Abstract
Despite the rapid and enormous progress in biotechnologies, the biochemical analysis of membrane proteins is still a difficult task. The presence of the large hydrophobic region buried in the lipid bilayer membrane (transmembrane domain) makes it difficult to analyze membrane proteins in standard assays developed for water-soluble proteins. To handle membrane proteins, the lipid bilayer membrane may be used as a platform to sustain their functionalities. Relatively slow progress in developing micro total analysis systems (μTAS) for membrane protein analysis directly reflects the difficulty of handling lipid membranes, which is a common problem in bulk measurement technologies. Nonetheless, researchers are continuing to develop efficient and sensitive analytical microsystems for the study of membrane proteins. Here, we review the latest developments, which enable detection of events caused by membrane proteins, such as ion channel current, membrane transport, and receptor/ligand interaction, by utilizing microfabricated structures. High-throughput and highly sensitive detection systems for membrane proteins are now becoming a realistic goal. Although most of these systems are still in the early stages of development, we believe this field will become one of the most important applications of μTAS for pharmaceutical and clinical screenings as well as for basic biochemical research.
Keywords: Microtechnology, Membrane protein, Lipid bilayer, Ion channel, Transporter
Introduction
Significant progress has been made in the field of micro total analysis systems (μTAS) and lab-on-a-chip in the last several decades [1, 2]. Microfabrication and microfluidic technologies enabled the development of miniaturized analytic and diagnostic tools in biology, including DNA/protein array chips [3, 4], microchannel electrophoresis [5–7], and micro immunoassay systems [5, 8]. Reactions of biomolecules contained in small volumes of buffer are efficiently controlled in sophisticated microfluidic systems, on the premise that the molecules are water-soluble.
Membrane proteins are a class of proteins present in the plasma membrane of cells. They perform specific roles in the cell boundary, enabling cells to sense, communicate, and regulate in response to fluctuations in the outside world. The hydrophobic regions in the polypeptide chain of membrane proteins penetrate into the inner hydrophobic layer of the lipid bilayer. Thus, the presence of the large hydrophobic part makes the membrane proteins notoriously difficult to separate from the membrane and handle in water-based assay systems. To create a microanalysis system for studying membrane proteins, it is necessary to construct an efficient and easily useable platform to handle the plasma membrane or the lipid bilayer.
In this review, we discuss the recent developments in μTAS and biochip technologies that are contributing to the functional analysis of membrane proteins. We first introduce the technologies for the measurement of membrane protein functions expressed on intact cell membranes, and then introduce the technologies which utilize reconstituted (artificial) membranes. We have classified the use of reconstituted membranes into three major subcategories based on their forms: (1) suspended lipid bilayers, planar membranes suspended over a small aperture, (2) supported lipid bilayers, planar membranes spanning a solid substrate, and (3) liposomes, spherical lipid vesicles. Here we especially focus on developments in the area of suspended bilayer systems, whereas applications of the other two categories are diverse and are discussed in other excellent reviews (e.g., [9–11]). The membrane proteins to be discussed are categorized by function into three groups: regulation of ion flow across the membrane by ion channel proteins, regulation of other molecules by transporter proteins, and association/dissociation of ligand molecules onto the receptor proteins. Like other applications, μTAS architectures for the study of membrane protein are oriented toward either or both of two directions: highly sensitive detection, ideally down to the single-protein level, and array systems for high-throughput screening applications.
Cell-membrane-based systems
In nature, membrane proteins function in intact cell membranes. The most straightforward way is to directly examine the membrane proteins on isolated or cultured cells. In this section, we review the use of microstructures for the analysis of membrane proteins expressed on the plasma membrane.
Ion channel recording
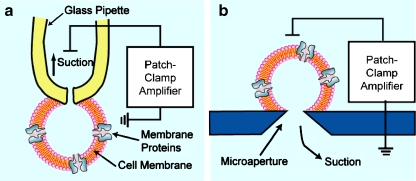
The patch-clamping technique, which was introduced by Neher and Sakmann [12] in 1976, is the most prominent technique in the history of membrane protein (ion channel) research. In this method, a sharp glass capillary (patch-clamp electrode) is pressed against the plasma membrane of the cell, and suction is applied to form a tight seal (gigaohm seal) between the membrane and the electrode. In the case of whole-cell recording, further suction is applied to rupture the membrane inside the capillary tip, providing electrical access to the inner space of the cell. The electric current and voltage across the membrane can be precisely controlled and measured using a patch-clamp amplifier (Fig. 1a). By using other variations of this technique, one can record the ion flow through a single ion channel protein.
