Abstract
Malignant pleural mesothelioma (MPM) is a rapidly fatal tumor with increasing incidence worldwide responsible for many thousands of deaths annually. Although there is a clear link between exposure to asbestos and mesothelioma, and asbestos is known to be both clastogenic and cytotoxic to mesothelial cells, the mechanisms of causation of MPM remain largely unknown. However, there is a rapidly emerging literature that describes inactivation of a diverse array of tumor suppressor genes (TSGs) via promoter DNA CpG methylation in MPM, although the etiology of these alterations remains unclear. We studied the relationships among promoter methylation silencing, asbestos exposure, patient demographics and tumor histology using a directed approach; examining six cell cycle control pathway TSGs in an incident case series of 70 MPMs. Promoter hypermethylation of APC, CCND2, CDKN2A, CDKN2B, HPPBP1 and RASSF1 were assessed. We observed significantly higher lung asbestos body burden if any of these cell cycle genes were methylated (P < 0.02), and there was a significant trend of increasing asbestos body counts as the number of methylated cell cycle pathway genes increased from 0 to 1 to >1 (P < 0.005). This trend of increasing asbestos body count and increasing number of methylated cell cycle pathway genes remained significant (P < 0.05) after controlling for age, gender and tumor histology. These data suggest a novel tumorigenic mechanism of action of asbestos and may contribute to the understanding of precisely how asbestos exposure influences the etiology and clinical course of malignant mesothelioma.
Introduction
Malignant pleural mesothelioma (MPM) is a highly lethal neoplasm linked with asbestos exposure in ∼70–80% of patients. Worldwide, the incidence of MPM is rising with ∼3000 cases per year reported in the USA, 2000 cases per year in Great Britain and 500 cases per year in Japan (1–5). The costs associated with compensation for asbestos-related disease and asbestos remediation have been estimated at $265 billion in the USA alone over the next 40 years (6). In addition, serious attention has recently been given to the potential contribution of exposure to the dust from the collapse of the World Trade Center towers in New York City to an increased risk for multiple serious conditions including MPM (7).
Since asbestos use has been on the decline for first world nations for 20–30 years, the overall incidence of MPM is expected to peak in the next few years in both USA and Europe (8,9). At the same time, asbestos continues to be mined, exported and widely used in many third world countries (10). Many nations including China, India and some Latin American countries are still importing vast amounts of asbestos (11,12). This fact, combined with the long 20–50 years latency of MPM, virtually assures that the MPM epidemic will continue for decades to come. This necessitates continuing research into the molecular genetic consequences of exposure to asbestos in an effort to better understand MPM pathogenesis, hopefully translating to prevention strategies and improved patient outcomes.
The pathogenic mechanisms of asbestos contributing to the development of MPM have long been studied, though they remain incompletely characterized. Many in vitro studies have demonstrated both clastogenic and cytotoxic effects of asbestos fibers (13,14). Phagocytosis of fibers by macrophages and oxidoreduction reactions on fiber surfaces are known to generate genotoxic reactive oxygen species that are capable of inducing DNA damage (15–17) and leading to genetic alterations in MPM (18). In addition to genetic alterations, the rapidly emerging literature indicates that epigenetic tumor suppressor gene (TSG) silencing via promoter methylation occurs in MPM (19–31). Methylation of cytosines in the context of promoter CpG islands of TSGs is a well-established mechanism of stable gene silencing in human cancers (32,33). However, the precise mechanisms underlying the induction of TSG methylation and the factors that influence tumor-specific methylation profiles are incompletely understood. Exposure to carcinogens has been associated with TSG methylation