Abstract
The twin-arginine translocation (Tat) system transports folded proteins across the bacterial plasma membrane, including FeS proteins that receive their cofactors in the cytoplasm. We have studied two Escherichia coli Tat substrates, NrfC and NapG, to examine how, or whether, the system exports only correctly folded and assembled FeS proteins. With NrfC, substitutions in even one of four predicted FeS centres completely block export, indicating an effective proofreading activity. The FeS mutants are rapidly degraded but only if they interact with the Tat translocon; they are stable in a tat deletion strain and equally stable in wild-type cells if the signal peptide twin-arginine motif is removed to block targeting. Basically similar results are obtained with NapG. The Tat apparatus thus proofreads these substrates and directly initiates the turnover of rejected molecules. Turnover of mutated FeS substrates is completely dependent on the TatA/E subunits that are believed to be involved in the late stages of translocation, and we propose that partial translocation triggers substrate turnover within an integrated quality control system for FeS proteins.
Keywords: FeS protein, protein transport, signal peptide Tat, twin-arginine
Introduction
The twin-arginine translocation (Tat) pathway has the unusual ability to transport folded proteins across tightly sealed, energy-transducing membranes, notably the bacterial plasma membrane and the chloroplast thylakoid membrane (reviewed by Robinson and Bolhuis, 2004; Müller, 2005). It operates alongside the well-characterised Sec pathway and appears to transport two broad classes of protein: globular proteins that fold too rapidly for the Sec system to handle, and proteins that are obliged to fold before transport. In bacteria, the latter class includes periplasmic proteins that bind any of a series of redox cofactors, such as FeS centres, molybdopterin centres and others (Berks, 1996; Santini et al, 1998; Weiner et al, 1998). These cofactors are inserted enzymatically in the cytoplasm, presumably necessitating export in a largely, if not completely, folded form. This raises the key question of how the Tat pathway assesses whether substrates are indeed correctly folded.
Studies on the Tat mechanism have suggested that the core translocation apparatus is membrane bound, at least in Escherichia coli and plant thylakoids. The key Tat components are the TatABC subunits in Gram-negative bacteria and the homologous Tha4, Hcf106 and cpTatC subunits in plants (Settles et al, 1997; Bogsch et al, 1998; Sargent et al, 1998; Weiner et al, 1998). TatA and TatB are single-span membrane proteins that are homologous but functionally distinct (Sargent et al, 1999), whereas TatC contains six transmembrane spans (Behrendt et al, 2004). These subunits form two distinct complexes in the membrane: a TatABC complex of about 400 kDa in E. coli (Bolhuis et al, 2001), and a separate, homo-oligomeric TatA complex, which is highly heterogeneous in size (Gohlke et al, 2005; Oates et al, 2005). Cross-linking studies (Cline and Mori, 2001; Alami et al, 2003) have shown that substrates first bind to the TatBC complex (Hcf106–cpTatC complex in plants), and this is believed to trigger the recruitment of the TatA complex (or plant Tha4 complex) to generate the full translocation system (Mori and Cline, 2002). However, the subsequent translocation process is not understood in any detail at the present time.
The question of ‘proofreading'—how the Tat pathway determines whether a substrate is correctly folded with any cofactors in place—has been addressed in several studies. First, it seems that the system has an inherent ability to preferentially transport folded proteins (at least with some types of proteins). In one case, it was shown that a heterologous protein, PhoA, was only transported if the native disulphide bonds had been formed to generate the correctly folded molecule (DeLisa et al, 2003), and a more recent study has suggested that the Tat system has an inbuilt tendency to transport globular proteins that do not possess exposed hydrophobic patches (Richter et al (2007). However, there is also evidence for the participation of other factors in some cases. Several studies on the related molybdoproteins TMAO reductase (TorA) and DMSO reductase (DmsA) (Oresnik et al, 2001; Sargent, 2007) have shown that the tor and dms operons encode chaperone molecules (TorD and DmsD) that bind to the newly synthesised apo-forms of TorA and DmsA and specifically prevent their export until the cofactors are in place. These proteins bind to the signal peptides of their cognate substrates and appear to be important only for the assembly and export of these specific proteins. For example, DmsD is essential for the efficient export of DmsA reductase (Oresnik et al, 2001) but a fusion comprising the DmsA signal peptide linked to a heterologous protein does not require DmsD for Tat-dependent export to the periplasm (Ray et al, 2003).
In this study, we have investigated the export of FeS proteins by the Tat pathway in E. coli, with the twin aims of testing whether the pathway senses the folded state of selected substrates, and determining the fate of substrate molecules that are unable to assemble correctly. We show that mutations in a single FeS cluster are sufficient to block export of two FeS proteins, and we further show that the Tat apparatus directly initiates the degradation of the rejected molecules.