Fig. 1.
a The conventional patch-clamping system with a glass capillary. b The planar patch-clamping system with a microaperture
Since ion channels are extremely important as a drug target, a high-throughput screening system on drug compounds is strongly desired especially in the pharmaceutical industry [13–15]. However, using the conventional patch-clamping technique involves laborious and time-consuming processes. One has to manipulate the tiny capillary to aim cells one by one under a microcope. To realize automated and high-throughput processing, a technique called “the planar patch clamping” has been proposed, and has been one of the most successful applications of microsystems (Fig. 1b). In this technique, individual cells are sucked into the small apertures etched into a substrate, instead of into glass capillaries, to form a cell patch. Since detection of extremely low electric current (picoamperes to nanoamperes) is required for ion channel recording, the choice of the substrate material is critical. Materials that have a low dielectric constant are generally used to avoid the noise incurred by stray capacitance. Fertig et al. [16] performed whole-cell patch clamping on a 1-μm-diameter aperture in a quartz substrate created by ion-track etching. They successfully recorded the respective ion channel currents through the K+ channel and the Ca2+ channel from Chinese hamster ovary (CHO) cells and neuroblastoma cells. By forming a cell-membrane patch in the cell attach mode, the same group succeeded in recording single K+ channel activity using the same glass aperture [17]. Klemic et al. [18] used polydimethylsiloxane (PDMS) elastomer as a substrate. They formed apertures as small as a few micrometers in diameter by casting PDMS on glass microcapillaries or a pyramidal mold, and recorded the K+ channel expressed in Xenopus oocytes. Use of PDMS should reduce the cost of device manufacturing. The lateral patch-clamping configuration with the PDMS microchannel was proposed by Lee’s group [19, 20], in which cells are trapped into lateral recording capillaries by negative pressure; they demonstrated the recording of a voltage-dependent K+ channel expressed on CHO cells. This configuration should make manufacturing and operation straightforward. Silicon microdevices, which offer precise and complicated manufacturing though well-established silicon-based microfabrication resources, provide facile fabrication of precisely-defined microapertures as well as integration of microfluidic systems; thus, they are expected to be advantageous for mass production. However, successful ion channel recording was not reported in early attempts [21, 22], probably owing to the low yield and large electric noise. The electric capacitance of the device, which is the main cause of the stray noise, was reduced by depositing a thick SiO2 layer (0.5–1 μm), and recording of ion channel current in a diverse group of cell types was presented [23, 24]. Recently, Zhang et al. [25] proposed the use of a silicon-on-insulator substrate for reduction of noise, and demonstrated the ligand-gated activation of TRPV1 receptor. All of the examples mentioned above are based on the single-cell patch, but the most problematic issue for realization of a high-throughput system is the reproducibility of the electrical sealing. At the single-cell level, the success rate of gigaohm seal formation is between 30 and 50% [16]. To overcome this inconsistency, Finkel et al. [26] proposed population patch-clamp technology, in which cells are patch-clamped into multiple apertures in a single recording well. Recording the ensemble signal of multiple whole-cell clamping events increased the success rate of ion channel recording to more than 95%. These planar patch-clamping technologies are now applied in commercially available automated patch-clamping systems [27, 28]. In addition to these efforts of improving the cell-patch configuration, Orwar’s group proposed the use of microfluidic channel system to control the concentration of ligand molecules for the study of sensitivity of GABAA receptors [29], and to increase the seal resistance of cell-patch [30] in regular glass-pipet based assay. Lee et al. [31] constructed PDMS based microfluidic device for planar patch clamping with integrated planar Ag/AgCl electrodes. These peripheral technologies will also be necessary for integrated recording systems.
Measurement of membrane transport
Transporters are the class of membrane proteins responsible for transportation of numerous kinds of molecules (e.g., polypeptides, nucleic acids, lipids, ions, and other chemicals, including drugs). Unlike ion channels, in which 106–107 electrons flow through a single channel per second, the turnover number of transporter proteins is generally considered to be in the range 1–103 s-1. Thus, reliable quantification of the membrane transport of nonionic molecules has been difficult owing to the small amount of substrate transported. Biologists have been examining the uptake of substrates into cells and proteoliposomes, using a radioactive substrate to increase sensitivity [32–34]. Nonetheless, the measurement error due to the nonspecific binding of substrates onto the membrane has not been negligible.
