Abstract
Background
Leifsonia xyli is a xylem-inhabiting bacterial species comprised of two subspecies: L. xyli subsp. xyli (Lxx) and L. xyli subsp. cynodontis (Lxc). Lxx is the causal agent of ratoon stunting disease in sugarcane commercial fields and Lxc colonizes the xylem of several grasses causing either mild or no symptoms of disease. The completely sequenced genome of Lxx provided insights into its biology and pathogenicity. Since IS elements are largely reported as an important source of bacterial genome diversification and nothing is known about their role in chromosome architecture of L. xyli, a comparative analysis of Lxc and Lxx elements was performed.
Results
Sample sequencing of Lxc genome and comparative analysis with Lxx complete DNA sequence revealed a variable number of IS transposable elements acting upon genomic diversity. A detailed characterization of Lxc IS elements and a comparative review with IS elements of Lxx are presented. Each genome showed a unique set of elements although related to same IS families when considering features such as similarity among transposases, inverted and direct repeats, and element size. Most of the Lxc and Lxx IS families assigned were reported to maintain transposition at low levels using translation regulatory mechanisms, consistent with our in silico analysis. Some of the IS elements were found associated with rearrangements and specific regions of each genome. Differences were also found in the effect of IS elements upon insertion, although none of the elements were preferentially associated with gene disruption. A survey of transposases among genomes of Actinobacteria showed no correlation between phylogenetic relatedness and distribution of IS families. By using Southern hybridization, we suggested that diversification of Lxc isolates is also mediated by insertion sequences in probably recent events.
Conclusion
Collectively our data indicate that transposable elements are involved in genome diversification of Lxc and Lxx. The IS elements were probably acquired after the divergence of the two subspecies and are associated with genome organization and gene contents. In addition to enhancing understanding of IS element dynamics in general, these data will contribute to our ongoing comparative analyses aimed at understanding the biological differences of the Lxc and Lxx.
Background
The Gram-positive, coryneform, fastidious, xylem-inhabiting bacteria Leifsonia xyli comprises two subspecies: L. xyli subsp. xyli (Lxx) and L. xyli subsp. cynodontis (Lxc). In its unique natural host, Lxx causes ratoon-stunting disease, a malady that affects sugarcane commercial fields worldwide, promoting losses of up to 30% in susceptible varieties [1]. Sequencing of the Lxx genome has provided important insights into the biology and pathogenicity of this bacterium [2]. Lxc is an endophyte of Bermuda grass (Cynodo dactylon) and, when artificially inoculated, can grow in and colonize the xylem of agriculturally important grasses (including sugarcane, corn and rice), causing no (or mild) symptoms of disease [3,4]. Some studies suggested that Lxc may be a potential vector for expressing heterologous proteins in plants [5-10,4]. We have initiated a genome-based approach to compare Lxc and Lxx by sample sequencing the Lxc genome. Our goal is to comprehensively assess gene content and genomic organization of these two closely related bacteria to enhance understanding of the differences in their pathogenicity and host range. Here, we present the in silico characterization of insertion sequence (IS) elements, the most abundant type of mobile genetic element found in L. xyli [2] and their involvement in Lxc and Lxx genome diversification.
IS elements are small transposable DNA fragments ranging from 0.7 to 3.5 kbp, comprising a transposase-encoding gene and terminal inverted repeats (IR) [11]. Close to 1,500 different IS elements have been reported in the chromosomes and plasmids of nearly all bacteria studied [12]. IS elements may inactivate genes upon insertion or activate and/or enhance the expression of nearby genes. Some are known to recognize specific sites of the genome that are duplicated after IS insertion, resulting in direct repeats (DR). IS elements may provide the structural basis necessary to enable the rearrangement of genomic fragments and the incorporation of foreign DNA either by active transposition process or indirectly, mediating homologous recombination between multiple copies present in a given genome [13]. They are believed to undergo frequent horizontal transfer and cycles of expansion and extinction within a given species, most likely as a consequence of transfer between genomes and plasmids [14]. Their expansion, genome location and composition may differ among related bacteria, representing an important source of genomic diversity [15-20,12]. Because its effects have a direct impact on cell survival, control of transposition is tightly regulated. Intrinsic regulation is basically at the transcriptional and translational level [21]. In addition, several host proteins have been identified as part of the transpososome, the assembly of which may be controlled by host factors, thus integrating transposition activity and host physiology [22].
Previously, 50 copies of five distinct IS elements (ISLxx1–5) in the Lxx genome were reported, along with 47 other transposase-related genes from uncharacterized elements [2,23]. In Lxc, three IS elements have previously been identified: IS1237 [24], ISLxc1 and ISLxc2 [25], located both on the chromosome and on the cryptic plasmid pCXC100 that is present in some isolates. A detailed characterization of two new Lxc IS elements (ISLxc3 and ISLxc4) and one new Lxx element (ISLxx6) is presented, as well as a comparative review of all elements found in both genomes, their distribution among families [11], and their comparative localization. The results support the hypothesis that IS elements are components involved in the diversification of Lxx and Lxc, with their influence most likely occurring after the divergence of the two subspecies from their common ancestor.
Results
The dataset of Lxc genomic DNA
The characterization of the IS elements were realized within the dataset of Lxc DSMZ46306 genomic DNA. The dataset is comprised of 9,766 reads, of which 5,854 were derived from the shotgun library and 3,912 from sequencing BAC ends, sub-cloning of inserts and primer-walking. All sequences were assembled into 1,064 contigs accounting for 1,368,731 non-redundant bases, representing approximately 50% of the Lxc chromosome. Comparing all the sequences of Lxc with the complete genome of Lxx, we found that nearly 70% share more than 80% nucleotide sequence identity considering a continuous segment of at least 200 bases. Two other genomes of related subspecies Clavibacter michiganensis subsp. michiganensis and C. michiganensis subsp. sepedonicus that were recently sequenced [26,27] have nearly 80% of nucleotide identity using the same criteria. These figures were derived from the Artemis Comparative Tool [28].
Lxc and Lxx each have their own set of IS elements
Among all the contigs, 70 shared sequence similarity to transposase-encoding genes. Fifty-six of these represented copies of five distinct IS elements that were characterized based on the criteria proposed (Table 1). The remaining 14 contigs correspond to uncharacterized IS elements, since no IR, site of insertion and structural limits were identified within the available sequences. They were classified only tentatively based on homology searches with Lxx genome, and were placed within IS families (Table 2): IS3 family (eleven elements); IS256 family (two elements); and IS481 family (one element). Elements of the same families were not characterized in Lxx as well [2,23]. These uncharacterized transposases may represent degenerated forms of old insertions, however they may also be used as site for rearrangements within Lxc genome.
Table 1.