Results
Mutations in FeS assembly lead to a complete block in NrfC export
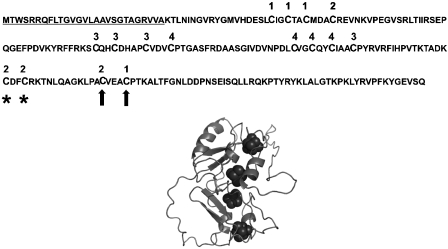
The aim of this study was to analyse the relationship between FeS cluster assembly and protein export in selected Tat substrates, and we first studied NrfC as a candidate substrate. This protein is part of the formate-dependent nitrite reductase complex and is predicted to contain four separate [4Fe-4S] centres (reviewed by Cole, 1996). It is part of the CooF family of iron–sulfur proteins, all which are involved in anaerobic bacterial metabolism. NrfC is strongly predicted to be a Tat substrate and has a typical twin-arginine signal peptide, which has been shown to direct the export of heterologous proteins by the Tat pathway (Tullman-Ercek et al, 2007). Figure 1 shows the primary sequence of pre-NrfC, with the signal peptide underlined and the Cys residues assigned to FeS clusters 1, 2, 3, or 4 on the basis of strong homology to formate dehydrogenase-N subunit β, the structure of which was reported by Jormakka et al (2002). Also shown is a homology model for the three-dimensional structure of NrfC, based on the sequence homology with this subunit.
Figure 1.
Predicted FeS ligands targeted for mutagenesis in NrfC. The top section shows the primary structure of E. coli pre-NrfC with the signal peptide underlined. The diagram also shows Cys residues predicted to be involved in formation of four [4Fe–4S] centres, and these have been denoted as belonging to FeS centres 1–4 (numbering above the residues) on the basis of strong homology to formate dehydrogenase-N, another member of this family of proteins. Residues substituted in mutants M1 and M2 are indicated by asterisks and arrows, respectively. The lower part of the figure shows a predicted three-dimensional structure for NrfC from homology modelling, using the primary sequence of NrfC and the known structure of formate dehydrogenase-N subunit β (Jormakka et al, 2002).
Pre-NrfC was expressed using the arabinose-inducible pBAD24 vector and we appended a C-terminal 6-His tag to aid in the identification of the protein. We first tested whether NrfC is indeed a Tat substrate, by expressing the precursor protein in wild-type MC4100 E. coli cells and a tat-null mutant strain, ΔtatABCDE. The results are shown in the top panel of Figure 2. When expressed in wild-type cells, immunoblotting shows the presence of two forms (of 25 and 22 kDa) in total cell extracts (lane ‘TC'). Fractionation of the cells shows that the 25 kDa form is in the membrane fraction (M), whereas the smaller form is in the periplasm (P). Very little protein is detected in the cytoplasm (C). Only the larger protein is visible after expression in the tat mutant strain, and no NrfC is detected in the periplasm. These data clearly show that, in wild-type cells, the 25 kDa precursor form has been exported to the periplasm and processed to the mature size. The complete absence of export in the tat mutant cells confirms that pre-NrfC is a Tat substrate.
Figure 2.
Mutagenesis of a single FeS cluster blocks the export of pre-NrfC by the Tat pathway. Pre-NrfC was expressed in wild-type E. coli MC4100 cells (WT) or ΔtatABCDE cells. After induction with arabinose for 2 h, cells were harvested and total cell contents (TC) were analysed together with cytoplasm, membrane and periplasm samples (C, M, P) after osmotic shock and fractionation. Precursor and mature forms of NrfC are indicated (pre, mat.). Mutated versions of NrfC (M1 and M2) were analysed in an identical manner (as indicated); only the precursor forms are detected (pre). Mobilities of molecular mass markers (in kDa) are shown on the left.
The question of proofreading was addressed by expressing two mutated versions of pre-NrfC. In mutant M1, 2 of the 16 Cys residues involved in FeS formation were substituted by Ala; the mutations were designed to affect the assembly of one of the four FeS centres (FeS centre 2; the substituted Cys are indicated by asterisks in the sequence shown in Figure 1). In mutant M2, two Cys residues from two separate FeS clusters were mutated in the same manner (arrowed in Figure 1). Expression of these mutants (Figure 2) shows that the substitutions have a dramatic inhibitory effect on export, with no periplasmic protein apparent even with the M1 mutant that contains mutations in a single FeS cluster. These data provide strong evidence that the Tat pathway effectively assesses (‘proofreads') NrfC's folding state and completely blocks the export of the misfolded/misassembled forms. To our knowledge, it is not known how the structure of any of this class of proteins is affected by substitutions of FeS ligands, so we are unable to gauge the extent of the structural changes that are evidently detected.
NrfC and NapG FeS mutants are rapidly degraded in wild-type cells but fully stable in a tat-null mutant strain
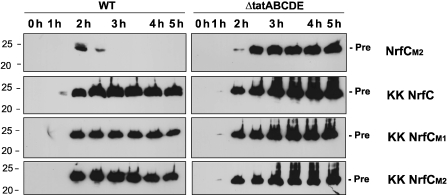
The data shown in Figure 2 were obtained after a 2-h induction of expression using arabinose, and in other experiments we noted that longer induction times resulted in a much-reduced signal strength with the NrfC mutants, or no signal at all. We therefore analysed the expression and processing of the NrfC forms over time, and the data are shown in Figure 3. Expression of non-mutated pre-NrfC in wild-type and ΔtatABCDE cells over a 5-h period (after addition of arabinose to induce synthesis) is illustrated in the upper panel. In wild-type cells, the precursor protein is first detected after 2-h and mature NrfC is detected (at low levels) after 3-h, with the mature protein becoming the dominant form thereafter. These data show that the export of NrfC is a relatively slow process, occurring over an hour or more, and this presumably reflects the time needed to assemble and insert the FeS centres before export.
