Abstract
Hypoxia-inducible factor (HIF-1) regulates the expression of genes that facilitate tumor cell survival by making them more resistant to therapeutic intervention. Recent evidence suggests that the activation of other transcription factors, in cooperation with HIF-1 or acting alone, is involved in the upregulation of hypoxia-inducible genes. Here we report that high cell density, a condition that might mimic the physiologic situation in growing tumor and most probably representing nutritional starvation, upregulates hypoxia-inducible genes. This upregulation can occur in HIF-independent manner since hypoxia-inducible genes carbonic anhydrase 9 (CA9), lysyloxidase like 2 (LOXL2) and n-myc-down regulated 1 (NDRG1)/calcium activated protein (Cap43) can be upregulated by increased cell density under both normoxic and hypoxic conditions in both HIF-1α-proficient and -deficient mouse fibroblasts. Moreover, cell density upregulates the same genes in 1HAEo− and A549 human lung epithelial cells. Searching for other transcription factors involved in the regulation of hypoxia-inducible genes by cell density, we focused our attention on ETS1. As reported previously, members of v-ets erythroblastosis virus E26 oncogene homolog (ETS) family transcription factors participate in the upregulation of hypoxia-inducible genes. Here, we provide evidence that ETS1 protein is upregulated at high cell density in both human and mouse cells. The involvement of ETS1 in the upregulation of hypoxia-inducible genes was further confirmed in a luciferase reporter assay using cotransfection of ETS1 expression vector with NDRG1/Cap43 promoter construct. The downregulation of ETS1 expression with small interfering RNA (siRNA) inhibited the upregulation of CA9 and NDRG1/Cap43 caused by increased cell density. Collectively, our data indicate the involvement of ETS1 along with HIF-1 in regulating hypoxia-inducible genes.
Introduction
Insufficient blood supply in growing tumors results in a lack of oxygen (hypoxia). This change in cellular environment provokes adaptive processes that are mediated by the elevated expression of hypoxia-inducible genes. The expression of these genes enables tumor cells to adapt and survive under harsh conditions. Inevitably, the upregulation of hypoxia-inducible genes makes tumor cells more resistant to therapeutic interventions, representing a serious clinical problem. The hypoxia-inducible factor (HIF-1) transcription factor is the master regulator of hypoxia-inducible genes (1), making it a tumor marker and an attractive target for pharmacologic interventions to block tumor progression (2). A number of HIF-inducible genes often upregulated in tumors, such as (CA9) and n-myc-down regulated 1 (NDRG1)/calcium activated protein (Cap43), are also used as tumor markers (3–5). CA9 was first identified as a marker protein expressed in several types of human carcinomas but not in the corresponding non-cancerous tissues (6). Additionally, its overexpression has been identified in a number of solid tumors, including renal carcinoma, particularly renal clear cell carcinoma (7,8), non-small cell lung carcinoma (4,8), head and neck squamous cell carcinoma (8,9) and cervix carcinoma (10). CA9 is strongly induced by hypoxia and correlates with a poor response to classic chemo- and radiotherapies, thus representing an interesting target for anticancer drug development (11). NDRG1/Cap43, which we originally cloned as a nickel-inducible gene (12), was found to be induced by hypoxia in HIF-dependent manner (13). NDRG1/Cap43 protein is expressed at low levels in normal tissues; however, its level increases in a variety of cancers, including lung, brain, melanoma, liver, prostate, breast and renal cancers (5,14), and in some cases, it serves as an indicator of poor prognosis (15). Another gene strongly induced by hypoxia is lysyloxidase (LOX), which codes for the extracellular matrix protein lysyl oxidase. It was shown recently that the elevation of LOX correlates with metastatic disease and is essential for hypoxia-induced metastasis (16). The inhibition of LOX expression eliminates metastasis in mice with orthotopically grown breast cancer tumors. This makes LOX a good therapeutic target for preventing and treating metastases.
Since these genes are potential targets for therapeutic intervention, the questions are whether HIF-1 is solely responsible for their activation in tumors and will the pharmacologic inhibition of HIF-1 also decrease their expression or are there any other factors that might play a role in the activation of these genes in growing tumors? It was shown recently that ELK1 is involved in hypoxic induction of the HIF-2α-dependent genes (17,18).
In the experiments reported in this manuscript, we used high cell density as a condition that might mimic the pathophysiologic situation in growing tumor, most probably representing nutritional starvation. We observed that high cell density, under normoxic conditions, upregulates CA9, NDRG1/Cap43 and lysyloxidase like 2 (LOXL2) genes in human and mouse cells in both an HIF-dependent and an HIF-independent manner, suggesting that, in addition to HIF-1, another transcription factor is involved. Previously, we have shown that HIF-dependent, but not HIF-independent, gene upregulation by cell density was sensitive to ascorbate (19). In this set of studies, we found that the activation of the ETS1 transcription factor, which appeared to be ascorbate independent, is critical for the upregulation of CA9, NDRG1/Cap43 and LOXL2 genes in conditions of high cell density. Thus, we can conclude that several signaling pathways could control the expression of hypoxia-inducible genes. Exploration of these pathways is critical for designing of new approaches directed at overcoming tumor resistance to therapeutic interventions.