silencing, and recently, simultaneous examination of multiple TSGs involved in different cellular pathways and processes has suggested that genes are phenotypically selected for silencing. Initial studies demonstrated that there is a dose response for methylation silencing of CDKN2A by tobacco smoke in lung cancer (34,35). Indeed, in lung adenocarcinoma, methylation of TSGs CDKN2A and APC also was significantly associated with exposure to tobacco smoke (36). Dammann et al. (37) have shown that asbestos exposure is significantly associated with methylation at CDKN2A in non-small cell lung cancer. Suzuki et al. (21) reported that methylation of RRAD, APPBP1, CCND2, RASSF1 and TMS1 was significantly more prevalent in SV40-positive MPM. Furthermore, in a recent study of 28 TSG loci in MPM, Tsou et al. (31) found a significant association between methylation of two TSGs; MT1A and MT2A with self-reported asbestos exposure. Taken together, these data strongly suggest that asbestos exposure may act to induce methylation silencing of TSGs. However, it remains unclear if this is a direct or indirect selection for TSG inactivation across phenotypically important pathways; if the process is stochastic and less phenotypically driven or whether a dose response exists between exposure and methylation extent. To examine this question, we have focused our efforts upon TSGs in the cell cycle control and proliferation pathway. We studied the APC, CCND2, CDKN2A, CDKN2B, HPPBP1 and RASSF1 genes for promoter hypermethylation in 70 incident cases of MPM. These genes were chosen as both a part of a larger pathway-based group of genes studied in our laboratory—in this and other types of human cancers—and because they are generally considered among the most important cell cycle control TSGs known to be inactivated via methylation in cancer (38–40). We examined whether methylation of specific genes, methylation at any of these loci or methylation of an increasing number of genes was associated with asbestos exposure, patient demographic variables or tumor histology. In this process, we were fortunate to have quantitative asbestos burden data to explore the relationship between exposure and epigenetic gene inactivation in MPM.
Materials and methods
Study population
Tumor material was obtained following surgical resection at Brigham and Women’s Hospital through the support of the International Mesothelioma Program. All patients provided informed consent under the approval of the appropriate Institutional Review Boards. Clinical information, including pathological diagnosis, was obtained from medical record review. Each patient was assessed for history of exposure to asbestos as well as additional demographic and environmental data by obtaining their medical and occupational history with an in-person questionnaire or interview. Patients were followed up for survival using the death index and last known clinic visit.
Methylation analysis
Tumor DNA was extracted from frozen tissue using the QIAamp DNA mini kit according to the manufacturer’s protocol (Qiagen, Valencia, CA). Tumor DNA was modified by sodium bisulfite to convert unmethylated cytosines to uracil using the EZ DNA Methylation Kit (Zymo Research, Orange, CA) according to the manufacturer’s protocol. Methylation-specific polymerase chain reaction (PCR) analysis was conducted with modified template DNA as described previously (41). PCR was performed with 50 ng of modified DNA in a mixture with 1x PCR buffer (Applied Biosystems, Foster City, CA), 0.2 mM dNTPs, 0.5 μM primers and 1.25 U of Ampli Taq Gold (Applied Biosystems) in a total volume of 25 μl. PCR products were analyzed by electrophoresis in 3% agarose gel. Sodium bisulfite-modified DNAs from circulating blood lymphocytes of healthy control subjects, untreated and treated with SssI DNA methylase, were used as negative and positive controls, respectively, in each run. In addition, no template-negative controls were also present in each run. All methylation-specific PCR reactions are optimized to detect ∼5% methylation in the sample, consistent with the cutoff values generally utilized in quantitative assays of methylation (42,43).