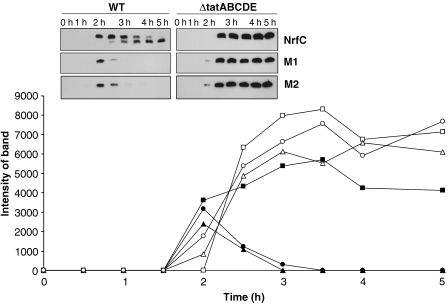
Figure 3.
Mutated forms of NrfC and NapG are rapidly degraded in wild-type cells but stable in ΔtatABCDE cells. Pre-NrfC, M1 and M2 were expressed in WT cells (black squares, triangles and circles, respectively) and ΔtatABCDE cells (white squares, triangles and circles, respectively). Cells were analysed at times (in hours) after induction of synthesis with arabinose, and the graph shows the amounts of protein present (by quantitative immunoblotting) as a function of time. The immunoblots are shown in the inset, with precursor and mature forms of the protein indicated (pre, mat).
Interestingly, the levels of the M1 and M2 forms also peak at 2-h when expressed in wild-type cells, but the protein levels then decline rapidly. Quantitation of the bands (graph) shows that virtually no M1/M2 protein can be detected by the 3-h time point, clearly showing that these mutated forms are subjected to rapid turnover. Surprisingly, the picture is completely different when these mutants are expressed in the Δtat background (right-hand-side panels of immunoblot inset). In these cases, pre-NrfC, M1 and M2 all accumulate stably throughout the induction period. The M1 and M2 forms are equally stable under these conditions and able to accumulate to massive levels when compared with the levels detected in wild-type cells. Note that each sample in this experiment (and others) was derived from equal numbers of identically grown/processed cells, with all immunoblots exposed for identical times to make them directly comparable. As a control to monitor protein synthesis rates, we expressed a Tat substrate comprising the signal peptide of TorA linked to green fluorescent protein; this fusion protein is an excellent Tat substrate (Thomas et al, 2001). TorA–GFP was expressed under identical conditions using the same vector (pBAD24) and we found that similar levels of TorA–GFP accumulated in the wild-type and ΔtatABCDE cells (data not shown; a more extensive version of this experiment is shown in Figure 8). This confirms that the higher signal levels observed in the mutant strain did not stem from enhanced synthesis rates. These data provide the first indication that the Tat system not only senses the folding state of this precursor, but directly initiates the degradation of mutated forms.
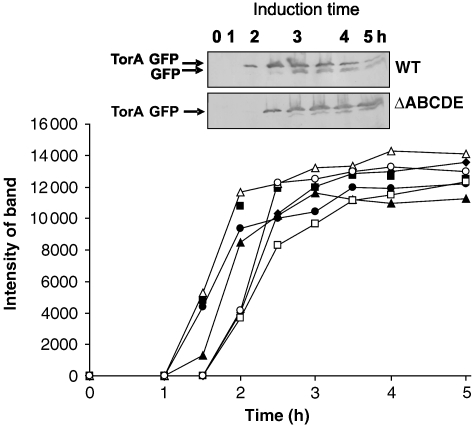
Figure 8.
A non-FeS Tat substrate, TorA–GFP, is synthesised at similar rates in strains disrupted for single or multiple tat genes. TorA–GFP was expressed using the pBAD24 vector as described above for NrfC constructs. The protein was expressed in the range of single/multiple tat disruptions under the conditions described for NrfC in Figure 6, and the GFP immunoblots were quantitated and expressed as shown in the graph. Data are shown after expression of TorA–GFP in wild-type cells (black squares), ΔtatABCDE (black triangles), ΔtatA (black circles), ΔtatB (black diamonds), ΔtatC (white squares), ΔtatE (white triangles) and ΔtatAE (white circles). The inset shows the immunoblots of TorA–GFP expression in two of the strains (wild type and ΔtatABCDE). Mobilities of TorA–GFP and GFP are shown.
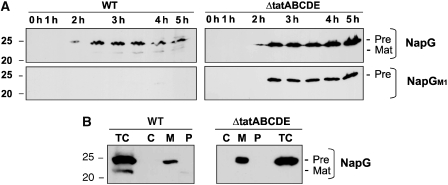
We also analysed NapG, a 4 × [4Fe–4S] component of periplasmic nitrate reductase (reviewed by Cole, 1996), which is known to be synthesised with a Tat signal peptide (Tullman-Ercek et al, 2007). Again, we expressed the protein using the pBAD24 promoter and we tested for export to the periplasm after expression for varying induction periods. Figure 4A shows that the precursor form of NapG again appears after about 2–2.5 h, and the data also show that a faster-migrating band, presumed to be the mature protein, is evident in wild-type cells but not in ΔtatABCDE cells. We fractionated cells after 3.5 h induction (Figure 4B), and the data show that the precursor form is found almost exclusively in the membrane fraction (M), suggesting a non-specific interaction with the membrane surface. A faint mature-size band is in the periplasmic fraction of wild-type cells but not ΔtatABCDE cells. NapG is thus exported in wild-type cells but with rather low efficiency. We did not co-express other genes from the nap operon, and it is possible that the cells do not correctly export NapG under these conditions.
Figure 4.
