Abstract
Heterocyclic and aromatic amine carcinogens are thought to lead to tumor initiation via the formation of DNA adducts, and bioactivation to arylhydroxylamine metabolites is necessary for reactivity with DNA. Carcinogenic arylhydroxylamine metabolites are cleared by a microsomal, NADH-dependent, oxygen-insensitive reduction pathway in humans, which may be a source of inter-individual variability in response to aromatic amine carcinogens. The purpose of this study was to characterize the identity of this reduction pathway in human liver. Based on our findings with structurally similar arylhydroxylamine metabolites of therapeutic drugs, we hypothesized that the reductive detoxication of arylhydroxylamine carcinogens was catalyzed by NADH cytochrome b5 reductase (b5R) and cytochrome b5 (cyt b5). We found that reduction of the carcinogenic hydroxylamines of the aromatic amine 4-aminobiphenyl (4-ABP; found in cigarette smoke) and the heterocyclic amine 2- amino-1-methyl-6-phenylimidazo [4,5-b] pyridine (PhIP; found in grilled meats) was indeed catalyzed by a purified system containing only human b5R and cyt b5. Specific activities were 56 to 346-fold higher in the purified system compared to human liver microsomes (HLM), with similar Michaelis-Menten constants (Km values) in both systems. The stoichiometry for b5R and cyt b5 that yielded the highest activity in the purified system was also similar to that found in native HLM (∼1:8 to 1:10). Polyclonal antisera to either b5R or cyt b5 significantly inhibited N-hydroxy-4-aminobiphenyl (NHOH-4-ABP) reduction by 95 and 89%, respectively, and immunoreactive cyt b5 protein content in individual HLM was significantly correlated with individual reduction of both NHOH-4-ABP and N-hydroxy-PhIP (NHOH-PhIP). Finally, titration of HLM into the purified b5R/cyt b5 system did not enhance the efficiency of reduction activity. We conclude that b5R and cyt b5 are together solely capable of the reduction of arylhydroxylamine carcinogens, and we further hypothesize that this pathway may be a source of individual variability with respect to cancer susceptibility following 4-ABP or PhIP exposure.
Introduction
Heterocyclic and arylamine carcinogens such as 2-amino-1-methyl-6-phenylimidazo[4,5-b] pyridine (PhIP,1 generated in cooked meats) and 4-aminobiphenyl (4-ABP, a component of cigarette smoke) are established rodent carcinogens that have been epidemiologically linked to human cancers of the colon, breast, and other organs. (1-4) Carcinogenesis from these compounds is thought to be initiated by DNA adducts; however, generation of arylhydroxylamine metabolites is necessary for DNA adducts to occur. (5) Carcinogenic arylhydroxylamines are generated by P450 1A2, 1A1, or 1B1, lactoperoxidase, or myeloperoxidase, (6-9) and are substrates for further activation through O-acetylation or O-sulfonation prior to adduct formation. (5,10,11)
Arylhydroxylamines are eliminated by a reduction pathway in human liver that is NADH-dependent, microsomal, and oxygen-insensitive. (12) This arylhydroxylamine reduction pathway is much more efficient than the oxidative formation of carcinogenic arylhydroxylamines, and the balance of reduction to forward oxidation can vary more than 20-fold among individuals. (12) Structurally similar arylhydroxylamine metabolites generated from drugs such as sulfamethoxazole and dapsone are metabolically reduced to their parent amines by an apparently similar NADH-dependent system, (13) which we have shown to be comprised solely of NADH cytochrome b5 reductase (b5R) and cytochrome b5 (cyt b5). (14)
In this report we provide evidence for the necessary and sufficient roles of b5R and cyt b5 in the reduction of arylhydroxylamine carcinogens. We show that purified b5R and cyt b5 together efficiently reduce the hydroxylamine metabolites of both 4-ABP (NHOH-4-ABP) and PhIP (NHOH-PhIP), and that other microsomal proteins do not enhance activity. We further show that the apparent Michaelis-Menten constants (Km) for hydroxylamine reduction are essentially identical in the purified system and in human liver microsomes, with 56 to 346-fold higher specific activities in the purified system. In addition, arylhydroxylamine carcinogen reduction activities correlate significantly with individual human liver microsomal cyt b5 content, and are strongly inhibited by antisera to either b5R or cyt b5. We conclude that b5R and cyt b5 are necessary and sufficient for direct reduction of arylhydroxylamine carcinogens in human liver, and further hypothesize that variability in the expression of either of these proteins may influence cancer risk following exposure to carcinogenic heterocyclic and aromatic amines.
Experimental Procedures
Expression of Soluble Human NADH Cytochrome b5 Reductase (b5R) and Soluble Human Cytochrome b5 (Cyt b5)
Human recombinant b5R and cyt b5 were expressed in Escherischia coli (E. coli) using a His-tag expression system, as previously described. (14,15) Briefly, the full length cDNA of human soluble b5R (kindly provided by Dr. Komei Shirabe, Oita Medical University, Japan) was inserted into an N-terminal histidine tag expression system (pCR T7/NT-TOPO®; Invitrogen; Carlsbad, CA) to allow purification from E. coli lysate, using nickel affinity chromatography. Induction with 1 mM IPTG was performed for 4 hours at room temperature. Stepwise elution of contaminants followed by elution of His-tagged b5R was accomplished with 20, 100 and 350 mM imidazole, each at pH 8.0 in binding buffer. The eluent was collected in 1 mL fractions, which were tested for purity by gel electrophoresis and silver staining, for identity using immunoblotting, and for activity using potassium ferricyanide reduction. (13) The fractions containing a single 36 kD band on silver stain (32 kD soluble protein plus 4 kD His tag) were pooled and dialyzed against binding buffer pH 8.0, through 10,000 Mr cut-off cassettes (Pierce Biotechnology, Rockford IL). The dialyzed sample was again bound to a nickel column and re-purified and pooled, as described for the first round of purification. The purified b5R was dialyzed against PBS, pH 7.4 and concentrated to a final protein concentration of 2−3 mg/mL using 10,000 Mr cut-off filtration devices (Centricon®, Millipore).
For cyt b5, the full length cDNA of human soluble cyt b5 was expressed in E. coli as described for b5R, except for the use of pCR T7/CT-TOPO® vector, stepwise elution with 20, 40 and 100 mM imidazole, and hemin loading after protein purification. (15) Fractions containing a single 15 kD band (12 kD soluble protein plus 3 kD His tag) on silver stain were pooled, dialyzed, and re-purified as described for b5R, with identity verified by immunoreactivity with antibody to human cyt b5. (14) Purified soluble proteins retained maximal activity at 4°C for approximately 3 weeks.
Arylhydroxylamine Carcinogen Reduction. Caution
NHOH-PhIP, NHOH-4-ABP, PhIP and 4-ABP are extremely hazardous, potentially mutagenic and carcinogenic compounds. Appropriate precautions should be taken when handling these compounds, i.e. gloves, lab coat, and a face mask should be worn, and transfer of the solid powders should be performed within a ventilated hood. NHOH-PhIP (Midwest Research Institute, Kansas City, MO; in 100% methanol, 2.5 % final concentration) or NHOH-4-ABP (Toronto Research Chemicals, Toronto, Ontario, Canada; in 50% DMSO, 1.25 % final concentration) were incubated at varying concentrations with purified soluble human recombinant b5R and cyt b5. The optimal stoichiometry of b5R : cyt b5 was evaluated by measuring reduction activity using different ratios of b5R to cyt b5 while keeping the total nanomol of protein constant. (16) Specifically, the molar ratio of b5R : cyt b5 was adjusted from 14:1, 10:1, 8:1, 2:1, 1:1, 1:2, 1:8, 1:10 and 1:14 while the total number of mol in the reaction remained constant at 1.1 nmol. Reactions were performed in PBS, pH 7.4, with 1 mM NADH. Ascorbic acid (17) (3 mM) stabilized both arylhydroxylamines from further oxidation over the incubation times of the assays (up to 30 min), and was included in all reactions.