Details of Lxc-IS elements.
| Lxc-IS | IS length (bp) | Tpase length (aa)a | IRs | DR (host sequence) | Minimal no of copies | IS familyb | Most related Lxx-IS |
| IS1237 | 899/798 | 218 | 5' GAGGTGGTTTCAGTAGTCG 3' GAGATGGTTTCAGTAGTCGc |
TTA (TAA) | 26 | IS5 | ISLxx6 |
| ISLxc1 | 2631 | 262 (istB) | 5' TGTCTGTGTCACTGTTAGCGGGCTCCG GGTTGTCGGAGTTGTCTGGCCCCGGCG 3' TGACTATGTCAAGGCTGGCGGGGACCG GGTTGTCGAAGTTAGTTGGCCCCAGCG |
fATCGCC | 2 | IS21 | ISLxx3 |
| ISLxc2 | 1105 | 332 | 5' TGTGTTTCTCAGGGACGTTGGT 3' TGTGCTGCTCAGGGACGTTGGT |
NCTAGN | 16 | IS481 | ISLxx4 |
| ISLxc3 | 1511 | 351 | 5' CTTGGGTTCTAACGGTCGTTGCAACA 3' GGCGGATTCTAGTGGTCGTTGCAACA |
- | 8 | IS30 | ISLxx5 |
| ISLxc4 | 1311/896 | 380 | 5' GGGTGGGTTCTAACAGTGGTCGCAACAC 3' GGGTGGGTTCTAACAGTGGTCGCAACAC |
- | 5 | IS30 | ISLxx5 |
N – not complementary nucleotides
a – assuming predicted features for regulation of transposition at translational level are overcome.
b – according to Chandler & Mahillon family classification [11].
c – three IS1237 copies have a G instead of A in the 3' IR, making perfectly complementary IRs.
Table 2.
Classification of Lxc and Lxx transposase-encoding genes into IS families [11].
| Lxc | Lxx | |||||
| IS Family | characterized | non-characterized | % Total | characterized | non-characterized | %Total |
| 5 | 26 | 0 | 37 | 1 | 4 | 5 |
| 21 | 1 | 0 | 1 | 5 | 3 | 8 |
| 481 | 16 | 1 | 24 | 26 | 6 | 33 |
| 30 | 13 | 0 | 19 | 15 | 0 | 16 |
| 110 | 0 | 0 | 0 | 4 | 1 | 5 |
| 3 | 0 | 11 | 16 | 0 | 19 | 20 |
| 256 | 0 | 2 | 3 | 0 | 8 | 8 |
| undefined | 0 | 0 | 0 | 0 | 5 | 5 |
| Total | 56 | 14 | 100 | 51 | 46 | 100 |
Columns in bold represent the percentage from the total number of predicted transposases in the Lxx genome and Lxc datasets. The other values are the numbers found within each genome.
The transposases of characterized IS elements shared less than 78% amino acid sequence identity with transposases of Lxx elements (Additional file 1). To be considered as iso-forms, transposases of a given element must share more than 95% identical amino acids [11]; therefore, Lxc and Lxx have different sets of IS elements. Despite of that, most of the elements found belong to the same families (Table 2), with the exception of IS110 (ISLxx2), which was not identified in our Lxc dataset.
Description of Lxc IS elements and comparison with related elements in Lxx
In addition to the three IS elements previously sequenced for Lxc: IS1237 (GenBank ID: X75973); ISLxc1 (GenBank ID: EF437436); and ISLxc2 (GenBank ID: EF176596), we identified two new elements, named ISLxc3 (GenBank ID: EF421582) and ISLxc4 (GenBank ID: EF433175) (Table 1, Fig. 1 and Additional file 2). A detailed characterization of all these Lxc-IS elements, and a comparison with related elements in Lxx, is presented below.
Figure 1.
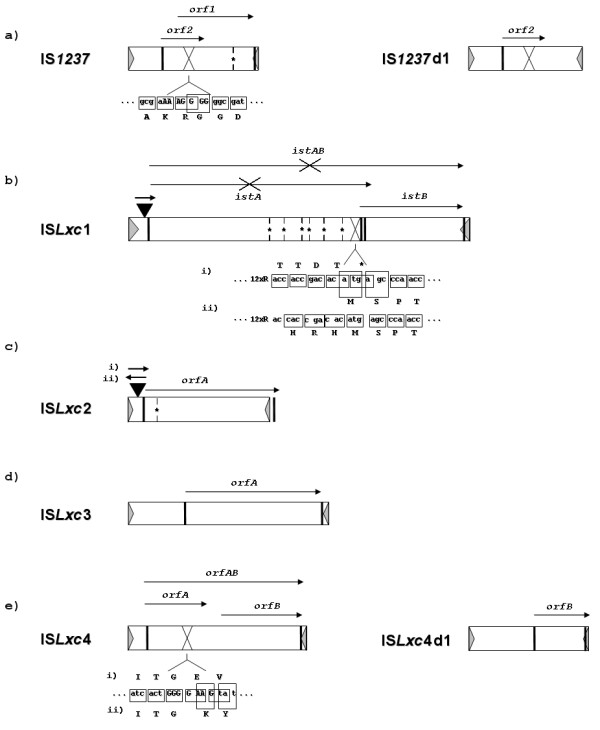
Characterization of IS elements of Lxc dataset. Each characterized IS element is represented by a rectangle, with IRs indicated by gray triangles. Black vertical bars indicate start and stop codons of the deduced transposases, and their transcriptional orientation is indicated by arrows. Crossed arrows represent truncated ORFs. Regions with slippery codons for frameshift are represented by X and the corresponding nucleotides are depicted in capital letters, with other nucleotides in lowercase letters. Amino acid sequences are shown beneath boxed codons. Vertical bars with asterisks represent premature stop codons. Inverted black triangles indicate the location of an IS1237 insertion within other IS elements, and the arrows above indicate the orientation of transcription of its transposase. In (b) we presented the nucleotides and deduced amino acid sequences immediately after the purine rich region that may promote either: (i) independent translation of istA and istB, or (ii) translation of istAB, depending on ribosomal slippage. In (c) copies of IS1237 were found inserted in both orientations (i and ii) when considering transcription of its transposase within ISLxc2. (e) in (i) indicates independent translation of orfA and (ii) shows translation of the same nucleotides after a +1 frameshift (translation of orfAB).
IS1237 and ISLxx6, IS5 family members
IS1237 was first identified in the plasmid pCXC100 detected in some Lxc isolates [24]. Recently, 13 upstream and 10 downstream flanking sequences of IS1237 were described [25]. We identified 26 copies of IS1237 (Additional files 3 and 4) represented by two variant forms: 24 copies were 899-bp long and almost identical, with a single polymorphism in the 3'-IR; two copies were 798 bp long (Table 1 and Fig. 1). Four copies were inserted within other IS elements: one inserted within ISLxc1 and three inserted within copies of ISLxc2. There was no read-through ORF encoding an entire transposase within IS1237 [24]. If functional, the coding sequence, starting and ending at nucleotide positions 232 and 889, respectively, would have to change frame (-1) and overcome a premature stop codon (Additional file 2). Since the coding sequence of all 899-bp long copies sequenced so far were identical, it is reasonable to assume that such translational features could be involved in the negative control of transposition [21]. The putative transposase has the expected catalytic domain containing a DDE motif [25], but the number of amino acids separating these residues does not follow the pattern of the IS5 family, and the glutamic acid is located downstream of the premature stop codon. Analyzing Lxx genome sequence, we identified ISLxx6, a single copy of which is represented within the genomic island LxxGI3 [2]. In Lxx, LxxGI3, is a depository of IS elements encompassing seventeen putative transposase genes [2,23]. ISLxx6 shows the highest nucleotide sequence similarity with an Lxc-IS element, the 899 bp-long IS1237 (Additional file 1). Interestingly, however, contrary to IS1237, the ISLxx6 putative transposase gene (Lxx22320.1) is read-through (Additional file 5). Both elements were classified as belonging to group IS427 within IS5 family based on multiple alignment using Tribe-MCl (Patricia Siguier, personal communication). However, they share conserved amino acids with elements of group IS1031 (Additional file 5). In addition, ISLxx6 presents a single ORF and recognize three nucleotides as site of insertion, also common features of elements of IS1031 group. This may indicate the existence of another group within IS5 family.