Mutagenesis of a single FeS cluster in pre-NapG leads to rapid degradation in wild-type cells but not in ΔtatABCDE cells. The precursor form of NapG was expressed in wild-type and ΔtatABCDE cells using the arabinose-inducible pBAD24 vector as described above for NrfC. A mutated version (preNapG-M1) in which two Cys FeS ligands have been substituted by Ala was analysed simultaneously. (A) Samples were taken at the indicated times after induction of synthesis with arabinose and immunoblotted. The precursor and mature forms are denoted by ‘Pre' and ‘Mat', respectively. (B) Cells were taken after 3.5 h induction and fractionated into membrane, cytoplasm, and periplasm (M, C, P) as shown in Figure 1 for NrfC. Mobility of a 25 kDa molecular weight marker is shown on the left of each panel.
We analysed the stability of pre-NapG in wild-type and ΔtatABCDE cells, and we also analysed a mutated form (M1), in which 2 Cys ligands for a single FeS centre were substituted by Ala. The results (Figure 4A) are striking. The non-mutated protein is relatively stable in both wild-type and ΔtatABCDE cells, accumulating steadily throughout the induction period. In contrast, the M1 mutated version is very rapidly degraded in wild-type cells (barely detectable at any point), yet fully stable in ΔtatABCDE cells. As with the NrfC mutants, NapG-M1 degradation is strongly correlated with Tat activity, and the data again suggest that the NapG mutant is interacting with the Tat system and being targeted for degradation when transport is prohibited.
The data in Figures 3 and 4 show that rapid turnover of mutated NrfC/NapG proteins is only observed in transport-competent cells, which suggests a key role for the Tat system in triggering their turnover. However, we considered it important to confirm the apparent importance of the substrate–translocase interaction using an alternative approach, in part to exclude the (unlikely) possibility that the Δtat phenotype may be indirectly responsible for the stabilisation of the mutated precursors. We therefore blocked the interaction between NrfC and the Tat system by mutating the NrfC signal peptide rather than the translocation apparatus. Various studies have confirmed the importance of the twin-arginine motif in Tat signal peptides (e.g., Chaddock et al, 1995; Stanley et al, 2001) and we substituted both arginines by lysine in the NrfC, M1 and M2 signal peptides, reasoning that such ‘KK' mutated forms would be completely blocked in Tat-dependent translocation. Figure 5 shows that, as expected, the KK variant of otherwise-wild-type pre-NrfC is not exported to any detectable extent, with no mature-size protein generated even over extended time points. The precursor form accumulates in a manner that resembles that observed during the expression in tatABCDE cells. The remaining panels of Figure 5 show the expression of the KK variants of M1 and M2 in both wild-type and ΔtatABCDE cells. The key point is that the KK forms of the M1 and M2 mutants are all highly stable over 5 h, even in wild-type cells. Stability is at least as high as that of M1/M2 when expressed in the tat-null mutant strain, and this confirms that the previously observed rapid turnover of M1 and M2 is totally dependent on interaction with the Tat apparatus. For direct comparison, we expressed one of the mutated NrfC forms bearing an intact signal peptide under the same conditions (M2, as used in Figures 1 and 2). As found previously, this mutated form is stable in ΔtatABCDE cells but rapidly turned over in wild-type cells. We conclude from these data that degradation of the mutated proteins is totally dependent on interaction with the membrane-bound Tat apparatus.
Figure 5.
Removal of the twin-arginine motif in the signal peptide blocks turnover of the NrfC M1/M2 mutants. The twin-arginine motifs in the pre-NrfC, M1 and M2 signal peptides were substituted by twin-lysine by site-specific mutagenesis, and the constructs were expressed and analysed exactly as shown in Figure 3, with time points shown after induction with arabinose. The M2 construct containing an intact signal peptide was also expressed for reference purposes (as shown in Figure 3). Total cell samples were analysed and the mobilities of the precursor proteins are shown.
TatA/E subunits have important functions in turnover of M1 and M2, indicating partial translocation before degradation
Which particular elements of the Tat apparatus are important for turnover of NrfC mutants? This is a key question because different components are believed to have different roles, with the TatA complex becoming involved only at the later stages of the translocation process. In E. coli, this analysis is complicated by the presence of a TatA paralogue, TatE, which is expressed from a separate gene. TatE is expressed at much lower levels than TatA, and as a result, ΔtatA cells exhibit a much more severe export defect than do ΔtatE cells. A complete block in translocation is observed only when both tatA and tatE are disrupted, and disruption of either tatB or tatC also results in a complete block in translocation (Sargent et al, 1998, 1999).
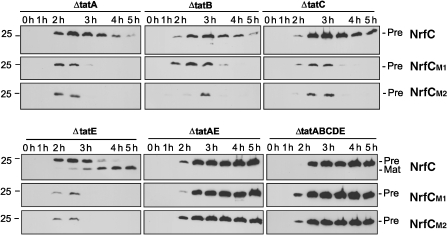
We expressed pre-NrfC and the M1/M2 mutants in a range of tat mutant strains, and the data are shown in Figure 6. The first, and most important, point is that the rapid turnover of M1 and M2 is completely abolished in the ΔtatAE double mutant. Both mutated NrfC proteins are as stable as when expressed in the full ΔtatABCDE strain, and this demonstrates a critical role for TatA/E in the turnover process. If current models for the E. coli Tat system are correct, this result indicates that coalescence of the TatABC and TatA complexes to form the full translocon is essential to trigger the degradation of M1 and M2. The clear implication is that these proteins are rejected, and perhaps concomitantly designated for disposal, at a very late stage in translocation.
