. Clinicopathologic data showed that decreased TCF4 expression occurred significantly more frequently in intestinal-type (22/37, 59%) than in diffuse-type (7/37, 19%) gastric cancers (P = 0.0004) and likewise more frequently in early (12/18, 67%) than in advanced (17/59, 29%) gastric cancers (P = 0.004). CpG methylation markedly increased with patient age among normal-appearing tissues, suggesting that CpG methylation in gastric mucosa may be one of the earliest events in carcinogenesis of intestinal-type gastric cancers. Furthermore, ectopic expression of TCF4 decreased cell growth in a gastric cancer cell line, and the knock down of TCF4 using small interfering RNA increased cell migration. Based on these results, we propose that the observed frequent epigenetic-mediated TCF4 silencing plays a role in tumor formation and progression.
Introduction
The basic helix-loop-helix (bHLH) family of transcription factors is categorized into distinct classes on the basis of biochemical and functional criteria and each member protein contains an HLH domain composed of two amphipathic helixes separated by a loop and a basic DNA-binding domain (1–3). These proteins can form homodimers and heterodimers with other classes of bHLH proteins through the HLH domain to facilitate binding to DNA (4,5). This basic DNA-binding domain is located N-terminal to the HLH domain and makes specific contacts with consensus DNA sequences known as E-boxes (CANNTG) (6). E-box sequences have been found in the promoters of a wide variety of genes, driving their specific activation (7,8). Among the several classes of bHLH families, the class I transcription factors (also called E proteins) are critical regulators in a diverse array of biological processes such as cell growth, differentiation, tissue-specific gene expression and programmed cell death (9–11).
The Transcription factor 4 (TCF4; also known as ITF2, E2-2, ME2 or SEF2) gene product is a member of the class I bHLH family together with the TCF12 (HEB) gene product and the alternatively spliced products E12 and E47 of TCF3 (ITF1, E2A) (9). This gene should not be confused with T-cell transcription factor 4 on human chromosome 10q25.3, which was previously termed TCF4, but is now designated as TCF7L2. Dimerization of TCF4 with other classes of bHLH proteins regulates tissue-specific gene expression through E-box sites and this process, in part, controls differentiation and proliferation in a wide range of cell types such as myocytes (12), osteoblasts (13), B and T lymphocytes (14) and neuronal cells (15). A previous study showed that TCF4 is a downstream target of the WNT/β-catenin/TCF pathway and, like cMYC and cyclin D1, functions as an oncogene when deregulated in human colon cancers (16). In contrast, it has been shown that the enforced expression of TCF4 suppresses the colony-forming efficiency of cells in several cell lines, suggesting that the gene acts as a negative regulator of cell proliferation (11). Very recently, genetic studies demonstrated that loss of one copy of TCF4 causes Pitt-Hopkins syndrome (17–19), a neurodevelopmental disease characterized by mental retardation, seizures and hyperventilation (20–21), suggesting that TCF4 is also critical for human nervous system development.
Epigenetic alterations such as DNA methylation and modification of chromatin structure often occur in neoplasia. It has been firmly established that aberrant methylation of CpG islands in the promoter regions and in the initial exons of many genes occurs in the early stages of carcinogenesis and results in suppressed expression of a variety of genes in a diverse array of cancers (22,23). Many reports have also shown that aberrant methylation of CpG islands leads to inactivation of many genes, particularly in gastric cancers (24–28). Although gastric cancer is the fourth most frequent human cancer and the second leading cause of cancer death in almost every country (29), it is still too often not diagnosed until at an advanced stage. Therefore, identification of effective biomarkers for early stage detection of gastric cancers is an urgent matter.
In this study, we identify TCF4 as a hypermethylated gene in gastric cancers using restriction landmark genomic scanning (RLGS) analysis. We demonstrate prominent hypermethylation of CpG dinucleotides in TCF4 exon 1, which significantly correlates with gene inactivation in early stage gastric cancers and in intestinal-type gastric cancers. Further, the effect of TCF4 on cell growth and migration in gastric cancer cells is investigated.
Materials and methods
Cell lines and tissue samples
Eleven human gastric cancer cell lines, SNU-001, -005, -016, -216, -484, -520, -601, -620, -638, -668, and -719, were obtained from the Korean Cell Line Bank (http://cellbank.snu.ac.kr/index.htm). These different cell lines were maintained at 37°C in humidified air containing 5% CO2 in RPMI 1640 medium (Gibco BRL, Gaithersburg, MD) supplemented with 10% fetal bovine serum. We obtained tissue samples from the Tissue Bank Program started at Chungnam National University Hospital, Daejeon, Korea, in 2001. Specimens from gastric cancer patients were originally obtained from tumors immediately after resection and adjacent normal mucosa specimens were obtained at least 3 cm away from the tumor edge. When the fresh specimens were resected, a portion of the tumor specimen was processed in a formalin-fixed paraffin block for pathologic observation and the remaining specimen was stored in a −80°C deep freezer in the Tissue Bank. Within 3 months, a portion of each frozen specimen was moved to a molecular biology laboratory for isolation of DNA and RNA from the frozen tissues. The purified DNAs and RNAs were stored in ethanol solution at −80°C until use. For this study, DNAs and RNAs from 77 gastric tumors and paired adjacent normal mucosa tissues were retrospectively identified from the surgical pathology files of Chungnam National University Hospital from 2001 to 2002. All specimens were obtained with informed consent and their use was approved by the Hospital's internal review board.
Formalin-fixed paraffin samples
Thirty-five archival samples of surgically resected gastric carcinomas, 35 archival samples of endoscopically resected gastric adenomas and 70 archival samples of endoscopically obtained non-neoplastic gastric mucosa (35 intestinal metaplasia and 35 chronic gastritis) were obtained from Seoul National University Hospital. After identifying the samples as gastric carcinoma, adenoma, intestinal metaplasia or chronic gastritis on hematoxylin and eosin-stained slides, a region corresponding to the identified lesion was scraped from 20 μm thick paraffin sections. The materials collected were dewaxed by washing in xylene and then by rinsing in ethanol. The dried tissues were digested with proteinase K and subjected to the standard method of DNA extraction using phenol/chloroform/isoamyl alcohol and ethanol precipitation.
RLGS assays and cloning of a methylated NotI locus
High molecular weight DNA was extracted from cultured cell lines or frozen clinical samples as described (30). RLGS was performed using paired tumor and mucosa DNA samples as described (31). For cell lines, RLGS was also performed in pairs consisting of cell line DNA alone and a cell line DNA sample mixed with gastric mucosa DNA. Differences between RLGS profiles from the paired samples were examined as described (32). Once a difference in spot intensity was detected, the spot position was compared with our Standard RLGS profile (32) or the Master RLGS profile (33) to determine the DNA fragment identity and access its sequence.