Materials and methods
Reagents
L-Ascorbic acid was obtained from Sigma (St Louis, MO). The complete mini protease inhibitor cocktail was purchased from Roche (Indianapolis, IN).
Cell culture
The 1HAEo− cell line was obtained from Dr D.C.Gruenert (University of Vermont) (20). Cells were grown on the mixture of bovine serum albumin (Invitrogen Corporation, Carlsbad, CA) and collagen (Cohesion, Palo Alto, CA) in Minimum Essential Medium with Earle’s modified salts (Invitrogen) containing 10% fetal calf serum, 2 mM L-glutamine, 100 μg/ml streptomycin and 100 U/ml penicillin. The A549 human lung adenocarcinoma cells were purchased from American Type Culture Collection (Manassas, VA) and maintained in F12K medium containing 10% fetal calf serum, 2 mM L-glutamine, 100 μg/ml streptomycin and 100 U/ml penicillin. Mouse embryo fibroblasts (HIF-1α+/+) and cells with HIF-1α knockout (HIF-1α−/−) were described previously (13). Mouse fibroblast-like SP1−/− cells were kindly provided by Dr J.Boss (Emory University) and were cultured as recommended by the supplier (21).
Cell density experiments were carried out as follows: 1HAEo− and A549 cells were plated at low density (LD) (2 × 104 cells/cm2) or high density (HD) (8 × 104 cells/cm2); since HIF-1α+/+ and HIF-1α−/−or SP1−/− cells grew faster than human cells, they were plated at LD (104 cells/cm2), intermediate (2.6 × 104 cells/cm2) and HD (4.3 × 104 cells/cm2). After 30 h, cells were collected for protein or RNA extraction. In some instances, cells were plated at LD and allowed to grow for 3–4 days. No differences were found between these two plating techniques.
Histological samples
The preparation and staining of normal and tumor tissues for histological evaluation have been described previously (22).
Real-time polymerase chain reaction
Total RNAs were extracted with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions, and 1 μg total RNA was reverse transcribed in 50 μl reaction using Taqman Reverse Transcription Reagents (Applied Biosystems, Foster City, CA). The expression was analyzed by real-time polymerase chain reaction (PCR) assays using Taqman assay-on-demand predesigned set with β-actin as a reference gene (cat#4326315E) (Applied Biosystems). Following primers were used for expression analyses of hypoxia-inducible genes: CA9 (Hs00154208_ml and Mm00519870_ml), ETS1 (Hs00428287 and Mm00468970), NDRG1/Cap43 (Hs00608389) and LOXL2 (Hs00158757_ml and Mm00804740). All primers spanned exon–exon junctions and were therefore messenger RNA/complementary DNA specific. Expression of each gene was analyzed in triplicates and repeated at least twice. Statistical analysis of individual gene expression was performed with Student’s t-test (two-tailed, unpaired).
Gene profiling analyses with microarrays
Cancer Profiling Arrays II, purchased from BD Biosciences Clontech (Palo Alto, CA), were used to analyze the expression of CA9 and NDRG1/Cap43 hypoxia-inducible genes in normal and tumor tissues. The arrays were hybridized with specific probes for CA9 or NDRG1/Cap43 labeled with 32P-α-deoxycytidine triphosphate. The images were obtained using the Packard Cyclone Storage Phosphor System (PerkinElmer, Shelton, CT).
Western blot analysis
Nuclei for nuclear extracts preparation were obtained as described earlier (23). Nuclei were resuspended in 50–150 μl of extraction buffer [0.42 M NaCl, 25% glycerol, 2 mM MgCl2, 0.2 mM ethylenediaminetetraacetic acid, 1 mM dithiothreitol and 0.5 mM phenylmethylsulfonyl fluoride; antipain 10 μg/ml, aprotinin 1.5 μg/ml, leupeptin 3 μg/ml and pepstatin 3 μg/ml; a cocktail of phosphatase inhibitors (Upstate Cell Signaling Solutions, Lake Placid, NY) and 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.9] and incubated on ice for 15 min, followed by maximum speed and 12 000g centrifugation for 15 min at 4°C. Supernatants were aliquoted and stored at −70°C. Fifteen micrograms of nuclear extracts were used for western blots. Western blot analysis for the hypoxia-inducible NDRG1/Cap43 and CA9 protein levels was performed with total cell extracts using rabbit polyclonal antibody, as described previously (24). CA9 antibodies were kindly provided by Dr E.Stanbridge; HIF-1α antibodies were purchased from BD Biosciences (Palo Alto, CA) and ETS1 and PEA3 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against β-actin were purchased from Abcam (Cambridgeshire, UK). Immunoreactive bands were detected using horseradish peroxidase-linked secondary antibody and enhanced chemiluminescence (PerkinElmer).
Reporter assays
A 730 bp promoter region (–330 to +400) of NDRG1/Cap43 was obtained with PCR using genomic DNA isolated from A549 cells. PCR was carried out under standard conditions using forward primer 5′-ACTCGGAGGGACTGCA-3′ and reverse primer 5′-GGAGACGACGCCAGGAA-3′ and the annealing temperature of 57°C. The obtained PCR product was cloned in TOPO TA cloning vector (Invitrogen) and verified by sequencing. The DNA fragment was recloned into pGL3 basic vector (Promega, Madison, WI). This fragment contained three potential ETS1-binding sites. The HRE-Luc construct was described earlier (25). The pCMV-Sport6-ETS1 expression vector was from American Type Culture Collection and was kindly provided by Dr Dechun Li (Saint Louis University), and the reference pGL4.73 Renilla vector with SV40 promoter was obtained from Promega. The transient transfection experiments were carried out as described previously (23).