Arylhydroxylamine reduction activities were determined by HPLC separation over a C18 column (Beckman ODS, 4.6 mm × 25 cm; Beckman Coulter Inc, Fullerton, CA), followed by UV detection at 254 nm. Solvent A was 0.1% triethylamine (TEA) /1% acetic acid in water, and solvent B was acetonitrile; the linear gradient for 4-ABP detection was 20−80% B over 15 min, at 2 mL/min. 4-ABP and NHOH-4-ABP eluted at 8.4 and 7.6 min, respectively. For PhIP detection, solvent A was 0.2% TEA in water and solvent B was methanol/0.2% TEA. The linear gradient was 50−80% B over 14 min at 1.5 mL/min; PhIP and NHOH-PhIP eluted at 9.4 and 10.5 min, respectively. Both assays were linear from 0.3 μM through 1 mM, with a limit of detection of < 0.3 μM, and inter-assay coefficients of variation for both assays of 0.3−9.6% over the range of concentrations used in kinetic determinations. During method development, parent and arylhydroxylamine peaks were analyzed by HPLC-MS, and were found to generate ions of the expected masses. Negative controls included reactions with human serum albumin (HSA) instead of b5R/cyt b5, and systems lacking NADH or substrate. Estimates for apparent Km and maximal velocity (Vmax) for the two substrates were determined by nonlinear curve fitting to a one site Michaelis Menten equation using commercial software (Prism 3; GraphPad Software, Inc., San Diego, CA). For comparison of specific activities and apparent Km values between purified and native systems, kinetics were also determined in pooled human liver microsomes (250 μg; BD Gentest, Woburn, MA). To assess the possible involvement of additional microsomal enzymes in the reduction pathway, reduction of NHOH-4-ABP or NHOH-PhIP (500 μM) by purified b5R and cyt b5 was also determined following the addition of increasing amounts of pooled human liver microsomes (0, 250, or 1000 μg), with preincubation of the proteins for 10 min prior to addition of NADH.
Determination of Native Stoichiometry Between b5R and Cyt b5 in Human Liver Microsomes
Purified human b5R and cyt b5 were each used to immunize individual rabbits using standard protocols (Panigen Inc., Blanchardville, WI). To approximate the molar ratio of b5R and cyt b5 as expressed in native human liver, these polyclonal antibodies were used to probe immunoblots made from pooled human liver microsomes (40 μg; BD Gentest, Woburn, MA). (14) Since b5R and cyt b5 are expressed only as holoenzymes, blots were also loaded with standards of purified human b5R (0.18, 0.45 and 0.72 pmol) and cyt b5 (1.2, 2.94 and 8.82 pmol), in order to generate standard curves relating densitometry to pmol protein. These curves were used for relative quantitation of immunoreactive b5R vs. cyt b5 in HLM.
Inhibition of Microsomal Arylhydroxylamine Reduction
To further confirm the importance of b5R and cyt b5 in arylhydroxylamine reduction in human liver, anti-cyt b5 and anti-b5R antibodies were used for immunoinhibition experiments in HLM. Our anti-cyt b5 and anti-b5R antibodies each bound to a single protein in full length immunoblots, and were inhibitory in a dose-dependent manner to both hydroxylamine reduction by purified b5R and cyt b5, and cytochrome c reduction (a marker activity for b5R/cyt b5 (18)) in HLM (data not shown). b5R and cyt b5 antisera (150 μl each, based on optimization for other hydroxylamine substrates (14)) were then each pre-incubated with pooled human liver microsomes (250 μg) for 30 min at room temperature, followed by quantitation of arylhydroxylamine reduction as described for the purified system. Similar inhibition experiments were performed in the presence of the chemical b5R inhibitor p-hydroxy-mercuribenzoate (19) (PHMB; Sigma-Aldrich; 1 mM in 50% DMSO). Preimmune rabbit serum or HSA instead of serum, and PHMB vehicle, were used as negative controls.
Immunocorrelation Studies
In order to determine the relationship between b5R and cyt b5 expression and arylhydroxylamine reduction in human liver, antisera to b5R or cyt b5 were also used to probe immunoblots prepared from individual human liver microsomes (40 μg; BD Gentest, Woburn, MA; samples H3, H13, H32, H37, H47, H56, H74, H77, H1, H91, and H95). PVDF membranes (Immobilon-P; Millipore Corporation) were blocked with 2.5% nonfat dry milk in PBS/0.1% Tween-20 and washed with PBS/0.1% Tween-20. Primary antisera were diluted 1:10,000 for immunoblotting; horseradish peroxidase-linked secondary antibody (donkey anti-rabbit immunoglobulin G; Amersham Biosciences, Inc., Piscataway, NJ) was diluted 1:10,000. An enhanced chemiluminescence system was used for signal detection (Amersham Biosciences, Inc.), and was quantified using a Biospectrum AC Imaging System (UVP, Inc.). For each of the individual human liver microsome samples (250 μg), b5R and cyt b5 immunoreactivity were correlated with microsomal reduction of both substrates, as determined by HPLC, using a correlation z test (Prism; GraphPad Software, Inc., San Diego CA).
Extra-Hepatic Expression of b5R and Cyt b5, and Extrahepatic Arylhydroxylamine Reduction Activity
Normal human breast and colon tissues were obtained from the Cooperative Human Tissue Network (Midwestern Division, Ohio State University, Department of Pathology). Microsomes were prepared from breast or colon from 3 to 5 different individuals, and were used to prepare immunoblots that were probed with b5R and cyt b5 antisera. In addition, reduction activities for NHOH-4-ABP and NHOH-PhIP were determined in pooled human breast and colon microsomes and cytosol, as described for liver microsomes.
Statistical Analyses
All data were analyzed using a commercial statistical software program (Prism, GraphPad Software Inc, San Diego, CA). Comparisons between groups were performed using unpaired t test. All data are reported as mean ± SD, with P < 0.05 considered significant.
Results
Expression and Purification of Human b5R and Cyt b5
Expression of histidine-tagged human soluble b5R in E. coli and purification using two rounds of nickel affinity chromatography yielded an average of 3 to 4 mg of purified protein per liter of culture. A single 36-kDa protein (32-kDa protein plus 4-kDa tag) was obtained with consistent purity on silver staining, and strong immunoreactivity with anti-b5R antibody. (14) Expression and purification of histidine-tagged human soluble cyt b5 yielded an average of 5 mg of purified protein per liter of culture. A single 15-kDa protein (12-kDa protein plus 3-kDa tag) was obtained with consistent purity on silver staining and strong immunoreactivity with anti-cyt b5 antibody. (14)
Reduction Kinetics of Arylhydroxylamine Carcinogens
Purified recombinant b5R and cyt b5 catalyzed the efficient reduction of the hydroxylamines of both the aromatic amine 4-ABP and heterocyclic amine PhIP. Optimal reduction activity by the purified system was obtained with cyt b5 in excess of b5R, with maximal activity observed with a ratio of b5R to cyt b5 of approximately 1:8 (Figure 1). As this is similar to what we had found for arylhydroxylamine metabolites of therapeutic drugs (1:8 to 1:10)(14), all subsequent experiments were conducted with cyt b5 in excess of b5R (b5R: cyt b5 = 1:10). The kinetics of NHOH-4-ABP and NHOH-PhIP reduction in the purified system and in HLM (Figure 2) were each fit to a one-site Michaelis Menten equation with similar apparent Km values for both the microsomal and purified systems (Table 1). The specific activities (Vmax) for NHOH-4-ABP and NHOH-PhIP reduction in the purified system containing only b5R and cyt b5 were 55 and 346 times higher than for HLM, respectively (Table 1). No activity was seen in pooled human liver cytosol, with HSA instead of enzymes, in the presence of 3 mM ascorbate without enzymes, or in the absence of NADH, and minimal activity was observed with either b5R or cyt b5 alone (data not shown).