ISLxc1, an IS21 family member
ISLxc1 [25] encompasses two ORFs similar to the IstA and IstB transposases found in elements of the IS21 family [11]. In our Lxc dataset, we identified one copy of ISLxc1, which was invaded by an IS1237 element. The IS1237 insertion is located 58 bp upstream of the first ORF (istA) (figure 1) and has duplicated the "TAA" site of insertion within ISLxc1 (Additional file 2). A second version of ISLxc1 not carrying a copy of IS1237 was amplified in DSMZ46306 with inwardly oriented primers complementary to the 5' and 3' IRs. However this element was not mapped on Lxx genome because we did not have the flanking sequences. ISLxc1 is 2,631 bp long and 99% identical to that previously sequenced [25]. Within both versions istA is truncated and the DDE motif was not identified, therefore istA disruption was prior to the IS1237 insertion. istB may encode a 262-amino-acid protein containing the predicted nucleoside triphosphate-binding domain. It is preceded by a purine reach site, which in other elements is an indicative of ribosome frameshifting and coupled translation of IstA and IstB [11] (Fig. 1 and additional file 2).
ISLxc2, an IS481 family member
ISLxc2 is a 1,105-bp-long element (Table 1 and Fig. 1) [25]. The 16 copies identified here all contain at least one nucleotide polymorphism in different positions, none of which located in crucial sites such as start and stop codons or the DDE motif. The putative transposase contains 332 amino acids and is interrupted by an in-frame stop-codon located 111 nucleotides downstream of the initiation codon (Additional file 2). Interestingly, the transposases of ISLxc2 and also ISLxx4 of Lxx genome [2] have stop codons within the host insertion site. The NCTAGN sequence is duplicated after element insertion and flanks both ends indicating that the translational end of the transposase may impose the structural limits on these elements, as described for other elements of the same family [29,30].
Three copies of ISLxc2 were invaded by IS1237, in all the cases the insertion was located 13 bp upstream of the putative transposase gene, duplicating the TTA site of recognition. Generally, genes invaded by IS elements become non-functional, but the IS1237 insertions were all at the same position within ISLxc2, located upstream of the transposase gene (Fig. 1). IS elements located upstream of the coding region may create a hybrid promoter between the IS element sequence and the gene and favor transcription of the later, which in this case is ISLxc2 [11,21]. However, this should be addressed experimentally.
ISLxc3 and ISLxc4, IS30 family members
Two novel elements were identified: ISLxc3 and ISLxc4 (Table 1, Fig. 1 and Additional file 2). Eight copies of ISLxc3 were found, six of which being 1,511 bp long, and two copies with 3' end deletions at different positions. One of these defective variants (occurrence number 8 – additional file 3) is associated with a reorganization event in Lxc compared to Lxx because one of the flanking regions is common to Lxx and the other one is specific to Lxc. This is an example showing that an IS element may serve as a subject for homologous recombination, independent of its integrity. An imperfect IR of ISLxc3 was defined as a twenty-six bp long sequence starting with three non-complemented nucleotides, "CTT", at the 5'-end and "GCC" at the 3'-end, which were also detected for ISLxx5 [2]. The proposed IRs were based on the experimentally proved structural limits of IS1655, an IS30 family element of Neisseria meningitides [31]. In fact, the first nine nucleotides of 3'-IR are conserved among IS1655, ISLxc3 and ISLxx5. We failed in detect a conserved target site for insertion in multiple alignment considering 20 bases upstream and downstream of the element limits, but the duplication of NTG sequence was detected flanking two copies of ISLxc3. The putative transposase was the only one described in Lxc that was not interrupted by a premature stop codon or by a purine repeat associated with a frameshift.
ISLxc4 was found in five copies represented by two variant forms. Two copies represented longer versions (1,311 bp) and the other three copies (896 bp) showed the same deletion within the core region, which were considered shorter versions of the ISLxc4 (ISLxc4d1 – GeneBank ID: EF494674). The IRs are twenty-eight bp long, and no DR was identified flanking the occurrences. Regardless of been unusual for IS30 family elements, the long version contain a cluster of purines (nucleotide position 431–437), resembling a slippery codon for the frameshift (Additional file 2). Transposases of the IS30 family encompass the helix-turn-helix (HTH) DNA-binding domain at the N-terminal part and the well-conserved D-(54–61 aa)-D-(33 aa)-E motif at the C-terminal part [30,32]. In ISLxc4, if the +1 frameshift does occur, the transposase (OrfAB) is 380 amino acids long and contains two domains. If the frameshift does not occur, the transposase (OrfA) is 115 amino acids long, carrying only the DNA-binding domain. Truncated transposases with only the DNA-binding domain may function as negative regulators of transposition when interacting with IRs, resulting in either repression of the transposase gene promoter or competition with full-length transposases [33,21]. Neither the DNA-binding domain nor DDE motif following the rule above were detected within the ORF of the shorter variant.
Analysis of IS element loci in the Lxc and Lxx genomes
The average nucleotide sequence identity between homologous fragments of Lxc and Lxx is 93%, as calculated on 80 kbp of 25 continuous Lxc-contigs, which were physically linked based on scaffold orientation. Comparing the adjacent regions of all 56 Lxc-IS element with the Lxx sequence, 44 elements could be mapped onto the Lxx genome (Additional file 3). All the flanking sequences of IS elements were submitted to GenBank and the accession number is available in the additional file 3. Four of these (occurrences 8, 14, 15 and 38, additional file 3), which are three copies of ISLxc2 and one copy of ISLxc3, have only one of the adjacent sequences homologous to the Lxx genome. The other end is specific to Lxc genome, indicating an association with genomic rearrangements. Although some IS elements of both genomes belong to the same families and may recognize the same target site for insertion, none of the Lxc-IS insertion loci were the same as for elements found in the Lxx genome (Fig. 2 and Additional file 3). The 12 remaining Lxc-elements were inserted in specific regions of its chromosome (Additional file 4).
Figure 2.
