Abstract
Background
We identified two 3p21.3 regions (LUCA and AP20) as most frequently affected in lung, breast and other carcinomas and reported their fine physical and gene maps. It is becoming increasingly clear that each of these two regions contains several TSGs. Until now TSGs which were isolated from AP20 and LUCA regions (e.g.G21/NPRL2, RASSF1A, RASSF1C, SEMA3B, SEMA3F, RBSP3) were shown to inhibit tumour cell growth both in vitro and in vivo.
Methodology/Principal Findings
The effect of expression HYAL1 and HYAL2 was studied by colony formation inhibition, growth curve and cell proliferation tests in vitro and tumour growth assay in vivo. Very modest growth inhibition was detected in vitro in U2020 lung and KRC/Y renal carcinoma cell lines. In the in vivo experiment stably transfected KRC/Y cells expressing HYAL1 or HYAL2 were inoculated into SCID mice (10 and 12 mice respectively). Tumours grew in eight mice inoculated with HYAL1. Ectopic HYAL1 was deleted in all of them. HYAL2 was inoculated into 12 mice and only four tumours were obtained. In 3 of them the gene was deleted. In one tumour it was present but not expressed. As expected for tumour suppressor genes HYAL1 and HYAL2 were down-expressed in 15 fresh lung squamous cell carcinomas (100%) and clear cell RCC tumours (60–67%).
Conclusions/Significance
The results suggest that the expression of either gene has led to inhibition of tumour growth in vivo without noticeable effect on growth in vitro. HYAL1 and HYAL2 thus differ in this aspect from other tumour suppressors like P53 or RASSF1A that inhibit growth both in vitro and in vivo. Targeting the microenvironment of cancer cells is one of the most promising venues of cancer therapeutics. As major hyaluronidases in human cells, HYAL1 and HYAL2 may control intercellular interactions and microenvironment of tumour cells providing excellent targets for cancer treatment.
Introduction
We have performed a deletion survey of 3p on more than 400 lung, renal, breast, cervical and ovarian carcinomas using a defined set of markers, combining conventional LOH (loss of heterozygosity) with quantitative real-time PCR (qPCR), comparative genomic and NotI microarrays hybridisations [1]–[5]. We identified two most frequently affected 3p21.3 regions, LUCA at the centromeric and AP20 at the telomeric border of 3p21.3. Aberrations of either region were detected in more than 90% of the studied tumours. Homozygous deletions (HD) were detected in 10%–18% of all tumours at both the LUCA and AP20 sites [4], [5]. The frequent chromosome losses in these regions suggest that they harbor multiple tumour suppressor genes (TSG) [6]–[8]. It was suggested that aberrations in both the LUCA and AP20 region could be functionally linked [5], [9], [10].
The definition of a TSG is based on the demonstration of its regular inactivation by mutation or epigenetic silencing in tumour samples. It is also important to obtain supportive evidence from functional studies. We have previously found (in collaboration with Stefan Imreh et al.) non-random losses of human 3p21-p22 fragments from mouse-human microcell hybrids following progressive growth in SCID mice [11]. In order to test whether a known suppressor gene, RB, would behave in a similar way, wild type and mutated RB genes were introduced into the pETE (Elimination Test Episomal) vector that permitted the expression of the gene in the absence but not in the presence of tetracycline. The expression of the gene could be modulated by tetracycline both in vivo and in vitro. When the transfectants were passaged as tumours in immunodeficient SCID mice, the wild type RB gene was deleted or functionally inactivated already after the first passage in all 20 tumours tested. In contrast, a non-functional mutant RB gene was maintained in all 10 tumours studied. In similar experiments with wt P53, the exogenous P53 gene was maintained and expressed in all 6 tumours tested, but in a mutated form. On the basis of these experiments we have developed the gene inactivation test (GIT) for a functional definition of TSG. It is based on the comparison of cell growth in vitro and tumour growth in vivo when the gene is/is not expressed. The main idea of the test is that a gene inhibiting growth of tumour cells should be inactivated in growing tumours by genetic or epigenetic mechanisms.
Using GIT and growth analysis under cultural conditions we have shown that FUS1, SEMA3B, G21/NPRL2, RASSF1A, RASSF1C, RBSP3 (genes from AP20 and LUCA regions) and other TSGs inhibit tumour cell growth both in vitro and in vivo [7], [9], [11]–[15].
However other genes from 3p21.3 (e.g. TCEA1, MLH1, RHOA, 3PK, PL6, 101F6, BLU, TGFBR2) did not show any effect in the tested cell lines [11].
Here we describe the functional analysis of two additional genes from the 3p21.3 LUCA region, hyaluronidases -1 and -2 (HYAL1 and HYAL2).
The HYAL1 gene (hyaluronoglucosaminidase 1) is located in the LUCA region. It contains 6 exons producing a 2.6 kb mRNA (coding for 436 aa protein). It is well expressed in all analysed normal human tissues including lung and kidney. It was not expressed in 18 out of 20 lung cancer cell lines [6].