Fig. 1.
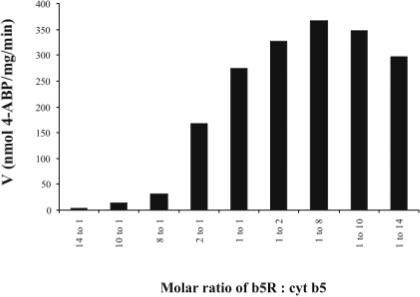
Arylhydroxylamine reduction activities at various molar ratios of b5R : cyt b5. A Job Plot (16) indicates maximal activity at a stoichiometry of 1:8 to 1:10 in the purified recombinant system (1.1 nmol protein total for each ratio). Activity for NHOH-4-ABP reduction (500 μM) is shown.
Fig. 2.
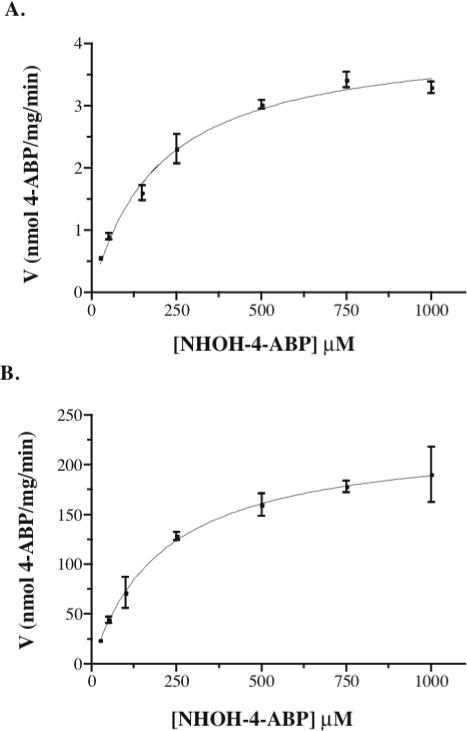
Velocity versus substrate concentrations for reduction of NHOH-4-ABP by (A) pooled human liver microsomes (250 μg), and (B) purified human recombinant b5R and cyt b5 (1:10 stoichiometry). All reactions were in PBS, pH 7.4, with 1 mM NADH and 3 mM ascorbate for arylhydroxylamine stabilization. Data for both enzyme sources were each fit to a one-site Michaelis Menten equation.
Table 1.
Kinetics of arylhydroxylamine carcinogen reduction by human liver microsomes or purified soluble NADH cytochrome b5 reductase (b5R) and cytochrome b5 (cyt b5).a
| Substrate | Enzyme Source | Km (μM) | Vmax (nmol/mg protein/min) |
|---|---|---|---|
| NHOH-4-ABP | HLM | 197 ± 1 b | 4.13 ± 0.05 |
| b5R/cyt b5 | 220 ± 89 b | 233 ± 36 d | |
| NHOH-PhIP | HLM | 229 ± 11 c | 1.57 ± 0.01 |
| b5R/cyt b5 | 225 c | 544 e |
The maximal velocity (Vmax) of arylhydroxylamine carcinogen reduction by purified b5R/cyt b5 was 56-fold
and 346-fold
higher than in HLM for NHOH-4-ABP and NHOH-PhIP, respectively. All reactions were performed in PBS, pH 7.4, with 1 mM NADH and 3 mM ascorbate, under conditions approximating linear kinetics. In the purified system, b5R and cyt b5 were used at a 1:10 stoichiometry. Velocity data are expressed in nmol per total mg protein per min. Data represent two to three experiments performed in duplicate for each condition, except for NHOH-PhIP by b5R/cyt b5, for which data are reported for one experiment performed in duplicate. Results are given as mean ± SD.
Km values were not significantly different between HLM and b5R/cyt b5 for either substrate
P = 0.75;
P = 0.65, one sample t test.
P = 0.01
P < 0.0001; one sample t test
Because other investigators have proposed the involvement of a third, P450-like protein in the reduction of therapeutic amidoximes and hydroxylamine metabolites of arylamine drugs, (20) we also evaluated the effect of titrated amounts of microsomal protein on the reduction activity of the purified system. These experiments were designed to determine whether the addition of other microsomal proteins enhanced the efficiency of reduction compared to the system containing b5R and cyt b5 alone. However, no increase in the efficiency of reduction, other than the expected additive effect (since b5R and cyt b5 are present in human liver microsomes), was seen with the addition of 250 or 1000 μg of pooled HLM to the recombinant system (Table 2).
Table 2.
Effect of addition of human liver microsomal protein to a purified system of b5R and cyt b5, on reduction capacity for NHOH-4-ABP and NHOH-PhIP.a
| Protein Source | Substrate (Reduction velocities; nmol/min) | |
|---|---|---|
| NHOH-4-ABP | NHOH-PhIP | |
| HLM | 1.02 ± 0.02 | 1.28 ± 0.001 |
| b5R/cyt b5 | 2.88 ± 0.01 | 2.88 ± 0.08 |
| Predicted additive HLM + b5R/cyt b5 | 3.90 b | 4.16 c |
| Observed HLM + b5R/cyt b5 | 4.15 ± 0.04 b | 3.72 ± 0.11 c |
All reactions performed as described in Table 1, using 250 μg HLM (for NHOH-4-ABP) or 1.0 mg HLM (for NHOH-PhIP), and 1.1 nmol b5R/cyt b5 (1:10 stoichiometry), or both enzyme systems in combination (observed activity). Predicted additive activity was derived by adding mean activities for each system alone. Note that total activities are not normalized to mg of protein, in order to more easily detect any enhancement of activity in the purified system following addition of microsomal protein. These results represent data run in duplicate for each condition. Similar results were observed for addition of 1.0 mg of HLM with NHOH-4-ABP as a substrate.
Not significantly different by one sample t-test.
Not significantly different by one sample t-test.
Immunoinhibition
Antisera to b5R and cyt b5 significantly inhibited NHOH-4-ABP reduction (500 μM) in pooled human liver microsomes by 95% (P = 0.004) and 89% (P = 0.002), respectively, compared with pre-immune sera (Figure 3). NHOH-PhIP reduction by pooled human liver microsomes (250 μg) was also inhibited by antisera to b5R (64%) and cyt b5 (75%). The chemical b5R inhibitor PHMB inhibited both NHOH-4-ABP and NHOH-PhIP reduction in pooled human liver microsomes by 84% (P = 0.026; Figure 3) and 93% (P = 0.004), respectively.
Fig. 3.