Asbestos body burden
Quantification of asbestos bodies was done using the protocol of Churg et al. (44). Portions of normal lung tissue (1–4 g) obtained from surgery were blotted to remove excess liquid, weighed, minced and digested with sodium hypochlorite. This was mixed, vented and then sealed for 48 h. Following digestion, samples were pelleted, resuspended in 25 ml of 50% ethanol and 10 ml of chloroform, vortexed and 15 ml of chloroform was added. Samples were then gently centrifuged for 10 min, supernatant was aspirated, pellets were resuspended in 25 ml of 25% ethanol and then mixed well and filtered through a 0.45 μm Millipore filter (Millipore, Billerica, MA). Sample tubes were washed twice with 25 ml of 25% ethanol and filtered. Similarly, the sides of the filter funnel were washed with 25 ml of 25% ethanol and filtered. Filters were dehydrated, cleared twice for 1 min each in 95% ethanol, 100% ethanol and then xylene, cut in half, recleared in xylene for another minute, mounted on microscope slides with a counting grid using Permount™ Mounting Medium (Fisher Scientific, Hampton, NH) and dried flat. Asbestos bodies were then counted, and the asbestos bodies per (wet weight) gram of lung were calculated with the following equation: number of asbestos bodies/(squares counted × 100.74 × weight in grams of the digested tissue sample).
Statistical analysis
Univariate tests for association between methylation at each of the cell cycle genes and patient demographic, tumor characteristic and exposure variables were carried out with the appropriate statistical tests using SAS analysis software. Similarly, tests for association between methylation at zero, one or greater than one gene, and patient demographic, tumor characteristic and exposure variables also were performed. Simple linear regression was used to test for association between the number of methylated cell cycle genes and asbestos body count. Finally, an ordered logistic model (SAS PROC PROBIT) predicting the number of methylated cell cycle genes was used to control for potential confounders and evaluate the contribution of asbestos body levels to cell cycle gene methylation.
Results
A total of 83 cases had available asbestos body burden data. Among the cases with available asbestos body counts, there were 3 extreme outliers (14 870, 19 681 and 303 852 compared with the median count 158) and 10 cases with zero counts. As pleural mesothelioma arising without detectable asbestos exposure may have a distinct etiology and biology, and in an effort to avoid an analysis anchored by extreme values, we did not include tumors with zero asbestos body counts or the extreme outliers in the analysis, restricting it to the remaining 70 cases. We investigated the methylation status of six cell cycle control-associated genes: CDKN2A, CDKN2B, RASSF1, CCND2, APC and APPBP1. Exposure, demographic and tumor characteristic data for these 70 cases are in presented in Table I.
Table I.
Mesothelioma patient demographics and tumor characteristics
| Gender, n (%) | |
| Female | 14 (20) |
| Male | 56 (80) |
| Patient age | |
| Range | 30–80 |
| Mean (SD) | 62 (9.1) |
| Histology, n (%) | |
| Epithelioid | 54 (77) |
| Mixed | 14 (20) |
| Sarcomatoid | 2 (3) |
| Asbestos exposurea, n (%) | |
| Yes | 53 (76) |
| No | 17 (24) |
| Asbestos body count | |
| Range | 6–6211 |
| Mean (SD) | 1000 (1529) |
Self-reported.
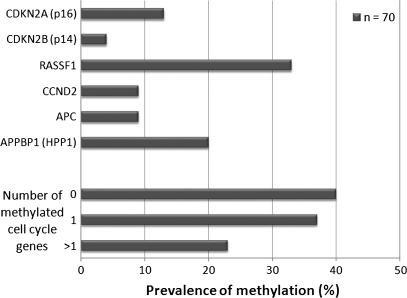
The prevalence of methylation among the cell cycle control genes varied; RASSF1 was methylated in 33% (n = 22) of cases, APPBP1 in 20% (n = 14), CDKN2A in 13% (n = 9), APC and CCND2 in 9% (n = 6) and finally CDKN2B was methylated in 4% (n = 3) of cases (Figure 1). We found no significant associations between patient gender or tumor histology and methylation at any of the six individual loci examined. However, patients with RASSF1 methylation were significantly older (65 ± 6.6 years) than patients without RASSF1 methylation (61 ± 9.5 years) (P < 0.05). We observed a similar relationship between methylation of CCND2 and older age (methylated, 69 ± 8.2 years; unmethylated, 61 ± 8.9 years) (P < 0.05). We then asked whether this relationship with age was a more general phenomenon and found that methylation at any (one or more gene) of the six TSGs was significantly associated with increased age (methylated, 64 ± 8.1 years; unmethylated, 58 ± 9.4 years) (P < 0.01). Since all these genes are involved in the process of cell cycle control, we grouped cases into three categories; cases with no genes methylated, cases with one gene methylated and cases with more than one cell cycle control gene methylated. Figure 1 displays the prevalence of methylation of zero (40%, n = 28), one (37%, n = 26) or more than one (23%, n = 16) cell cycle pathway gene.