RNA isolation and real-time quantitative reverse transcription–polymerase chain reaction
Total RNA was extracted from each sample using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions and treated with DNase I (Promega, Madison, WI). DNase-treated total RNA (1 μg) was reverse transcribed using the Superscript II Reverse Transcriptase kit (Invitrogen) according to the manufacturer's instructions and a portion of the resulting cDNAs were amplified by standard polymerase chain reaction (PCR) to verify the presence of TCF4 messenger RNAs (mRNAs) in gastric cancer cell lines. The reverse transcription (RT) product (100 ng) was amplified in a 15 μl reaction using f-Taq polymerase (SolGent, Daejeon, Korea) on a GeneAmp PCR System 9700 (PerkinElmer). The following RT–PCR primers were designed to anneal to TCF4 cDNA and produce a product of 378 bp using the Primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi): forward primer, 5′-TGAGAACCTGCAAGACACGA-3′ and reverse primer, 5′-GGAGGCTCTGAGGACACCTT-3′. All PCRs were performed using the following conditions: 95°C for 2 min, 30 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 30 s, with a final extension at 72°C for 5 min. PCR products were subjected to 2% agarose gel electrophoresis and visualized by ethidium bromide staining. β-Actin was used as an internal control. Next, real-time RT–PCR analysis was performed using an Exicycler Quantitative Thermal Block (BiONEER, Daejeon, Korea) to quantitatively measure TCF4 mRNA levels from cell line samples or from clinical samples. About 100 ng of each RT reaction was amplified in a 15 μl reaction containing forward and reverse primers as above and 2× SYBR Premix EX Taq (Takara, Shiga, Japan) according to the manufacturer's instructions. The real-time RT–PCR was analyzed under the same conditions for RT–PCR, but for 45 cycles instead of 30 cycles for RT–PCR as amplification step. To minimize errors arising from variation in the amount of starting RNA among samples, β-actin mRNA was used as an internal reference against which other RNA values could be normalized. The Ct value of internal reference β-actin in gastric tissue samples and cell lines is shown in supplementary Figure 1 (available at Carcinogenesis Online). Normalized results were expressed as the ratio of the copy number of each gene to the copy number of the β-actin gene.
Methylation-specific polymerase chain reaction
Genomic DNA (1 μg) from cancer cells or clinical samples was modified by sodium bisulfite using the EZ DNA Methylation kit (ZYMO Research, Orange, CA) according to the manufacturer's instructions. Two regions of genomic DNA were targeted for methylation-specific polymerase chain reaction (MSP) analysis: the NotI-linked region in TCF4 intron 8 and the CpG-clustered region in TCF4 exon 1, as shown in Figure 1A. The following MSP primer sets were designed with the MethPrimer program (http://www.urogene.org/methprimer/index.html): for the methylated DNA in TCF4 intron 8, the forward primer 5′-TTAATTTTAGAGTGGAGAACGTGC-3′ and the reverse primer 5′-AAATAACAATACGACCCGCC-3′ were designed to yield a 198 bp product; for the unmethylated DNA in TCF4 intron 8, the forward primer 5′-TTTTAGAGTGGAGAATGTGTGT-3′ and the reverse primer 5′-AAACAAAATAACAATACAACCCACC-3′ were designed to yield a 199 bp product; for the methylated DNA in TCF4 exon 1, the forward primer 5′-GAATTTGTAATTTCGTGCGTTTC-3′ and the reverse primer 5′-AAAAAAAACTCTCCGTACACCG-3′ were designed to yield a 258 bp product and for the unmethylated DNA in TCF4 exon 1, the forward primer 5′-TGAATTTGTAATTTTGTGTGTTTTG-3′ and the reverse primer 5′-AAAAAAAACTCTCCATACACCACC-3′ were designed to yield a 259 bp product. MSPs were performed with 25 ng bisulfite-modified genomic DNA as follows: 94°C for 5 min, 35 cycles of 94°C for 45 s, 58°C for 30 s and 72°C for 60 s for TCF4 intron 8 or 94°C for 45 s, 59°C for 30 s and 72°C for 60 s for TCF4 exon 1, followed by 72°C for 7 min. PCR products were subjected to 2% agarose gel electrophoresis and visualized by ethidium bromide staining.
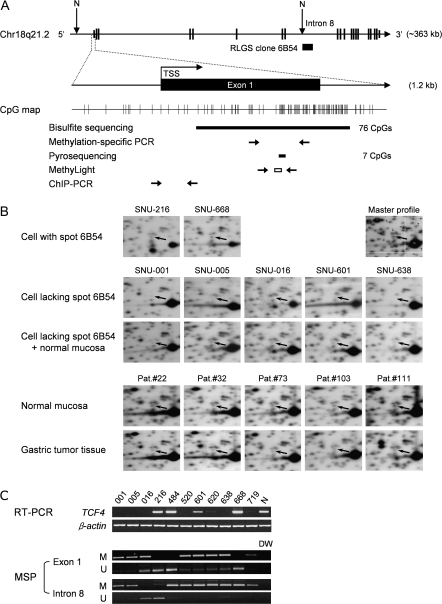
Fig. 1.
RLGS analysis of human TCF4 in gastric tumors and gastric cancer cell lines. (A) Schematic diagram of the TCF4 structure at human chromosome 18q21.2 derived from the UCSC Genome Browser (http://genome.ucsc.edu). TCF4 consists of 20 exons (exons 1 and 20 are non-coding) and spans 363 kb. The previously cloned NotI-linked 6B54 sequence (GenBank accession no. CG464927) was found in intron 8. The regions of CpG dinucleotides in exon 1 selected for bisulfite sequencing, MSP, pyrosequencing and MethyLight analysis are indicated below the CpG map. The location of the DNA fragment amplified by the ChIP assay is also indicated. N, NotI enzyme site; TSS, transcription start site. (B) Representative examples of decreased spot 6B54 (TCF4 intron 8) intensities from NotI–EcoRV–HinfI RLGS profiles. The uppermost panels show two cell lines in which the intensity of spot 6B54 relative to its neighboring spots was not different from the master profile. The arrow indicates the position of spot 6B54 in each RLGS profile; the upper right panel shows the Master RLGS profile (31). The middle set of panels show gastric cancer cell lines with a decreased intensity of spot 6B54 relative to its neighboring spots. ‘Cells lacking spot 6B54 + normal mucosa’ indicates samples in which DNA from cells lacking spot 6B54 were mixed with DNA from normal mucosa. The lower set of panels show RLGS profiles of gastric tumor tissues compared with those of adjacent normal mucosa tissues from five patients. (C) RT–PCR and methylation-sensitive PCR (MSP) analyses of TCF4. In the upper set of panels, TCF4 mRNA expression was analyzed in the 11 gastric cancer cell lines indicated using RT–PCR. β-Actin was used as an internal control. N, normal mucosa control. In the lower set of panels, TCF4 methylation in exon 1 or intron 8 was analyzed using MSP analysis. M, methylated DNA; U, unmethylated DNA, amplified by specific primers. DW, distilled water used as a negative control.
Bisulfite sequencing
Sequence analysis of bisulfite-modified DNA was performed on 76 CpG sites covering parts of exon 1 and intron 1 of TCF4, as shown in Figure 1A. Bisulfite-modified DNA (25 ng) was amplified by PCR in a 20 μl reaction containing a forward primer (5′-TTTAGGTTTTAGATTGTAATTGA-3′) and a reverse primer (5′-AAAAAAAATATCTCTTCTAAAAAC-3′) designed to yield a 617 bp product. All amplification reactions were performed as follows: 95°C for 1 min, 40 cycles of 95°C for 45 s, 51°C for 45 s, 72°C for 1 min, followed by 72°C for 5 min. PCR products were cloned into the pGEM-T Easy Vector (Promega), and four to seven clones were randomly chosen for sequencing.
Pyrosequencing
Within TCF4 exon 1, seven CpG sites were selected for quantitative determination of methylation status (Figure 1A). Bisulfite-modified DNA (100 ng) was amplified by PCR in a 25 μl reaction containing a forward primer (5′-GAAGAGAGTTGGTGTTAAGAGTTAG-3′) and a biotinylated reverse primer (5′-CCACCAAAAAAAACTCTCC-3′) designed to yield a 192 bp product. All amplification reactions were performed as follows: 95°C for 1 min, 50 cycles of 95°C for 30 s, 56°C for 40 s, 72°C for 40 s, followed by 72°C for 5 min. Pyrosequencing was performed according to the manufacturer′s instructions using the sequencing primer 5′-TGTGTGTTTGAGGATTTG-3′ on the PSQ HS 96A System (Biotage AB, Kungsgatan, Sweden).