ETS1 suppression with small interfering RNA
ETS1 small interfering RNA (siRNA) (SMARTpool Plus) and non-targeting control siRNA were purchased from Dharmacon (Lafayette, CO). A549 cells were transfected with HiPerFect Transfection Reagent (Qiagen, Valencia, CA) according to the manufacturer’s reverse transfection protocol. Briefly, 1.2 × 106 cells per 60 mm plate were combined with the premade siRNA (final concentration 100 nM) and HiPerFect reagent complexes. After 48 h, cells were counted and split into new 60 mm dishes at a density of 2 × 104 cells/cm2 (LD) or 8 × 104 cells/cm2 (HD). Thirty hours later, RNA was purified using TRIzol method, and the expression of ETS1, CA9 or NDRG1/Cap43 was analyzed by real-time PCR using TaqMan Gene Expression Assays (Applied Biosystems).
Results
Expression of CA9 and NDRG1/Cap43 is elevated in human tumors
The elevated expression of CA9 and NDRG1/Cap43 in human tumors has been documented previously (5,8). However, a comparative analysis of the expression of the same genes in the same tumor samples and respective normal tissues has not been reported. Using Cancer Profiling Array II, we showed that both CA9 and NDRG1/Cap43 were significantly upregulated in tumors, as compared with their expression in normal tissues (Figure 1A and B). Both CA9 and NDRG1/Cap43 were upregulated in tumors of colon, kidney, lung, ovary, rectum, cervix, uterus, breast and pancreas, with the strongest induction in tumors of kidney, colon and lung for CA9 and kidney, lung and ovary for NDRG1/Cap43. In many cases, the same tumor samples displayed strong expression of both genes. For example, in lung tumors, samples 2, 3, 5 and 10 had strong expression of both NDRG1/Cap43 and CA9 genes (Figure 1A and B), suggesting similar regulation in these tumors. However, in some tumors of kidney, pancreas and colon, distinct patterns of NDRG1/Cap43 and CA9 expression were also found (Figure 1A and B), suggesting that in addition to hypoxia other mechanisms could be involved in the expression regulation of these genes.
Fig. 1.
Overexpression of CA9 (A) and NDRG1/Cap43 (B) in tumors in comparison with matched normal tissues. Cancer Profiling Arrays II was purchased from BD Biosciences Clontech. They represent RNA samples from 19 tumor types (T) and matched normal tissues (N) that are spotted onto membranes and arranged in columns from 1 to 10 from top to bottom. Probes corresponding to CA9 or NDRG1/Cap43 were hybridized with the membranes as indicated in Materials and Methods.
The upregulation of both tumor markers also occurs at the protein level. As an example, the immunohistochemical staining of NDRG1/Cap43 protein is shown in Figure 2. NDRG1/Cap43 protein expression showed an intense staining of tumor cells, but not the neighboring tissue, in several lung adenocarcinomas (Figure 2). At higher magnification, the plasma membrane location of this protein is clearly seen. The detailed analysis of CA9 protein expression in different tumors has been reported previously by one of us (8).
Fig. 2.
Expression of NDRG1/Cap43 protein in human normal lungs and lung tumors. Tissue sections were stained with antibodies against NDRG1/Cap43 protein as shown in Materials and Methods. Two upper panels represent normal human lungs. Middle and bottom panels represent lung tumors. Overexpression of NDRG1/Cap43 protein is clearly seen in tumor cells. The magnification is shown below each image.
HIF-independent induction of hypoxic genes
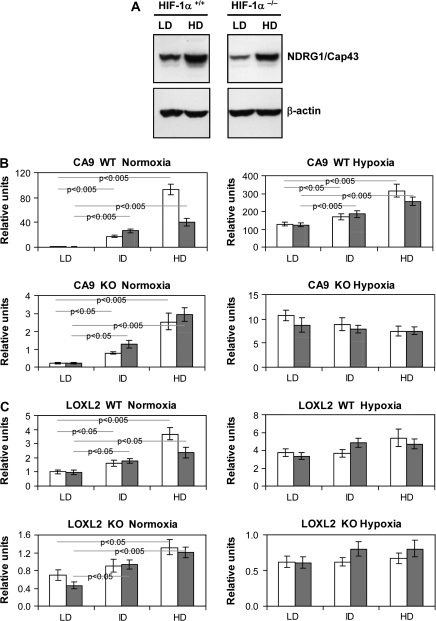
Both our group and others have shown the dependence of CA9 and NDRG1/Cap43 expression on the von hippel-lindau/HIF pathway (13,26,27). CA9 and NDRG1/Cap43 can also be upregulated by high cell density under normoxic conditions (19,27,28). In order to understand whether HIF-1 transcription factor is involved in this upregulation, we first investigated the effect of cell density on the expression of these genes in HIF-1α+/+ and HIF-1α−/− mouse fibroblasts. The basal level of NDRG1/Cap43 was lower in HIF-1α−/− cells, but in both cell lines, NDRG1/Cap43 protein expression was induced by high cell density (Figure 3A).