Fig. 1.
Prevalence of cell cycle control gene methylation in pleural mesothelioma. Prevalence of methylation-positive cell cycle control genes among pleural mesotheliomas as measured by methylation-specific PCR, and prevalence of tumors with zero, one or more than one methylation-positive cell cycle control gene.
Next, we examined the relationship between cell cycle control gene methylation and exposure to asbestos using both self-reported and quantitative asbestos body counts as exposure variables. While we found no significant associations between methylation at any one of the six genes and self-reported asbestos exposure, cases with RASSF1 methylation had significantly higher asbestos body counts (mean count = 698) compared with cases without RASSF1 methylation (mean count = 409) (P < 0.01, Wilcoxon test). Similarly, there was no significant relationship between methylation of any cell cycle gene (comparing samples with no genes methylated to those with any genes methylated) and self-reported asbestos exposure. Notably, although we were unable to detect an association between self-reported asbestos exposure and methylation of cell cycle control-related genes, we observed a significant association between reported asbestos exposure and elevated asbestos body count (P < 0.005). We also examined the relationships between asbestos body count and patient age, gender and tumor histology. Although we did not find any association between asbestos body count and age or histology (data not shown), we did observe a significant difference in asbestos body count in males (mean count = 1218) compared with females (mean count = 213) (P < 0.001, Wilcoxon test).
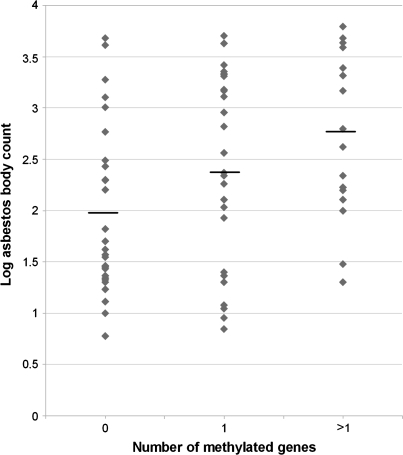
Figure 2 shows that log transformed asbestos body counts are significantly correlated with the number of cell cycle control genes methylated (linear regression F-test, P < 0.005). In order to control for potential confounders of this relationship and to better represent the discreet ordinal nature of the methylation count, we modeled the data using an ordered logistic regression that predicts zero, one or more than one methylated cell cycle pathway gene (Table II). This model indicates that when controlling for gender and tumor histology, both age and asbestos body count are significant predictors (P = 0.04 and P < 0.05, respectively) of an increased number of methylated cell cycle control genes.
Fig. 2.
Asbestos body count versus cell cycle gene methylation. Log transformed asbestos body count (y-axis) is plotted versus the number of methylated cell cycle control genes (x-axis). Using simple linear regression, there is a significant association between increasing asbestos burden and increasing number of methylated cell cycle control genes (P < 0.005, R2 = 0.12).
Table II.
Ordered logistic regression model predicting increasing number of methylated cell cycle control genes in pleural mesothelioma (n = 70)
| Predictor | Estimate | P-value |
| Age | 0.67 | 0.04 |
| Gender | ||
| Male | 1.0 | Referent |
| Female | −0.73 | 0.32 |
| Histology | ||
| Epithelioid | 1.0 | Referent |
| Mixed and sarcomatoid | 0.77 | 0.21 |
| Asbestos body counta | 0.33 | <0.05 |
Scaled to: (asbestos body count per 1000).