5-aza-2′-deoxycytidine and trichostatin A treatment
Gastric cancer cells (SNU-601, -620 and -638) were seeded in 10 cm dishes at a density of 1 × 106 cells 1 day before drug treatment. The cells were treated with either 1 μM 5-aza-2′-deoxycytidine (5-aza-dC; Sigma, St Louis, MO) every 24 h for 3 days and then harvested or with 250 nM trichostatin A (TSA; Sigma) for 1 day and then harvested. To test the combined effect of 5-aza-dC and TSA, cells were treated with 1 μM 5-aza-dC every 24 h for 3 days followed by treatment with 250 nM TSA for 1 day. Total RNA was prepared and TCF4 expression was examined by real-time RT–PCR. The average relative mRNA levels were calculated from three independent experiments and from a total of six independent PCR analyses.
MethyLight analysis
DNAs from lesions of gastric carcinomas, gastric adenomas, intestinal metaplasia or chronic gastritis from hematoxylin and eosin-stained slides were modified by sodium bisulfite using the EZ DNA Methylation kit and analyzed according to the previously published MethyLight procedure (34). Briefly, bisulfite-modified DNA (100 ng) was amplified in a 30 μl reaction containing 0.3 μM locus-specific PCR primers (forward, 5′-TCGGAGAAGAGAGTTGGTGTT-3′; reverse, 5′-CTCCCGCGCCTACTACCT-3′), 0.2 μM oligonucleotide probe (6FAM5′-CGCCGCCACTACAAATCCGC-3′TAMRA), with 200 μM dNTPs, 3.5 mM MgCl2, 0.01% Tween 20, 0.05% gelatin and 0.5 U AmpliTaq Gold polymerase (Applied Biosystems, Foster City, CA). The reporter's fluorescence was detected with an Exicycler Quantitative Thermal Block. A standard curve for the Alu repeat control reaction was generated from 1:25 serial dilutions of bisulfite-converted, M. SssI-treated DNA for the methylated MethyLight reactions. The data for methylated DNA were expressed as percent of methylated reference values. The percent of methylated reference value was calculated by dividing the TCF4:Alu ratio of a sample by the TCF4:Alu ratio of M. SssI-treated human genomic DNA and multiplying by 100.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed using a ChIP assay kit (Upstate Biotechnology, Lake Placid, NY) according to the manufacturer's protocol with some modifications. Briefly, proteins were cross-linked to DNA by addition of formaldehyde directly to the culture medium and the reaction was quenched with glycine. Cells were then collected and washed twice with ice-cold phosphate-buffered saline containing proteinase inhibitors and resuspended in sodium dodecyl sulfate lysis buffer. To produce DNA fragments ranging from 200 to 500 bp, lysates were sonicated 21 times for 5 s with 30 s intervals on ice at power setting 3 using a Fisher Sonicator Dismembrator 100 and centrifuged at 13 000 r.p.m. for 10 min at 4°C. The supernatants were diluted in nine volumes of ChIP dilution buffer and precleared with salmon sperm DNA/protein A agarose beads and immunoprecipitated with either 5 μg anti-acetyl-histone H3 antibody (K9, K14) (Upstate Biotechnology, catalog no. 06-599), 5 μg anti-acetyl-histone H4 antibody (K5, K8, K12, K16) (Upstate Biotechnology, catalog no. 06-866) or 5 μg anti-trimethyl-histone H3 (K9) antibody (Upstate Biotechnology, catalog no. 07-442) or with no antibody, according to the manufacturer's recommendations. Immunoprecipitated DNA was recovered using the QIAquick PCR Purification kit (Qiagen) and amplified in 15 μl reactions containing SYBR Premix EX Taq (Takara) using the primers 5′-TAAACTTGCTTTGCCGTGTG-3′ and 5′-TCGAGCACCTCATTTTTCCT-3′ (located in TCF4 exon 1, product size 183 bp). Real-time PCR was performed as follows: 95°C for 1 min and 45 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 30 s. The amount of immunoprecipitated DNA was normalized to the input DNA. The average relative amount of each amplified product was calculated from two independent ChIP experiments and a total of four independent PCR analyses.
Generation of TCF4 expression constructs
Full-length TCF4 cDNA was amplified by PCR from the pBluescriptR-TCF4 plasmid (KUGI clone ID no. hMU003024) provided by Korean UniGene Information (http://kugi.kribb.re.kr/KUGI/index.html) and then cloned into the pGEM-T Easy vector (Promega). The resulting TCF4 cDNA clone was confirmed by sequence analysis and then subcloned into the EcoRI sites of the pEGFP-C1 vector (Clontech). The expression of green fluorescence protein (GFP)-TCF4 from the resulting pEGFP-C1-TCF4 plasmid was confirmed by western blotting using an anti-GFP antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Colony-forming assay
To analyze anchorage-dependent growth, a colony formation assay was performed on cell monolayers. pEGFP-C1-TCF4 or empty control vector (pEGFP-C1) was transfected into SNU-638 cells in six-well plates using Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's protocol. The cells were selected with G418 (300 μg/ml) for 2 weeks and then plated into fresh six-well plates at a density of 4 × 104 cells per well. After 2 weeks of incubation with G418, colonies were then stained with crystal violet and counted. To analyze anchorage-independent growth, colony formation assays in soft agar were also performed. SNU-638 cells were transfected and selected as above. The cells were suspended in top agar medium containing G418 (300 μg/ml) and 0.35% (wt/vol) agarose and 4 × 104 cells were overlaid into each well of a six-well plate containing bottom agar[0.6% (wt/vol) agarose]. Every 5 days, 500 μl fresh medium with G418 was applied to the top layer. Two weeks after seeding, the size of the colonies was measured in four randomly chosen microscopic fields. The value of each point was calculated as the average ± standard deviation from three independent experiments performed in duplicate.
Small interfering RNA and transfection
The following small interfering RNAs (siRNAs) were designed to inhibit expression of TCF4: sense sequence (5′-CAAGCACUGCCGACUACAAUAGGdGdA-3′) and antisense sequence (5′-UCCCUAUUGUAGUCGGCAGUGCUUGUU-3′). The 27 nucleotide synthetic siRNA duplex was synthesized and purified by Samchully Pharmaceutical (Seoul, Korea). The day before transfection, SNU-484 and SNU-668 cells were seeded at a density of 5 × 105 cells per well in six-well plates. TCF4-siRNA was then transfected into the cells using Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's instructions. Total RNA was extracted 48 h after transfection and TCF4 mRNA expression was analyzed via RT–PCR as described above.
Cell migration assay
Cell migration was examined using transwell chambers (Corning Costar, Corning, NY) with an 8 μm pore diameter membrane coated with fibronectin (F2006, Sigma) at a concentration of 25 mg/ml. SNU-484 and SNU-638 cells transfected with TCF4-siRNA or mock transfected were seeded into 24-well transwell plates (5 × 104 cells per chamber) and incubated for 20 and 26 h, respectively, at 37°C in a humidified atmosphere under 5% CO2. After removing the cells from the upper chamber, the cells that had migrated to the lower chamber were fixed with methanol for 1.5 min and stained with crystal violet for over 1 h. The membrane was mounted onto glass slides for viewing. The number of cells in four randomly chosen microscopic fields was counted. The value of each point was calculated as the average ± standard deviation from three independent experiments performed in duplicate.