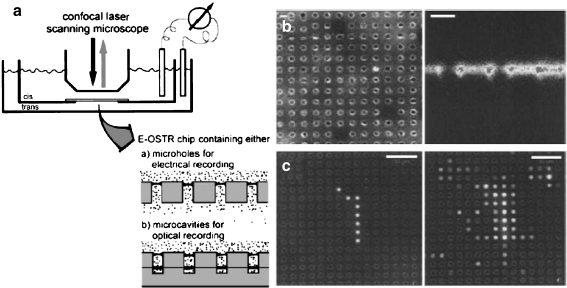
Over the last decade, Peter’s group [35] has been developing an excellent measurement technology for the optical (fluorescence-based) detection of membrane transport using a microwell array. In their optical single transporter recording (OSTR) system, membranes were firmly attached to a polycarbonate sheet with homogeneous distributions of cylindrical pores. The flux of the fluorescent substrate across the membrane was quantified by measuring the increase in fluorescence intensity inside the pores using confocal laser scanning microscopy. Transport of the fluorescent protein B-phycoerythrin through the membrane of erythrocytes [36] and that of the nuclear transport receptors NTF2 and GFP through the nuclear pore complex [37] were measured quantitatively. Since the transported substrate was concentrated in a tiny volume (1 aL–1 pL), it became possible to detect the activity of single transporters.
Measurement of receptor activity
In the examples mentioned above, patches of the cell membrane mounted over micropores were utilized to measure the transport of molecules across the membrane. Danelon et al. [38] proposed the use of similar cell membrane patches for the study of receptor proteins. In this study, a Si3N4 membrane, in which holes with a diameter of 50–600 nm were created, was pressed on adhering live cells. This technique provides the fluidic accessibility to both extracellular and intracellular surfaces of the membrane, enabling the side-specific assay of 5HT3 receptors.
Reconstituted membrane-based systems
A lipid bilayer membrane can be artificially reconstituted in various forms from purified or synthesized lipid molecules. In an aqueous environment, phospholipids spontaneously self-assemble into bilayer membranes through hydrophobic interactions between their acyl chains. The utilization of artificial lipid membranes for the analysis of membrane protein has several advantages over intact cell-based assays. First, it makes it possible to analyze the proteins in the membrane that are difficult to access with a glass pipette or microapertures, including proteins located in intracellular organelles. Second, the measurement conditions, such as the composition of the lipid membrane and buffer, can be precisely defined for delicate analysis. Thus, experiments in conditions far from physiological conditions are possible. Third, unlike in experiments with live cells, where protein expression levels fluctuate widely in each cell, the population of membrane proteins in reconstituted membranes can be controlled. In the following sections, we introduce approaches to use microtechnologies for the controlled reconstitution of three different major classes of artificial bilayer membranes.
Suspended lipid bilayer systems
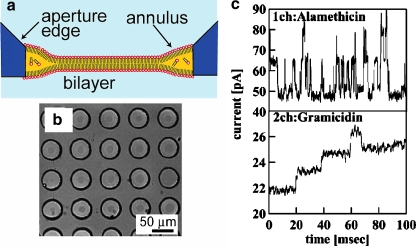
A planar bilayer lipid membrane (also called a black lipid membrane, BLM) has been used for the reconstitution and electrophysiological analysis of ion channel proteins (Fig. 2a). A layer of nonpolar solvent containing phospholipids is painted over a tiny aperture and spontaneously becomes a biofunctional lipid bilayer as it thins down. Ion channel proteins are typically incorporated into the bilayer membrane by fusing the proteoliposome (liposome or membrane fraction containing membrane proteins) by adding the suspension into the buffer bath. A BLM generally has a much better electrical seal than the cell membrane patch, which helps in the detection of single ion channel current. Since the 1970s, many types of ion channels have been studied in this platform [39–42]. However, in these conventional BLM-forming methods (i.e., the painting method and the Langmuir–Blodgett method), bilayer formation apertures have been fabricated by manually shaving the indentation made on the polymer sheet [43]. Thus, the aperture diameter is nonuniform and relatively large (50–200 μm). Because the lipid membrane is only 5–10-nm thick, the large planar membrane is mechanically fragile and easily broken by small mechanical and electrical disturbances. Thus, the stability and reproducibility of the reconstituted BLM are expected to be improved by using precisely defined micromachined apertures.