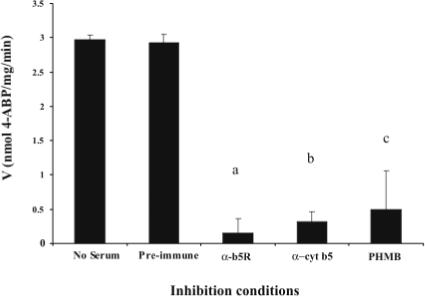
Inhibition of NHOH-4-ABP reduction. Antisera to b5R or cyt b5 significantly inhibited NHOH-4-ABP reduction (500 μM) in HLM by 95% (a P = 0.004) and 89% (b P = 0.002), respectively, compared with pre-immune serum incubation. The chemical b5R inhibitor p-hydroxymercuribenzoate (PHMB), also significantly inhibited NHOH-4-ABP reduction by 84% (c P = 0.026) compared to microsomes incubated with vehicle. Microsomes (250 μg) were pre-incubated with HSA (no serum), pre-immune sera or anti-sera (150 μl each), or PHMB (1 mM) or its vehicle for 30 min at room temperature prior to initiation of the reaction as described in Fig. 1. Data shown are from two experiments, each performed in duplicate.
Native Stoichiometry of b5R to Cyt b5 in Human Liver Microsomes
b5R and cyt b5 expression were semi-quantified in pooled human liver microsomes by immunoblotting, along with b5R and cyt b5 purified standards. Membranes were first probed for cyt b5, then stripped and probed again for b5R to directly compare b5R and cyt b5 immunoreactive protein concentrations within the same blot. Four separate experiments all indicated a molar ratio of 1:8 to 1:10 for b5R:cyt b5 in native HLM (representative blot shown in Figure 4). This estimate of native stoichiometry for b5R : cyt b5 in pooled human liver microsomes was consistent with the stoichiometry that yielded maximal activity for arylhydroxylamine reduction in the purified system (Figure 1).
Fig. 4.
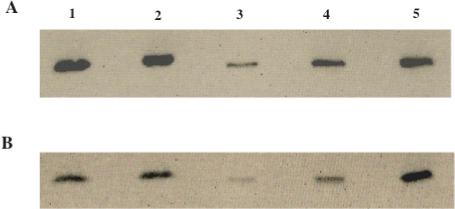
Immunoblot of pooled human liver microsomes, probed for both NADH cytochrome b5 reductase (A) and cytochrome b5 (B) content. Lane 1: pooled human liver microsomes, lane 2: duplicate pooled human liver microsomes, lanes 3−5: increasing concentrations of purified recombinant b5R (Panel A; 0.18, 0.45, and 0.72 pmol) or cyt b5 (Panel B; 1.2, 2.94, and 8.82 pmol) to establish standard curves (pmol b5R = 0.0166 * HLM b5R density + 0.2198; pmol cyt b5 = 0.6455 * HLM cyt b5 density + 0.6409). (Note: this cyt b5 antibody has a lower affinity for cyt b5 (less dense bands), compared to the b5R antibody for b5R; therefore, more pmol of cyt b5 have been loaded as standards to give a linear range for quantitation). Results from four immunoblots (representative shown) indicated a native b5R : cyt b5 molar ratio of between 1:8 to 1:10 in HLM.
Immunocorrelation
Reduction of NHOH-4-ABP or NHOH-PhIP did not correlate with microsomal content of immunoreactive b5R protein in 11 individual human liver microsomes; however, b5R content in these commercially available samples differed only by 1.8-fold (Figure 5A). On the other hand, microsomal cyt b5 content was significantly correlated with reduction activities for both NHOH-4-ABP (r = 0.79; P =0.004; (Figure 5B)) and NHOH-PhIP (r=0.70; P =0.016) in these individual human liver samples.
Fig. 5.
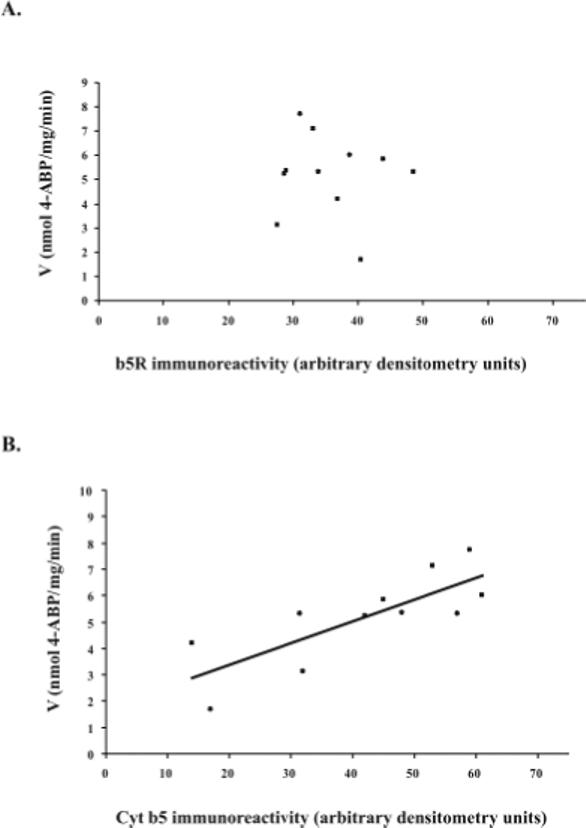
(A) Microsomal content of NADH cytochrome b5 reductase (as measured by immunoreactivity to b5R antibodies) did not correlate with NHOH-4-ABP reduction activity (500 μM substrate) in 11 individual human liver microsomes (r = −0.11). However, there was only a 1.8-fold range in b5R content in this sample of microsomes. (B) Microsomal cyt b5 content (as measured by immunoreactivity to anti-cyt b5 antibody) did correlate significantly with NHOH-4-ABP reduction in 11 individual human liver microsomes (r = 0.79; P = 0.004). Activity results are representative of three experiments each performed in duplicate. Reactions included human liver microsomes (250 μg), NHOH-4-ABP (500 μM), 1 mM NADH, and ascorbate (3 mM), in PBS, pH 7.4.
Extra-Hepatic Expression of b5R and Cyt b5 and Arylhydroxylamine Reduction Activity
Immunoreactive b5R and cyt b5 were both readily detected in pooled human breast and colon microsomes (Figure 6). Importantly, breast and colon microsomes showed reduction activity towards NHOH-4-ABP and NHOH-PhIP, respectively, at levels approximately 20%−40% of those seen in HLM. Cytosolic reduction activity was absent in both breast and colon, as found for liver cytosol (Table 3).
Fig. 6.
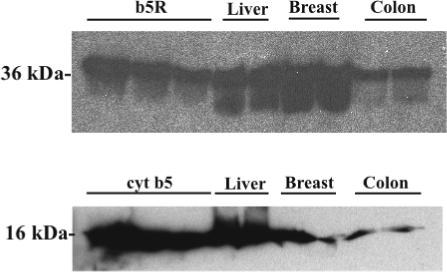
(A) Immunoblot of pooled microsomes prepared from human liver, breast, and colon (25 μg per lane), and probed with polyclonal antibody to b5R. First 3 lanes of panel A: 0.32, 0.16, and 0.0.08 pmol human recombinant b5R. (B) Immunoblot of pooled microsomes prepared from human liver (10 μg per lane), breast (50 μg), and colon (50 μg), and probed with polyclonal antibody to cyt b5. First 3 lanes of panel B: 0.42, 0.21, and 0.11 pmol human recombinant cyt b5. 12% polyacrylamide gels.
Table 3.