Statistical methods for analysis
The Student's paired t-test was performed to measure the differences in TCF4 expression or TCF4 CpG methylation between primary gastric tumors and adjacent normal tissues. The clinicopathologic factors in various groups of patients with negative or positive TCF4 expression were compared by means of χ2 test or Student's t-test. Results with P values of <0.05 were considered statistically significant.
Results
Identification of a previously unidentified epigenetic target in gastric cancers by RLGS analysis
We identified in RLGS analysis a spot of NotI-linked genomic DNA with significantly decreased intensity in 8 of 11 (73%) gastric cell lines examined (Figure 1B, arrows, upper and middle panels). The same spot was also decreased in samples from primary gastric tumors compared with those from adjacent normal tissues (Figure 1B, bottom panel) and corresponded to spot number 5C23 from the Master RLGS profile (33) or to spot number 6B54 from the Standard RLGS profile (32) (Figure 1B, upper panel). The sequence corresponding to this DNA spot (GenBank accession no. CG464927) was mapped to intron 8 of TCF4 on human chromosome 18q21.1 in our previous RLGS study (32). To further examine epigenetic control of TCF4 expression, RT–PCR and MSP analysis of gastric cancer cell lines were used in tandem to compare TCF4 mRNA expression and TCF4 CpG methylation near the NotI sites in intron 8 (spot 6B54). Additional MSP analysis (Figure 1A and C) was also performed to examine methylation of the CpG dinucleotides in exon 1 of the gene because no CpG islands were found within 1 kb of the TCF4 transcription start site in the UCSC Genome Browser. We found little or no TCF4 expression in 8 of 11 gastric cancer cell lines by RT–PCR (Figure 1C, upper panel) and this observed silencing correlated with the pattern of increased CpG methylation observed in TCF4 exon 1, but not in intron 8 (Figure 1C, lower panel).
CpG methylation and histone acetylation regulate transcriptional silencing of TCF4 in gastric cancer cells
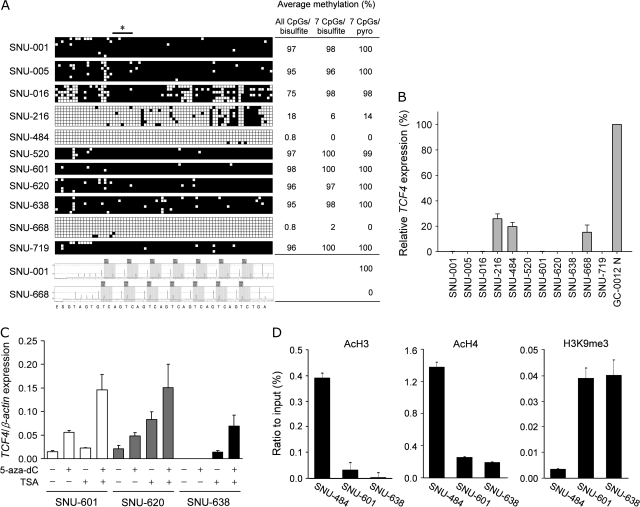
Bisulfite sequencing in exon 1 and intron 1 revealed extensive hypermethylation (75–98%) in the eight cell lines with little or no TCF4 expression, whereas hypomethylation (0.8–18%) was detected in the three TCF4-expressing cell lines (Figure 2A). Pyrosequencing analysis of seven CpG dinucleotides within exon 1 of TCF4 was used to confirm the bisulfite sequencing results (Figure 2A, right panel). The methylation status of the seven CpG sites examined by pyrosequencing analysis was representative of the 76 CpG sites examined by bisulfite sequencing. Real-time RT–PCR analysis also revealed that silencing of TCF4 expression correlated with the methylation status of exon 1 determined by pyrosequencing (Figure 2B), suggesting that CpG methylation at exon 1 is associated with the inactivation of TCF4 in human gastric cancer cell lines. We next determined that TCF4 mRNA expression was restored in SNU-601, -620 and -638 cells after 5-aza-dC and/or TSA treatment (Figure 2C). This result suggests that TCF4 mRNA expression in gastric cancer cells may be regulated by both CpG methylation and histone acetylation. We therefore examined histone acetylation and methylation in chromatin associated with TCF4 exon 1 using ChIP analysis. Histone-associated DNA fragments immunoprecipitated with antibodies against acetyl-H3 (acetylated at residues K9 and K14), acetyl-H4 (acetylated at residues K5, K8, K12 and K16) or H3K9me3 (acetylated at residue K9 and methylated at residue 3) were subjected to PCR analysis using primers designed to amplify a region of TCF4 exon 1. The acetylation of histones H3 and H4 within TCF4 exon 1 was elevated in SNU-484 cells, in which TCF4 is unmethylated and transcriptionally active, compared with SNU-601 or -638 cells, in which TCF4 is hypermethylated and transcriptionally silent (Figure 2D). In contrast, anti-H3K9me3 immunoprecipitation enriched amplification of TCF4 exon 1 in SNU-601 and -638 cells (Figure 2D). These results clearly indicate that histone modification is also likely involved in transcriptional silencing of TCF4 in gastric cancers.
Fig. 2.
TCF4 CpG methylation and histone modifications correlate with TCF4 silencing in gastric cancer cell lines. (A) Quantitative measurement of CpG dinucleotide methylation in TCF4 exon 1. In the upper panels, each horizontal row of boxes represents a single clone analyzed by bisulfite sequencing. Each small square box represents a CpG site. Filled and open square boxes indicate methylated and unmethylated CpG sites, respectively. The bar with an asterisk indicates the seven CpG sites used for pyrosequencing analysis. Representative pyrograms for SNU-001 and SNU-668 cells are shown in the lower set of panels. Numerical values at right represent the average percent methylation of the indicated CpG sites determined by bisulfite sequencing (bisulfite) or by pyrosequencing (pyro). (B) Relative TCF4 expression in gastric cancer cell lines. Relative TCF4 expression was obtained using real-time RT–PCR, and results from three independent analyses are expressed relative to β-actin mRNA levels. (C) Restoration of TCF4 expression in gastric cancer cells after treatment with the indicated drugs. Three gastric cancer cell lines, SNU-601, SNU-620 and SNU-638, were treated with 5-aza-dC, TSA or both. For each cell line, TCF4 expression was measured using real-time RT–PCR and results from three independent analyses are expressed relative to β-actin mRNA levels. (D) ChIP assays of the TCF4 exon 1. Chromatin DNA was immunoprecipitated with antibodies specific for acetyl-H3 (AcH3), acetyl-H4 (AcH4) or trimethyl-H3-K9 (H3K9me3). DNA fragments corresponding to 183 bp in the TCF4 exon 1 (see Figure 1A) were amplified by PCR. The amount of immunoprecipitated DNA was normalized to the input DNA.