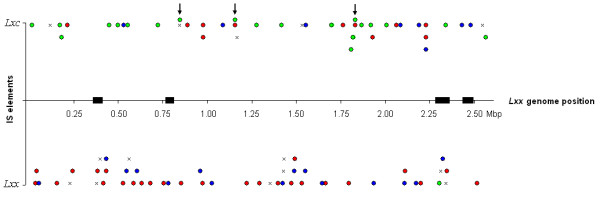
Distribution of Lxc and Lxx IS elements across the complete nucleotide sequence of Lxx genome. Mapping of Lxc elements was based on similarity of flanking sequences. Elements located less than 20 kbp apart are shown on different levels for ease of visualization. Elements sharing the same family assignment and possibly the same target site of insertion are represented by the same symbol in Lxc and Lxx: IS1237 and ISLxx6 (green circles); ISLxc2 and ISLxx4 (red circles); ISLxc3 and ISLxx5 (blue circles). The remaining IS elements are marked as "X". Arrows indicate insertions of IS1237 copies within other IS elements. Black boxes indicate four genomic islands described for Lxx genome.
IS elements were randomly distributed throughout Lxx genome (Fig. 2), 25% of all insertions were located in what has been described as genomic islands [2,23], in particular within genomic island LxxGI3. Comparatively the distribution of the elements in Lxc also seems to be random; however our approach so far does not allow us to make the same inferences about islands in Lxc genome.
To assess the impact of IS elements in gene disruption, they were classified into three categories: IS insertions within predicted genes; insertions in non-coding sequences; and insertions in non-coding sequences, but with one or two truncated genes nearby (less than 100 nucleotides distant) (Fig. 3). Truncated genes were defined as those ORFs that have a disrupted coding sequence based on BlastX results. Most of the elements were found inserted within intergenic regions for both genomes. In the Lxx genome, disrupted genes were associated mainly with degradation of polysaccharides, transport and regulatory functions, while in the Lxc genome they were linked to cell structure, regulatory functions and hypothetical genes (Additional file 3 and 4). Five of the putative genes truncated in Lxc have truncated orthologs in the Lxx genome however, in Lxx no IS insertions were detected: three of them probably encoded hypothetical proteins, Lxx05470 (Lxx genome position: 552,768–555,523), Lxx17040 (1,767,422–1,768,564) and Lxx21675 (2,229,230–2,230,675); another one, Lxx01850 (180,918–182,055), presented a conserved acyltransferase domain, associated with lipopolysaccharide modification; and the last one, Lxx14740 (1,531,157–1,531,354), presented a DNA-binding domain. Within Lxc, those genes were invaded by IS1237, ISLxc2, ISLxc3, IS1237 and ISLxc4, respectively. No DRs were detected within Lxx orthologous that could be an indicative of a prior invasion and excision event. Therefore is more likely that these genes were truncated prior to the IS elements insertion in Lxc.
Figure 3.
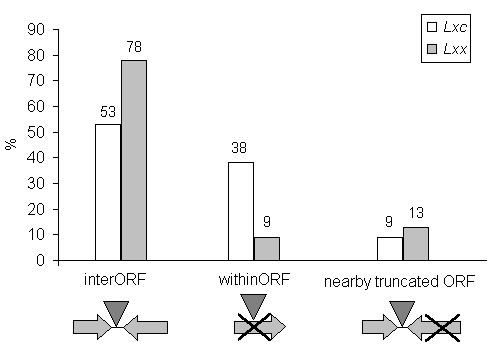
Classification of IS insertions according to the effect on proximal genes in Lxc and Lxx: inter ORF (intergene); within ORF; and nearby truncated ORFs. The orientation of adjacent genes or genes invaded by IS elements were not considered. Percent-values are relative to all IS elements present in Lxx genome and in our Lxc dataset.
Distribution of L. xyli-IS-related elements throughout Actinobacteria
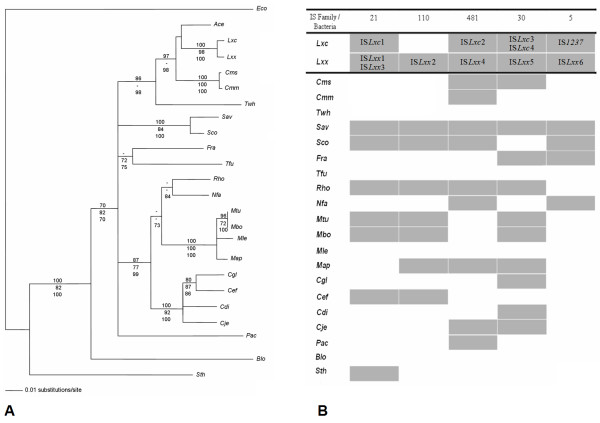
According to the ISfinder database, elements of the IS5 family are common in both Archaea and Eubacteria [12]. Considering all IS families found in Leifsonia xyli, examining ISfinder submission reports and using sequence similarity searches, we found that IS5 elements were the least represented in the sequenced genomes of Actinobacteria, despite being the most highly represented IS element in the Lxc genome. As observed in L. xyli genomes, transposases from the IS30 and IS481 families are abundant in most Actinobacteria (Fig. 4). Each putative transposase of characterized IS elements of Leifsonia xyli genomes was used as query and compared to transposases of Actinobacteria genomes, and the best BlastX hit result was considered. Most of the transposases shared around 70% of amino acid identity. The exception was IS1237, which related elements of Actinobacteria genomes did not share more than 50% identical amino acids. Actinobacteria is one of the largest taxonomic units in terms of number and variety of species [34] and members of IS elements of most characterized families are represented within this group [12]. As probably expected no correlation was found between phylogenetic relatedness and IS family distribution in Actinobacteria genomes (Fig. 4). Not even among subspecies of Leifsonia xyli and Clavibacter michiganensis, which are much related to each other. This observation is in concert with the cycles of expansion and extinction of IS elements proposed before [14].
Figure 4.
Comparative analysis of Leifsonia xyli IS elements with putative transposases of Actinobacteria. (A) Actinobacteria were arranged based on their phylogenetic relatedness. Consensus neighbor-joining phylogenetic tree was constructed using SSU rDNA(16S) sequences of: twenty-two completely sequenced genomes; in addition to Agromyces sp., which was reported as forming a coherent cluster to Leifsonia [44,45]; and E. coli as a outgroup. Numbers at the branches represent bootstrap values for parsimony (top, 2000 replicates), maximum likelihood (middle, 100 replicates) and neighbor-joining (bottom, 2000 replicates). Only bootstrap values above 70 are shown. (B) IS elements characterized from Lxc and Lxx genomes were used as in silico probes for the identification of distribution of IS families belonging to Leifsonia xyli genomes in other Actinobacteria. The ISfinder database was also searched for the presence of IS families in the Actinobacteria assayed here. Species names and accession numbers are listed: Eco-Escherichia coli K12 (NC_000913); Ace – Agromyces sp. (AM410680); Lxc – Leifsonia xyli subsp. cynodontis DSMZ46306 (unpublished); Lxx – Leifsonia xyli subsp. xyli CTCB07 (NC_006087); Cms – Clavibacter michiganensis subsp. sepedonicus ATCC33113 (NC_010407); Cmm – Clavibacter michiganensis subsp. michiganensis NCPPB 382 (NC_009480); Twh – Tropheryma whipplei str. Twist (NC_004572) and TW08/27 (NC_004551); Sav – Streptomyces avermitilis MA-4680 (NC_003155); Sco – Streptomyces coelicolor A3(2) (NC_003888); Fra – Frankia sp. CcI3 (NC_007777); Tfu – Thermobifida fusca YX (NC_007333); Rho – Rhodococcus sp. RHA1 (NC_008268); Nfa – Nocardia farcinica IFM 10152 (NC_006361); Mtu – Mycobacterium tuberculosis H37Rv (NC_000962) and CDC1551 (NC_002755); Mbo – Mycobacterium bovis AF2122/97 (NC_002945); Mle – Mycobacterium leprae TN (NC_002677); Map – Mycobacterium avium subsp. paratuberculosis str. k10 (NC_002944); Cgl – Corynebacterium glutamicum ATCC 13032 (NC_006958); Cef – Corynebacterium efficiens YS-314 (NC_004369); Cdi – Corynebacterium diphtheriae NCTC 13129 (NC_002935); Cje – Corynebacterium jeikeium K411 (NC_007164); Pac – Propionibacterium acnes KPA171202 (NC_006085); Blo – Bifidobacterium longum NCC2705 (NC_004307); Sth – Symbiobacterium thermophilum IAM 14863 (NC_006177).