The HYAL2 (hyaluronoglucosaminidase 2, located in LUCA region) contains 4 exons producing a 2 kb mRNA (that encode 473 aa). It is well expressed in all analysed human tissues including lung, kidney and many lung cancer cell lines. The protein is attached to the membrane by the glycosylphosphatidyl-inositol-anchor (GPI-anchor) [16]. The HYAL2 protein was identified as a receptor for the sheep lung cancer retrovirus, JSRV, and a sequestration mechanism inactivating HYAL2 protein was demonstrated. The env gene of JSRV was shown to transform human bronchial epithelial cells in vitro and sequesters the HYAL2 protein. The absence of HYAL2 (mediated either by a putative virus or mutational inactivation) leads to ligand-independent activation of the RON receptor tyrosine kinase and its downstream AKT and MAPK signaling pathways [17].
These two genes contribute to intracellular and extracellular catabolism of hyaluronic acid (HA) in a CD44-dependent manner [18]. HA has a great number of biological functions: it mediates cell-cell and cell-matrix interactions and plays an important role in cell migration, tumour growth and progression.
Thus analysis of HYAL1 and HYAL2 can have a great importance not only for better understanding of HA catabolism and human carcinogenesis but could provide therapeutic targets for cancer treatment. In this study we have found that the expression of either gene suppressed tumour growth in vivo but not in vitro. These findings are consistent with earlier somatic hybrid studies by Henry Harris, George Klein and Francis Wiener, showing that somatic hybridisation of normal and malignant cells can suppress tumorigenicity in vivo but not cell growth in vitro [19], [20]. The mechanisms of the in vivo suppressive effects have not been clarified, but at least part of them may have acted at the level of tumour-host interactions. The finding of a similar phenomenon with the two genes involved in the present study is a step towards the analysis of this important phenomenon.
Results
Colony formation inhibition by HYAL1 and HYAL2 in vitro
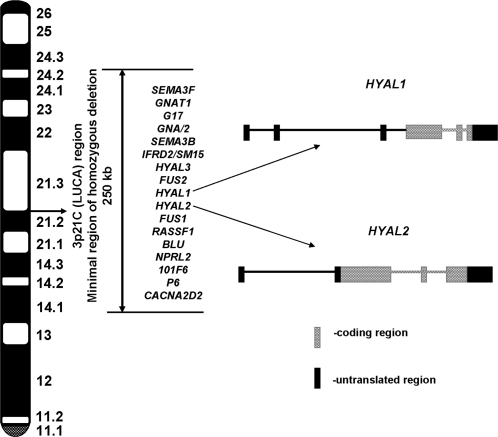
In this study we performed initial functional analysis of HYAL1 and HYAL2. These genes are located in the minimally deleted region of LUCA (Figure 1). The work was a continuation of our previous study to characterize different genes, located in the same region, e.g. RASSF1A [13], RASSF1C [14] and G21/NPRL2 [15]. These studies showed that these genes have strong growth inhibiting activity both in vitro and in vivo.
Figure 1. Schematic map of the LUCA region picturing the HYAL1 and HYAL2 genes.
Expression of HYAL1 and HYAL2 was almost undetectable in the KRC/Y renal and U2020 lung carcinoma lines (data not shown), which were used in our growth suppression experiments for testing RASSF1A and G21/NPRL2 [13]–[15]. As both HYAL1 and HYAL2 are well expressed in normal kidney and lung (see for example http://www.genecards.org/index.shtml) we have chosen KRC/Y and U2020 for our study.
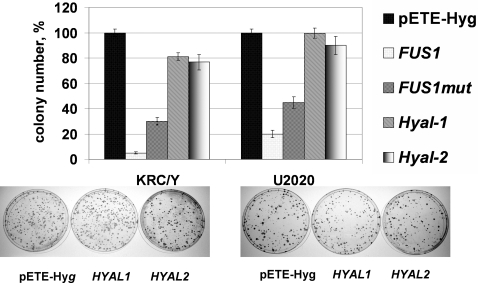
HYAL1 and HYAL2 were cloned into the episomal tetracycline-regulated pETE-Hyg vector [21] and used for transfection of the KRC/Y and U2020 cells (for colony formation experiments, Figure 2). The empty pETE vector was used as a negative control. HYAL1 and HYAL2 showed almost no inhibition of colony formation. When HYAL1 and HYAL2 were expressed the cloning efficiency was 77–100%, compared to the empty vector. As a positive control we used FUS1 gene, a strong TSG cloned in pETE [7]. Colony formation efficiency of the KRC/Y cells expressing FUS1 was less than 5% and for U2020 cells it was less than 25% compared to controls.
Figure 2. Effect of HYAL1 and HYAL2 transgenes on colony formation efficiency in KRC/Y and U2020 cells compared to empty pETE vector (negative control) and wild type or mutated FUS1 transgenes.
Graphical representation summarizing three independent experiments and photographic images of Petri dishes stained with methylene blue. Values are the mean ±s.d. of three separate experiments each calculated from triplicate plates.