Carcinogenic arylhydroxylamine reduction by microsomes and cytosol prepared from pooled human breast, liver, and colon tissues.a
| NHOH-4-ABP reduction (nmol/mg/min) | NHOH-PhIP reduction (nmol/mg/min) | |||
|---|---|---|---|---|
| Breast | Liver | Colon | Liver | |
| Microsomes | 0.23 | 1.25 ± 0.08 | 0.38 | 0.92 ± 0.05 |
| Cytosol | * | * | * | NP |
All reactions performed in PBS, pH 7.4, with 1 mM NADH, 3 mM ascorbate, and 100 μM substrate, with 1 mg breast or colon microsomes or cytosol, and 0.5 mg human liver microsomes or cytosol.
Not detected. NP = not performed. Data represent the mean of 2 experiments run in duplicate, except for breast and colon microsomes, one experiment, each run in duplicate.
Discussion
NADH cytochrome b5 reductase (b5R) is an FAD-containing protein expressed in soluble and membrane bound forms, which are both the products of the same gene. (21,22) These isoforms have identical catalytic domains, but differ only in a ∼ 4 kDa hydrophobic anchor at the N terminus, which targets the membrane-bound enzyme to the endoplasmic reticulum and outer mitochondrial membranes of somatic cells. (23) The hemoprotein cytochrome b5 (cyt b5), is also expressed in soluble and membrane-bound forms, with identical catalytic domains that are encoded by alternative splicing of a single gene; (24) for membrane-bound cyt b5, the hydrophobic anchor is located at the C- terminus of the protein. (25) In liver and other tissues, b5R and cyt b5 together mediate electron transfer from NADH to fatty acid desaturases or P450 oxidases. (26,27) The soluble forms of b5R and cyt b5 are expressed primarily in erythrocytes, where they function to maintain hemoglobin in its reduced state. (28)
The results of these studies indicate that the complex of b5R and cyt b5 also has a direct role in the metabolism of arylhydroxylamine carcinogens. This enzyme complex efficiently catalyzed the reduction of the hydroxylamines of the aromatic amine 4-ABP and the heterocyclic aromatic amine PhIP. The apparent Km values for reduction by the purified system were nearly identical to those found in native human liver microsomes, which is consistent with, but not proof of, the involvement of the same enzyme system. Microsomal activity was inhibited by PHMB, a chemical inhibitor of b5R, and although PHMB has not been fully evaluated for its specificity for b5R, antibodies to b5R, as well as to cyt b5, dramatically inhibited activity. Further, arylhydroxylamine reduction correlated strongly with microsomal content of cyt b5. These data together provide strong evidence that the enzyme complex of b5R and cyt b5 is capable of the direct reduction of arylhydroxylamine carcinogens, and that these two enzymes alone very likely comprise this pathway in human liver.
The finding that addition of microsomal protein to the recombinant system did not enhance activity more than additively is also supportive of the view that additional microsomal proteins, other than b5R and cyt b5, are not required for efficient activity. However, these data should be interpreted cautiously, in that soluble proteins in the purified system may not have interacted optimally with microsomal enzymes in HLM under the conditions of these experiments.
The reduction of arylhydroxylamines represents an intriguing direct role for the b5R / cyt b5 complex in xenobiotic metabolism. Although b5R has been shown to reduce some chemotherapeutic agents to their cytotoxic forms, (29,30) direct xenobiotic detoxication by this enzyme complex had not been described. We recently reported a direct role for b5R and cyt b5 in the reduction of hydroxylamines of sulfamethoxazole and dapsone, and in the reduction of two amidoximes, the antihypertensive agent benzamidoxime and the antiprotozoal drug DB289. (14,31) The nature of the enzymes involved in the reduction of hydroxylamines has been somewhat controversial. Kadlubar and Ziegler, in their initial characterization of this azide and carbon-monoxide insensitive microsomal pathway in pig liver,(32) concluded b5R and cyt b5 were necessary, but that a third protein was involved in final electron transfer to the hydroxylamine substrate.(33) This third protein was not isolated, but was thought not to contain any heme or chromophores. Clement et al., also working in pig liver, reported that b5R and cyt b5 were cofactors in amidoxime and hydroxylamine reduction, but concluded that a third P450 protein was essential for efficient reduction. (20,34,35) This third protein has been reported to share homology with P450 2D, (20) although the protein fractions used in reconstituted activity assays have not been purified to homogeneity. (34) In addition, specific activities with this reconstituted system were more than 400-fold lower than those observed by our group for the same arylhydroxylamines, using only b5R and cyt b5.(14) Finally, a third group, investigating amidoxime reduction, has reported a role for stearoyl CoA desaturase in amidoxime and hydroxylamine reduction in rodent adipose.(36) However, a role for stearoyl CoA desaturase in hydroxylamine reduction in human liver was not established.
Our data indicate that a purified recombinant system, containing only b5R, cyt b5, and NADH, is capable of efficient and complete reduction of arylhydroxylamines in human liver. We found that an excess of cyt b5 was optimal for activity, and that the highest activities were seen at a ratio of 1:8 for the purified system containing b5R and cyt b5. This is similar to the findings of Kadlubar and Ziegler, who, although they invoked the involvement of a third unidentified protein in electron transfer, found maximal reduction activity when the ratio of b5R to cyt b5 was 1:9. (33) Further, we found this same approximate molar ratio in native human liver microsomes, which is consistent with the stoichiometry of 1:10 that has been reported for b5R and cyt b5 in rat and rabbit liver microsomes. (37,38) Previous studies implicating the need for other proteins in this pathway in liver have incorporated b5R in excess of cyt b5, (20,35) which may be non-physiologic. These experiments may have underestimated the capacity of b5R and cyt b5 alone to reduce arylhydroxylamines and amidoximes.
Although b5R is necessary for arylhydroxylamine reduction, b5R immunoreactive protein did not correlate with arylhydroxylamine carcinogen reduction activity in individual human liver microsomes. This may be due to the relatively narrow range of b5R content in the small number of livers evaluated. Studies are underway in a much larger group of individual liver human livers to determine whether outliers in b5R content will correlate with arylhydroxylamine reduction activities for various substrates. An alternative explanation for the lack of correlation with b5R content could be that electron transfer from cyt b5, but not from b5R, is rate limiting for hydroxylamine reduction. This is consistent with the finding that electron transfer from b5R to cyt b5 is extremely rapid. (39,40) Indeed, in our studies, cyt b5 content significantly correlated with arylhydroxylamine reduction, and appears to vary more widely in expression in human liver than does its reductase.(14) Cyt b5 may therefore be a significant source of toxicogenetic variability in the metabolism of arylhydroxylamine carcinogens in humans.
In addition to liver, we also found that cyt b5 and b5R expression, and arylhydroxylmine reduction activities, are readily detectable in both human breast and colon, which are suspected targets of the carcinogenic effects of 4-ABP and PhIP. (41,42) A number of studies have demonstrated DNA adducts, arising from heterocyclic and aromatic amine carcinogens, in human breast and colon, indicating that these tissues are directly exposed to these environmental carcinogens. (41,43-46) While previous studies have investigated the relationships between the formation of carcinogenic DNA adducts and several enzyme pathways, to include P450 1A1, (47,48) glutathione-S-transferase, (49-52) sulfotransferases, (53) and N-acetyltransferases, (54-59) the role of reductive detoxication of arylhydroxylamines has not been evaluated, either in the liver or locally in tissues affected by arylamine carcinogenesis.