Fig. 3.
Upregulation of hypoxia-inducible genes by cell density in mouse fibroblasts. (A) Western blot analysis of NDRG1/Cap43 expression in HIF-1α+/+ and HIF-1α−/− mouse fibroblasts grown at low density (LD) and high density (HD). Real-time PCR was performed using CA9 primers (B) or LOXL2 primers (C), as described in Materials and Methods. For real-time PCR, HIF-1α+/+ and HIF-1α−/− mouse fibroblasts were grown at LD, intermediate density (ID) or HD in the absence (open bars) or presence (gray bars) of 50 μM ascorbate. Total RNA was extracted and reverse transcribed for real-time PCR analysis. The results are expressed as relative units and represent means ± SDs from three replicates. In case of significant differences, P values are shown on the graphs. For each gene, upper panel represents data for HIF-1α+/+ cells and lower panel represents data for HIF-1α−/− cells.
To determine whether high cell density affects expression of other HIF-dependent genes, such as CA9 and LOXL2, we utilized real-time PCR. The results, shown in Figure 3B and C, indicate that these genes are also upregulated by the cell density in an HIF-independent manner. It is important to note that upregulation of these genes by cell density was much stronger in HIF-1α+/+ cells. In these cells, CA9 that was upregulated under hypoxic culture conditions was further upregulated by high cell density. These data indicate that HIF-1 plays an important role in this upregulation, but another transcription factor is activated by high cell density independent of HIF-1.
In previously reported studies, we showed that HIF-1α and HIF-2α were induced by increased cell density, and this upregulation could be alleviated by the addition of ascorbate (19). Therefore, we tested the comparative effect of ascorbate on CA9 and LOXL2 induction by increased cell density using HIF-1α+/+ and HIF-1α−/− cells. The addition of ascorbate to cultures decreased the induction of CA9 and LOXL2 in HIF-1α+/+ cells but had little effect in HIF-1α−/− cells (Figure 3B and C). Since ascorbate downregulates the HIF-1 activity by activating HIF-prolyl hydroxylases, it was conceivable that cell density activated another transcription factor, which is not dependent on hydroxylation. Collectively, these data indicated the activation of another transcription factor by cell density; this other transcription factor may cooperate with HIF-1 and may be involved in the upregulation of hypoxia-inducible genes.
ETS1 is involved in the upregulation of hypoxic genes
A number of candidate transcription factors were proposed to be involved in upregulating hypoxia-inducible genes by cell density. For example, it has been reported that upregulation of CA9 by cell density is dependent on SP1 activity (28). All three promoters, CA9, LOXL2 and NDRG1/Cap43, contain clusters of SP1-binding sites. To determine if SP-1 is involved, we analyzed the upregulation of NDRG1/Cap43 in SP1−/− mouse cells. Figure 4B shows density-dependent upregulation of NDRG1/Cap43 in these cells. The upregulation of NDRG1/Cap43 was similar to that observed in HIF-1α+/+ mouse fibroblasts. Thus, the absence of the SP1 transcription factor did not preclude NDRG1/Cap43 activation by cell density. Additionally, we were not able to detect any difference in the amounts of SP1 protein in sparse and dense cultures of 1HAEo− cells (data not shown). These data suggest that SP1 is neither activated by cell density nor a transcription factor involved in upregulating hypoxia-inducible genes by cell density.
Fig. 4.
Transcription factor involved in upregulation of hypoxic genes by cell density. (A) Schematic representation of NDRG1/Cap43 and CA9 promoter regions. Potential transcription factor-binding sites are underlined. TSS, transcription start site. (B) Western blot analysis of density-dependent NDRG1/Cap43 expression in SP1−/− mouse cells. Cells were plated at initial density of 104 cells/cm2. Total cell extracts isolated after 5 days were resolved over sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane and probed with antibodies against NDRG1/Cap43. (C) Density-dependent expression of NDRG1/Cap43 promoter–luciferase construct in 1HAEo− cells. 1HAEo− cells were transfected with the NDRG1/Cap43 promoter–luciferase construct and pGL4 Renilla. Next day, cells were plated at different densities, shown in the figure. Luciferase expression levels were determined 24 h later. (D) ETS1 upregulates expression of NDRG1/Cap43 promoter–luciferase construct. NDRG1/Cap43 promoter–luciferase reporter was transiently cotransfected with vector only (1), ETS1 expression vector at the ratio of 1:0.5 (2) or ETS1 expression vector at the ratio of 1:1 (3). Luciferase expression levels were determined as in (C).
A previous study proposed that family members of HIF are involved with members of v-ets erythroblastosis virus E26 oncogene homolog (ETS) transcription factors in the regulation of hypoxia-inducible genes (18). To search for a potential binding site for this transcription factor, we used Genomatix software (http://www.genomatix.de) to analyze the promoter regions of three hypoxia-inducible genes, NDRG1/Cap43, CA9 and LOXL2, and found several potential ETS1-binding sites in the promoters of all these genes. The consensus sequence of the binding site for the ETS1 transcription factor, PuCC/a-GGAA/T-GCPy, was reported earlier (29). Figure 4A shows promoter sequences for NDRG1/Cap43 and CA9 genes with HIF-1- and ETS1-binding sites. The closest position of ETS1- and HIF-1-binding sites was found in the promoter of CA9 gene (Figure 4A).