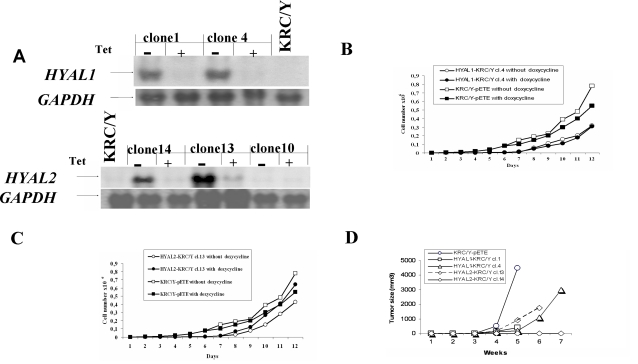
We performed also growth curve inhibition experiments using KRC/Y cells stably transformed with HYAL1 and HYAL2 transgenes. The pETE-HYAL1 and pETE-HYAL2 vectors carrying wild type alleles of the tested genes were transfected into KRC/Y expressing tTA and clonal cell lines were selected. Expression of the transgenes was tested by Northern hybridization. Two of the best tetracycline regulated clones for each gene were then used in further experiments (see for example Figure 3A). For HYAL1 clones 1 and 4 were selected (HYAL1-KRC/Ycl.1 and HYAL1-KRC/Ycl.4) and for HYAL2 clones 13 and 14 (HYAL2-KRC/Ycl.13 and HYAL2-KRC/Ycl.14). Growth curves for HYAL1-KRC/Ycl.4 and HYAL2-KRC/Ycl.13 are shown in Figure 3B and C. Only modest growth inhibition was seen like in the colony assays. Two other clones showed similar results.
Figure 3. Analysis of HYAL1 and HYAL2 stably transformed KRC/Y clones.
Northern analysis (A) of tetracycline regulated clones. (+), tetracycline (Tet) or doxycycline is present, gene is OFF. (−), tetracycline or doxycycline is absent, gene is ON. Growth inhibition of KRC/Y cells with HYAL1 (B) and HYAL2 (C) transgenes in vitro. Tumour growth inhibition of KRC/Y cells by HYAL1 and HYAL2 in vivo in SCID mice (D). Mice were drinking water with tetracycline (+Tet, gene is OFF) or without (−Tet, gene is ON) but for simplicity curves are shown only for mice when genes were ON (no tetracycline).
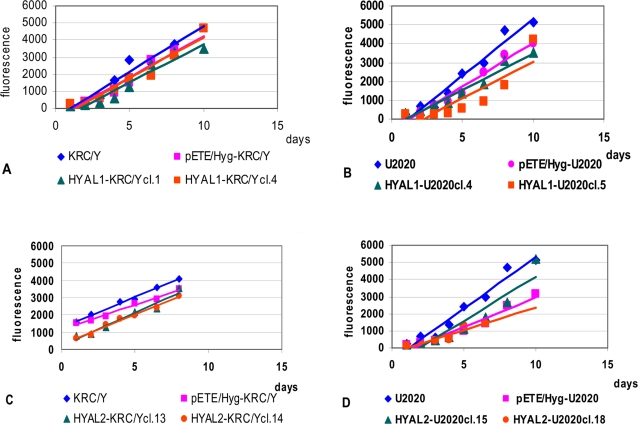
In the same way we selected U2020 cell clones expressing HYAL1 (HYAL1-U2020cl.4 and HYAL1-U2020cl.5) and HYAL2 (HYAL2-U2020cl.15 and HYAL2-U2020cl.18). All eight clones expressing either HYAL1 or HYAL2 in KRC/Y and U2020 cells were tested with CyQUANT NF Cell Proliferation Assay based on measurement of cellular DNA content via fluorescent dye binding (see Materials and Methods). Figure 4 shows that neither HYAL1 nor HYAL2 inhibited KRC/Y or U2020 cells to a significant extent.
Figure 4. CyQUANT Cell Proliferation Assay.
Effect of expression of HYAL1 transgene in KRC/Y (A) and in U2020 (B) cells. The same is shown for HYAL2 (KRC/Y in C and U2020 in D). Experiments were done in triplicates in the absence of doxycycline. The same experiments were done in the presence of doxycycline and showed similar results (data not shown). Plotted data points represent averages of triplicate samples, the plotted line is a linear regression fit of all data points. The assay is designed to produce a linear analytical response from at least 100–20,000 cells per well in most cell lines.
Gene inactivation test with HYAL1 and HYAL2 in SCID mice
We investigated HYAL1 and HYAL2 using the gene inactivation test (GIT) as described by Li et al., 1999 [22] and Protopopov et al., 2002 [21]. The test mimics the inactivation of TSGs during tumour growth in vivo. Clonal cell lines expressing transgenes were inoculated into SCID mice and the expression of the transgene was controlled by tetracycline administered ad libitum in the drinking water. In this setting TSGs suppressed tumour formation in SCID mice, unless they were eliminated or mutated. All grown tumours were analysed for the presence and expression of the transgene.
KRC/Y derived HYAL1 and HYAL2 cell clones were inoculated into 22 SCID mice (6 mice/HYAL1-KRC/Ycl.1; 4 mice/HYAL1-KRC/Ycl.4; 8 mice/HYAL2-KRC/Ycl.13 and 4 mice/HYAL2-KRC/Ycl.14). For each SCID mouse, 5×106 cells were inoculated (one inoculation per mouse). In control mice, KRC/Y cells transfected with empty pETE vector were used (twelve SCID mice were injected). Half of the SCID mice were then given drinking water containing 1mg/ml tetracycline. Results of these experiments are shown in Figure 3D (to make Figure more clear only curves for mice drinking water without tetracycline are shown). In many cases no tumour growth was observed at all (see Table 1). Strong inhibition of tumour growth was observed for all clones expressing HYAL1 or HYAL2. All grown 12 tumours (T1–T12) were explanted and tested for the presence of pETE-HYAL1 and pETE-HYAL2 constructs by PCR. Transgenes were detected only in one tumour (HYAL2) and in eleven tumours it was deleted. In tumour T10 where the HYAL2 gene was present by PCR, there was no detectable expression by Northern (data not shown).