Discussion
We evaluated promoter hypermethylation of six cell cycle control and progression pathway genes in an incident case series of 70 MPMs examining whether methylation of specific genes, methylation of any of these loci or methylation of an increasing number of genes was associated with patient demographic variables, tumor histology or asbestos exposure. We chose to study these genes in part because they have been studied by our laboratory as a part of a pathway-based approach to investigating TSG methylation in other human cancers. Further, these genes are known to be subject to inactivation by promoter hypermethylation in cancer and are recognized as important in cell cycle control and progression (38–40). We examined these genes for methylation using methylation-specific PCR. This technique is known to be sensitive to 5% of cells with methylation, and is therefore ample for detecting aberrant methylation events of phenotypic importance (45).
Methylation of RASSF1 has been observed in 32% of MPM (n = 66) and previously significantly associated with SV40 exposure (25). We observed essentially the same prevalence of RASSF1 methylation among our cases (33%), and we also found that RASSF1 methylation was significantly associated with increased asbestos body count. Furthermore, significant, independent associations between older patient age and methylation of CCND2 and RASSF1 were observed. The association between older age and methylation is not unexpected since it is known that CpG island hypermethylation often increases with age (46,47). Also, in another report of TSG methylation in MPM, Toyooka et al. (25) reported prevalences of methylation at the APC, CDKN2A and CDKN2B genes similar to ours.
One great advantage of this study was the availability of a quantitative measure of asbestos exposure. Ferruginous asbestos bodies form as a result of the interaction of macrophages with asbestos fibers, and presence of asbestos bodies is an indicator of past exposure to asbestos. By quantifying their level, we are able to estimate the degree of asbestos fiber burden in an individual (48). In our data, tumors with methylation have significantly higher asbestos exposure, using the asbestos body counts as a quantitative measure of burden. Furthermore, there was a significant trend between increasing number of methylated cell cycle TSGs (0 to 1 to >1) and increasing asbestos body count. Finally, an ordered logistic regression model controlling for gender, and tumor histology, showed that both age and an increasing asbestos body count are independent significant predictors of an increased number of methylated cell cycle pathway genes in MPM.
Hence, these data suggest that the induction of methylation in a phenotypically important pathway might occur as a result of physical interaction between asbestos fibers and the parietal pleura. However, precisely how any exposure selects TSGs for silencing has only recently begun to be explored. Maintenance of control over the cell cycle is critical to tumor suppression, but the relationship between dynamic carcinogen exposure and the targeting and induction of tumor-specific methylation profiles is likely to be highly complex. Asbestos exposure is associated with chronic inflammation (49), and the physical presence of asbestos fibers at the interface of the mesothelial membrane and the lung induces a dose-dependent cycle of death and regrowth of mesothelial cells in the area of fiber deposition (50). Additionally, persistent mitotic stimulation of mesothelial cells after direct physical insult, and reactive oxygen species generated by the fiber clearance-related cellular response, may induce a reaction by mesothelial cells akin to that of cells in culture subject to repeated cycles of growth. Repeated passaging of cells in tissue culture, similar to the process of aging, is associated with the induction of TSG silencing by promoter methylation (51). The known decades-long latency of MPM then suggests that there is ample time for appreciable fields of clonally altered cells to accumulate, perhaps leading to malignancy through a combination of acquired genetic and epigenetic alterations enhanced by repeated mitotic selection. Additionally, asbestos fibers are known to be clastogenic and lead to genotoxic damage, and tumors with higher asbestos fiber burden may be induced to grow faster, possibly leading to the preferential selection of clones with silenced cell cycle control TSGs. While these mechanisms of clonal selection for epigenetic silencing are consistent with our data, it does not necessarily imply any direct asbestos fiber interaction with the histone/DNA methylation machinery, but instead that the chronic inflammation response and/or accelerated tumor growth related to asbestos burden may select for cells capable of continued proliferation.