Figure 6.
The TatA/E subunits have an important function in the Tat-dependent turnover of the M1/M2 mutants. Pre-NrfC, M1 and M2 were expressed in the ΔtatABCDE strain and strains specifically disrupted in tatA, tatE, tatAE, tatB or tatC as indicated. Samples were analysed at times (in hours) after induction of NrfC/M1/M2 synthesis using arabinose and immunoblotted as described in Figure 3. Mobilities of the precursor proteins are shown.
Close inspection of the data obtained with the ΔtatE and ΔtatA single mutants does yield interesting results that bear consideration. Pre-NrfC is not exported at all in ΔtatA cells (no mature-size protein is apparent) but is exported rather efficiently in ΔtatE cells, as shown by the accumulation of mature size protein during the time course. These data are not surprising, as TatE is known to have a less important function in the translocation process, just because it is present in such low quantities. TatE is thus unable to support the export of NrfC in the absence of TatA, and the natural prediction is that TatE would be similarly unable to support M1/M2 turnover in the absence of TatA. However, this is not the case: these mutants are turned over very rapidly indeed in both ΔtatA and ΔtatE cells (as rapidly as when expressed in wild-type cells). We conclude that TatE can function efficiently in this proofreading/turnover system, apparently as efficiently as TatA.
Pre-NrfC, M1 and M2 were also expressed in ΔtatB and ΔtatC cells (Figure 6). The non-mutated precursor is relatively stable, but M1 and M2 were rapidly degraded over the 5-h time-course analysis. These data are surprising; heterodimeric TatBC units are believed to form the initial binding site for precursor, and we expected that the absence of either subunit would completely block substrate binding and hence M1/M2 turnover. The observed turnover of M1 and M2 in ΔtatB and ΔtatC cells suggests one of two scenarios: (i) that either subunit, on its own, can bind precursor and recruit the TatA complex, thereby stimulating M1/M2 turnover, or (ii) that the roles of Tat complexes/subunits in translocation are different from their roles in M1/M2 proofreading and turnover, and TatA/E can enhance turnover of the mutants even in the absence of substrate binding to TatBC.
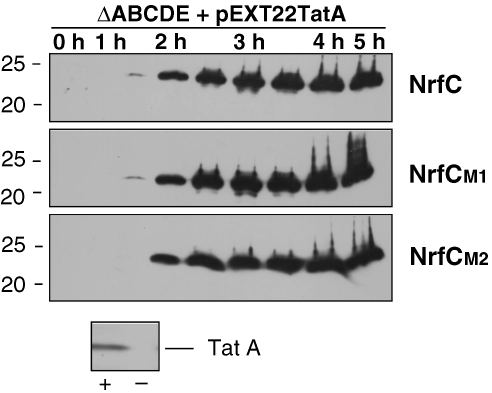
Given the key role of TatA/E in the turnover of NrfC mutants, we thought it important to test whether one of these subunits can support turnover of NrfC in the complete absence of TatBC. We therefore expressed TatA together with NrfC/M1/M2 in a ΔtatABCDE background, and the data are shown in Figure 7. The lower section of the figure shows a TatA blot in these cells (+ lane) and ΔtatABCDE control cells (−), and this confirms that TatA is indeed expressed as expected. The time-course analyses of NrfC/M1/M2 show that all of these forms are stable in the presence of TatA, and this confirms that turnover requires both TatA/E and, minimally, either TatB or TatC (as suggested by the data in Figure 6).
Figure 7.
TatA cannot direct the degradation of NrfC mutants in the absence of TatBC. TatA was expressed in ΔtatABCDE cells using the pEXT22 plasmid, together with NrfC and mutants M1 and M2. The NrfC forms were induced by addition of arabinose as shown in previous figures, and samples were analysed by immunoblotting at times (in hours) after induction. The lower part of the figure shows a TatA immunoblot of a sample from one of these strains (the TatA/NrfC-expressing cells) and ΔtatABCDE cells expressing NrfC but not TatA.
There is ample evidence that other, non-FeS Tat substrates do not exhibit such widely different stabilities in wild-type versus tat cells (see Discussion), but we considered it important to examine the synthesis and stability of a non-FeS substrate under identical conditions. For this purpose, we used the TorA–GFP substrate described earlier. The protein was expressed under identical conditions in all of the mutant strains used in Figure 6, and the quantitated data are shown in Figure 8, with the inset showing the immunoblot of two of the panels (TorA–GFP expression in wild-type and ΔtatABCDE cells). The protein is exported in wild-type cells as shown previously (Thomas et al, 2001), and the mature-size protein accumulates throughout the induction series. No export is observed in ΔtatABCDE cells, although some proteolytic clipping to a near-mature-size form is evident, and no export is observed in ΔtatAE, ΔtatB or ΔtatC cells either (data not shown). Comparison of the data sets shows that this substrate accumulates to broadly similar levels in all of these strains, and this shows that this Tat substrate is (i) synthesised at similar rates and (ii) equally stable in the presence or absence of the Tat apparatus.