The status of TCF4 expression in primary gastric tumors
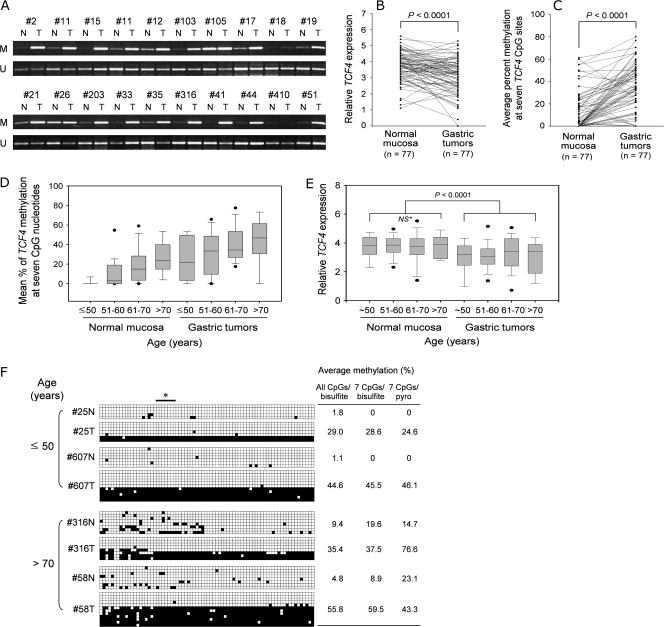
To test whether CpG methylation in TCF4 exon 1 is abnormally increased in primary tumors compared with the corresponding normal tissue, we next performed MSP analysis on clinical samples. Figure 3A shows that a single PCR product was amplified using primers for unmethylated CpG sequence from all gastric tumors, as well as from normal tissues, suggesting the presence of normal mucosa or stromal cells in the gastric tumor specimens that may have contributed to the amplification of unmethylated products. Nonetheless, a prominent PCR product for methylated CpG sequence was detected in all tumors except for 2 (samples 18 and 410) of 20 tumors, whereas no or weak bands were observed in 16 of the corresponding normal tissue specimens. This result suggests that CpG methylation in TCF4 exon 1 may be a feature of gastric tumors.
Fig. 3.
Reduced TCF4 expression correlates with CpG methylation in primary gastric tumors. (A) MSP analysis in TCF4 exon 1 of primary gastric tumors. Amplified PCR products for 20 gastric tumors and adjacent normal tissues from the indicated patients are shown. M, methylated DNA; U, unmethylated DNA amplified by specific primers. N, normal mucosa; T, gastric tumor. (B) Quantitative real-time RT–PCR analysis of TCF4 in 77 gastric tumors and adjacent normal tissues. TCF4 mRNA levels in primary gastric tumors (right) and in matched adjacent normal tissue control (left) are shown. Each sample was normalized to the β-actin internal control. (C) Pyrosequencing analysis at seven CpG sites in TCF4 exon 1 from 77 gastric tumors and adjacent normal tissues. (D) Average percent CpG methylation in TCF4 exon 1 with respect to age. Patients were subdivided into four groups by age (≤50 years, n = 17; 51–60 years, n = 22; 61–70 years, n = 26; >70 years, n = 12) and the average percent CpG methylation in gastric tumors or adjacent normal tissues within each age group was estimated. The box plot analysis shows the median, 25th and 75th percentiles and outliers. (E) Box plot of relative TCF4 expression with respect to age. Patients were subdivided into four groups by age as above, and the relative TCF4 expression in gastric tumors or adjacent normal tissues within each age group was plotted. NS with an asterisk indicates a statistically insignificant difference. (F) Quantitative measurement of CpG dinucleotide methylation in TCF4 exon 1 in gastric tumors and adjacent normal tissues by bisulfite sequencing analysis. The results from younger and older patients are presented in the upper and the lower panels, respectively. The open and filled boxes and numerical values at the right are the same as for Figure 2A. The bar with an asterisk indicates the seven CpG sites used for pyrosequencing analysis.
To elucidate the status of TCF4 expression during gastric tissue carcinogenesis, we investigated TCF4 mRNA expression in 77 paired clinical tissues using real-time quantitative RT–PCR and examined the associated clinicopathologic parameters. TCF4 mRNA expression in tumor tissues was significantly lower than in adjacent normal tissues (P < 0.0001) after normalization to β-actin (Figure 3B). Loss of expression (LOE) was assigned to tumors in which the expression of TCF4 mRNA was less than or equal to half of that in adjacent normal tissues. TCF4 LOE was detected in 38% (29 of 77) of primary tumors. Quantitative methyl-CpG analysis of the same seven CpG sites in TCF4 exon 1 described in Figure 1A was performed on the 77 paired clinical tissues via pyrosequencing. The mean percent methylation was 10.9, 13.8, 13.5, 15.1, 12.4, 13.3 and 13.3% for each of the seven CpG sites examined in TCF4 exon 1 from normal tissues. The overall mean percent methylation was calculated from the average of these individual values and was determined to be 13.2. In the same manner, the overall mean percent methylation in TCF4 exon 1 from tumor tissues was 34.7%, revealing a significant difference compared with normal tissues (P < 0.0001) (Figure 3C). As shown in Table I, the mean percent methylation was significantly higher in tumors with TCF4 LOE (43.0 ± 23.2%) than in those with TCF4 expression comparable with their matched normal tissue (29.8 ± 19.1%) (P = 0.006), indicating that increased CpG methylation correlated with decreased TCF4 expression in primary gastric tumors. Clinicopathologic characteristics of patients with respect to tumor TCF4 expression levels are also shown in Table I. TCF4 LOE was more frequent among females (14/26, 53.8%) than males (15/51, 29.4%) (P = 0.04). In particular, TCF4 LOE was significantly more common in intestinal-type than diffuse-type gastric cancers (P = 0.0004) and in early gastric cancers than in advanced gastric cancers (P = 0.004), suggesting that TCF4 LOE may be an early event in the multistep process leading to gastric carcinogenesis.
Table I.
Relative TCF4 mRNA expression in tumors with respect to clinicopathologic characteristics
| Clinicopathologic parameter | Gastric tumors with decreased relative TCF4 expression a |
P valueb | |
| ≥50% decrease (n = 29) | <50% decrease (n = 48) | ||
| Mean patient age (in years ± SD) | 61.1 ± 11.4 | 58.9 ± 11.5 | 0.41 |
| Gender | |||
| Male | 15 | 36 | 0.04 |
| Female | 14 | 12 | |
| Tumor size in cmc (mean ± SD) | 5.2 ± 2.4 | 5.7 ± 2.3 | 0.39 |
| Histology | |||
| Intestinal | 22 | 15 | 0.0004 |
| Diffuse | 7 | 30 | |
| Mixedd | 0 | 3 | |
| Tumor progressione | |||
| EGC | 12 | 6 | 0.004 |
| AGC | 17 | 42 | |
| Lymph node metastasis | |||
| Negative | 14 | 14 | 0.09 |
| Positive | 15 | 34 | |
| TCF4 methylationf (mean % ± SD) | 43.0 ± 23.2 | 29.8 ± 19.1 | 0.006 |
All tumors were classified into two subtypes: those in which the decrease in TCF4 mRNA expression was 50% or more compared with adjacent normal tissue (≥50% decrease) and those in which the decrease in TCF4 mRNA expression was <50% compared with adjacent normal tissue (<50% decrease).
Analyzed by Student's t-test or by χ2 test.
Size was calculated based on the largest diameter measured for each tumor.
Mixed type tumors were excluded from this analysis because of a small sample number (n = 3).
EGC, early gastric cancer; AGC, advanced gastric cancer.
The percent methylation at seven distinct CpG sites in TCF4 exon 1 was determined by pyrosequencing analysis. The percentages were then averaged to calculate TCF4 methylation.