Table 1. Growth of tumours in SCID mice.
| Genes | Clone | Tetracycline | Tumours | PCR | Northern |
| HYAL1 | clone1 | - | T1 | - | |
| + | T2 | - | |||
| - | No tumour | ||||
| + | No tumour | ||||
| - | T3 | - | |||
| + | T4 | - | |||
| clone4 | - | T5 | - | ||
| + | T6 | - | |||
| - | T7 | - | |||
| + | T8 | - | |||
| HYAL2 | clone13 | - | T9 | - | |
| + | T10 | + | NO EXPR | ||
| - | No tumour | ||||
| + | No tumour | ||||
| - | T11 | - | |||
| + | T12 | - | |||
| - | No tumour | ||||
| + | No tumour | ||||
| clone14 | - | No tumour | |||
| + | No tumour | ||||
| - | No tumour | ||||
| + | No tumour |
Expression of HYAL1 and HYAL2 genes is significantly decreased in lung and kidney cancer samples
As our data suggested that both HYAL1 and HYAL2 induce growth inhibition of tumours in SCID mice we tested expression of these genes in lung and renal tumours using qPCR (see Materials and Methods).
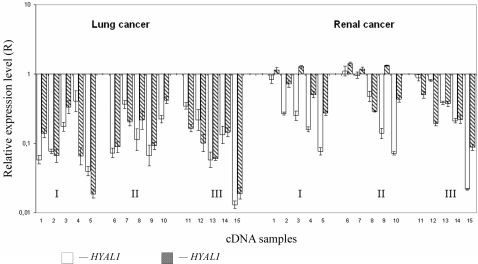
Fifteen SCC and fifteen RCC tumours were studied. We assumed that expression fell down if it was at least 2-fold lower than in matched control samples. Expression of both genes was drastically decreased in all lung samples (15/15, P<0.02, Figure 5). Average decrease level for HYAL1 was 7.8 (from 2.5 up to 77) and for HYAL2 8.8 (2.5–53) times . In seven cases expression of HYAL1 fell down more than 10-fold. For HYAL2 eight such cases were found. In six of fifteen lung biopsies we found more than 10-fold decrease of both HYAL1 and HYAL2 expression. In RCC samples expression of HYAL1 declined in 10 of 15 cases (67%, P<0.02) and average decrease was 6.5 times (from 2 to 46). For HYAL2 reduced expression was detected in 9 of 15 samples (60%, P<0.02) and average decrease was 4.9 times (2–11.4). Only in three RCC samples expression of HYAL1 decreased more than 10-fold and in one case we detected similar strong decline of HYAL2 expression. No statistically significant difference in cases with different tumour stages was observed both in SCC and RCC cases.
Figure 5. QPCR mRNA expression profile of HYAL1 and HYAL2 in SCC and RCC biopsies.
The Y axis indicates the values of relative expression level of target genes in log10 scale in tumour samples relative to control normal samples normalized to the reference gene GAPDH. The X axis shows the cDNA samples isolated from tumours at different stages (I–III). Open bars show HYAL1 and hatched bars HYAL2 expression.
Discussion
In this work we describe functional characterization of HYAL1 and HYAL2 genes. Using the GIT test we previously tested altogether 15 genes from 3p21.3 LUCA and AP20 regions. Nine genes (TCEA1, MLH1, VILL, RHOA, 3PK, PL6, 101F6, BLU, TGFBR2) did not show any effect in the tested cell lines. Six genes (RBSP3, NPRL2/G21, RASSF1A, RASSF1C, SEMA3B, SEMA3F) had strong growth inhibitory activity, both in vitro and in vivo.
For example, NPRL2/G21 demonstrated growth inhibition in vitro in small cell lung cancer cell line U2020 and renal carcinoma cell line KRC/Y cells almost 90%. RASSF1A gene showed the same results. In SCID mice both genes either did not permit tumour formation or in growing tumours they were inactivated (for NPRL2 it was 10 mice and for RASSF1A it was 8 mice).
In contrast, neither HYAL1 nor HYAL2 showed significant growth inhibition in colony formation and growth curve assays in vitro (see Figures 2–3). CyQUANT NF Cell Proliferation Assay (Figure 4) also demonstrated that neither HYAL1 nor HYAL2 inhibited KRC/Y or U2020 cells proliferation. Importantly, pETE vectors expressed mRNA of inserted genes in quantity comparable to physiologically normal levels [14]. However in SCID mice both genes had a very strong inhibiting effect: no tumours grew in 10 mice and in 12 mice where tumours grew the genes were inactivated (deletion or loss of expression, see for example Figure 3D and Table 1). In our SCID mice experiments HYAL1 and HYAL2 genes were inactivated (or tumours didn't grow) even when the genes were repressed (water with tetracycline). However we have already shown that gene expression leakage in vivo is stronger than in vivo. Moreover it is known that tetracycline is a weaker inhibitor of expression compared to doxycycline in tTA system [9], [14], [15].
Thus, results clearly showed that the expression of either gene has led to the inhibition of tumour growth in vivo, most likely by influencing some interaction between the tumour cells and the host, but without directly affecting tumour cell growth in vitro. HYAL1 and HYAL2 thus differ in this aspect from other tumour suppressors like P53 or RASSF1A that inhibit growth both in vitro and in vivo. Since the products of both genes have a potential to influence intercellular interactions, their impairment may inhibit microenvironmental controls that normally protect the host from tumour growth.