Discussion
Proofreading of substrates is likely to be a critical aspect of Tat functioning because the export of apo-forms of complex substrates would be a futile process, with possible adverse consequences. Emerging evidence suggests that the Tat system may indeed preferentially transport folded forms of globular proteins, perhaps through an inbuilt tendency to avoid proteins with exposed hydrophobic patches (Richter et al, 2007), but the pathway may need a more rigorous quality control system for some substrates, with additional safeguards. Studies on molybdoproteins such as TorA and DmsA have shown that soluble, substrate-specific chaperones have an important function, and these accessory factors appear to have exactly this function (reviewed by Sargent, 2007). Here, we have shown that mutation of only one FeS centre is sufficient to completely block export of the NrfC FeS protein, and these data represent the first evidence for the proofreading of FeS substrates. Our data also show why such a system may be vital for the cell; NrfC is exported very slowly in wild-type E. coli cells, and this would provide ample opportunity for the export of apo-protein in the absence of a rigorous quality control system.
Although this finding is of interest in itself, our study has shown that the quality control of FeS protein export involves much more than the proofreading of either substrate conformation or the status of FeS cluster assembly. FeS-deficient molecules are rapidly degraded by the cell, and this degradation is wholly dependent on interaction with the Tat apparatus. Thus, the mutated NrfC proteins are completely stable in a tat-null mutant background, or when non-functional signal peptides are used to block interaction with the Tat translocon (the KK mutants). Effectively, the Tat system is at the centre of an integrated quality control system that involves not only the proofreading of FeS protein substrates before transport, but also the targeted degradation of substrate molecules that are found to be inappropriately folded or assembled. Turnover is triggered only after productive interaction between the precursor and the Tat components.
Importantly, the TatA/E subunits have an important function in this quality control operation, and this points to an involvement of the TatA (or TatE) complex, which is believed to become involved only at a late stage in translocation. NrfC may, therefore, be tagged for degradation as a result of partial translocation by the membrane-bound Tat apparatus. The other FeS protein studied here, NapG, does not appear to be efficiently exported under these conditions, but it may nevertheless be subjected to the same form of quality control audit. Our data show that the mutated form of this protein (NapG-M1) is very rapidly turned over in wild-type cells, but highly stable in ΔtatABCDE cells. We believe that the wild-type NapG precursor protein may interact extensively with the Tat apparatus, but not to the extent that it is fully translocated and processed (for unknown reasons). Apparently, this partial translocation of the M1 NapG mutant is nevertheless sufficient to trigger very rapid degradation in wild-type cells, to the extent that the protein is virtually undetectable.
It is unsurprising that misfolded substrates are degraded (this is common practice in biological systems) but highly unusual for a protein transport system to make the vital decision about the fate of its substrates. Key questions are whether this mechanism applies to all FeS substrates of the Tat system, and whether a similar system operates for other, non-FeS Tat substrates. Further substrates need to be studied before this question can be properly answered, but previous studies on other (non-FeS) substrates suggest that the Tat apparatus is not involved in the degradation of its other substrates. In several cases it has been shown that other types of Tat substrate (whether native or misfolded) are destabilised, rather than stablilised, when transport is prevented by the absence of translocase components or mutagenesis of the signal peptide (Santini et al, 2001; Barrett et al, 2003; DeLisa et al, 2003; Fisher et al, 2006). None of these studies reported enhanced rates of substrate degradation in the presence of a functioning Tat system, and our data are therefore consistent with the idea that FeS proteins are subjected to a particular form of quality control analysis. Of course, further studies are required to determine whether there are parallels with other types of Tat substrate.
How might the Tat translocon initiate the disposal of rejected substrates? This is a key question, but we have very little information at present. One possibility is that ancillary ‘proofreading' factors remain associated with the substrate and prevent its correct translocation once bound to the TatABC components. In this scenario, the translocation event may actually serve to partially unfold the substrate and render it protease sensitive. Alternatively, the substrate may remain locked onto the translocon in a transport-incompetent state and the system may then recruit the necessary degradation machinery; this would represent a more active role for the Tat system in triggering turnover. These, and other, possibilities are experimentally testable, and the priorities will be to determine how the Tat translocon operates within this quality control pathway, and to identify other factors involved.
Materials and methods
Bacterial strains, plasmids and growth conditions
Escherichia coli strain MC4100 was the parental strain (Casadaban and Cohen, 1979); ΔtatA, ΔtatB, ΔtatC, ΔtatE, ΔtatAE and ΔtatABCDE have been described earlier (Bogsch et al, 1998; Sargent et al, 1998, 1999; Wexler et al, 2000). All of the constructs described below were expressed using the arabinose-inducible pBAD24 vector, and arabinose-resistant derivatives were used as described previously (Guzman et al, 1995; Bolhuis et al, 2001). E. coli was grown aerobically at 37 °C in Luria broth (LB) as described earlier (Bolhuis et al, 2001). Medium supplements were used at the following final concentrations: ampicillin, 100 μg/ml; arabinose, 100 μM unless otherwise stated (Table I).
Table 1.