In summary, using a directed pathway-based approach to methylation analysis and a quantitative measure of asbestos exposure, we observed that methylation silencing of cell cycle TSGs is associated with both older age and asbestos exposure in MPM. Our data, using a quantitative measure of asbestos exposure, demonstrate that epigenetic gene inactivation is a crucial and novel mechanism for asbestos action in the genesis of this rapidly fatal cancer.
Funding
International Mesothelioma Program at Brigham and Women’s Hospital (http://www.impmeso.org/); Mesothelioma Applied Research Foundation; National Institutes of Health/National Institute of Environmental Health Sciences training grant in environmental health sciences (T32ES007155); National Institutes of Health/National Institute of Environmental Health Sciences (P42ES05947); National Cancer Institute (CA126939).
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- MPM
malignant pleural mesothelioma
- PCR
polymerase chain reaction
- TSG
tumor suppressor gene
References
- 1.Morinaga K, et al. Asbestos-related lung cancer and mesothelioma in Japan. Ind. Health. 2001;39:65–74. doi: 10.2486/indhealth.39.65. [DOI] [PubMed] [Google Scholar]
- 2.Pelin K, et al. Expression of cell adhesion molecules and connexins in gap junctional intercellular communication deficient human mesothelioma tumour cell lines and communication competent primary mesothelial cells. Carcinogenesis. 1994;15:2673–2675. doi: 10.1093/carcin/15.11.2673. [DOI] [PubMed] [Google Scholar]
- 3.Price B. Analysis of current trends in United States mesothelioma incidence. Am. J. Epidemiol. 1997;145:211–218. doi: 10.1093/oxfordjournals.aje.a009093. [DOI] [PubMed] [Google Scholar]
- 4.Roushdy-Hammady I, et al. Genetic-susceptibility factor and malignant mesothelioma in the Cappadocian region of Turkey. Lancet. 2001;357:444–445. doi: 10.1016/S0140-6736(00)04013-7. [DOI] [PubMed] [Google Scholar]
- 5.Price B, et al. Mesothelioma trends in the United States: an update based on Surveillance, Epidemiology, and End Results Program data for 1973 through 2003. Am. J. Epidemiol. 2004;159:107–112. doi: 10.1093/aje/kwh025. [DOI] [PubMed] [Google Scholar]
- 6.Bhagavatula R, et al. Asbestos: a moving target. Best’s Review. 2001;102:85–90. [Google Scholar]
- 7.Landrigan PJ, et al. Health and environmental consequences of the world trade center disaster. Environ. Health Perspect. 2004;112:731–739. doi: 10.1289/ehp.6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kukreja J, et al. Malignant pleural mesothelioma: overview of the North American and European experience. Thorac. Surg. Clin. 2004;14:435–445. doi: 10.1016/j.thorsurg.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 9.Peto J, et al. The European mesothelioma epidemic. Br. J. Cancer. 1999;79:666–672. doi: 10.1038/sj.bjc.6690105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson BW, et al. Advances in malignant mesothelioma. N. Engl. J. Med. 2005;353:1591–1603. doi: 10.1056/NEJMra050152. [DOI] [PubMed] [Google Scholar]
- 11.Joshi TK, et al. Asbestos in developing countries: magnitude of risk and its practical implications. Int. J. Occup. Med. Environ. Health. 2004;17:179–185. [PubMed] [Google Scholar]