HYAL1 and HYAL2 are the major mammalian hyaluronidases in somatic tissues. They may act in concert to degrade high molecular weight hyaluronan (HA, a negatively charged, high molecular weight glycosaminoglycan) to the tetrasaccharide level [23]. HYAL2 hydrolyses high molecular weight hyaluronic acid (or hyaluronate or hyaluronan) to produce an intermediate-sized product which is further hydrolysed by HYAL1 to give small oligosaccharides. Hyaluronan is claimed to be involved in tumour invasion and metastatic spread. The levels of HA surrounding tumour cells are often correlated with tumour aggressiveness and poor outcome [24].
Overproduction of HA enhances anchorage-independent tumour cell growth [25], [26]. Loss of hyaluronidase activity, permitting accumulation of HA, may be one of several steps required by cells in the multi-step process of carcinogenesis [23].
In contrast to the tumour suppressor function of HYAL1 and HYAL2 in these studies, no frequent inactivating mutations were identified so far in these genes. Nevertheless it was reported that HYAL1 was inactivated in six of seven head and neck squamous cell carcinoma lines by illegitimate splicing [23], [27]. We found that the HYAL2 promoter was methylated in more than 50% of non- small cell lung cancer (NSCLC) cases with decreased expression of the HYAL2 (V. Senchenko, personal communication).
Suppressor activity of HYAL1 and HYAL2 was investigated in the study of Ji et al. [28]. The results showed that HYAL1 and HYAL2 did not significantly inhibit tumour cells growth neither in vitro nor in vivo. However in this work it was demonstrated that HYAL2 inhibited experimental lung metastases in nu/nu mice. It is difficult to compare this work with our study. They used non-small cell lung cancer cell lines and recombinant adenoviral vectors. Completely different methods were used to measure suppressor activity. Despite these differences main results are similar to our work: HYAL1 and HYAL2 do not have strong growth inhibiting activity in vitro and HYAL2 displayed some tumour suppressor activity in vivo.
Since we demonstrated tumour suppressor function of HYAL1 and HYAL2 in SCID mice, we suggested that in primary renal and lung cancers expression of these genes might also be distorted. mRNA quantification was done for these genes in fifteen lung (SCC) and fifteen kidney (RCC) tumours using qPCR. In the majority of the tumour samples mRNA level of both genes was significantly decreased (Figure 5), sometimes more than 50-fold. The difference in expression level in tumour and normal tissues for both SCC and RCC samples was statistically valid (P<0.02). Interestingly, the expression of both genes frequently declined in the same SCC samples. The correlation coefficient according to Spearmen's rank test between HYAL1 and HYAL2 mRNA level was 0.67 (P = 0.005) in lung cancer samples (i.e statistically valid). However in RCC samples it was only 0.4 (P = 0.14, i.e not statistically valid). In any case, the results indicated that these genes may play important role in the development of lung and renal malignancies.
What genes could be responsible for the suppression of tumour growth in vivo, without any inhibition of cell growth in vitro?
There are at least three conceivable categories: genes encoding products required for responding to differentiation inducing signals in vivo; products required for normal cellular responses to microenvironmental controls; or genes whose products inhibit angiogenesis [29].
Hyaluronidases are known to play an important role in tumour/host interactions. They may be examples of genes encoding interactive molecules that participate in microenvironmental growth control and carcinogenesis.
There may be many other, as yet unidentified suppressor genes with similarly asymmetric inhibitory properties. The large numbers of LOHs that occur in the major human tumours and are not accounted for by known tumour suppressor genes, as well as the existence of “tumour suppressor gene clusters”, as documented on human chromosome 3p [11], point to a fertile area of further investigation ahead.
Materials and Methods
Cell lines, tumour samples and general methods
Paired tumour/normal samples were obtained from Blokhin Cancer Research Center, Russian Academy of Medical Sciences. Altogether fifteen specimens of lung squamous cell carcinoma (SCC) and fifteen clear cell renal carcinoma (RCC) cases and adjacent morphologically normal tissues (conventional “normal” tissues) were obtained from patients after surgical resection of primary tumours and and stored in liquid nitrogen.. Top and bottom sections (3–5 µm thick) cut from frozen tumour tissues were examined histologically and only samples containing 70% or more tumour cells were used in the study. The samples were collected in accordance to the guidelines issued by the Ethic Committee of Blokhin Cancer Research Center, Russian Academy of Medical Sciences (Moscow). All patients gave written informed consent that is available upon request. The study was done in accordance with the principles outlined in the Declaration of Helsinki. All tumor specimens were characterized according to the International System of Clinico-Morphological Classification of Tumors (TNM).
SCLC (small cell lung carcinoma) cell line U2020 was described earlier [30] and RCC (renal cell carcinoma) cell line KRC/Y was obtained from the MTC-KI (Stockholm, Sweden) cell lines collection [1].
Molecular cloning of the human HYAL1, HYAL2 and FUS1 genes was described previously [6]. Spontaneous mutant FUS1mut containing Val33Met amino acid substitution was isolated from KRC/Y cell line transformed with wild type FUS1 (unpublished data). These genes were re-cloned into the pETE vector [21]. Recombinants were confirmed by sequencing on ABI310 Sequencer (Applied Biosystems, Foster City, USA) according to manufacturer's protocol.