Next, we cloned a promoter fragment spanning the region from −330 to +400 bp of the NDRG1/Cap43 gene containing ETS1- and HIF-1-binding sites into pGL3 luciferase reporter plasmid. This promoter responded to hypoxia mimetics such as nickel sulfate, dimethyloxallyl glycine and desferoxamine (data not shown). To confirm that this promoter fragment also responds to density-dependent changes, the reporter was transfected into 1HAEo− cells plated at three different densities: 104 cells/cm2, 3 × 104 cells/cm2 and 6 × 104 cells/cm2. The expression of the reporter increased with cell density, suggesting that the density-dependent response element was present in the reporter plasmid (Figure 4C). To test the involvement of the ETS1 transcription factor in the regulation of NDRG1/Cap43 gene, we cotransfected the NDRG1/Cap43 promoter–luciferase construct with the ETS1 expression vector. The cotransfection of ETS1 with the NDRG1/Cap43 promoter–luciferase construct resulted in strong activation of the luciferase reporter, whereas cotransfection with the empty vector had no effect (Figure 4D). Several dilutions of the ETS1 expression vector in cotransfection experiments resulted in dose-dependent response, suggesting the involvement of ETS1 in regulating the expression of the NDRG1/Cap43 gene.
To determine whether ETS1 itself is upregulated by cell density, real-time PCR and western blot analyses were carried out on RNA and nuclear extracts isolated from 1HAEo− and A549 cells, as well as from HIF-1α+/+ and HIF-1α−/− mouse fibroblasts grown at LD and HD (Figure 5A–D). The ETS1 messenger RNA levels were slightly upregulated in A549 and 1HAEo− cells (Figure 5A) and moderately downregulated in HIF-1α+/+ and HIF-1α−/− mouse fibroblasts (Figure 5C). The ETS1 protein level was increased at high cell density in all cell lines (Figure 5B and D); however, particularly strong ETS1 protein induction was observed in mouse fibroblasts. To confirm that upregulation by cell density was specific for ETS1 transcription factor, we examined the levels of another member of the ETS family, PEA3, at LD and HD (Figure 5B). No changes in PEA3 protein level were observed in 1HAEo− cells; moreover, in A549 cells, this protein was undetectable. These data indicate specific upregulation of the ETS1 transcription factor by cell density.
Fig. 5.
ETS1 expression is increased by cell density. (A) ETS1 RNA levels at low density (LD) and high density (HD) in A549 and 1HAEo− cells. Total RNA was extracted and reverse transcribed for real-time PCR. Real-time PCR was performed using ETS1 primers, as described in Materials and Methods. The results are expressed as relative units and represent means ± SDs from three replicates. In case of significant differences, P values are shown on the graphs. (B) Western blot analysis of ETS1 expression in A549 and 1HAEo− cells grown at LD and HD. (C) ETS1 RNA levels at LD and HD in HIF-1α+/+ and HIF-1α−/− mouse fibroblasts. (D) Western blot analysis of ETS1 expression in HIF-1α+/+ and HIF-1α−/− mouse fibroblasts grown at LD and HD.
Targeting ETS1 by siRNA alleviates induction of hypoxia-inducible genes by cell density
To confirm the involvement of the ETS1 transcription factor in the upregulation of hypoxia-inducible genes by cell density, we suppressed ETS1 expression using the siRNA technique. Following transfection of siRNA, the level of ETS1 RNA in A549 cells was 40% lower at LD and 60% lower at HD, as compared with the non-targeting control siRNA (Figure 6A). The real-time PCR showed that the suppression of ETS1 resulted in the decreased expression of LOXL2 (Figure 6B), CA9 (Figure 6C) and NDRG1/Cap43 (Figure 6D) at HD.
Fig. 6.
siRNA targeting ETS1 decreases density-dependent induction of hypoxic genes. A549 cells were transfected with non-targeting (NT) or ETS1-targeting (ETS) siRNA and plated at low density (LD) and high density (HD), as described in Materials and Methods. Total RNA was extracted and reverse transcribed for real-time PCR. Real-time PCR was performed using primers for ETS1 (A), LOXL2 (B), CA9 (C) and NDRG1/Cap43 (D) or β-actin. The results are expressed as relative units and represent means of three measurements ± SDs. The experiments were repeated twice. In case of significant differences, P values are shown on the graphs.
Discussion
Tumor hypoxia is a well-known phenomenon. Cells in hypoxic regions of a tumor are more resistant to the effects of radiation and chemotherapy (30). It is generally accepted that upregulation of the HIF-1 transcription factor and its target genes is an important part of this resistance (31). Therefore, HIF-1 has been considered a tumor marker related to poor prognosis (32,33) and targeting it for inactivation has a significant clinical value (34). However, mounting evidence suggests that, in some cases, upregulation of hypoxia-inducible genes is not linked to the hypoxia or HIF-1α expression (35), suggesting that other transcription factors may be involved in upregulating hypoxia-inducible genes in tumors. For example, in the head and neck squamous cell carcinoma, CA9 demonstrated a positive correlation with HIF-1α and HIF-2α, but the correlation coefficient was low (36).