Distribution of IS elements in Lxc isolates
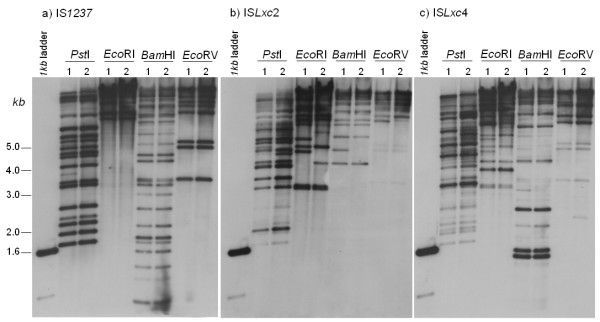
To further examine the distribution of IS elements between genomic DNA of two isolates of Lxc to check the involvement of these elements in diversification of strains, hybridization experiments were performed using fragments of three IS elements of different families as probes. There are two BamHI sites within IS1237, one within ISLxc4, and none within ISLxc2. There are no sites for PstI, EcoRV or EcoRI within these elements. IS1237 is the only element showing the same hybridization pattern between the two isolates for all tested enzymes (Fig. 5). However, variable DNA band patterns were detected for ISLxc2 and ISLxc4 (Fig. 5). We believe that partial digestions can not justify such differences because the same stripped membrane was used in each experiment, and we do not see partials in the assay with IS1237. Also, polymorphism at the enzymes restriction site could not explain the differences since they were found for all enzymes used when probed with ISLxc2 and ISLxc4, and they were not detected for IS1237, mainly considering the abundance of the later in Lxc genome. Finally, we assume that these differences were probably not the result of large-scale rearrangements; otherwise they would also have been detected in the distribution of IS1237. It is plausible to assume that the observed variation may result from homologous recombination between copies of these elements not interspersed by IS1237 copies, or that it may be associated with the presence of a variable number of elements, as a result of transposition of ISLxc2 and ISLxc4 to new locations. To assure any of these two hypotheses the bands should be isolated and the DNA sequenced. In any case, the difference exists and is probably originating diversification in the isolates of Lxc genomes. Consistent with this are the data generated by Young and collaborators where isolates of Lxc showed different band pattern in DNA fingerprinting [35].
Figure 5.
Southern hybridization of digested DNA of two Lxc isolates. The isolates used were (1) DSMZ46306 and (2) SB with probes (a) IS1237, (b) ISLxc2 and (c) ISLxc4. The DNA molecular marker used was a 1kb ladder (Invitrogen, Carlsbad, CA).
Discussion
Sequencing of approximately 50% of the Lxc genome identified 56 copies of five distinct IS elements, establishing that the Lxc chromosome contains a larger number of IS copies than the Lxx genome [2]. The IS elements of Lxc were subjected to a careful analysis and compared to the elements of Lxx and other Actinobacterial genomes, in an attempt to understand their contribution to the genome diversification of the two L. xyli subspecies. Also, taking into account that genome projects usually fail in a detailed characterization of these elements, we reviewed all the elements in both Lxc and Lxx genomes.
Apart from sharing the same IS families, there were no iso-forms in common between the Lxc and Lxx and the locations of insertions were distinct to each genome. Analysis of the impact of IS insertions revealed that, in both subspecies, most elements were inserted within intergenic regions. The higher percentage of this kind of insertion in Lxx (78%) as compared to the Lxc genome (53%) is probably due to the Lxx-genome decay process, since a higher percentage of genes were already non-functional, as previously proposed [2]. Insertions within genes or intergenic regions were independent of IS family assignment for both subspecies. The Lxc and Lxx genome comparative arrangement was assessed by mapping the flanking sequences of each Lxc-IS element in the complete Lxx sequence; some elements were associated with DNA rearrangements and others were inserted into specific fragments of the Lxc genome.
IS1237 is an unique element, with some similarity to elements of Streptomyces coelicolor A3(2) (NP_624431), S. avermitilis MA-4680 (NP_821293) and Frankia sp. CcI3 (YP_481128). An expansion of it was detected in the Lxc chromosome, whereas elements of the IS5 family were the least represented in Actinobacteria. Particularly intriguing is the presence of a single copy of an IS1237-related element within Lxx genome, the ISLxx6. Despite sharing 85% of nucleotide identity with IS1237, the transposase of ISLxx6 appears to be intact. One would probably expect a larger expansion of it within Lxx genome, but somehow its transposition may be controlled by mechanisms other than that regulated at translational level. This is probably an interesting model to be further analyzed. IS elements are very often plasmid-borne, and transfer events between genomes and plasmids are common [12], which may be the case of IS1237, first described in the cryptic plasmid pCXC100 present in some of the Lxc isolates [24]. The extended survey of all Lxc and Lxx-IS elements among Actinobacteria genomes showed no correlation between phylogenetic relatedness and distribution of IS families, which is probably in agreement with the hypothesis that genomes undergo repetitive extinction-reinfection cycles of different IS elements throughout bacterial evolution [14].
Most of the IS families assigned in L. xyli were characterized by maintaining transposition at low levels using mechanisms, such as regulation at the translational level [21], i.e., transposition occurs only when translation overcomes features of the sequences such as sites for ribosomal slippage and premature termination codons. So far, copies of Lxc and Lxx elements sequenced within each genome have proved almost identical, therefore, we have assumed that these features are part of the mechanism to regulate transposition, rather than being indicative of defective IS elements [21].
To assess the involvement of L. xyli IS elements carrying these types of transposases in promoting genome reorganization, the distribution of three IS elements of different families was analyzed in two strains of Lxc. The observed band polymorphism raised the hypothesis that ISLxc2 and ISLxc4 are involved in diversification, either by being active in transposition or by promoting homologous recombination. Variation was not observed for IS1237 hybridization, supporting previous experimental data where mobilization in vitro was not achieved [25]. Conversely, the elevated number of identical copies within the Lxc chromosome, the identical positioning in different isolates, and insertions within other elements suggest recent expansion of IS1237.