All molecular, cell biology and microbiology procedures were performed as described previously [21], [31].
Construction of U2020 and KRC/Y cell lines producing tetracycline trans-activator tTA was described in [21].
Transfection, positive clone selection and Northern blotting
Plasmid DNAs containing HYAL1, HYAL2, FUS1 and FUS1mut genes were purified using R.E.A.L. Prep kit (Qiagen, Valencia, CA). Transfections were performed using Lipofectamine PLUS Reagent (Life Technologies, Rockville, MD) according to the manufacturer's protocol.
After transfection, cells were selected with 200U/ml Hygromycin and 200ng/ml doxycycline for four weeks.
PCR positive clones from each recombinant were grown in Iscove's cell culture medium supplemented with 10% heat-inactivated FBS, 100 µg/ml penicillin, 100 µg/ml streptomycin, 200 U/ml hygromycin and 200ng/ml doxycycline. Each clone was split into two parallel flasks, in one of them cells were grown without doxycycline. After one week, 106 cells were collected and total RNA was isolated with Trizol® reagent (Life Technologies, Rockville, MD).
Northern blotting and hybridization were performed as described before [9], [14], [15]. HYAL1 and HYAL2 probes were purified using electrophoresis and the Jetquick Gel Purification kit (Saveen, Germany). The probes were labeled with α-P32 dCTP by random labeling.
All growth inhibition experiments in vivo and in vitro were done as described previously [9], [13], [14], [21]. Work with SCID mice has been approved by North Stockholm Ethical Committee, decision No. 150/08.
Briefly, U2020 or KRC/Y cells were transfected with plasmid DNA using Lipofectamine/Plus reagent (Invitrogen, Germany). Transfected cells were stripped and plated on 100 mm cell culture plates. After selection with 400 µg/ml Hygromycin for 2 weeks, Giemsa-stained colonies were photographed and counted.
The tumorigenicity of each cell line was tested by subcutaneous injection. In total, 5×106 cells were injected into 4-week old female SCID mice. Each mouse received only 1 injection. Twelve control mice were injected with empty pETE vector (six mice were drinking water with DOX and six without). Twenty two mice were used for the experiments (see Table 1). Tumour growth in animals was checked twice a week, if tumour formation was observed, tumours were measured using calipers.
Cell proliferation assay
Cell proliferation rate was determined using CyQUANT NF Cell Proliferation Assay (Invitrogen) according to the manufacturer's protocol. Briefly, cells were plated at density of 100–500 cells per well in a 96-well plate (totally 8–12 identical wells). Number of cells in wells was counted every 24 hours: growth medium was removed, 50 µl of green-fluorescent CyQUANT GR dye (which exhibits strong fluorescence enhancement when bound to cellular nucleic acid) was added to the well and incubated for 30 min at 37°C. The fluorescence intensity of each sample was measured using a fluorescence microplate reader with excitation at 485nm and emission detection at 530nm. Plotted data points represent averages of triplicate samples, the plotted line is a linear regression fit of all data points. The assay is designed to produce a linear analytical response from at least 100–20,000 cells per well in most cell lines.
Quantitative real-time PCR (qPCR)
RNA isolated from primary tumour samples was reverse transcribed using the GeneAmp® RNA PCR Kit (Applied Biosystems) according to the manufacturer's protocol. The relative mRNA level of target genes and GAPDH was assessed using qPCR (ABI PRISM ® 7000 Sequence Detection System, Applied Biosystems). Ratio of mRNA level of target genes in all analysed tumours was measured relative to the reference “normal” samples (paired morphologically normal tissues obtained from the same patient).
The primers and probes for transcripts were as follows:
HYAL1:
Forward primer, 5′- TTTCTGCCCCTGGATGAGC-3′;
Reverse primer, 5′-CTCACCCAGAGCACCACTCC-3′;
Probe, 5′-FAM-CCCAGGCTGTGCTCCAGCTCA-[RTQ1]-3′;
(amplicon size 80 bp);
HYAL2:
Forward primer, 5′- CACCACAAGCACGGAGACCT-3′;
Reverse primer, 5′-CAGGCACTAGGCGGAAACTG-3′;
Probe 5′-FAM-CCTTCCTGCATCTCAGCACCAACAG-[RTQ1]-3′;
(amplicon size 197 bp);
GAPDH:
Forward primer, 5′- CGGAGTCAACGGATTTGGTC– 3′;
Reverse primer, 5′- TGGGTGGAATCATATTGGAACAT– 3′;
Probe, 5′-FAM-CCCTTCATTGACCTCAACTACATGGTTTACAT–[RTQ1]-3′;
(amplicon size 141 bp);
The optimal primer and probe concentrations for the target and control genes were as follows:
HYAL1 primers, 300 nM, probe, 300 nM (for lung cancer cDNA samples); primers, 200 nM, probe, 200 nM (for RCC cDNA samples);
HYAL2 primers, 300 nM, probe, 300 nM (for lung cancer cDNA samples); primers, 200 nM, probe, 200 nM (for RCC cDNA samples);
GAPDH primers, 300 nM; probe, 150 nM.
The thermocycler conditions were 10 min at 95°C, then 50 two-step cycles 15sec at 95°C and 60sec at 60° C. qPCR amplification was carried out in triplicate in 25-µl reaction volume using TaqMan Universal Master Mix (Applied Biosystems) and 10ng template cDNA. The sequences of the amplicons were verified by sequencing in 3730 DNA Analyzer automated sequencer (Applied Biosystems).