Fig. 2.
a The planar bilayer lipid membrane (BLM) formed over an aperture. b Array formation of 25 BLMs in a Parylene sheet device. c Simultaneous recording of alamethicin and gramicidin peptide channels in the addressable parallel recording chip [52]
Schmidt et al. [44] used a micromachined aperture to reconstitute a BLM. They formed a lipid membrane by attracting a giant liposome on a 0.6–7-μm aperture opened in the Si3N4/SiO2 diaphragm and demonstrated a recording of the alamethicin ion channel. Peterman et al. [45] also utilized a silicon-based microaperture for BLM experiments. Fertig et al. [46] performed the recording of a single gramicidin A ion channel current using a BLM in a glass device with a track-etched aperture (diameter approximately 1 μm), which was also used for the patch-clamping experiments [16, 17]. Pantoja et al. [47] successfully recorded a single channel current of maxi-K channels reconstituted in the BLM formed over the silicon microaperture (diameter 200 μm). Mayer et al. [48] thoroughly examined the size effect of the apertures on the electric noise and the stability of a BLM with the apertures opened in a Teflon sheet. Although electric noise was expected to decrease with smaller BLMs, they found that the noise from other contributions became dominant with apertures smaller than 40 μm. Nonetheless, noise was reduced to less than 0.4 pA rms in practice, and the BLM was stable at voltages up to 460 mV. They also formed multiple parallel BLMs in a single sheet to indicate the possibility of high-throughput screening.
These early studies showed that the use of micromachined apertures efficiently reduces electric noise and stabilizes the BLM. However, the method of forming the BLM relied on conventional manual techniques. Recently, several groups have proposed the integration of microfluidic platforms to improve the reproducibility of BLM formation to help develop parallel and addressable systems. Our group proposed forming BLMs in a microfluidic platform by alternately inserting the lipid solution and buffer through a microchannel [49]. Since the amount of lipid solution remaining over the aperture is automatically adjusted in the microfluidic systems, the reproducibility of the BLM formation improved dramatically (approximately 90%) [50, 51]. In these studies, poly(methyl methacrylate) was used as a substrate to reduce electric noise. However, spatial resolution in the manufacturing of microapertures was low, because we used mechanical machining. Recently, we found that the use of Parylene film improved the formation efficiency of a lipid bilayer, on which apertures of micrometer resolution can be easily fabricated. Using this technology, we realized a parallel recording in multiple independent wells in a single chip [52] (Fig. 2b,c). Other groups have also developed methods for the reproducible formation of BLMs. Ide and Ichikawa [53] showed that the thinning of the lipid layer, which generally takes a long time, can be accelerated by pressing the lipid layer on an agarose layer. Microchannel-based BLM formation was also performed by Morgan’s group [54, 55], who successfully demonstrated the recording of KcsA ion channels in a microfluidic platform [56]. In all of the studies mentioned above, BLMs were formed over microapertures in a manner analogous to conventional BLM methods. Several groups have recently demonstrated the efficient formation of a BLM across a microfluidic channel. We have developed a “contact method” in which the BLM is formed by contacting the two water interfaces separated by a solvent containing phospholipids [57]. This method is extremely simple and forms planar lipid bilayers reproducibly, and is thus suitable for multiple bilayer formation in microfluidic channels. A similar approach was described by Malmstadt et al. [58], showing the formation of a BLM in a PDMS microchannel. They utilized the characteristics of PDMS to absorb the solvent, to drive the thinning of solvent layer between the two monolayer interfaces. Holden et al. [59] carried out the formation of planar membrane networks by joining multiple droplets in hexadecane containing phospholipids, and demonstrated the modulation of membrane current with bacteriorhodopsin. These contact-method-based approaches can increase the flexibility in designing automated BLM-based systems for membrane protein assays. In addition, this method also has the potential to enable the formation of asymmetric bilayers, as two different lipid monolayers are brought into contact.
The BLM system has not yet been applied for the assay of transporter proteins because of the technical issue that small amounts of transported substrate should diffuse and be diluted in a relatively large volume of buffer bath. However, Peter’s group [60] also developed a BLM-based system to detect membrane transport phenomena using the same principle as the OSTR system mentioned above (Fig. 3). They determined the flux of fluorescent molecules through a single α-hemolysin peptide pore. This technology is very promising for a transportation assay of functional transporter proteins at the single-molecule level, which will be a significant breakthrough in the study of transporters.
Fig. 3.
a Optical single transporter recording (OSTR) system based on the BLM for the membrane transport assay. b Confocal images of the bilayer membranes formed over the microwells. c Increase of fluorescence intensity due to the transport (diffusion) of the fluorescent substrate across the BLM and accumulation in the microwells [60]
Supported lipid bilayer systems
A supported lipid bilayer (SLB or supported BLM) is a reconstituted membrane that is formed on the surface of a solid substrate, which is formed by incubating a liposome suspension on a silicon oxide or polymer surface [61, 62]. This class of artificial membranes has the excellent quality of being easily micropatterned by various techniques, such as patterning of hydrophilic/phobic surfaces [63], microcontact printing [64, 65], lift-off techniques [66], and photopolymerization [67]. Because they are easily formed, stable, and versatile, many researchers expect SLBs to be applied to biosensor applications based primarily on the interaction between membrane proteins and ligands on the membrane surface. Some examples of applications are in binding assays of bacterial toxin [68], in immunological assays [69, 70], in detection of ligand/receptor interaction [71, 72], and other membrane/molecule interactions [73]. Generally, SLBs are not suitable for electrophysiological measurements of ion channels owing to their large electrical permeability. However, because SLBs are much more stable than BLMs (SLBs lasts for many days, while BLMs only last for hours) the effort to develop electrochemical sensors is ongoing [74–76]. For further information on SLB-based assay systems, readers should refer to the excellent reviews by Sackmann and Tanaka [10], Groves [9], Groves and Boxer [77], Groves and Dustin [78], and Cooper [79].