Plasmids and strains used in the study
| Strain/plasmid | Description | Source/reference |
|---|---|---|
| DH5α | F_ _80lacZ_M15 _(lacZYA-argF) U169 deoR recA1 endA1 hsdR17 (rk_, mk_) gal-phoA supE44 thi-1 gyrA96 relA1 | Invitrogen, Carlsbad, CA, USA |
| MC4100AR | AraR, FaraD139ΔlacU169 rpsL150 relA1 flB5301 deoC1 ptsF25 rbsR | Casadaban et al (1979); Bolhuis et al (2000) |
| ΔtatABCDE | As MC4100AR; ΔtatABCDE | Wexler et al (2000); Bolhuis et al (2000) |
| ΔtatA | As MC4100AR; ΔtatA | Sargent et al (1998); Bolhuis et al (2000) |
| ΔtatB | As MC4100AR; ΔtatB | Sargent et al (1999); Bolhuis et al (2000) |
| ΔtatC | As MC4100AR; ΔtatC | Bogsch et al (1998); Bolhuis et al (2000) |
| ΔtatE | As MC4100AR; ΔtatE | Sargent et al (1998); Bolhuis et al (2000) |
| ΔtatAE | As MC4100AR; ΔtatAE | Sargent et al (1998); Bolhuis et al (2000) |
| pJDT1 | PBAD24 expressing ssTorA–GFP under control of pBAD promoter | Thomas et al (2001) |
| pCM1 | PBAD24 expressing pre-NrfC with a hexa-Histag under control of pBAD promoter | This study |
| pCM2 | As pCM1 but point mutations C152A and C155A in the nrfC gene | This study |
| pCM3 | As pCM1 but point mutations C168A and C172A in the nrfC gene | This study |
| pCM4 | PBAD24 expressing pre-NapG with a hexa-Histag under control of pBAD promoter | This study |
| pCM5 | As pCM4 but point mutations C107A and C111A in the napG gene | This study |
| pCM6 | As pCM1 but point mutations R5K and R6K in the nrfC gene | This study |
| pCM7 | As pCM2 but point mutations R5K and R6K in the nrfC gene | This study |
| pCM8 | As pCM3 but point mutations R5K and R6K in the nrfC gene | This study |
Constructs and cloning
To construct a plasmid for the overproduction of His-tagged NrfC, a DNA fragment containing the nrfC gene was amplified with the primers CM_nrfC_Fw (5′-CGTAGCCAGAATTCATGACCTGGTCTCGTCG-3′) and CM_nrfC_Rev (5′-CGTAGCCACTGCAGTTAATGGTGATGGTGATGGTGTTGGCTCACCTCCCCG-3′) using E. coli MC4100 chromosomal DNA as template. The resulting product was digested with EcoRI and PstI and cloned into plasmid pBAD24 (Guzman et al, 1995). The resulting plasmid was denoted by pCM1.
To construct a plasmid for the overproduction of His-tagged NapG, a DNA fragment containing the napG gene was amplified with the primers CM_napG_Fw (5′-CGTAGCCAGAATTCATGTCCCGGTCAGCGAA-3′) and CM_napG_Rev (5′-CGTAGCCACTGCAGTTAATGGTGATGGTGATGGTGCGATTTGCCATTGTTCCC-3′) using E. coli MC4100 chromosomal DNA as template. The resulting product was digested with EcoRI and PstI and cloned into plasmid pBAD24. The resulting plasmid was denoted by pCM4. For co-expression of TatA with above-mentioned plasmids, the coding sequence for E. coli TatA was cloned into pEXT22 using the XbaI site, generating pEXT-TatA.
Site-specific mutagenesis was used to generate the mutants nrfC M1, nrfC M2 on pCM1 background and the mutant napG M2 on pCM4 background. The resulting plasmids were denoted by pCM2, pCM3 and pCM5, respectively. To obtain these, the point mutations were made using the Quickchange™ mutagenesis system (Stratagene) according to the manufacturer's instructions. In the case of nrfC M1 (pCM2), the Cys residues at positions 152 and 155 were mutated to Ala with the primers CM_nrfC_C152A_C155A_Fw (5′-GACGGCGGACAAAGCCGATTTCGCCCGTAAGACCAATTTGC-3′) and CM_nrfC_C152A_C155A_R (5′-GCAAATTGGTCTTACGGGCGAAATCGGCTTTGTCCGCCGTC-3′) using pCM1 as a template. In the case of nrfC M2 (pCM3), the Cys residues at position 168 and 172 were mutated to Ala with the primers CM_nrfC_C168A_C172A_Fw (5′-GTTGCCCGCGGCCGTTGAAGCTGCCCCGACCAAG-3′) and CM_nrfC_C168A_C172A_Rev (5′-CTTGGTCGGGGCAGCTTCAACGGCCGCGGGCAAC-3′) using pCM1 as a template. In the case of napG M2 (pCM5), the Cys residues at position 107 and 111 were mutated to Ala with the primers CM_napG_C107A_C111A_Fw (5′-GGACATTCCGGCCGCCAAAGTGGCCCCAAGCGGTG-3′) and CM_napG_C107A_C111A_Rev (5′-CACCGCTTGGGGCCACTTTGGCGGCCGGAATGTCC-3′) using pCM4 as a template.
Site-specific mutagenesis was used again to generate the mutants KK nrfC, KK nrfC M1 and KK nrfC M2 of pCM1, pCM2 and pCM3, respectively. The resulting plasmids were denoted by pCM6, pCM7 and pCM8, respectively. To obtain these, the point mutations were made using the Quickchange mutagenesis system (Stratagene) according to the manufacturer's instructions. To produce pCM6, pCM7 and pCM8, both arginine residues of NrfC at positions 5 and 6 were mutated to lysine residues with the primers CM_KK_nrfC_ F (5′-GACCTGGTCTAAAAAGCAGTTTCTCACCGGCGTCGGCGTGCTGGC-3′) and CM_KK_nrfC_R (5′-GCCAGCACGCCGACGCCGGTGAGAAACTGCTTTTTAGACCAGGTC-3′) using pCM1, pCM2 and pCM3, respectively, as a template.