Age-related TCF4 CpG methylation in gastric tumors and adjacent normal mucosa
No mean age differences were found between patients with tumors having TCF4 LOE and those having non-LOE tumors. When patient ages were subdivided into four age groups, however, the mean percent TCF4 methylation determined by pyrosequencing of the seven CpG sites in exon 1 in age group 1 (≤50 years, n = 17) was ∼1.7 and 24.5% in gastric mucosa and tumors, respectively. The mean percent TCF4 methylation in gastric mucosa and tumors was 9.5 and 30.9%, respectively, in age group 2 (51–60 years, n = 22), 18.2 and 41.0%, respectively, in age group 3 (61–70 years, n = 26) and 25.3 and 42.5%, respectively, in age group 4 (>70 years, n = 12), as shown in Figure 3D. Thus, CpG methylation was significantly higher in tumors than in gastric mucosa within every age group and gradually increased with increasing age in both gastric mucosa and primary tumors, suggesting that increased CpG methylation in exon 1 of TCF4 correlates with both age and tumorigenesis. Interestingly, CpG methylation was not detected in any of the normal gastric mucosa samples taken from patients younger than age 50 (n = 14), whereas TCF4 CpG methylation in normal gastric mucosa from patients over age 70 (n = 12) was 25.3%, very similar to that observed in tumors from patients younger than age 50 (24.5%, n = 17). To determine whether TCF4 CpG methylation in normal gastric mucosa affects TCF4 expression in normal tissue from older patients, we compared the relative TCF4 expression levels between the four age groups and between normal mucosa and gastric tumors (Figure 3E). No significant difference in TCF4 expression was detected between age groups for the normal mucosa, suggesting that methylation at the seven CpG sites of TCF4 tested may not be critical in regulating TCF4 expression in normal-appearing tissues.
In addition, Figure 3F shows that the gastric tumors used in this study consisted of two types of clones based on the proportion of CpG methylation: one clone type is a cell with extensive hypermethylation (over 95% of the 76 CpG sites were methylated) and the other was methylation free at almost all CpG sites. Given that the bisulfite sequencing analysis revealed extensive hypermethylation in the gastric cancer cell lines expressing little or no TCF4, the extensively hypermethylated clone may be responsible for reduced TCF4 expression in gastric tumors from both younger and older patients and in some gastric cancer cell lines. Although TCF4 CpG methylation at the seven CpG sites in normal tissues of older patients is similar to that in tumors of younger patients, we suggest that a significant reduction in TCF4 mRNA levels could not be detected in normal tissues of older patients because these tissues do not contain the extensively hypermethylated clone.
TCF4 CpG methylation during gastric carcinogenesis
To elucidate the methylation status of TCF4 during gastric carcinogenesis, we performed MethyLight analysis on the CpG sites in exon 1 (shown in Figure 1A) on 35 sets of paraffin-embedded tissues of chronic gastritis, intestinal metaplasia, gastric adenomas and gastric carcinomas. No CpG methylation in TCF4 exon 1 was found in the chronic gastritis samples but a gradual increase in CpG methylation from intestinal metaplasia (mean percent of methylated reference 0.5%, range of 0.0–11.5%) to gastric adenomas (mean 2.4%, range of 0.0–31.8%) or gastric carcinomas (mean 3.3%, range of 0.0–49.7%). This result suggests that CpG methylation in TCF4 exon 1 is initiated at an early stage such as intestinal metaplasia during gastric carcinogenesis and tends to accumulate along the multistep carcinogenesis.
In vitro effects of TCF4 on cell growth and migration in gastric cancer cells
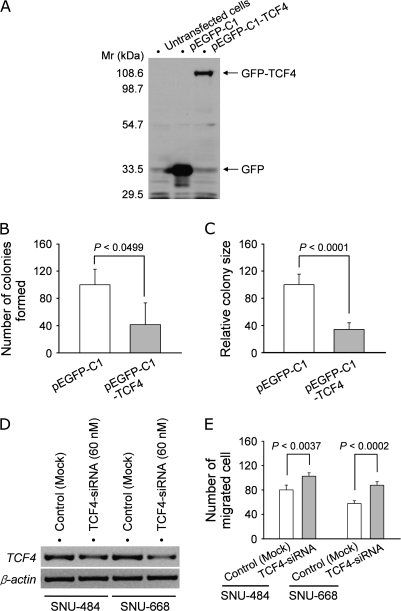
To determine whether TCF4 is involved in cell growth in gastric cancer cells, we introduced a TCF4 expression vector into SNU-638 cells, where methyl-CpG-associated endogenous TCF4 silencing was demonstrated (see Figure 2). TCF4 expression in cells transfected with the pEGFP-C1-TCF4 expression plasmid, but not in the parental SNU-638 cells or in cells transfected with the control pEGFP-C1 vector, was confirmed by western blotting using a monoclonal anti-GFP antibody (Figure 4A). In the monolayer assay used to evaluate anchorage-dependent growth, the number of colonies from pEGFP-C1-TCF4 transfectants was much smaller than that of control pEGFP-C1 transfectants (P < 0.0499) (Figure 4B). A soft agar assay for anchorage-independent growth also showed decreased colony size in pEGFP-C1-TCF4 transfectants compared with control pEGFP-C1 transfectants (P < 0.0001) (Figure 4C). We also addressed whether transfection with pEGFP-C1-TCF4 resulted in any morphological alterations but did not note any morphological change or apoptosis at 24 h after transfection. We further examined whether reducing TCF4 expression by RNA interference altered cell migration in gastric cancer cells. RT–PCR analysis of SNU-484 and SNU-668 cells transfected with a broad range of TCF4-siRNA concentrations indicated that 60 nM TCF4-siRNA reduced TCF4 transcript levels in both cell lines compared with mock-transfected controls (Figure 4D). Transwell assays also demonstrated increased cell migration in SNU-484 (P < 0.0037) and SNU-668 cells (P < 0.0002) transfected with TCF4-siRNA compared with mock-transfected cells (Figure 4E). These results suggest that TCF4 silencing may be associated with cell growth and migration in gastric cancer cells.
Fig. 4.
Ectopic expression or siRNA knockdown of TCF4. (A) Immunoblot analysis of exogenous TCF4 expression in gastric cancer cells. SNU-638 cells were transfected with pEGFP-C1-TCF4 or control pEGFP-C1 vector and GFP-TCF4 or GFP expression was confirmed by western blotting using an anti-GFP antibody. Mr, molecular mass markers. (B) Anchorage-dependent colony formation assay in monolayer cultures. SNU-638 cells selected for transfection with pGFP-C1-TCF4 or control plasmid were cultured for 25 days and stained with crystal violet to visualize colony formation. The bar graph indicates the number of colonies formed. Values represent the mean ± SD from three separate experiments performed in duplicate. (C) Anchorage-independent colony formation assay in soft agar. SNU-638 cells transfected with pGFP-C1-TCF4 or control plasmid were seeded in soft agar and examined for colony formation after 2 weeks. The bar graph indicates the relative size of the colonies formed with average size of the control colonies arbitrarily set to 100%. Values represent the mean ± SD from three separate experiments performed in duplicate. (D) TCF4 knockdown by TCF4-siRNA in two TCF4-expressing cell lines, SNU-484 and SNU-668. Cells transfected with 60 nM TCF4-siRNA were analyzed for TCF4 expression using RT–PCR. β-Actin was analyzed as an internal control. (E) Transwell cell migration assay. Cells transfected with TCF4-siRNA or mock-transfected cells were seeded in fibronectin-coated transwell plates and allowed to migrate for 20 h (SNU-484 cells) or 26 h (SNU-668 cells) (left panel). The bar graph indicates the number of migrated cells. Values represent the mean ± SD from three independent experiments performed in duplicate. Scale bar, 65 μm.