Liposome-based systems
Liposomes or lipid vesicles are spherical lipid membranes. They have been widely used as a model cell membrane system and as a cell-sized container [11, 80]. Since liposomes form as a result of the self-assembly of lipids, it is difficult to control their properties, such as size, lamellarity, and entrapment efficiency. Various formation methods have been under investigation for decades with the goal of forming unilamellar liposomes, large or giant liposomes, and high encapsulation efficiency, and these efforts have been discussed in numerous papers and books [11, 80]. Reconstitution of membrane proteins into liposomes is an important research field in molecular biology and, thus, is beyond the scope of this review. In this section, we focus on current efforts to develop microsystems designed for liposome-based membrane protein assays.
In conventional analysis methods, characteristics of liposomes are studied as an average of the bulk suspension, although liposomes vary greatly in size, shape, and lamellarity. This diversity raises difficulties in quantitatively determining the physical constants associated with phenomena such as encapsulation efficiency, internal reaction, and membrane transport. Monodisperse and unilamellar liposomes can be prepared by passing polydisperse liposomes through a polycarbonate membrane with nanoscale holes (extrusion method) [81]. However, the resulting size is restricted to less than 1 μm in diameter, which is not suitable for handling and optical observation of individual liposomes. Many researchers expect microsystems to greatly aid in the formation of monodisperse giant liposomes for single-vesicle assays. Tan et al. [82] recently proposed a method of producing uniform-sized lipid vesicles by transferring the uniform water-in-oil emulsion into the water phase. By carefully designing the chemical composition of reagents, the oil phase was removed from the vesicle shell, resulting in solvent-free liposomes. Giant unilamellar liposomes can also be efficiently formed by electroformation, although there is no size control in the conventional method [11]. Paunov’s group [83, 84] demonstrated the electroformation of giant liposomes from micropatterned dry lipids on an indium tin oxide substrate. The liposomes had a narrow size distribution (5–20 μm), since the size was determined by the number of lipids in each patterned grid. Our group demonstrated the electroformation of liposomes in microfluidic structures [85], and realized nearly monodisperse liposomes from lipids patterned by a lift-off process using a Parylene sheet [86]. Another issue which should be overcome in electroformation is that this technology is usually not applicable in buffers with a high ionic concentration, which limits its use for membrane protein reconstitution. However, Estes and Mayer [87] used electroformation to form unilamellar giant liposomes and exchanged the inner solution with high-ionic strength buffer in a flow chamber, and demonstrated the binding assay of protein annexin V. Estes et al. [88] fused electroformed liposomes induced by a fusogenic peptide in the microfluidic platform. Many expect these technologies to be applied to membrane protein assays to determine the physical constants in the phenomena originating from membrane proteins, which is only possible in single-vesicle assays [89, 90].
Conclusion
Like the microsystems designed for the assay of water-soluble biomolecules, microfabricated structures and microfluidic platforms are expected to improve many aspects of membrane protein assays, making them superior to the conventional bulk methods in areas such as reduction of sample volume and improvement in sensitivity, reproducibility, and throughput. Owing to the difficulties in handling membranes and membrane proteins, advances in μTAS technologies in this field have been relatively slow in coming. However, it is certain that handy, low-cost, reliable diagnostic tools for membrane proteins are now strongly desired for clinical and pharmacological applications. Planar patch-clamping systems for drug screening are commercially available. In addition, high-throughput technologies based on artificial lipid membranes are now rapidly progressing toward the realization of sensitive and reproducible assay systems. Other than the study of ion channels, the assay of transporter proteins is currently the most important avenue in cancer studies, since transporters expressed in tumor cells export drugs out of the membrane. The assay of receptor/ligand interaction is important for screening sensory and immunological reaction represented by the G-protein-coupled receptors. We believe that in the near future the μTAS and biochip assay technologies for membrane proteins will be used therapeutically as well as in the pharmaceutical industry for rapid and/or high-throughput screening systems.
Contributor Information
Hiroaki Suzuki, Email: suzuki@ist.osaka-u.ac.jp.