The comparative CT method was used as described previously [4], [5].
Statistical analysis
Nonparametric Wilcoxon test was used to compare mRNA expression differences of HYAL1 and HYAL2 and reference gene for the same SCC and RCC patients. The evaluation of statistical significance of mRNA level was tested for all studied cases. Nonparametric Spearmen's rank test was used to calculate the coefficient of correlation between the level of mRNA decrease for HYAL1 and HYAL2 genes. P-values <0.05 were considered statistically significant. All statistical procedures were performed using BioStat software [32].
Acknowledgments
Authors are grateful to Dr. L.L. Kisselev for his useful recommendations and discussions and to Dr. G.S. Krasnov for help with figure preparations.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by research grants from the Swedish Cancer Society, the Swedish Research Council, the Swedish Foundation for International Cooperation in Research and Higher Education (STINT), the Swedish Institute, the Royal Swedish Academy of Sciences, INTAS and Karolinska Institute. VNS, EAA and AVK received grants from the Russian Federal Agency of Science and Innovations (State Contract no 02.512.11.21.01 and 02.512.11.2241) and the Russian Foundation for Basic Research (Project no. 008-04-01577). EVG was recipient of fellowship from the Concern Foundation in Los Angeles and the Cancer Research Institute in New York. TVP was supported by fellowship from The Swedish Institute. MIL and DA were supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Alimov A, Kost-Alimova M, Liu J, Li C, Bergerheim U, et al. Combined LOH/CGH analysis proves the existence of interstitial 3p deletions in renal cell carcinoma. Oncogene. 2000;19:1392–1399. doi: 10.1038/sj.onc.1203449. [DOI] [PubMed] [Google Scholar]
- 2.Braga E, Senchenko V, Bazov I, Loginov W, Liu J, et al. Critical tumour-suppressor gene regions on chromosome 3P in major human epithelial malignancies: allelotyping and quantitative real-time PCR. Int J Cancer. 2002;100:534–541. doi: 10.1002/ijc.10511. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Protopopov A, Wang F, Senchenko V, Petushkov V, Vorontsova O, et al. NotI subtraction and NotI-specific microarrays to detect copy number and methylation changes in whole genomes. Proc Natl Acad Sci U S A. 2002;99:10724–10729. doi: 10.1073/pnas.132271699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Senchenko V, Liu J, Braga E, Mazurenko N, Loginov W, et al. Deletion mapping using quantitative real-time PCR identifies two distinct 3p21.3 regions affected in most cervical carcinomas. Oncogene. 2003;22:2984–2992. doi: 10.1038/sj.onc.1206429. [DOI] [PubMed] [Google Scholar]
- 5.Senchenko VN, Liu J, Loginov W, Bazov I, Angeloni D, et al. Discovery of frequent homozygous deletions in chromosome 3p21.3 LUCA and AP20 regions in renal, lung and breast carcinomas. Oncogene. 2004;23:5719–5728. doi: 10.1038/sj.onc.1207760. [DOI] [PubMed] [Google Scholar]
- 6.Lerman MI, Minna JD. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumour suppressor genes. The International Lung Cancer Chromosome 3p21.3 Tumour Suppressor Gene Consortium. Cancer Res. 2000;60:6116–6133. [PubMed] [Google Scholar]
- 7.Zabarovsky ER, Lerman MI, Minna JD. Tumour suppressor genes on chromosome 3p involved in the pathogenesis of lung and other cancers. Oncogene. 2002;21:6915–6935. doi: 10.1038/sj.onc.1205835. [DOI] [PubMed] [Google Scholar]
- 8.Protopopov A, Kashuba VI, Zabarovska VI, Muravenko OV, Lerman MI, et al. An Integrated Physical and Gene Map of the 3.5-Mb Chromosome 3p21.3 (AP20) Region Implicated in Major Human Epithelial Malignancies. Cancer Research. 2003;63:404–412. [PubMed] [Google Scholar]
- 9.Kashuba VI, Li J, Wang F, Senchenko VN, Protopopov A, et al. RBSP3 (HYA22) is a tumour suppressor gene implicated in major epithelial malignancies. Proc Natl Acad Sci U S A. 2004;101:4906–4911. doi: 10.1073/pnas.0401238101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shivakumar L, Minna J, Sakamaki T, Pestell R, White MA. The RASSF1A tumour suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol Cell Biol. 2002;22:4309–4318. doi: 10.1128/MCB.22.12.4309-4318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imreh S, Klein G, Zabarovsky ER. Search for unknown tumour-antagonizing genes. Genes Chromosomes Cancer. 2003;38:307–321. doi: 10.1002/gcc.10271. [DOI] [PubMed] [Google Scholar]