Because hydroxylamine generation is required for DNA adduct formation by arylamine carcinogens, (5) the balance of arylhydroxylamine metabolites is likely to be very important in determining the outcome of mutagenic DNA adduct formation in tissues susceptible to these compounds. In this report we demonstrate that NADH cytochrome b5 reductase and cytochrome b5 are sufficient for the reduction of arylhydroxylamine carcinogens in human liver. We further hypothesize that b5R and cyt b5 comprise the primary pathway catalyzing the reductive detoxication of carcinogenic arylhydroxylamines in target tissues of arylamine carcinogens, such as human breast and colon. Further work is underway to fully characterize the role of b5R and cyt b5 in carcinogenic arylhydroxylamine detoxication in these tumor target tissues.
Supplementary Material
Acknowledgements
The authors would like to acknowledge Dr. Komei Shirabe, Medical College of Oita, Japan, for kindly providing human b5R cDNA, and Dr. Grank Mauk, University of British Columbia, for the gift of cyt b5 cDNA used in initial experiments; Ms. Jacquie Leighton at Panigen Inc. for generating rabbit antisera, and Dr. Scott Jewell at the Cooperative Human Tissue Network, Midwest Division, Ohio State University, for coordinating the provision of human breast and colon tissue. This project was performed, in part, using compounds provided by the National Cancer Institute's Chemical Carcinogen Reference Standards Repository, operated under contract by Midwest Research Institute, NO. NO2-CB-07008. This work was supported by the National Institutes of Health, grant GM61753. JRK was supported by training grant T32 ES07015 from NIEHS.
Footnotes
Abbreviations used: b5R: NADH cytochrome b5 reductase; cyt b5: cytochrome b5; 4-ABP: 4-aminobiphenyl; PhIP: 2-amino-1-methyl-6-phenylimidazo[4,5-b] pyridine; NHOH-4-ABP: N-hydroxy-4-aminobiphenyl; NHOH-PhIP: N-hydroxy-2-amino-1-methyl-6-phenylimidazo[4,5-b] pyridine; HLM: human liver microsomes; PBS: phosphate buffered saline (8.0 g/L NaCl, 0.2 g/L KCl, 0.2 g/L KH2PO4 (anhyd), 1.15 g/L Na2HPO4 (anhyd), pH 7.4); HSA: human serum albumin; TEA: triethylamine; PVDF: polyvinylidenedifluoride; PHMB: p-hydroxy-mercuribenzoate; Mr: relative molecular mass (molecular weight).
Supporting Information Available: Supplemental figures depicting full length immunoblots of human liver microsomes probed with antibodies to both b5R and cyt b5 (showing specificity of each antibody for a single protein); and demonstrating dose dependent inhibition by these antibodies of cytochrome c reduction in human liver microsomes (a specific marker of the b5R/cyt b5 pathway in microsomes) is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Nakagama H, Nakanishi M, Ochiai M. Modeling human colon cancer in rodents using a food-borne carcinogen, PhIP. Cancer Sci. 2005;96:627–636. doi: 10.1111/j.1349-7006.2005.00107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shan L, Yu M, Snyderwine EG. Gene expression profiling of chemically induced rat mammary gland cancer. Carcinogenesis. 2005;26:503–509. doi: 10.1093/carcin/bgh330. [DOI] [PubMed] [Google Scholar]
- 3.Ito N, Hasegawa R, Sano M, Tamano S, Esumi H, Takayama S, Sugimura T. A new colon and mammary carcinogen in cooked food, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP). Carcinogenesis. 1991;12:1503–1506. doi: 10.1093/carcin/12.8.1503. [DOI] [PubMed] [Google Scholar]
- 4.Hecht SS. Tobacco smoke carcinogens and breast cancer. Environ. Mol. Mutagen. 2002;39:119–126. doi: 10.1002/em.10071. [DOI] [PubMed] [Google Scholar]
- 5.Fan L, Schut HA, Snyderwine EG. Cytotoxicity, DNA adduct formation and DNA repair induced by 2-hydroxyamino-3-methylimidazo[4,5-f]quinoline and 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine in cultured human mammary epithelial cells. Carcinogenesis. 1995;16:775–779. doi: 10.1093/carcin/16.4.775. [DOI] [PubMed] [Google Scholar]
- 6.Culp SJ, Roberts DW, Talaska G, Lang NP, Fu PP, Lay JO, Jr., Teitel CH, Snawder JE, Von Tungeln LS, Kadlubar FF. Immunochemical, 32P-postlabeling, and GC/MS detection of 4-aminobiphenyl-DNA adducts in human peripheral lung in relation to metabolic activation pathways involving pulmonary N-oxidation, conjugation, and peroxidation. Mutat. Res. 1997;378:97–112. doi: 10.1016/s0027-5107(97)00101-2. [DOI] [PubMed] [Google Scholar]
- 7.Crofts FG, Sutter TR, Strickland PT. Metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by human cytochrome P4501A1, P4501A2 and P4501B1. Carcinogenesis. 1998;19:1969–1973. doi: 10.1093/carcin/19.11.1969. [DOI] [PubMed] [Google Scholar]
- 8.Butler MA, Iwasaki M, Guengerich FP, Kadlubar FF. Human cytochrome P-450PA (P-450IA2), the phenacetin O-deethylase, is primarily responsible for the hepatic 3-demethylation of caffeine and N-oxidation of carcinogenic arylamines. Proc. Natl. Acad. Sci. U S A. 1989;86:7696–7700. doi: 10.1073/pnas.86.20.7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorlewska-Roberts KM, Teitel CH, Lay JO, Jr., Roberts DW, Kadlubar FF. Lactoperoxidase-catalyzed activation of carcinogenic aromatic and heterocyclic amines. Chem. Res. Toxicol. 2004;17:1659–1666. doi: 10.1021/tx049787n. [DOI] [PubMed] [Google Scholar]
- 10.Williams JA, Stone EM, Fakis G, Johnson N, Cordell JA, Meinl W, Glatt H, Sim E, Phillips DH. N-Acetyltransferases, sulfotransferases and heterocyclic amine activation in the breast. Pharmacogenetics. 2001;11:373–388. doi: 10.1097/00008571-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 11.Sadrieh N, Davis CD, Snyderwine EG. N-acetyltransferase expression and metabolic activation of the food-derived heterocyclic amines in the human mammary gland. Cancer Res. 1996;56:2683–2687. [PubMed] [Google Scholar]
- 12.King R, Teitel C, Shaddock J, Casciano D, Kadlubar F. Detoxification of carcinogenic aromatic and heterocyclic amines by enzymatic reduction of the N-hydroxy derivative. Cancer Lett. 1999;143:167–171. doi: 10.1016/s0304-3835(99)00119-6. [DOI] [PubMed] [Google Scholar]
- 13.Cribb AE, Spielberg SP, Griffin GP. N4-hydroxylation of sulfamethoxazole by cytochrome P450 of the cytochrome P4502C subfamily and reduction of sulfamethoxazole hydroxylamine in human and rat hepatic microsomes. Drug Metab. Disp. 1995;23:406–414. [PubMed] [Google Scholar]