Shoji Takeuchi, Email: takeuchi@iis.u-tokyo.ac.jp.
References
- 1.Whitesides GM (2006) Nature 442:368–373 [DOI] [PubMed]
- 2.Dittrich PS, Tachikawa K, Manz A (2006) Anal Chem 78:3887–3908 [DOI] [PubMed]
- 3.DeRisi JL, Iyer VR, Brown PO (1997) Science 278:680–686 [DOI] [PubMed]
- 4.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, Mitchell T, Miller P, Dean RA, Gerstein M, Snyder M (2001) Science 293:2101–2105 [DOI] [PubMed]
- 5.Auroux PA, Iossifidis D, Reyes DR, Manz A (2002) Anal Chem 74:2637–2652 [DOI] [PubMed]
- 6.Kaji N, Tezuka Y, Takamura Y, Ueda M, Nishimoto T, Nakanishi H, Horiike Y, Baba Y (2004) Anal Chem 76:15–22 [DOI] [PubMed]
- 7.Tabuchi M, Ueda M, Kaji N, Yamasaki Y, Nagasaki Y, Yoshikawa K, Kataoka K, Baba Y (2004) Nat Biotechnol 22:337–340 [DOI] [PubMed]
- 8.Sato K, Tokeshi M, Kimura H, Kitamori T (2001) Anal Chem 73:1213–1218 [DOI] [PubMed]
- 9.Groves JT (2002) Curr Opin Drug Discov Dev 5:606–612 [PubMed]
- 10.Sackmann E, Tanaka M (2000) Trends Biotechnol 18:58–64 [DOI] [PubMed]
- 11.Luisi PL, Walde P (2000) Giant vesicles. Wiley, New York
- 12.Neher E, Sakmann B (1976) Nature 260:799–802 [DOI] [PubMed]
- 13.Xu J, Wang X, Ensign B, Li M, Wu L, Guia A, Xu J (2001) Drug Discov Today 6:1278–1287 [DOI] [PubMed]
- 14.Gonzalez JE, Oades K, Leychkis Y, Harootunian A, Negulescu PA (1999) Drug Discov Today 4:431–439 [DOI] [PubMed]
- 15.Denyer J, Worley J, Cox B, Allenby G, Banks M (1998) Drug Discov Today 3:323–332 [DOI]
- 16.Fertig N, Blick RH, Behrends JC (2002) Biophys J 82:3056–3062 [DOI] [PMC free article] [PubMed]
- 17.Fertig N, Klau M, George M, Blick RH, Behrends JC (2002) Appl Phys Lett 81:4865–4867 [DOI]
- 18.Klemic KG, Klemic JF, Reed MA, Sigworth FJ (2002) Biosens Bioelectron 17:597–604 [DOI] [PubMed]
- 19.Ionescu-Zanetti C, Shaw RM, Seo J, Jan YN, Jan LY, Lee LP (2005) Proc Natl Acad Sci USA 102:9112–9117 [DOI] [PMC free article] [PubMed]
- 20.Lau AY, Hung PJ, Wu AR, Lee LP (2006) Lab Chip 6:1510–1515 [DOI] [PubMed]
- 21.Lehnert T, Gijs MAM, Netzer R, Bischoff U (2002) Appl Phys Lett 81:5063–5065 [DOI]
- 22.Pandey S, Mehrotra R, Wykosky S, White MH (2004) Solid-State Electron 48:2061–2066 [DOI]
- 23.Pantoja R, Nagarah JM, Starace DM, Melosh NA, Blunck R, Bezanilla F, Heath JR (2004) Biosens Bioelectron 20:509–517 [DOI] [PubMed]
- 24.Sordel T, Garnier-Raveaud S, Sauter F, Pudda C, Marcel F, De Waard M, Arnoult C, Vivaudou M, Chatelain F, Picollet-D’hahan N (2006) J Biotechnol 125:142–154 [DOI] [PubMed]
- 25.Zhang ZL, Asano T, Uno H, Tero R, Suzui M, Nakao S, Kaito T, Shibasaki K, Tominaga M, Utsumi Y, Gao YL, Urisu T (2007) Thin Solid Films (in press). DOI 10.1016/j.tsf.2007.04.104
- 26.Finkel A, Wittel A, Yang N, Handran S, Hughes J, Costantin J (2006) J Biomol Screen 11:488–496 [DOI] [PubMed]
- 27.Brueggemann A, George M, Klau M, Beckler M, Steindl J, Behrends JC, Fertig N (2004) Curr Drug Discov Technol 1:91–96 [DOI] [PubMed]
- 28.Wood C, Williams C, Waldron GJ (2004) Drug Discov Today 9:434–441 [DOI] [PubMed]