Discussion
In this study, we identified epigenetic silencing of TCF4 in human gastric cancers and demonstrated that this process is linked to cell growth and migration. Genetic and epigenetic alterations in tumor suppressor genes are a prerequisite of cancer development and progression. In particular, epigenetic alterations within tumors are inevitable and often promote further genetic modifications (35). Similar to the findings in this study, CpG methylation of promoter regions or the first exon of tumor suppressor genes has been established as an important mechanism for gene silencing (36). Moreover, silencing of TCF4 was relieved in several gastric cancer cell lines upon treatment with a DNA methyltransferase and/or a histone deacetylase inhibitor. Thus, our data demonstrate epigenetic regulation of TCF4 in gastric cancers.
Although our understanding of the molecular mechanisms of the pathology of sporadic gastric cancers is increasing, the cellular events that trigger initiation of carcinogenesis in human gastric mucosa cells remain unclear (37). It is widely accepted, however, that epigenetic alterations are a prerequisite of virtually all tumors and that epigenetic alterations facilitate the accumulation of further genetic modifications that result in cancer progression through clonal expansion of cells with a resulting proliferative advantage (35). In this study, we demonstrated that 38% of the gastric tumors examined displayed TCF4 silencing and we significantly correlated this effect with CpG dinucleotide methylation in TCF4 exon 1. Gastric cancers are histologically classified into two main types, intestinal and diffuse (38). Intestinal-type gastric cancers are thought to be derived from gastric mucosa cells, and patients progress through well-characterized sequential stages such as chronic gastritis, atrophy, intestinal metaplasia and dysplasia, whereas diffuse-type gastric cancers are presumed to develop from gastric epithelial cells through a shorter, less well-characterized sequence of events (39). In this study, examination of clinicopathologic characteristics of tumors showed that TCF4 silencing was significantly more prevalent in intestinal type and in early stage gastric cancers than in diffuse type or in advanced stage gastric cancers. Recent results have shown that promoter hypermethylation in many genes occurs in the non-cancerous tissues adjacent to gastric cancer tumors (40,41) and in non-neoplastic gastric mucosa of patients without gastric cancer (42), indicating that an overall deregulation of the DNA methylation machinery may be an early event in the formation of gastric cancers and may become more severe as carcinogenesis progresses.
Because the normal gastric mucosa and tumor tissues used in this study were not dissected by a technique such as laser-captured microdissection, the normal gastric mucosa may have actually been normal-appearing precancerous gastric mucosa, and likewise, the tumor tissues may have included some normal mucosa or precancerous cells. Although our samples may have contained these possible mixtures, our data showed that CpG methylation in TCF4 exon 1 gradually increased in an age-dependent manner in all normal-appearing gastric mucosa samples examined—the average percent of methylated CpG sites ranging from 1.7% in age group 1 (<50 years) to 25.3% in age group 4 (over 70 years). These data are highly relevant to gastric neoplasia because the incidence of sporadic gastric tumors is strongly age related (43). In agreement with this, it is worthwhile to note that the mean percent TCF4 CpG methylation in normal-appearing gastric mucosa of older patients (25.3% for patients over age 70) was similar to that in gastric tumor tissues from younger patients (24.5% for patients younger than age 50), suggesting that gastric mucosa from patients over age 70 may be predisposed to neoplasia. However, it is unlikely that TCF4 CpG methylation contributes to decreased TCF4 expression in normal gastric mucosa in older patients based on our observation that normal gastric mucosa from older patients contains no extensively hypermethylated clone, which may be correlated with TCF4 expression, in contrast to its presence in gastric tumors from younger and older patients and in some gastric cancer cell lines. In fact, no significant difference in TCF4 expression in the normal mucosa was detected between age groups. Although CpG methylation in TCF4 exon 1 was not found in chronic gastritis, a gradual increase in CpG methylation from intestinal metaplasia to neoplastic gastric mucosa, such as gastric adenomas or gastric carcinomas, was noted. Therefore, it is reasonable to propose that increased TCF4 CpG methylation in normal-appearing gastric mucosa may be due to local methylation of CpG sites initiated at an early stage during gastric carcinogenesis (such as in intestinal metaplasia), rather than due to extensive CpG methylation as proposed in the ‘field cancerization effect’ in an epithelial carcinogenesis model in which the development of a field of genetically altered cells plays a central role (44).
TCF4 consists of 20 exons (exons 1 and 20 are non-coding), spans 363 kb, and encodes at least two isoforms of the TCF4 protein that differ with respect to the presence of four amino acids (RSRS) located 17 residues N-terminal to the HLH domain (19). Although previous studies have presented controversial cellular roles for TCF4 (11,16), our data show that overexpression of TCF4 reduced colony formation in an anchorage-dependent and -independent manner, supporting a role for TCF4 as a negative regulator of cell proliferation (16). Because it has also been shown that the functional patterns of TCF4 are very similar to those of TCF3, another member of the bHLH transcriptional factor family, and that TCF3 acts act as a tumor suppressor in several cancer cell lines (11), TCF4 may be associated with the induction of apoptosis. Furthermore, no significant correlation between β-catenin mutations and expression of TCF4 in hepatoblastomas has been demonstrated (45), although β-catenin-binding proteins, such as ICAT or Chibby, may regulate β-catenin function and its ability to activate TCF7L2-regulated transcription (46,47). A possible explanation for the observed variation in TCF4 expression is that the expression may be controlled by other signaling pathways in a cell type-specific manner. We showed that knock down of TCF4 using TCF4-siRNA significantly increased cell migration in vitro, also suggesting an inhibitory effect of TCF4 on cell migration. Thus, our results suggest that TCF4 has a possible negative effect on both cell growth and migration in gastric cancer cells. In this study, however, because no correlation was found between lymph node metastasis and TCF4 silencing in clinical samples and because we found that TCF4 silencing was significantly more frequent in early stage gastric cancers than in advanced stage gastric cancers, further analysis is needed to resolve the role of TCF4 silencing in carcinogenesis. However, CpG methylation of TCF4 may prove to be a useful molecular biomarker for assessing the risk of gastric cancer development and drug therapies targeting TCF4 expression in gastric cancers may improve prognosis for gastric cancer patients.
Supplementary material
Supplementary Figure 1 can be found at http://carcin.oxfordjournals.org/
Funding
21C Frontier Functional Human Genome Project (FG08-11-01) from the Ministry of Science and Technology of Korea.