- 12.Burbee DG, Forgacs E, Zochbauer-Muller S, Shivakumar L, Fong K, et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93:691–699. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreijerink K, Braga E, Kuzmin I, Geil L, Duh FM, et al. The candidate tumour suppressor gene, RASSF1A, from human chromosome 3p21.3 is involved in kidney tumourigenesis. Proc Natl Acad Sci U S A. 2001;98:7504–7509. doi: 10.1073/pnas.131216298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Wang F, Protopopov A, Malyukova A, Kashuba V, et al. Inactivation of RASSF1C during in vivo tumour growth identifies it as a tumour suppressor gene. Oncogene. 2004;23:5941–5949. doi: 10.1038/sj.onc.1207789. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Wang F, Malyukova A, Protopopov A, Duh F-M, et al. Functional characterization of the candidate tumour suppressor gene NPRL2/G21 located in 3p21.3C. Cancer Research. 2004;64:6438–6443. doi: 10.1158/0008-5472.CAN-03-3869. [DOI] [PubMed] [Google Scholar]
- 16.Rai SK, Duh FM, Vigdorovich V, Danilkovitch-Miagkova A, Lerman MI, et al. Candidate tumour suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc Natl Acad Sci U S A. 2001;98:4443–4448. doi: 10.1073/pnas.071572898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danilkovitch-Miagkova A, Duh FM, Kuzmin I, Angeloni D, Liu SL, et al. Hyaluronidase 2 negatively regulates RON receptor tyrosine kinase and mediates transformation of epithelial cells by jaagsiekte sheep retrovirus. Proc Natl Acad Sci U S A. 2003;100:4580–4585. doi: 10.1073/pnas.0837136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harada H, Takahashi M. CD44-dependent intracellular and extracellular catabolism of hyaluronic acid by hyaluronidase-1 and -2. J Biol Chem. 2007;282:5597–5607. doi: 10.1074/jbc.M608358200. [DOI] [PubMed] [Google Scholar]
- 19.Harris H, Miller OJ, Klein G, Worst P, Tachibana T. Suppression of malignancy by cell fusion. Nature. 1969;223:363–368. doi: 10.1038/223363a0. [DOI] [PubMed] [Google Scholar]
- 20.Harris H, Wiener F, Klein G. The analysis of malignancy by cell fusion. 3. Hybrids between diploid fibroblasts and other tumour cells. J Cell Sci. 1971;8:681–92. doi: 10.1242/jcs.8.3.681. [DOI] [PubMed] [Google Scholar]
- 21.Protopopov AI, Li J, Winberg G, Gizatullin RZ, Kashuba VI, et al. Human cell lines engineered for tetracycline-regulated expression of tumour suppressor candidate genes from a frequently affected chromosomal region, 3p21. J Gene Med. 2002;4:397–406. doi: 10.1002/jgm.283. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Protopopov AI, Gizatullin RZ, Kiss C, Kashuba VI, et al. Identification of new tumour suppressor genes based on in vivo functional inactivation of a candidate gene. FEBS Lett. 1999;451:289–294. doi: 10.1016/s0014-5793(99)00598-0. [DOI] [PubMed] [Google Scholar]
- 23.Csoka AB, Frost GI, Stern R. The six hyaluronidase-like genes in the human and mouse genomes. Matrix Biol. 2001;20:499–508. doi: 10.1016/s0945-053x(01)00172-x. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Underhill CB, Chen L. Hyaluronan on the surface of tumour cells is correlated with metastatic behavior. Cancer Res. 1995;55:428–433. [PubMed] [Google Scholar]
- 25.Kosaki R, Watanabe K, Yamaguchi Y. Overproduction of hyaluronan by expression of the hyaluronan synthase Has2 enhances anchorage-independent growth and tumourigenicity. Cancer Res. 1999;59:1141–1145. [PubMed] [Google Scholar]
- 26.Liu N, Gao F, Han Z, Xu X, Underhill CB, et al. Hyaluronan synthase 3 overexpression promotes the growth of TSU prostate cancer cells. Cancer Res. 2001;61:5207–5214. [PubMed] [Google Scholar]
- 27.Frost GI, Mohapatra G, Wong TM, Csoka AB, Gray JW, et al. HYAL1LUCA-1, a candidate tumour suppressor gene on chromosome 3p21.3, is inactivated in head and neck squamous cell carcinomas by aberrant splicing of pre-mRNA. Oncogene. 2000;19:870–877. doi: 10.1038/sj.onc.1203317. [DOI] [PubMed] [Google Scholar]
- 28.Ji L, Nishizaki M, Gao B, Burbee D, Kondo M, et al. Expression of several genes in the human chromosome 3p21.3 homozygous deletion region by adenovirus vector results in tumor suppressor activities in vitro and in vivo. Cancer Res. 2002;62:2715–2720. [PMC free article] [PubMed] [Google Scholar]
- 29.Klein G, Imreh S, Zabarovsky ER. Why do we not all die of cancer at an early age? Adv. Cancer Research. 2007;98:1–16. doi: 10.1016/S0065-230X(06)98001-4. [DOI] [PubMed] [Google Scholar]
- 30.Heppell-Parton AC, Nacheva E, Carter NP, Rabbitts PH. A combined approach of conventional and molecular cytogenetics for detailed karyotypic analysis of the small cell lung carcinoma cell line U2020. Cancer Genet Cytogenet. 1999;108:110–119. doi: 10.1016/s0165-4608(98)00130-7. [DOI] [PubMed] [Google Scholar]
- 31.Zabarovsky ER, Boldog F, Thompson T, Scanlon D, Winberg G, et al. Construction of a human chromosome 3 specific NotI linking library using a novel cloning procedure. Nucleic Acids Res. 1990;18:6319–6324. doi: 10.1093/nar/18.21.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glantz SA. NY: McGraw-Hill, Inc; 1994. Primer of biostatistics. 4th edition. pp. 1–459. [Google Scholar]