- 29.Granfeldt D, Sinclair J, Millingen M, Farre C, Lincoln P, Orwar O (2006) Anal Chem 78:7947–7953 [DOI] [PubMed]
- 30.Sinclair J, Olofsson J, Phil J, Orwar O (2003) Anal Chem 75:6718–6722 [DOI] [PubMed]
- 31.Li X, Klemic KG, Reed MA, Sigworth FJ (2006) Nano Lett 6:815–819 [DOI] [PubMed]
- 32.Borths EL, Poolman B, Hvorup RN, Locher KP, Rees DC (2005) Biochemistry 44:16301–16309 [DOI] [PubMed]
- 33.van Aubel RA, van Kuijck MA, Koenderink JB, Deen PM, van Os CH, Russel FG (1998) Mol Pharmacol 53:1062–1067 [PubMed]
- 34.Dong M, Penin F, Baggetto LG (1996) J Biol Chem 271:28875–28883 [DOI] [PubMed]
- 35.Peters R (2003) Annu Rev Biophys Biomol Struct 32:47–67 [DOI] [PubMed]
- 36.Tschodrich-Rotter M, Peters R (1998) J Microsc 192:114–125 [DOI] [PubMed]
- 37.Kiskin NI, Siebrasse JP, Peters R (2003) Biophys J 85:2311–2322 [DOI] [PMC free article] [PubMed]
- 38.Danelon C, Perez JB, Santschi C, Brugger J, Vogel H (2006) Langmuir 22:22–25 [DOI] [PubMed]
- 39.Miller C, Racker E (1976) J Membr Biol 30:283–300 [DOI] [PubMed]
- 40.White MM, Miller C (1979) J Biol Chem 254:10161–10166 [PubMed]
- 41.Labarca P, Coronado R, Miller C (1980) J Gen Physiol 76:397–324 [DOI] [PMC free article] [PubMed]
- 42.Latorre R, Vergara C, Hidalgo C (1982) Proc Natl Acad Sci USA 79:805–809 [DOI] [PMC free article] [PubMed]
- 43.Wonderlin WF, Finkel A, French RJ (1990) Biophys J 58:289–297 [DOI] [PMC free article] [PubMed]
- 44.Schmidt C, Mayer M, Vogel H (2000) Angew Chem Int Ed 39:3137–3140 [DOI] [PubMed]
- 45.Peterman MC, Ziebarth JM, Braha O, Bayley H, Fishman HA, Bloom DM (2002) Biomed Microdev 4:231–236 [DOI]
- 46.Fertig N, George M, Klau M, Meyer C, Tilke A, Sobotta C, Blick RH, Behrends JC (2003) Receptors Channels 9:29–40 [DOI] [PubMed]
- 47.Pantoja R, Sigg D, Blunck R, Bezanilla F, Heath JR (2001) Biophys J 81:2389–2394 [DOI] [PMC free article] [PubMed]
- 48.Mayer M, Kriebel JK, Tosteson MT, Whitesides GM (2003) Biophys J 85:2684–2695 [DOI] [PMC free article] [PubMed]
- 49.Suzuki H, Tabata K, Kato-Yamada Y, Noji H, Takeuchi S (2004) Lab Chip 4:502–505 [DOI] [PubMed]
- 50.Suzuki H, Tabata KV, Noji H, Takeuchi S (2006) Langmuir 22:1937–1942 [DOI] [PubMed]
- 51.Suzuki H, Tabata KV, Noji H, Takeuchi S (2007) Biosens Bioelectron 22:1111–1115 [DOI] [PubMed]
- 52.Pioufle BL, Suzuki H, Tabata KV, Noji H, Takeuchi S (2008) Anal Chem 80:328–332 [DOI] [PubMed]
- 53.Ide T, Ichikawa T (2005) Biosens Bioelectron 21:672–677 [DOI] [PubMed]
- 54.Sandison ME, Morgan H (2005) J Micromech Microeng 15:S139–S144 [DOI]
- 55.Sandison ME, Zagnoni M, Morgan H (2007) Langmuir 23:8277–8284 [DOI] [PubMed]
- 56.Zagnoni M, Sandison ME, Marius P, Lee AG, Morgan H (2007) Lab Chip 7:1176–1183 [DOI] [PMC free article] [PubMed]
- 57.Funakoshi K, Suzuki H, Takeuchi S (2006) Anal Chem 78:8169–8174 [DOI] [PubMed]
- 58.Malmstadt N, Nash MA, Purnell RF, Schmidt JJ (2006) Nano Lett 6:1961–1965 [DOI] [PubMed]