- 12.Kazan-Allen L. Asbestos and mesothelioma: worldwide trends. Lung Cancer. 2005;49(suppl. 1):S3–S8. doi: 10.1016/j.lungcan.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Jaurand MC. Mechanisms of fiber-induced genotoxicity. Environ. Health Perspect. 1997;105(suppl. 5):1073–1084. doi: 10.1289/ehp.97105s51073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelsey KT, et al. The in vitro genetic effects of fibrous erionite and crocidolite asbestos. Br. J. Cancer. 1986;54:107–114. doi: 10.1038/bjc.1986.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang NS, et al. The interactions between asbestos fibers and metaphase chromosomes of rat pleural mesothelial cells in culture. A scanning and transmission electron microscopic study. Am. J. Pathol. 1987;126:343–349. [PMC free article] [PubMed] [Google Scholar]
- 16.Okayasu R, et al. Asbestos and DNA double strand breaks. Cancer Res. 1999;59:298–300. [PubMed] [Google Scholar]
- 17.Xu A, et al. Role of oxyradicals in mutagenicity and DNA damage induced by crocidolite asbestos in mammalian cells. Cancer Res. 1999;59:5922–5926. [PubMed] [Google Scholar]
- 18.Xu A, et al. Genotoxic mechanisms of asbestos fibers: role of extranuclear targets. Chem. Res. Toxicol. 2007;20:724–733. doi: 10.1021/tx600364d. [DOI] [PubMed] [Google Scholar]
- 19.He B, et al. Secreted frizzled-related protein 4 is silenced by hypermethylation and induces apoptosis in beta-catenin-deficient human mesothelioma cells. Cancer Res. 2005;65:743–748. [PubMed] [Google Scholar]
- 20.Lee AY, et al. Expression of the secreted frizzled-related protein gene family is downregulated in human mesothelioma. Oncogene. 2004;23:6672–6676. doi: 10.1038/sj.onc.1207881. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki M, et al. Aberrant methylation profile of human malignant mesotheliomas and its relationship to SV40 infection. Oncogene. 2005;24:1302–1308. doi: 10.1038/sj.onc.1208263. [DOI] [PubMed] [Google Scholar]
- 22.Shivapurkar N, et al. Aberrant methylation of trail decoy receptor genes is frequent in multiple tumor types. Int. J. Cancer. 2004;109:786–792. doi: 10.1002/ijc.20041. [DOI] [PubMed] [Google Scholar]
- 23.Ohta Y, et al. Thrombospondin-1 expression and clinical implications in malignant pleural mesothelioma. Cancer. 1999;85:2570–2576. [PubMed] [Google Scholar]
- 24.Tsou JA, et al. Distinct DNA methylation profiles in malignant mesothelioma, lung adenocarcinoma, and non-tumor lung. Lung Cancer. 2005;47:193–204. doi: 10.1016/j.lungcan.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 25.Toyooka S, et al. Aberrant methylation and simian virus 40 tag sequences in malignant mesothelioma. Cancer Res. 2001;61:5727–5730. [PubMed] [Google Scholar]
- 26.Murthy SS, et al. Expression of GPC3, an X-linked recessive overgrowth gene, is silenced in malignant mesothelioma. Oncogene. 2000;19:410–416. doi: 10.1038/sj.onc.1203322. [DOI] [PubMed] [Google Scholar]
- 27.Wong L, et al. Inactivation of p16INK4a expression in malignant mesothelioma by methylation. Lung Cancer. 2002;38:131–136. doi: 10.1016/s0169-5002(02)00178-2. [DOI] [PubMed] [Google Scholar]
- 28.Hirao T, et al. Alterations of the p16(INK4) locus in human malignant mesothelial tumors. Carcinogenesis. 2002;23:1127–1130. doi: 10.1093/carcin/23.7.1127. [DOI] [PubMed] [Google Scholar]
- 29.Toyooka S, et al. Progressive aberrant methylation of the RASSF1A gene in simian virus 40 infected human mesothelial cells. Oncogene. 2002;21:4340–4344. doi: 10.1038/sj.onc.1205381. [DOI] [PubMed] [Google Scholar]
- 30.Shigematsu H, et al. Aberrant methylation of HIN-1 (high in normal-1) is a frequent event in many human malignancies. Int. J. Cancer. 2005;113:600–604. doi: 10.1002/ijc.20622. [DOI] [PubMed] [Google Scholar]