Supplementary Material
Glossary
Abbreviations
- 5-aza-dC
5-aza-2′-deoxycytidine
- bHLH
basic helix-loop-helix
- ChIP
chromatin immunoprecipitation
- GFP
green fluorescence protein
- LOE
loss of expression
- mRNA
messenger RNA
- MSP
methylation-specific polymerase chain reaction
- PCR
polymerase chain reaction
- RLGS
restriction landmark genomic scanning
- RT
reverse transcription
- siRNA
small interfering RNA
- TCF4
transcription factor 4
- TSA
trichostatin A
References
- 1.Murre C, et al. Structure and function of helix-loop-helix proteins. Biochim. Biophys. Acta. 1994;1218:129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 2.Atchley WR, et al. A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl Acad. Sci. USA. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ledent V, et al. Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol. 2002;3:0030.1–0030.18. doi: 10.1186/gb-2002-3-6-research0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murre C, et al. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 5.Murre C, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 6.Saisanit S, et al. A novel enhancer, the pro-B enhancer, regulates Id1 gene expression in progenitor B cells. Mol. Cell. Biol. 1995;15:1513–1521. doi: 10.1128/mcb.15.3.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Church GM, et al. Cell-type-specific contacts to immunoglobulin enhancers in nuclei. Nature. 1985;313:798–801. doi: 10.1038/313798a0. [DOI] [PubMed] [Google Scholar]
- 8.Naya FJ, et al. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 9.Massari ME, et al. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004;5:226. doi: 10.1186/gb-2004-5-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pagliuca A, et al. Class A helix-loop-helix proteins are positive regulators of several cyclin-dependent kinase inhibitors’ promoter activity and negatively affect cell growth. Cancer Res. 2000;60:1376–1382. [PubMed] [Google Scholar]
- 12.Lassar AB, et al. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 13.Beck GR, Jr, et al. Gene array analysis of osteoblast differentiation. Cell Growth Differ. 2001;12:61–83. [PubMed] [Google Scholar]
- 14.Quong MW, et al. E protein function in lymphocyte development. Annu. Rev. Immunol. 2002;20:301–322. doi: 10.1146/annurev.immunol.20.092501.162048. [DOI] [PubMed] [Google Scholar]
- 15.Persson P, et al. HASH-1 and E2-2 are expressed in human neuroblastoma cells and form a functional complex. Biochem. Biophys. Res. Commun. 2000;274:22–31. doi: 10.1006/bbrc.2000.3090. [DOI] [PubMed] [Google Scholar]
- 16.Kolligs FT, et al. ITF-2, a downstream target of the Wnt/TCF pathway, is activated in human cancers with b-catenin defects and promotes neoplastic transformation. Cancer Cell. 2002;1:145–155. doi: 10.1016/s1535-6108(02)00035-1. [DOI] [PubMed] [Google Scholar]
- 17.Brockschmidt A, et al. Severe mental retardation with breathing abnormalities (Pitt-Hopkins syndrome) is caused by haploinsufficiency of the neuronal bHLH transcription factor TCF4. Hum. Mol. Genet. 2007;16:1488–1494. doi: 10.1093/hmg/ddm099. [DOI] [PubMed] [Google Scholar]
- 18.Amiel J, et al. Mutations in TCF4, encoding a class I basic helix-loop-helix transcription factor, are responsible for Pitt-Hopkins syndrome, a severe epileptic encephalopathy associated with autonomic dysfunction. Am. J. Hum. Genet. 2007;80:988–993. doi: 10.1086/515582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zweier C, et al. Haploinsufficiency of TCF4 causes syndromal mental retardation with intermittent hyperventilation (Pitt-Hopkins syndrome) Am. J. Hum. Genet. 2007;80:994–1001. doi: 10.1086/515583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pitt D, et al. A syndrome of mental retardation, wide mouth and intermittent overbreathing. Aust. Paediatr. J. 1978;14:182–184. doi: 10.1111/jpc.1978.14.3.182. [DOI] [PubMed] [Google Scholar]
- 21.Peippo MM, et al. Pitt-Hopkins syndrome in two patients and further definition of the phenotype. Clin. Dysmorphol. 2006;15:47–54. doi: 10.1097/01.mcd.0000184973.14775.32. [DOI] [PubMed] [Google Scholar]
- 22.Esteller M. CpG island hypermethylation and tumour suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 23.Herman JG, et al. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 24.Grady WM, et al. Methylation of the CDH1 promoter as the second genetic hit in hereditary diffuse gastric cancer. Nat. Genet. 2000;26:16–17. doi: 10.1038/79120. [DOI] [PubMed] [Google Scholar]
- 25.Shim YH, et al. Correlation of p16 hypermethylation with p16 protein loss in sporadic gastric carcinomas. Lab. Invest. 2000;80:689–695. doi: 10.1038/labinvest.3780072. [DOI] [PubMed] [Google Scholar]
- 26.Iida S, et al. Alterations and hypermethylation of the p14(ARF) gene in gastric cancer. Int. J. Cancer. 2000;87:654–658. doi: 10.1002/1097-0215(20000901)87:5<654::aid-ijc6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 27.Oue N, et al. Promoter hypermethylation of MGMT is associated with protein loss in gastric carcinoma. Int. J. Cancer. 2001;93:805–809. doi: 10.1002/ijc.1403. [DOI] [PubMed] [Google Scholar]
- 28.Kim SK, et al. The epigenetic silencing of LIMS2 in gastric cancer and its inhibitory effect on cell migration. Biochem. Biophys. Res. Commun. 2006a;349:1032–1040. doi: 10.1016/j.bbrc.2006.08.128. [DOI] [PubMed] [Google Scholar]
- 29.Parkin DM, et al. Global cancer statistics, 2002. CA Cancer J. Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 30.Birren B, et al. Genome Analysis: A Laboratory Manual. Vol. 1. New York: Cold Spring Harbor Laboratory Press; 1997. pp. 1–15. [Google Scholar]
- 31.Hatada I, et al. A genomic scanning method for higher organisms using restriction sites as landmarks. Proc. Natl Acad. Sci. USA. 1991;88:9523–9527. doi: 10.1073/pnas.88.21.9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim JH, et al. Cloning of NotI-linked DNA detected by restriction landmark genomic scanning of human genome. Genomics Inform. 2006b;4:1–10. [Google Scholar]
- 33.Costello JF, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat. Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 34.Eads CA, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grady WM. Epigenetic events in the colorectum and in colon cancer. Biochem. Soc. Trans. 2005;33:684–688. doi: 10.1042/BST0330684. [DOI] [PubMed] [Google Scholar]
- 36.Baylin SB. DNA methylation and gene silencing in cancer. Nat. Clin. Pract. Oncol. 2005;2(suppl 1):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 37.Zheng L, et al. Molecular basis of gastric cancer development and progression. Gastric Cancer. 2004;7:61–77. doi: 10.1007/s10120-004-0277-4. [DOI] [PubMed] [Google Scholar]
- 38.Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol. Microbiol. Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 39.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process—First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52:6735–6740. [PubMed] [Google Scholar]
- 40.Kang GH, et al. CpG island methylation in premalignant stages of gastric carcinoma. Cancer Res. 2001;61:2847–2851. [PubMed] [Google Scholar]
- 41.Leung WK, et al. Concurrent hypermethylation of multiple tumor-related genes in gastric carcinoma and adjacent normal tissues. Cancer. 2001;91:2294–2301. [PubMed] [Google Scholar]
- 42.Chan AO, et al. Promoter methylation of E-cadherin gene in gastric mucosa associated with Helicobacter pylori infection and in gastric cancer. Gut. 2003;52:502–506. doi: 10.1136/gut.52.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee H, et al. Gastric cancer in Korea. Gastric Cancer. 2002;5:177–182. doi: 10.1007/s101200200031. [DOI] [PubMed] [Google Scholar]
- 44.Braakhuis BJ, et al. A genetic explanation of Slaughter's concept of field cancerization: evidence and clinical implications. Cancer Res. 2003;63:1727–1730. [PubMed] [Google Scholar]
- 45.Koch A, et al. Elevated expression of Wnt antagonists is a common event in hepatoblastomas. Clin. Cancer Res. 2005;11:4295–4304. doi: 10.1158/1078-0432.CCR-04-1162. [DOI] [PubMed] [Google Scholar]
- 46.Tago K, et al. Inhibition of Wnt signaling by ICAT, a novel beta-catenin-interacting protein. Genes Dev. 2000;14:1741–1749. [PMC free article] [PubMed] [Google Scholar]
- 47.Takemaru K, et al. Chibby, a nuclear beta-catenin-associated antagonist of the Wnt/Wingless pathway. Nature. 2003;422:905–909. doi: 10.1038/nature01570. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.