Transposon activity is regarded as an important source of bacterial diversity not only in promoting gene inactivation and genome reorganization but also in acquisition of new sequences [12]. Consequently, there must be some equilibrium between the impact of those events in successful maintenance of the element and host viability. How the differential expansion happens even among close related species and strains remains to be fully understood.
Conclusion
The impact of IS elements on genomic organization and gene content among closely related species has previously been described [16,17], and our data further support such analyses. Although the approach used to sequence the genome of Lxc was limited, an extensive study using the data gathered was sufficient to show that the genomic diversification of Lxc and Lxx is also and perhaps primarily associated with the presence of distinct types of IS elements. In addition to that, we have made a detailed characterization of IS elements present in each genome, which is often missing in analysis of fully sequenced genomes. The set of IS elements being unique to each genome, their specific location in combination with rearrangements and horizontal gene transfer are probably the major forces of genome evolution in Leifsonia xyli and consequently should have an impact on its biology. The same is probably true for isolates of Lxc as determined by our hybridization experiments. Our study also provided information in concert with the concept that distribution of IS elements in a given genome happens in evolutionary recent events due to cycles of expansion and extinction.
These data will probably contribute to our ongoing comparative analyses aimed at understanding the biological differences of the Lxc and Lxx genomes.
Methods
Bacterial strains and growth conditions
The bacterial strains used were: Lxx CTCB07 (NC_006087) [2] (Brazil); Lxc DSMZ46306 (Taiwan); and Lxc SB (Australia). The DSMZ46306 strain was grown in a MSC New modified liquid medium [3,2] at 28°C for 5–10 days, under agitation at 300 rpm. The SB genomic DNA was kindly provided by Dr. Steven Brumbley (Bureau of Sugar Experiment Station/Queensland, Australia).
Genomic library construction, DNA sequencing and assembly
Two DSMZ46306 genomic libraries were prepared. Genomic DNA extracted as previously described [2] was mechanically sheared and the resulting fragments cloned into pUC19 (Q-Biogene, Carlsbad, CA). A BAC clone library was prepared with 25 kbp-inserts obtained by partial digestion with BamHI [36] cloned into pIndigoBAC-5 (Epicentre®). Both ends of each insert were sequenced using an automated sequencer (model 3700, ABI Prism, Applied Biosystems, Foster City, CA). Results were analyzed by ABI sequencing analysis software, and assembled using the phred/Phrap/consed package [37,38]. All consensus sequences were generated with phred quality ≥ 20.
Characterization of IS elements
Inserts containing end-sequences similar to transposases were subcloned or primer-walked. Homology searches were performed using BlastN and BlastX [39] at GenBank [40] and IS finder [41]. Identities were considered significant only when the E-value was less than 10-05. Positive matches for transposase/integrase were manually verified to determine the presence of the following determinants of a given family: element size; presence of terminal inverted repeats (IR) and conserved terminal based pairs; target site of insertion and number of bases associated to direct repeats (DR); number of ORFs; distance among amino acids of DDE; as well as comparisons with related elements [11]. We also analyzed the presence of variants, domains, frameshifts and premature stop codons.
Transposases sharing more than 95% of amino acid identity were grouped. Their nucleotide sequences in addition to 300 bases up and downstream were aligned using ClustalW [42]. For each cluster of sequences, the first sixty bases of the 5'-end was aligned with the reverse complement of the first sixty bases of the 3'-end. Best matches were considered as IR candidates and compared to IRs of other characterized IS elements. IRs were selected when the minimum number of non complemented nucleotides followed the pattern of a given family. In an attempt to identify the target sites of insertion and the DR, an alignment of up to 20 bases flanking each IS element was done and the sequence was compared to the already described target sites of other IS elements. Features such as stop codon in frame and purine rich sites related to ribosome frameshift were sought in ORFs whose alignment partially matches transposases, based on BlastX results. Domains were identified within the Blast results at the CDD database [40]. DDE motifs were identified following IS finder pattern [41]. Minimal numbers of occurrences were determined based on flanking sequences.
Comparing Lxx and Lxc sequences
Lxc-IS element loci were compared to those of the Lxx genome using adjacent sequences as anchors and a cross_match program, which is part of the swat/cross_match/phrap package [38], with default parameters. Lxc-specific loci were annotated using SABIA [43].
Analysis of IS element distribution within Actinobacteria
Amino acid sequences of putative transposases were used as queries within completely sequenced genomes of Actinobacteria. Genomes were downloaded from GenBank (IDs in Fig. 4). Searches were performed using BlastX and tBlastN [39].
Southern blot hybridization
Lxc genomic DNA (1 μg; DSMZ46306 and SB) was digested completely with PstI, EcoRI, BamHI and EcoRV. Fragments were separated in a 0.8% (w/v) agarose gel and transferred to a Hybond N+ membrane (Amersham, Piscataway, NJ). Probe labeling, hybridization and detection were performed with an ECL kit (Amersham) according to the manufacturer's guidelines and under high stringency conditions. Inserts of shotgun clones containing the entire fragments of three Lxc-IS elements (IS1237, ISLxc2 and ISLxc4) were amplified and used as probes. The same membrane was stripped and re-used according to the manufacturer's recommendations.
Authors' contributions
MMZ: carried out the molecular genetic studies, prepared the in silico analysis and drafted the manuscript. M–AVS: Data interpretation and analysis and helped to draft the manuscript. LEAC: Data interpretation and analysis and helped to draft the manuscript. CBM–V: conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Comparative analysis of Lxc-IS elements and the most related IS element of Lxx genome.
Characterization of IS elements of Leifsonia xyli subsp. cynodontis.
Mapping of Lxx (gray lines) and Lxc (white lines) IS elements within the Lxx genome based on their flanking sequences.
IS elements inserted in specific fragments of Lxc genome.
Details of ISLxx6 and IS1237 sequence alignment and putative transposase amino acid sequences.
Acknowledgments
Acknowledgements
We thank: Steven Brumbley from the Bureau of Sugar Experiment Station (Queensland) for kindly providing genomic DNA of Lxc isolated in Australia; Maria Cristina Rodrigues Rangel and Daniela Truffi from the Laboratório de Genética Molecular (ESALQ/Brazil) for technical assistance; and Mariana Cabral de Oliveira from the Laboratório de Biologia Molecular de Plantas (USP/Brazil) for assistance in constructing the consensus phylogenetic tree. This work was supported by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) to CBMV, MAVS and LEAC, and a scholarship to MMZ.
Contributor Information
Marcelo M Zerillo, Email: mzerillo@usp.br.
Marie-Anne Van Sluys, Email: mavsluys@usp.br.
Luis Eduardo A Camargo, Email: leacamar@esalq.usp.br.
Claudia B Monteiro-Vitorello, Email: cbmvitor@lncc.br.