- 14.Kurian J, Bajad S, Miller J, Chin N, Trepanier L. NADH cytochrome b5 reductase and cytochrome b5 catalyze the microsomal reduction of xenobiotic hydroxylamines and amidoximes in humans. J. Pharmacol. Exp. Ther. 2004;311 doi: 10.1124/jpet.104.072389. [DOI] [PubMed] [Google Scholar]
- 15.Trepanier L, Bajad S, Kurian J. Evaluation of the cytochrome b5 / cytochrome b5 reductase pathway. In: Hodgson E, editor. Current Protocols in Toxicology. John Wiley & Sons; Hoboken NJ: 2005. [DOI] [PubMed] [Google Scholar]
- 16.Huang C. Methods in Enzymology. Academic Press Inc.; New York: 1982. Determination of binding stoichiometry by the continuous variation method: the Job plot. pp. 509–525. [DOI] [PubMed] [Google Scholar]
- 17.Trepanier L, Miller J. NADH-dependent reduction of sulphamethoxazole hydroxylamine in dog and human liver microsomes. Xenobiotica. 2000;30:1111–1121. doi: 10.1080/00498250010013908. [DOI] [PubMed] [Google Scholar]
- 18.Fitzsimmons S, Workman P, Grever M, Paull K, Camalier R, Lewis A. reductase enzyme expression across the NCI tumor cell line panel: correlation with sensitivity to mitomycin C and EO9. J. Natl. Cancer Inst. 1996;88:259–269. doi: 10.1093/jnci/88.5.259. [DOI] [PubMed] [Google Scholar]
- 19.Barham HM, Inglis R, Chinje EC, Stratford IJ. Development and validation of a spectrophotometric assay for measuring the activity of NADH: cytochrome b5 reductase in human tumour cells. Br. J. Cancer. 1996;74:1188–1193. doi: 10.1038/bjc.1996.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clement B, Lomb R, Moller W. Isolation and characterization of the protein components of the liver microsomal O2-insensitive NADH-benzamidoxime reductase. J. Biol. Chem. 1997;272:19615–19620. doi: 10.1074/jbc.272.31.19615. [DOI] [PubMed] [Google Scholar]
- 21.Tomatsu S, Kobayashi Y, Fukumaki Y, Yubisui T, Orii T, Sakaki Y. The organization and the complete nucleotide sequence of the human NADH-cytochrome b5 reductase gene. Gene. 1989;80:353–361. doi: 10.1016/0378-1119(89)90299-0. [DOI] [PubMed] [Google Scholar]
- 22.Leroux A, Vieira L, Kahn A. Transcriptional and translational mechanisms of cytochrome b5 reductase isozyme generation in humans. Biochem. J. 2001;355:529–535. doi: 10.1042/0264-6021:3550529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozols J, Carr S, Strittmatter P. Identification of the NH2-terminal blocking group of NADH cytochrome b5 reductase as myristic acid and the complete amino acid sequence of the membrane binding domain. J. Biol. Chem. 1984;259:13349–13354. [PubMed] [Google Scholar]
- 24.Giordano S, Steggles A. The human liver and reticulocyte cytochrome b5 mRNA's are products from a single gene. Biochim. Biophys. Res. Commun. 1991;178:38–44. doi: 10.1016/0006-291x(91)91776-9. [DOI] [PubMed] [Google Scholar]
- 25.Borgese N, D'Arrigo A, DeSilvestris M, Pietrini G. NADH cytochrome b5 reductase and cytochrome b5 isoforms as a model for the study of post-translational targeting to the endoplasmic reticulum. FEBS. Letters. 1993;325:70–75. doi: 10.1016/0014-5793(93)81416-w. [DOI] [PubMed] [Google Scholar]
- 26.Oshino N, Imai Y, Sato R. A function of cytochrome b5 in fatty acid desaturation by rat liver microsomes. J. Biochem. (Tokyo) 1971;69:155–167. doi: 10.1093/oxfordjournals.jbchem.a129444. [DOI] [PubMed] [Google Scholar]
- 27.Hildebrandt A, Estabrook R. Evidence for the participation of cytochrome b5 in hepatic microsomal mixed-function oxidation reactions. Arch. Biochem. Biophys. 1971;143:66–79. doi: 10.1016/0003-9861(71)90186-x. [DOI] [PubMed] [Google Scholar]
- 28.Hultquist D, Passon P. Catalysis of methaemoglobin reduction by erythrocyte cytochrome b5 and cytochrome b5 reductase. Nature New Biol. 1971;229:252–254. doi: 10.1038/newbio229252a0. [DOI] [PubMed] [Google Scholar]
- 29.Mahmutoglu I, Kappus H. Redox cycling of bleomycin-Fe(III) and DNA degradation by isolated NADH cytochrome b5 reductase: involvement of cytochrome b5. Mol. Pharm. 1988;34:578–583. [PubMed] [Google Scholar]
- 30.Hodnick W, Sartorelli A. Reductive activation of mitomycin C by NADH:cytochrome b5 reductase. Cancer Res. 1993;53:4907–4912. [PubMed] [Google Scholar]
- 31.Saulter JY, Kurian JR, Trepanier LA, Tidwell RR, Bridges AS, Boykin DW, Stephens CE, Anbazhagan M, Hall JE. Unusual dehydroxylation of antimicrobial amidoxime prodrugs by cytochrome b5 and NADH cytochrome b5 reductase. Drug Metab. Dispos. 2005;33:1886–1893. doi: 10.1124/dmd.105.005017. [DOI] [PubMed] [Google Scholar]
- 32.Kadlubar FF, McKee EM, Ziegler DM. Reduced pyridine nucleotide-dependent N-hydroxy amine oxidase and reductase activities of hepatic microsomes. Arch. Biochem. Biophys. 1973;156:46–57. doi: 10.1016/0003-9861(73)90339-1. [DOI] [PubMed] [Google Scholar]
- 33.Kadlubar FF, Ziegler DM. Properties of a NADH-dependent N-hydroxy amine reductase isolated from pig liver microsomes. Arch. Biochem. Biophys. 1974;162:83–92. doi: 10.1016/0003-9861(74)90107-6. [DOI] [PubMed] [Google Scholar]
- 34.Clement B, Behrens D, Amschler J, Matschke K, Wolf S, Havemeyer A. Reduction of sulfamethoxazole and dapsone hydroxylamines by a microsomal enzyme system purified from pig liver and pig and human liver microsomes. Life Sci. 2005;77:205–219. doi: 10.1016/j.lfs.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 35.Clement B, Demesmaeker M. Microsomal catalyzed N-hydroxylation of guanfacine and reduction of N-hydroxyguanfacine. Arch. Pharm. Pharm. Med. Chem. 1997;330:303–306. doi: 10.1002/ardp.19973300907. [DOI] [PubMed] [Google Scholar]
- 36.Johanssen I, Thelin A, Hofmann Y, Andersson S, Nordling A, Li Q, Carlsson S, Andersson T, Ingelman-Sundberg M. Identification of stearoyl CoA desaturase as the enzyme responsible for the reduction of ximelagatran/N-hydroxyximelagatran and benzamidoxime in adipocytes. Drug Metab. Rev. 2005;37:48. (abstr) [Google Scholar]
- 37.Spatz L, Strittmatter P. A form of cytochrome b5 that contains an additional hydrophobic sequence of 40 amino acid residues. Proc. Natl. Acad. Sci U.S.A. 1971;68:1042–6. doi: 10.1073/pnas.68.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang M-X, Cederbaum A. Interaction of ferric complexes with NADH-cytochrome b5 reductase and cytochrome b5: lipid peroxidation, H20 2 generation, and ferric reduction. Arch. Biochem. Biophys. 1996;331:69–78. doi: 10.1006/abbi.1996.0284. [DOI] [PubMed] [Google Scholar]
