Abstract
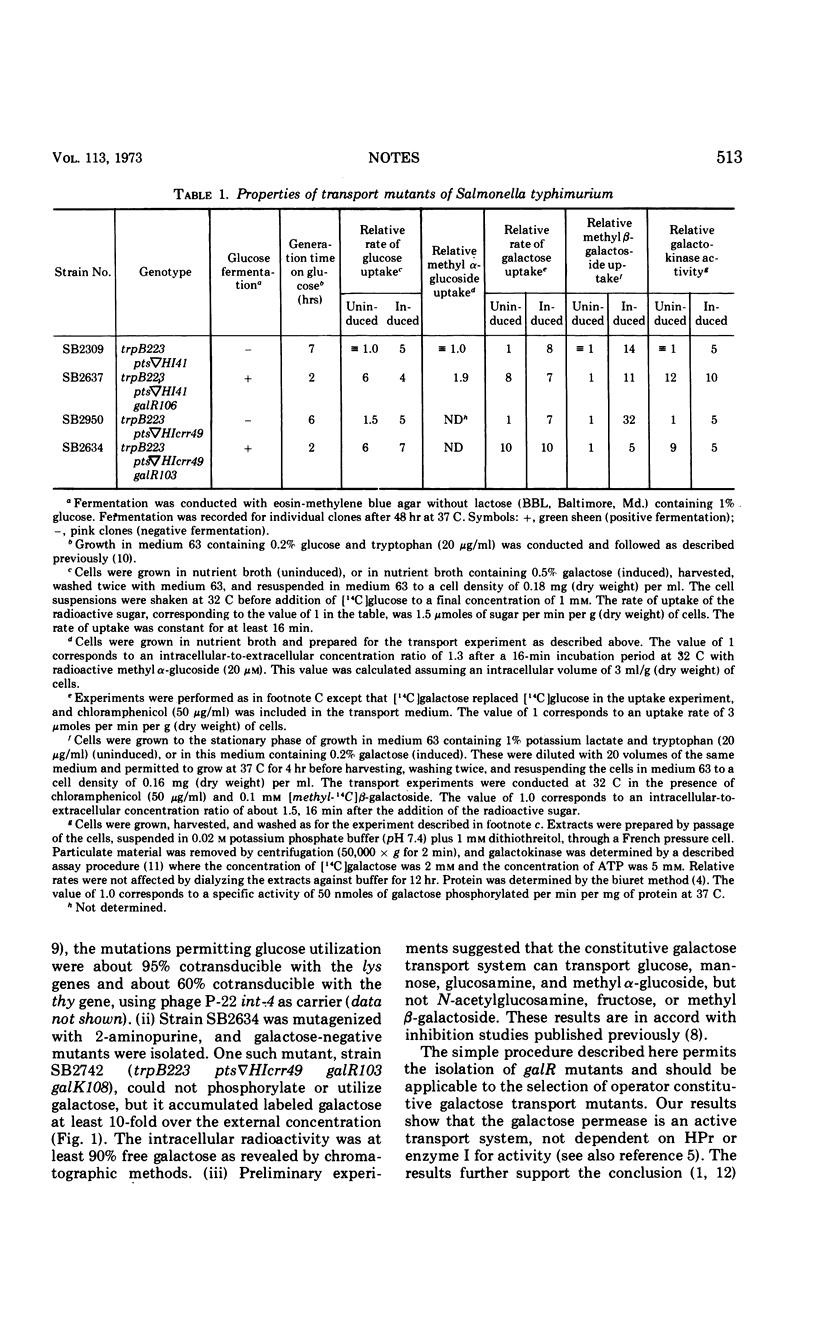
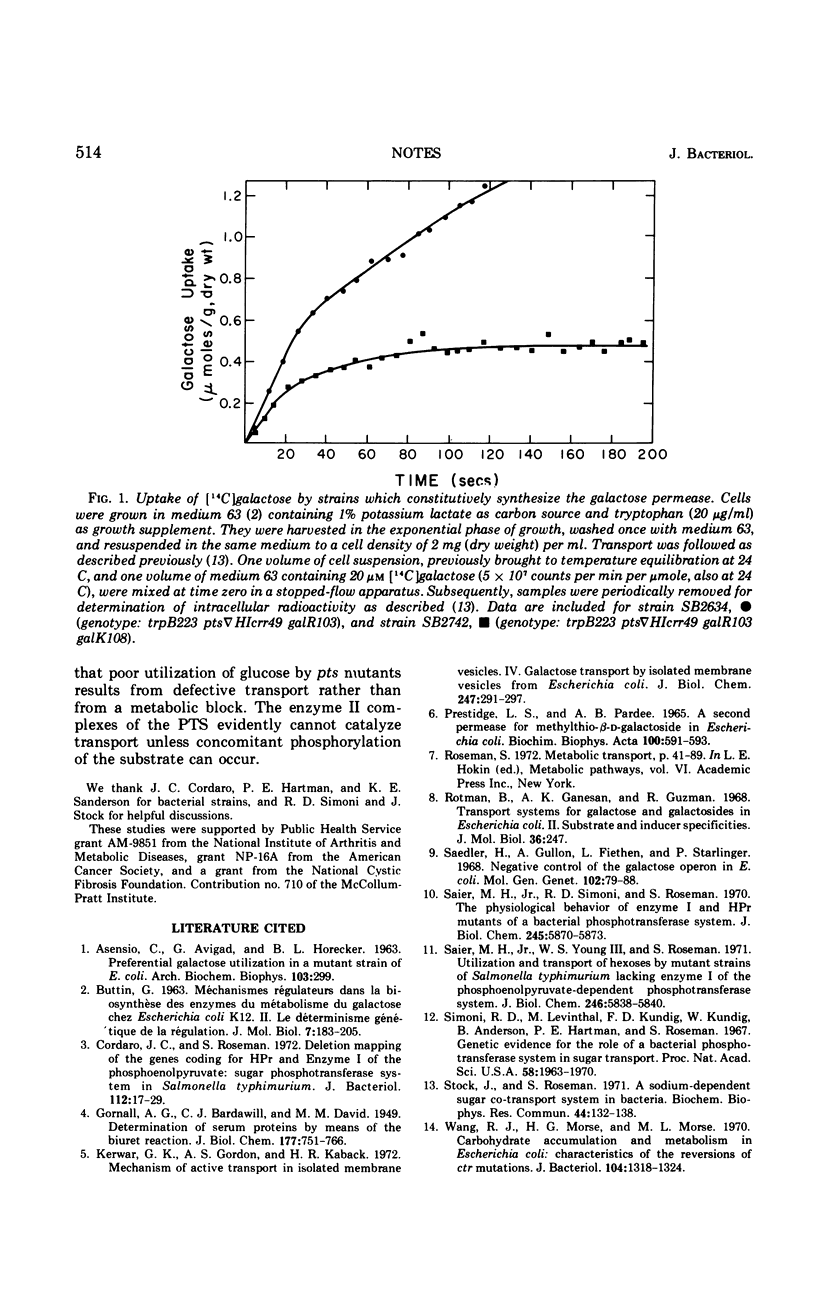
Salmonella typhimurium strains, lacking both enzyme I and the phosphocarrier protein, HPr, of the phosphoenolpyruvate-sugar phosphotransferase system, cannot transport or metabolize glucose and other sugar substrates of this enzyme system. Mutants which regain the ability to specifically utilize glucose were found to constitutively synthesize a galactose permease by virtue of a mutation in the galR gene. This permease, shown to be an active transport system, does not require HPr or enzyme I for activity.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ASENSIO C., AVIGAD G., HORECKER B. L. PREFERENTIAL GALACTOSE UTILIZATION IN A MUTANT STRAIN OF E. COLI. Arch Biochem Biophys. 1963 Dec;103:299–309. doi: 10.1016/0003-9861(63)90419-3. [DOI] [PubMed] [Google Scholar]
- BUTTIN G. M'ECANISMES R'EGULATEURS DANS LA BIOSYNTH'ESE DES ENZYMES DU M'ETABOLISME DU GALACTOSE CHEZ ESCHERICHIA COLI K12. II. LE D'ETERMINISME G'EN'ETIQUE DE LA R'EGULATION. J Mol Biol. 1963 Aug;7:183–205. doi: 10.1016/s0022-2836(63)80045-5. [DOI] [PubMed] [Google Scholar]
- Cordaro J. C., Roseman S. Deletion mapping of the genes coding for HPr and enzyme I of the phosphoenolpyruvate: sugar phosphotransferase system in Salmonella typhimurium. J Bacteriol. 1972 Oct;112(1):17–29. doi: 10.1128/jb.112.1.17-29.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerwar G. K., Gordon A. S., Kaback H. R. Mechanisms of active transport in isolated membrane vesicles. IV. Galactose transport by isolated membrane vesicles from Escherichia coli. J Biol Chem. 1972 Jan 10;247(1):291–297. [PubMed] [Google Scholar]
- PRESTIDGE L. S., PARDEE A. B. A SECOND PERMEASE FOR METHYL-THIO-BETA-D-GALACTOSIDE IN ESCHERICHIA COLI. Biochim Biophys Acta. 1965 May 4;100:591–593. doi: 10.1016/0304-4165(65)90029-2. [DOI] [PubMed] [Google Scholar]