In normal tissues, cell growth is tightly regulated by mechanisms that depend on growth factors and cell–cell or cell–substrate interactions, and most of these mechanisms are lost or disordered in tumor-forming cell population. Some of these tumor cell-related abnormalities can be studied in cell lines in vitro. The best example is contact inhibition of growth. Another interesting example is the upregulation of hypoxia-inducible genes in dense cell culture. The mechanism of this upregulation is not fully understood. It was suggested that in dense culture in vitro, local (pericellular) hypoxia could be responsible for upregulation of hypoxia-inducible genes (37). However, local hypoxia is unlikely a main factor responsible for this effect in dense culture. Cell culture, in contrast to living tissues that are exposed to ∼4% of oxygen, is exposed to 21% of oxygen in the air. The number of cells per given volume even in dense culture is less than in the tissue. The oxygen is easily diffusing through 5 mm of the medium layer. Additionally, Kaluz et al. (37) recognized that the changes in oxygen concentration near the cell surface of dense culture ‘are relatively small and are not as low as those seen in experimentally induced hypoxic responses’. Hence, high cell density in vitro represents an interesting model for investigating the mechanisms of gene regulation that may occur in a growing tumor.
In this manuscript, we report that the expression of the CA9, NDRG1/Cap43 and LOXL2 hypoxia-inducible genes can be increased by cell density under normal oxygen tension. Previously, we have found that high cell density stabilizes, although not strongly, HIF-1α and HIF-2α (19). This stabilization can be alleviated by the addition of ascorbate. Here, using HIF-1α-proficient mouse fibroblasts, we found both ascorbate-dependent and ascorbate-independent increases in the expression of CA9 and LOXL2 at HD. The ascorbate-sensitive increase in the expression of these genes was most probably due to the HIF-α proteins stabilization, since, in HIF-1α-deficient mouse fibroblasts, which have neither HIF-1α nor functional HIF-2α (38), only the ascorbate-independent component was present. This finding correlated well with the observed lower level of upregulation of hypoxia-inducible genes in HIF-1α-deficient mouse fibroblasts, as compared with HIF-1α-proficient mouse fibroblasts. Similarly, the upregulation of the CA9 promoter by cell density was found in HIF-1α-deficient chinese hamster ovary (CHO) cells, although it was weaker than that found in HIF-1α-proficient CHO cells (37). These data, along with our findings, suggest that the ascorbate-independent component may represent another transcription factor that is upregulated at high cell density.
Since the density-dependent upregulation of hypoxia-inducible genes occurs under normal oxygen tension, the involvement of a transcription factor other than HIF-1 was suggested, namely SP1 (28). However, we and others (28) were not able to detect any changes in the levels of SP1 in sparse versus dense cultures. The lack of modulation of this transcription factor by cell density, as well as the induction of NDRG1/Cap43 in SP1−/− mouse cells, suggested that SP1 was not the transcription factor that upregulates hypoxia-inducible genes in conditions of high cell density.
Searching for other candidates, we have identified putative binding sites for the ETS1 transcription factor in the promoters of CA9, NDRG1/Cap43 and LOXL2 genes. Our results suggest that ETS1 is indeed involved in the density-dependent upregulation of hypoxia-inducible genes. The following arguments support this involvement: first, the ETS1 protein was upregulated by cell density in both human lung epithelial cells and mouse fibroblasts. Another member of the ETS family, PEA3, was not changed by cell density. Second, cotransfection of the ETS1 expression vector with NDRG1/Cap43 promoter–luciferase reporter resulted in a strong and dose-dependent upregulation of the reporter in human 1HAEo− cells. Finally, targeting ETS1 expression with the siRNA resulted in inhibition of upregulation of CA9, NDRG1/Cap43 and LOXL2 genes in high cell density.
ETS1 is an oncoprotein. Its expression was shown in a variety of solid tumors, including epithelial tumors, sarcomas and astrocytomas, and it was increased in invasive, higher grade tumors. (39). High ETS1 levels correlated with poorer prognosis in lung, colorectal and squamous cell carcinomas. Moreover, ETS1 expression was associated with a higher incidence of lymph node metastasis (39). Additionally, it was shown that ETS1 was required for angiogenesis, in part by stimulating the proteases necessary for the endothelial activation program (40) and in part by stimulating vascular endothelial growth factor expression (39,41). Collectively, these data indicate that ETS1 is activated in vivo.
Using immunoprecipitation and chromatin immunoprecipitation assays, the physical interaction between another member of the ETS family, ELK1, and HIF-2α has been shown (17,18). Considering the close proximity of the ETS1- and HIF-1-putative binding sites on NDRG1/Cap43 promoter, it is conceivable that these transcription factors can interact with each other and cooperate in regulating NDRG1/Cap43 transcription. However, such interaction is not obligatory. Here we demonstrated that, in the absence of HIF-1α or HIF-2α, ETS1 alone is sufficient for the upregulation of hypoxia-inducible genes.
Collectively, our finding of the involvement of ETS1 along with HIF-1 in regulating hypoxia-inducible genes points out the necessity for fine-tuning a strategy to fight cancer through more precise targeting of transcription factors activated in a particular tumor. These data imply that more research is needed for better understanding the mechanisms of regulation of hypoxia-inducible genes.
Funding
Intramural Research Program of the National Institutes of Health; National Cancer Institute; Center for Cancer Research.