References
- Gillaspie AG, Teakle DS. Ratoon stunting disease. In: Ricaud C, Egan BT, Gillaspie AG Jr, Hughes CG, editor. Diseases of Sugarcane: Major Diseases. Amsterdam: Elsevier Science Publishers; 1989. pp. 58–80. [Google Scholar]
- Monteiro-Vitorello CB, Camargo LEA, Van Sluys MA, Kitajima JP, Truffi D, do Amaral AM, Harakava R, de Oliveira JCF, Wood D, de Oliveira MC, Miyaki C, Takita MA, da Silva AC, Furlan LR, Carraro DM, Camarotte G, Almeida NF, Jr, Carrer H, Coutinho LL, El-Dorry HA, Ferro MI, Gagliardi PR, Giglioti E, Goldman MH, Goldman GH, Kimura ET, Ferro ES, Kuramae EE, Lemos EG, Lemos MV, Mauro SM, Machado MA, Marino CL, Menck CF, Nunes LR, Oliveira RC, Pereira GG, Siqueira W, de Souza AA, Tsai SM, Zanca AS, Simpson AJ, Brumbley SM, Setúbal JC. The genome of the Gram-positive sugarcane pathogen Leifsonia xyli subsp. xyli. Mol Plant Microbe Interact. 2004;17:827–836. doi: 10.1094/MPMI.2004.17.8.827. [DOI] [PubMed] [Google Scholar]
- Davis MJ, Gillaspie AG, Vidaver AK, Harris RW. Clavibacter: A new genus containing some phytopathogenic coryneform bacteria, including Clavibacter xyli subsp. xyli sp. nov., subsp. nov. and Clavibacter xyli subsp. cynodontis subsp. nov., pathogens that cause ratoon stunting disease of sugarcane and Bermuda grass stunting disease. Int J Syst Bacteriol. 1984;34:107–117. [Google Scholar]
- Li TY, Zeng HL, Ping Y, Lin H, Fan XL, Guo ZG, Zhang CF. Construction of a stable expression vector for Leifsonia xyli subsp. cynodontis and its application in studying the effect of the bacterium as an endophytic bacterium in rice. FEMS Microbiol Lett. 2007;267:176–83. doi: 10.1111/j.1574-6968.2006.00551.x. [DOI] [PubMed] [Google Scholar]
- Metzler MC, Zhang YP, Chen TA. Transformation of Gram-positive bacterium Clavibacter xyli subsp. cynodontis by electroporation with plasmids from the IncP incompatibility group. J Bacteriol. 1992;174:4500–4503. doi: 10.1128/jb.174.13.4500-4503.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampel JS, Canter GL, Dimock MB, Anderson JJ, Uratani BB, Turner JT. Integrative cloning, expression and stability of the cryA (c) gene from Bacillus thuringiensis in a recombinant strain of Clavibacter xyli subsp. cynodontis. Appl Environ Microbiol. 1994;60:501–508. doi: 10.1128/aem.60.2.501-508.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uratani BB, Alcorn SC, Tsang BH, Kelly JL. Construction of secretion vectors and use of heterologous signal sequences for protein secretion in Clavibacter xyli subsp. cynodontis. Mol Plant Microbe Interact. 1995;8:892–898. doi: 10.1094/mpmi-8-0892. [DOI] [PubMed] [Google Scholar]
- Haapalainen M, Karp M, Metzler MC. Isolation of strong promoters from Clavibacter xyli subsp. cynodontis using a promoter probe plasmid. Biochim Biophys Acta. 1996;1305:130–134. doi: 10.1016/0167-4781(95)00203-0. [DOI] [PubMed] [Google Scholar]
- Haapalainen M, Kobet N, Piruzian E, Metzler MC. Integrative vector for stable transformation and expression of a β-1,3-glucanase gene in Clavibacter xyli subsp. cynodontis. FEMS Microbiol Lett. 1998;162:1–7. doi: 10.1111/j.1574-6968.1998.tb12971.x. [DOI] [PubMed] [Google Scholar]
- Li TY, Yin P, Zhou Y, Zhang Y, Zhang YY, Chen TA. Characterization of the replicon of a 51-kb native plasmid from the gram-positive bacterium Leifsonia xyli subsp. cynodontis. FEMS Microbiol Lett. 2004;236:33–39. doi: 10.1111/j.1574-6968.2004.tb09623.x. [DOI] [PubMed] [Google Scholar]
- Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P, Filée J, Chandler M. Insertion sequences in prokaryotic genomes. Curr Opin Microbiol. 2006;9:1–6. doi: 10.1016/j.mib.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Mahillon J, Leonard C, Chandler M. IS elements as constituents of bacterial genomes. Res Microbiol. 1999;150:675–687. doi: 10.1016/S0923-2508(99)00124-2. [DOI] [PubMed] [Google Scholar]
- Wagner A. Periodic Extinctions of Transposable Elements in Bacterial Lineages: Evidence from Intragenomic Variation in Multiple Genomes. Mol Biol Evol. 2006;23:723–733. doi: 10.1093/molbev/msj085. [DOI] [PubMed] [Google Scholar]
- Schneider D, Duperchy E, Depeyrot J, Coursange E, Lenski R, Blot M. Genomic comparisons among Escherichia coli strains B, K-12, and O157:H7 using IS elements as molecular markers. BMC Microbiol. 2002;2:1–8. doi: 10.1186/1471-2180-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson NR, Harris DE, Holden MT, Churcher CM, Bentley SD, Mungall KL, Cerdeño-Tárraga AM, Temple L, James K, Harris B, Quail MA, Achtman M, Atkin R, Baker S, Basham D, Bason N, Cherevach I, Chillingworth T, Collins M, Cronin A, Davis P, Doggett J, Feltwell T, Goble A, Hamlin N, Hauser H, Holroyd S, Jagels K, Leather S, Moule S, Norberczak H, O'Neil S, Ormond D, Price C, Rabbinowitsch E, Rutter S, Sanders M, Saunders D, Seeger K, Sharp S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Unwin L, Whitehead S, Barrell BG, Maskell DJ. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet. 2003;35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- Chain PS, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, Regala WM, Georgescu AM, Vergez LM, Land ML, Motin VL, Brubaker RR, Fowler J, Hinnebusch J, Marceau M, Medigue C, Simonet M, Chenal-Francisque V, Souza B, Dacheux D, Elliott JM, Derbise A, Hauser LJ, Garcia E. Insights into the evolution of Yersinia pestis through whole genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci USA. 2004;101:13826–13831. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugger K, Torarinsson E, Redder P, Chen L, Garrett RA. Shuffling of Sulfolobus genomes by autonomous and non-autonomous mobile elements. Biochem Soc Trans. 2004;32:179–183. doi: 10.1042/BST0320179. [DOI] [PubMed] [Google Scholar]
- Nascimento AL, Ko AI, Martins EA, Monteiro-Vitorello CB, Ho PL, Haake DA, Verjovski-Almeida S, Hartskeerl RA, Marques MV, Oliveira MC, Menck CF, Leite LC, Carrer H, Coutinho LL, Degrave WM, Dellagostin OA, El-Dorry H, Ferro ES, Ferro MI, Furlan LR, Gamberini M, Giglioti EA, Góes-Neto A, Goldman GH, Goldman MH, Harakava R, Jerônimo SM, Junqueira-de-Azevedo IL, Kimura ET, Kuramae EE, Lemos EG, Lemos MV, Marino CL, Nunes LR, de Oliveira RC, Pereira GG, Reis MS, Schriefer A, Siqueira WJ, Sommer P, Tsai SM, Simpson AJ, Ferro JA, Camargo LE, Kitajima JP, Setubal JC, Van Sluys MA. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J Bacteriol. 2004;186:2164–72. doi: 10.1128/JB.186.7.2164-2172.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro-Vitorello CB, Oliveira MC, Zerillo MM, Varani AM, Civerolo E, Van Sluys MA. Xylella and Xanthomonas Mobil'omics. OMICS. 2005;9:146–159. doi: 10.1089/omi.2005.9.146. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Chandler M. Regulation of transposition in bacteria. Res Microbiol. 2004;155:387–98. doi: 10.1016/j.resmic.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Gueguen E, Rousseau P, Duval-Valentin G, Chandler M. The transpososome: control of transposition at the level of catalysis. Trends Microbiol. 2005;13:543–549. doi: 10.1016/j.tim.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Monteiro-Vitorello CB, Zerillo MM, Van Sluys M-A, Camargo LEA. Genome sequence-based insights into the biology of the sugarcane pathogen Leifsonia xyli subsp. xyli. Plant Pathogenic Bacteria Genomics and Molecular Biology – Horizon Press.