- 39.Meyer TE, Shirabe K, Yubisui T, Takeshita M, Bes MT, Cusanovich MA, Tollin G. Transient kinetics of intracomplex electron transfer in the human cytochrome b5 reductase-cytochrome b5 system: NAD+ modulates protein-protein binding and electron transfer. Arch. Biochem. Biophys. 1995;318:457–64. doi: 10.1006/abbi.1995.1254. [DOI] [PubMed] [Google Scholar]
- 40.Tonegawa Y, Umeda N, Hayakawa T, Ishibashi T. Evaluation of data in terms of two-dimensional random walk model: interaction between NADH-cytochrome b5 reductase and cytochrome b5. Biomed. Res. 2005;26:207–12. doi: 10.2220/biomedres.26.207. [DOI] [PubMed] [Google Scholar]
- 41.Li D, Wang M, Dhingra K, Hittelman WN. Aromatic DNA adducts in adjacent tissues of breast cancer patients: clues to breast cancer etiology. Cancer Res. 1996;56:287–293. [PubMed] [Google Scholar]
- 42.Dingley KH, Curtis KD, Nowell S, Felton JS, Lang NP, Turteltaub KW. DNA and protein adduct formation in the colon and blood of humans after exposure to a dietary-relevant dose of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine. Cancer Epidemiol. Biomarkers Prev. 1999;8:507–512. [PubMed] [Google Scholar]
- 43.Zhu J, Chang P, Bondy ML, Sahin AA, Singletary SE, Takahashi S, Shirai T, Li D. Detection of 2-amino-1-methyl-6-phenylimidazo[4,5-b]-pyridine-DNA adducts in normal breast tissues and risk of breast cancer. Cancer Epidemiol. Biomarkers Prev. 2003;12:830–837. [PubMed] [Google Scholar]
- 44.Lightfoot TJ, Coxhead JM, Cupid BC, Nicholson S, Garner RC. Analysis of DNA adducts by accelerator mass spectrometry in human breast tissue after administration of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and benzo[a]pyrene. Mutat. Res. 2000;472:119–127. doi: 10.1016/s1383-5718(00)00134-0. [DOI] [PubMed] [Google Scholar]
- 45.Gorlewska-Roberts K, Green B, Fares M, Ambrosone CB, Kadlubar FF. Carcinogen-DNA adducts in human breast epithelial cells. Environ. Mol. Mutagen. 2002;39:184–192. doi: 10.1002/em.10060. [DOI] [PubMed] [Google Scholar]
- 46.DeBruin LS, Martos PA, Josephy PD. Detection of PhIP (2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine) in the milk of healthy women. Chem. Res. Toxicol. 2001;14:1523–1528. doi: 10.1021/tx015556u. [DOI] [PubMed] [Google Scholar]
- 47.Ambrosone CB, Freudenheim JL, Graham S, Marshall JR, Vena JE, Brasure JR, Laughlin R, Nemoto T, Michalek AM, Harrington A, et al. Cytochrome P4501A1 and glutathione S-transferase (M1) genetic polymorphisms and postmenopausal breast cancer risk. Cancer Res. 1995;55:3483–3485. [PubMed] [Google Scholar]
- 48.Ishibe N, Hankinson SE, Colditz GA, Spiegelman D, Willett WC, Speizer FE, Kelsey KT, Hunter DJ. Cigarette smoking, cytochrome P450 1A1 polymorphisms, and breast cancer risk in the Nurses' Health Study. Cancer Res. 1998;58:667–671. [PubMed] [Google Scholar]
- 49.Vogl FD, Taioli E, Maugard C, Zheng W, Pinto LF, Ambrosone C, Parl FF, Nedelcheva-Kristensen V, Rebbeck TR, Brennan P, Boffetta P. Glutathione S-transferases M1, T1, and P1 and breast cancer: a pooled analysis. Cancer Epidemiol. Biomarkers Prev. 2004;13:1473–1479. [PubMed] [Google Scholar]
- 50.Rundle A, Tang D, Zhou J, Cho S, Perera F. The association between glutathione S-transferase M1 genotype and polycyclic aromatic hydrocarbon-DNA adducts in breast tissue. Cancer Epidemiol. Biomarkers Prev. 2000;9:1079–1085. [PubMed] [Google Scholar]
- 51.Firozi PF, Bondy ML, Sahin AA, Chang P, Lukmanji F, Singletary ES, Hassan MM, Li D. Aromatic DNA adducts and polymorphisms of CYP1A1, NAT2, and GSTM1 in breast cancer. Carcinogenesis. 2002;23:301–306. doi: 10.1093/carcin/23.2.301. [DOI] [PubMed] [Google Scholar]
- 52.Egan KM, Cai Q, Shu XO, Jin F, Zhu TL, Dai Q, Gao YT, Zheng W. Genetic polymorphisms in GSTM1, GSTP1, and GSTT1 and the risk for breast cancer: results from the Shanghai Breast Cancer Study and meta-analysis. Cancer Epidemiol. Biomarkers Prev. 2004;13:197–204. doi: 10.1158/1055-9965.epi-03-0294. [DOI] [PubMed] [Google Scholar]
- 53.Tang D, Rundle A, Mooney L, Cho S, Schnabel F, Estabrook A, Kelly A, Levine R, Hibshoosh H, Perera F. Sulfotransferase 1A1 (SULT1A1) polymorphism, PAH-DNA adduct levels in breast tissue and breast cancer risk in a case-control study. Breast Cancer Res. Treat. 2003;78:217–222. doi: 10.1023/a:1022968303118. [DOI] [PubMed] [Google Scholar]
- 54.Lash TL, Bradbury BD, Wilk JB, Aschengrau A. A case-only analysis of the interaction between N-acetyltransferase 2 haplotypes and tobacco smoke in breast cancer etiology. Breast Cancer Res. 2005;7:R385–R393. doi: 10.1186/bcr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hunter DJ, Hankinson SE, Hough H, Gertig DM, Garcia-Closas M, Spiegelman D, Manson JE, Colditz GA, Willett WC, Speizer FE, Kelsey K. A prospective study of NAT2 acetylation genotype, cigarette smoking, and risk of breast cancer. Carcinogenesis. 1997;18:2127–32. doi: 10.1093/carcin/18.11.2127. [DOI] [PubMed] [Google Scholar]
- 56.Millikan RC, Pittman GS, Newman B, Tse CK, Selmin O, Rockhill B, Savitz D, Moorman PG, Bell DA. Cigarette smoking, N-acetyltransferases 1 and 2, and breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 1998;7:371–378. [PubMed] [Google Scholar]
- 57.Zheng W, Deitz AC, Campbell DR, Wen WQ, Cerhan JR, Sellers TA, Folsom AR, Hein DW. N-acetyltransferase 1 genetic polymorphism, cigarette smoking, well-done meat intake, and breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 1999;8:233–239. [PubMed] [Google Scholar]
- 58.Deitz AC, Zheng W, Leff MA, Gross M, Wen WQ, Doll MA, Xiao GH, Folsom AR, Hein DW. N-Acetyltransferase-2 genetic polymorphism, well-done meat intake, and breast cancer risk among postmenopausal women. Cancer Epidemiol. Biomarkers Prev. 2000;9:905–910. [PubMed] [Google Scholar]
- 59.Delfino RJ, Sinha R, Smith C, West J, White E, Lin HJ, Liao SY, Gim JS, Ma HL, Butler J, Anton-Culver H. Breast cancer, heterocyclic aromatic amines from meat and N-acetyltransferase 2 genotype. Carcinogenesis. 2000;21:607–615. doi: 10.1093/carcin/21.4.607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