- Rotman B., Ganesan A. K., Guzman R. Transport systems for galactose and galactosides in Escherichia coli. II. Substrate and inducer specificities. J Mol Biol. 1968 Sep 14;36(2):247–260. doi: 10.1016/0022-2836(68)90379-3. [DOI] [PubMed] [Google Scholar]
- Saedler H., Gullon A., Fiethen L., Starlinger P. Negative control of the galactose operon in E. coli. Mol Gen Genet. 1968;102(1):79–88. doi: 10.1007/BF00341872. [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr, Simoni R. D., Roseman S. The physiological behavior of enzyme I and heat-stable protein mutants of a bacterial phosphotransferase system. J Biol Chem. 1970 Nov 10;245(21):5870–5873. [PubMed] [Google Scholar]
- Saier M. H., Jr, Young W. S., 3rd, Roseman S. Utilization and transport of hexoses by mutant strains of Salmonella typhimurium lacking enzyme I of the phosphoenolpyruvate-dependent phosphotransferase system. J Biol Chem. 1971 Sep 25;246(18):5838–5840. [PubMed] [Google Scholar]
- Simoni R. D., Levinthal M., Kundig F. D., Kundig W., Anderson B., Hartman P. E., Roseman S. Genetic evidence for the role of a bacterial phosphotransferase system in sugar transport. Proc Natl Acad Sci U S A. 1967 Nov;58(5):1963–1970. doi: 10.1073/pnas.58.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J., Roseman S. A sodium-dependent sugar co-transport system in bacteria. Biochem Biophys Res Commun. 1971 Jul 2;44(1):132–138. doi: 10.1016/s0006-291x(71)80168-7. [DOI] [PubMed] [Google Scholar]
- Wang R. J., Morse H. G., Morse M. L. Carbohydrate Accumulation and Metabolism in Escherichia coli: Characteristics of the Reversions of ctr Mutations. J Bacteriol. 1970 Dec;104(3):1318–1324. doi: 10.1128/jb.104.3.1318-1324.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]