- 31.Tsou JA, et al. DNA methylation profile of 28 potential marker loci in malignant mesothelioma. Lung Cancer. 2007 doi: 10.1016/j.lungcan.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baylin SB, et al. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 33.Jones PA, et al. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 34.Kim DH, et al. p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res. 2001;61:3419–3424. [PubMed] [Google Scholar]
- 35.Toyooka S, et al. Dose effect of smoking on aberrant methylation in non-small cell lung cancers. Int. J. Cancer. 2004;110:462–464. doi: 10.1002/ijc.20125. [DOI] [PubMed] [Google Scholar]
- 36.Toyooka S, et al. Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int. J. Cancer. 2003;103:153–160. doi: 10.1002/ijc.10787. [DOI] [PubMed] [Google Scholar]
- 37.Dammann R, et al. CpG island methylation and expression of tumour-associated genes in lung carcinoma. Eur. J. Cancer. 2005;41:1223–1236. doi: 10.1016/j.ejca.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 38.Agathanggelou A, et al. Role of the Ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res. 2005;65:3497–3508. doi: 10.1158/0008-5472.CAN-04-4088. [DOI] [PubMed] [Google Scholar]
- 39.Kusy S, et al. p14ARF, p15INK4b and p16INK4a methylation status in chronic myelogenous leukemia. Leuk. Lymphoma. 2004;45:1989–1994. doi: 10.1080/10428190410001714025. [DOI] [PubMed] [Google Scholar]
- 40.Schulz WA. Understanding urothelial carcinoma through cancer pathways. Int. J. Cancer. 2006;119:1513–1518. doi: 10.1002/ijc.21852. [DOI] [PubMed] [Google Scholar]
- 41.Herman JG, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eads CA, et al. Fields of aberrant CpG island hypermethylation in Barrett’s esophagus and associated adenocarcinoma. Cancer Res. 2000;60:5021–5026. [PubMed] [Google Scholar]
- 43.Ogino S, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J. Mol. Diagn. 2006;8:209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Churg A, et al. Correlation of quantitative asbestos body counts and occupation in urban patients. Arch. Pathol. Lab. Med. 1977;101:629–634. [PubMed] [Google Scholar]
- 45.Marsit CJ, et al. Epigenetic inactivation of SFRP genes and TP53 alteration act jointly as markers of invasive bladder cancer. Cancer Res. 2005;65:7081–7085. doi: 10.1158/0008-5472.CAN-05-0267. [DOI] [PubMed] [Google Scholar]
- 46.Holliday R. The significance of DNA methylation in cellular aging. Basic Life Sci. 1985;35:269–283. doi: 10.1007/978-1-4899-2218-2_17. [DOI] [PubMed] [Google Scholar]
- 47.Issa JP. CpG-island methylation in aging and cancer. Curr. Top. Microbiol. Immunol. 2000;249:101–118. doi: 10.1007/978-3-642-59696-4_7. [DOI] [PubMed] [Google Scholar]
- 48.Dodson RF, et al. Stability of ferruginous bodies in human lung tissue following death, embalmment, and burial. Inhal. Toxicol. 2005;17:789–795. doi: 10.1080/08958370500240199. [DOI] [PubMed] [Google Scholar]
- 49.Sabo-Attwood T, et al. Gene expression profiles reveal increased mClca3 (Gob5) expression and mucin production in a murine model of asbestos-induced fibrogenesis. Am. J. Pathol. 2005;167:1243–1256. doi: 10.1016/S0002-9440(10)61212-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adamson IY, et al. Mesothelial cell proliferation after instillation of long or short asbestos fibers into mouse lung. Am. J. Pathol. 1993;142:1209–1216. [PMC free article] [PubMed] [Google Scholar]
- 51.Baylin SB. Mechanisms underlying epigenetically mediated gene silencing in cancer. Semin. Cancer Biol. 2002;12:331–337. doi: 10.1016/s1044-579x(02)00053-6. [DOI] [PubMed] [Google Scholar]