Acknowledgments
We thank R.Bare for excellent technical assistance and Dr J.Boss for providing Sp1−/− cells. Critical comments of Dr J.Phang are greatly appreciated. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products or organizations imply endorsement by the USA Government.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CA9
carbonic anhydrase 9
- Cap43
calcium activated protein
- ETS
v-ets erythroblastosis virus E26 oncogene homolog
- HD
high density
- HIF-1
hypoxia-inducible factor
- LD
low density
- LOX
lysyloxidase
- LOXL2
lysyloxidase like 2
- NDRG1
n-myc-down regulated 1
- PCR
polymerase chain reaction
- siRNA
small interfering RNA
References
- 1.Semenza GL. Expression of hypoxia-inducible factor 1: mechanisms and consequences. Biochem. Pharmacol. 2000;59:47–53. doi: 10.1016/s0006-2952(99)00292-0. [DOI] [PubMed] [Google Scholar]
- 2.Semenza GL. Development of novel therapeutic strategies that target HIF-1. Expert Opin. Ther. Targets. 2006;10:267–280. doi: 10.1517/14728222.10.2.267. [DOI] [PubMed] [Google Scholar]
- 3.Potter C, et al. Hypoxia inducible carbonic anhydrase IX, marker of tumour hypoxia, survival pathway and therapy target. Cell Cycle. 2004;3:164–167. [PubMed] [Google Scholar]
- 4.Vermylen P, et al. Carbonic anhydrase IX antigen differentiates between preneoplastic malignant lesions in non-small cell lung carcinoma. Eur. Respir. J. 1999;14:806–811. doi: 10.1034/j.1399-3003.1999.14d14.x. [DOI] [PubMed] [Google Scholar]
- 5.Cangul H, et al. Enhanced expression of a novel protein in human cancer cells: a potential aid to cancer diagnosis. Cell Biol. Toxicol. 2002;18:87–96. doi: 10.1023/a:1015376032736. [DOI] [PubMed] [Google Scholar]
- 6.Pastorek J, et al. Cloning and characterization of MN, a human tumor-associated protein with a domain homologous to carbonic anhydrase and a putative helix-loop-helix DNA binding segment. Oncogene. 1994;9:2877–2888. [PubMed] [Google Scholar]
- 7.McKiernan JM, et al. Expression of the tumor-associated gene MN: a potential biomarker for human renal cell carcinoma. Cancer Res. 1997;57:2362–2365. [PubMed] [Google Scholar]
- 8.Ivanov S, et al. Expression of hypoxia-inducible cell-surface transmembrane carbonic anhydrases in human cancer. Am. J. Pathol. 2001;158:905–919. doi: 10.1016/S0002-9440(10)64038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beasley NJ, et al. Carbonic anhydrase IX, an endogenous hypoxia marker, expression in head and neck squamous cell carcinoma and its relationship to hypoxia, necrosis, and microvessel density. Cancer Res. 2001;61:5262–5267. [PubMed] [Google Scholar]
- 10.Loncaster JA, et al. Carbonic anhydrase (CA IX) expression, a potential new intrinsic marker of hypoxia: correlations with tumor oxygen measurements and prognosis in locally advanced carcinoma of the cervix. Cancer Res. 2001;61:6394–6399. [PubMed] [Google Scholar]
- 11.Thiry A, et al. Targeting tumor-associated carbonic anhydrase IX in cancer therapy. Trends Pharmacol. Sci. 2006;27:566–573. doi: 10.1016/j.tips.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Zhou D, et al. Cap43, a novel gene specifically induced by Ni2+ compounds. Cancer Res. 1998;58:2182–2189. [PubMed] [Google Scholar]
- 13.Salnikow K, et al. Carcinogenic nickel induces genes involved with hypoxic stress. Cancer Res. 2000;60:38–41. [PubMed] [Google Scholar]
- 14.Ellen T, et al. NDRG1, a growth and cancer related gene: regulation of gene expression and function in normal and disease states. Carcinogenesis. 2008;29:2–8. doi: 10.1093/carcin/bgm200. [DOI] [PubMed] [Google Scholar]
- 15.Chua MS, et al. Overexpression of NDRG1 is an indicator of poor prognosis in hepatocellular carcinoma. Mod. Pathol. 2007;20:76–83. doi: 10.1038/modpathol.3800711. [DOI] [PubMed] [Google Scholar]
- 16.Erler JT, et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 17.Elvert G, et al. Cooperative interaction of hypoxia-inducible factor-2alpha (HIF-2alpha) and Ets-1 in the transcriptional activation of vascular endothelial growth factor receptor-2 (Flk-1) J. Biol. Chem. 2003;278:7520–7530. doi: 10.1074/jbc.M211298200. [DOI] [PubMed] [Google Scholar]
- 18.Aprelikova O, et al. Role of ETS transcription factors in the hypoxia-inducible factor-2 target gene selection. Cancer Res. 2006;66:5641–5647. doi: 10.1158/0008-5472.CAN-05-3345. [DOI] [PubMed] [Google Scholar]
- 19.Karaczyn A, et al. Ascorbate depletion mediates up-regulation of hypoxia-associated proteins by cell density and nickel. J. Cell. Biochem. 2006;97:1025–1035. doi: 10.1002/jcb.20705. [DOI] [PubMed] [Google Scholar]
- 20.Gruenert DC, et al. Culture and transformation of human airway epithelial cells. Am. J. Physiol. 1995;268:L347–L360. doi: 10.1152/ajplung.1995.268.3.L347. [DOI] [PubMed] [Google Scholar]