- 59.Holden MA, Needham D, Bayley H (2007) J Am Chem Soc 129:8650–8655 [DOI] [PubMed]
- 60.Hemmler R, Bose G, Wagner R, Peters R (2005) Biophys J 88:4000–4007 [DOI] [PMC free article] [PubMed]
- 61.Sackmann E (1996) Science 271:43–48 [DOI] [PubMed]
- 62.Richter RP, Berat R, Brisson AR (2006) Langmuir 22:3497–3505 [DOI] [PubMed]
- 63.Groves JT, Ulman N, Boxer SG (1997) Science 275:651–653 [DOI] [PubMed]
- 64.Hovis JS, Boxer SG (2001) Langmuir 17:3400–3405 [DOI]
- 65.Hovis JS, Boxer SG (2000) Langmuir 16:894–897 [DOI]
- 66.Orth RN, Kameoka J, Zipfel WR, Ilic B, Webb WW, Clark TG, Craighead HG (2003) Biophys J 85:3066–3073 [DOI] [PMC free article] [PubMed]
- 67.Morigaki K, Baumgart T, Offenhausser A, Knoll W (2001) Angew Chem Int Ed 40:172–174 [DOI] [PubMed]
- 68.Moran-Mirabal JM, Edel JB, Meyer GD, Throckmorton D, Singh AK, Craighead HG (2005) Biophys J 89:296–305 [DOI] [PMC free article] [PubMed]
- 69.Orth RN, Wu M, Holowka DA, Craighead HG, Baird BA (2003) Langmuir 19:1599–1605 [DOI]
- 70.Yang T, Jung S, Mao H, Cremer PS (2001) Anal Chem 73:165–169 [DOI] [PubMed]
- 71.Wu M, Holowka D, Craighead HG, Baird B (2004) Proc Natl Acad Sci USA 101:13798–13803 [DOI] [PMC free article] [PubMed]
- 72.Yamazaki V, Sirenko O, Schafer RJ, Nguyen L, Gutsmann T, Brade L, Groves JT (2005) BMC Biotechnol 5:18 [DOI] [PMC free article] [PubMed]
- 73.Majd S, Mayer M (2005) Angew Chem Int Ed 44:6697–6700 [DOI] [PubMed]
- 74.Bayley H, Cremer PS (2001) Nature 413:226–230 [DOI] [PubMed]
- 75.Jenkins ATA, Boden N, Bushby RJ, Evans SD, Knowles PF, Miles RE, Ogier SD, Schönherr H, Vancso GJ (1999) J Am Chem Soc 121:5274–5280 [DOI]
- 76.Andersson M, Keizer HM, Zhu C, Fine D, Dodabalapur A, Duran RS (2007) Langmuir 23:2924–2927 [DOI] [PubMed]
- 77.Groves JT, Boxer SG (2002) Acc Chem Res 35:149–157 [DOI] [PubMed]
- 78.Groves JT, Dustin ML (2003) J Immunol Methods 278:19–32 [DOI] [PubMed]
- 79.Cooper MA (2004) J Mol Recognit 17:286–315 [DOI] [PubMed]
- 80.Torchilin V, Wessig V (2003) Liposomes: a practical approach, 2nd edn. Oxford University Press, Oxford
- 81.Mayer LD, Hope MJ, Cullis PR (1986) Biochim Biophys Acta 858:161–168 [DOI] [PubMed]
- 82.Tan YC, Hettiarachchi K, Siu M, Pan YR, Lee AP (2006) J Am Chem Soc 128:5656–5658 [DOI] [PubMed]
- 83.Taylor P, Xu C, Fletcher PD, Paunov VN (2003) Chem Commun 1732–1733 [DOI] [PubMed]
- 84.Taylor P, Xu C, Fletcher PDI, Paunov VN (2003) Phys Chem Chem Phys 5:4918–4922 [DOI]
- 85.Kuribayashi K, Tresset G, Coquet P, Fujita H, Takeuchi S (2006) Meas Sci Technol 17:3121–3126 [DOI]
- 86.Kuribayashi K, Takeuchi S (2005) Proc μTAS 2:1455–1457
- 87.Estes DJ, Mayer M (2005) Biochim Biophys Acta 1712:152–160 [DOI] [PubMed]
- 88.Estes DJ, Lopez SR, Fuller AO, Mayer M (2006) Biophys J 91:233–243 [DOI] [PMC free article] [PubMed]
- 89.Tamba Y, Yamazaki M (2005) Biochemistry 44:15823–15833 [DOI] [PubMed]
- 90.Noireaux V, Libchaber A (2004) Proc Natl Acad Sci USA 101:17669–17674 [DOI] [PMC free article] [PubMed]