- Laine MJ, Zhang Y-P, Metzler MC. IS1237, a repetitive chromosomal element from Clavibacter xyli subsp. cynodontis, is related to insertion sequences from Gram-negative and Gram-positive bacteria. Plasmid. 1994;32:270–279. doi: 10.1006/plas.1994.1066. [DOI] [PubMed] [Google Scholar]
- Lin H, Li TY, Xie MH, Zhang Y. Characterization of the variants, flanking genes and promoter activity of Leifsonia xyli subsp. cynodontis insertion sequence IS1237. J Bacteriol. 2007;189:3217–27. doi: 10.1128/JB.01403-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartemann K-H, Abt B, Bekel T, Burger A, Engemann J, Flügel M, Gaigalat L, Goesmann A, Gräfen I, Kalinowski J, Kaup O, Kirchner O, Krause L, Linke B, McHardy A, Meyer F, Pohle S, Rückert C, Schneiker S, Zellermann E, Pühler A, Eichenlaub R, Kaiser O, Bartels D. The Genome Sequence of the Tomato-Pathogenic Actinomycete Clavibacter michiganensis subsp. michiganensis NCPPB382 Reveals a Large Island Involved in Pathogenicity. J Bacteriol. 2008;190:2138–2149. doi: 10.1128/JB.01595-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley SD, Corton C, Brown SE, Barron A, Clark L, Doggett J, Harris B, Ormond D, Quail MA, May G, Francis D, Knudson D, Parkhill J, Ishimaru CA. Genome of the actinomycete plant pathogen Clavibacter michiganensis subspecies sepedonicus suggests recent niche adaptation. J Bacteriol. 2008;190:2150–2160. doi: 10.1128/JB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artemis Comparative Tool http://www.webact.org/WebACT/home
- Tauch A, Zheng Z, Pühler A, Kalinowski J. Corynebacterium striatum chloramphenicol resistance transposon Tn5564: Genetic organization and transposition in Corynebacterium glutamicum. Plasmid. 1998;40:126–139. doi: 10.1006/plas.1998.1362. [DOI] [PubMed] [Google Scholar]
- Chandler M, Mahillon J. Insertion Sequence Nomenclature (letter) ASM NEWS. 2000;66:324. [Google Scholar]
- Kiss J, Nagy Z, Tóth G, Kiss GB, Jakab J, Chandler M, Olasz F. Transposition and target specificity of the typical IS30 family element IS1655 from Neisseria meningitidis. Mol Microbiol. 2007;63:1731–1747. doi: 10.1111/j.1365-2958.2007.05621.x. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Szabó M, Chandler M, Olasz F. Analysis of the N-terminal DNA binding domain of the IS30 transposase. Mol Microbiol. 2004;54:478–488. doi: 10.1111/j.1365-2958.2004.04279.x. [DOI] [PubMed] [Google Scholar]
- Stalder R, Caspers P, Olasz F, Arber W. The N-terminal domain of the insertion sequence 30 transposase interacts specifically with the terminal inverted repeats of the element. J Biol Chem. 1990;265:3757–3762. [PubMed] [Google Scholar]
- Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AJ, Petrasovits LA, Croft BJ, Gillings M, Brumbley SM. Genetic uniformity of international isolates of Leifsonia xyli subsp. xyli, causal agent of ratoon stunting disease of sugarcane. Aust Plant Pathol. 2006;35:503–511. doi: 10.1071/AP06055. [DOI] [Google Scholar]
- Chang N, Chui LA. Standardized Protocol for the Rapid Preparation of Bacterial DNA for Pulse-Field Gel Electrophoresis. Diagn Microbiol Infect Dis. 1998;31:227–279. doi: 10.1016/S0732-8893(98)00007-8. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Gordon D, Abajian C, Grenn P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- GenBank http://www.ncbi.nlm.nih.gov/
- IS finder http://www-is.biotoul.fr/
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida LG, Paixão R, Souza RC, Costa GC, Barrientos FJ, Santos MT, Almeida DF, Vasconcelos AT. A System for Automated Bacterial (genome) Integrated Annotation SABIA. Bioinform. 2004;20:2832–2833. doi: 10.1093/bioinformatics/bth273. [DOI] [PubMed] [Google Scholar]
- Suzuki KI, Suzuki M, Sasaki J, Park YH, Komagata K. Leifsonia gen. nov., a genus for 2,4 diaminouyric and acid-containing actinomycetes to accomodate Corynebacterium aquticum Leifson 1962 and Clavibacter xyli subsp. cynodontis Davis et al. 1984. J Gen Appl Microbiol. 1999;45:253–262. doi: 10.2323/jgam.45.253. [DOI] [PubMed] [Google Scholar]
- Evtushenko L, Dorofeeva LV, Subbbotin SA, Cole JR, Tiedje JM. Leifsonia poae gen. nov., sp. nov., isolated from nematode galls on Poa annua, and reclassification of Corynebacterium aquticum Leifson 1962 as Leifsonia aquatica (ex Leifson 1962) gen. nov., nom. rev., comb. nov. and Clavibacter xyli Davis et al. 1984 with two subspecies as Leifsonia xyli (Davis et al. 1984) gen. nov., comb. nov. Int J Syst Evol Microbiol. 2000;50:371–380. doi: 10.1099/00207713-50-1-371. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparative analysis of Lxc-IS elements and the most related IS element of Lxx genome.
Characterization of IS elements of Leifsonia xyli subsp. cynodontis.
Mapping of Lxx (gray lines) and Lxc (white lines) IS elements within the Lxx genome based on their flanking sequences.
IS elements inserted in specific fragments of Lxc genome.
Details of ISLxx6 and IS1237 sequence alignment and putative transposase amino acid sequences.