- 21.Ping D, et al. Sp1 binding is critical for promoter assembly and activation of the MCP-1 gene by tumor necrosis factor. J. Biol. Chem. 2000;275:1708–1714. doi: 10.1074/jbc.275.3.1708. [DOI] [PubMed] [Google Scholar]
- 22.Cangul H, et al. Enhanced overexpression of an HIF-1/hypoxia-related protein in cancer cells. Environ. Health Perspect. 2002;110(suppl. 5):783–788. doi: 10.1289/ehp.02110s5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salnikow K, et al. Depletion of intracellular ascorbate by the carcinogenic metals nickel and cobalt results in the induction of hypoxic stress. J. Biol. Chem. 2004;279:40337–40344. doi: 10.1074/jbc.M403057200. [DOI] [PubMed] [Google Scholar]
- 24.Salnikow K, et al. The regulation of hypoxic genes by calcium involves c-Jun/AP-1, which cooperates with hypoxia-inducible factor 1 in response to hypoxia. Mol. Cell. Biol. 2002;22:1734–1741. doi: 10.1128/MCB.22.6.1734-1741.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salnikow K, et al. Nickel-induced transformation shifts the balance between HIF-1 and p53 transcription factors. Carcinogenesis. 1999;20:1819–1823. doi: 10.1093/carcin/20.9.1819. [DOI] [PubMed] [Google Scholar]
- 26.Wykoff CC, et al. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res. 2000;60:7075–7083. [PubMed] [Google Scholar]
- 27.Ivanov SV, et al. Down-regulation of transmembrane carbonic anhydrases in renal cell carcinoma cell lines by wild-type von Hippel-Lindau transgenes. Proc. Natl Acad. Sci. USA. 1998;95:12596–12601. doi: 10.1073/pnas.95.21.12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaluz S, et al. Expression of the hypoxia marker carbonic anhydrase IX is critically dependent on SP1 activity. Identification of a novel type of hypoxia-responsive enhancer. Cancer Res. 2003;63:917–922. [PubMed] [Google Scholar]
- 29.Nye JA, et al. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992;6:975–990. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- 30.Vaupel P, et al. Hypoxia and anemia: effects on tumor biology and treatment resistance. Transfus. Clin. Biol. 2005;12:5–10. doi: 10.1016/j.tracli.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Acker T, et al. A role for hypoxia and hypoxia-inducible transcription factors in tumor physiology. J. Mol. Med. 2002;80:562–575. doi: 10.1007/s00109-002-0355-1. [DOI] [PubMed] [Google Scholar]
- 32.Kim SJ, et al. Expression of HIF-1alpha, CA IX, VEGF, and MMP-9 in surgically resected non-small cell lung cancer. Lung Cancer. 2005;49:325–335. doi: 10.1016/j.lungcan.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 33.Hui EP, et al. Coexpression of hypoxia-inducible factors 1alpha and 2alpha, carbonic anhydrase IX, and vascular endothelial growth factor in nasopharyngeal carcinoma and relationship to survival. Clin. Cancer Res. 2002;8:2595–2604. [PubMed] [Google Scholar]
- 34.Semenza GL. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 35.Mayer A, et al. Carbonic anhydrase IX expression and tumor oxygenation status do not correlate at the microregional level in locally advanced cancers of the uterine cervix. Clin. Cancer Res. 2005;11:7220–7225. doi: 10.1158/1078-0432.CCR-05-0869. [DOI] [PubMed] [Google Scholar]
- 36.Winter SC, et al. The relation between hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression with anemia and outcome in surgically treated head and neck cancer. Cancer. 2006;107:757–766. doi: 10.1002/cncr.21983. [DOI] [PubMed] [Google Scholar]
- 37.Kaluz S, et al. Lowered oxygen tension induces expression of the hypoxia marker MN/carbonic anhydrase IX in the absence of hypoxia-inducible factor 1 alpha stabilization: a role for phosphatidylinositol 3′-kinase. Cancer Res. 2002;62:4469–4477. [PubMed] [Google Scholar]
- 38.Park SK, et al. Hypoxia-induced gene expression occurs solely through the action of hypoxia-inducible factor 1alpha (HIF-1alpha): role of cytoplasmic trapping of HIF-2alpha. Mol. Cell. Biol. 2003;23:4959–4971. doi: 10.1128/MCB.23.14.4959-4971.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dittmer J. The biology of the Ets1 proto-oncogene. Mol. Cancer. 2003;2:29. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwasaka C, et al. Ets-1 regulates angiogenesis by inducing the expression of urokinase-type plasminogen activator and matrix metalloproteinase-1 and the migration of vascular endothelial cells. J. Cell. Physiol. 1996;169:522–531. doi: 10.1002/(SICI)1097-4652(199612)169:3<522::AID-JCP12>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 41.Hashiya N, et al. In vivo evidence of angiogenesis induced by transcription factor Ets-1: ets-1 is located upstream of angiogenesis cascade. Circulation. 2004;109:3035–3041. doi: 10.1161/01.CIR.0000130643.41587.DB. [DOI] [PubMed] [Google Scholar]