Cell fractionation
Starter cultures (5 ml) were grown overnight and used to inoculate 50 ml of LB (with appropriate antibiotic selection and 100 μM arabinose to induce the expression of NrfC and 500 μM arabinose to induce the expression of NapG) in a 250 ml flask to a starting OD600 of 0.01. Cells were grown to mid-exponential growth phase before fractionation as follows. Periplasm and spheroplasts were prepared by the EDTA/lysozyme/cold osmoshock procedure (Randall and Hardy, 1986). Spheroplasts were lysed by sonication, and intact cells and cellular debris were removed by centrifugation (5 min at 10 000 g). Membranes were separated from the cytoplasmic fraction by centrifugation (30 min at 250 000 g). Protein concentration was determined using a BCA-linked assay (Pierce). Protein fractions were separated by 15% SDS–polyacrylamide gel electrophoresis and immunoblotted with anti-His antibody (Invitrogen) and anti-GFP (Clontech). As secondary antibodies, horseradish peroxidase anti-mouse and anti-rabbit IgG conjugate were used, respectively, and detected by ECL detection system (Amersham Pharmacia Biotech). ImageQuant software (Molecular Dynamics) was used for quantitative analysis of immunoblots.
Time course during arabinose induction
Starter cultures (5 ml) were grown overnight and used to inoculate 50 ml of LB (with appropriate antibiotic selection and 100 μM arabinose to induce the expression of NrfC and 500 μM arabinose to induce the expression NapG) in a 250 ml flask to a starting OD600 of 0.01. Cells were grown at 37 °C and cell samples were taken every 30 min for a period of 5 h during arabinose induction. At each time point, 500 μl of cells were collected by centrifugation and immediately frozen. Total cell protein content was separated on a 15% SDS–polyacrylamide gel and immunoblotted as described earlier.
Generation of a structural model for NrfC
PDB file 1KQF for formate dehydrogenase was used as a template for generation of a homology model for NrfC structure. The primary sequence was superimposed on the amino-acid backbone ensuring that FeS ligands were positioned at equivalent points, and the model shown in Figure 1 was generated using PyMOL (http://pymol.sourceforge.net/).
Acknowledgments
This work was funded by a grant from the Biotechnology and Biological Sciences Research Council and grant LSHG-CT-2004-005257 from the EU to C.R.
References
- Alami M, Luke I, Deitermann S, Eisner G, Koch HG, Brunner J, Müller M (2003) Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol Cell 12: 937–946 [DOI] [PubMed] [Google Scholar]
- Barrett CML, Ray N, Thomas JD, Robinson C, Bolhuis A (2003) Quantitative export of a heterologous protein, GFP, by the twin-arginine translocation pathway in Escherichia coli. Biochem Biophys Res Comm 304: 279–284 [DOI] [PubMed] [Google Scholar]
- Behrendt J, Standar K, Lindenstrauss U, Bruser T (2004) Topological studies on the twin-arginine translocase component TatC. FEMS Microbiol Lett 234: 303–308 [DOI] [PubMed] [Google Scholar]
- Berks BC (1996) A common export pathway for proteins binding complex redox cofactors? Mol Microbiol 22: 393–404 [DOI] [PubMed] [Google Scholar]
- Bogsch EG, Sargent F, Stanley NR, Berks BC, Robinson C, Palmer T (1998) Anessential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J Biol Chem 273: 18003–18006 [DOI] [PubMed] [Google Scholar]
- Bolhuis A, Bogsch EG, Robinson C (2000) Subunit interactions in the twin-arginine translocase complex of Escherichia coli. FEBS Letts 472: 88–92 [DOI] [PubMed] [Google Scholar]
- Bolhuis A, Mathers JE, Thomas JD, Barrett CML, Robinson C (2001) TatB and TatC form a structural and functional unit of the twin-arginine translocase of Escherichia coli. J Biol Chem 276: 20213–20219 [DOI] [PubMed] [Google Scholar]
- Casadaban MJ, Cohen SN (1979) Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci USA 76: 4530–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaddock AM, Mant A, Karnauchov I, Brink S, Herrmann RG, Klosgen RB, Robinson C (1995) A new type of signal peptide: central role of a twin-arginine motif in transfer signals for the delta pH-dependent thylakoidal protein translocase. EMBO J 14: 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K, Mori H (2001) Thylakoid ΔpH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. J Cell Biol 154: 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J (1996) Nitrate reduction to ammonia by enteric bacteria: redundancy, or a strategy for survival during oxygen starvation? FEMS Microbiol Lett 136: 1–11 [DOI] [PubMed] [Google Scholar]
- Delisa MP, Tullman D, Georgiou G (2003) Folding quality control in the export of proteins by the bacterial twin-arginine translocation pathway. Proc Natl Acad Sci USA 100: 6115–6120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AC, Kim W, DeLisa MP (2006) Genetic selection for protein solubility enabled by the folding quality control feature of the twin-arginine translocation pathway. Protein Sci 15: 449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohlke U, Pullan L, McDevitt CA, Porcelli I, de Leeuw E, Palmer T, Saibil HR, Berks BC (2005) The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter. Proc Natl Acad Sci USA 102: 10482–10486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177: 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jormakka M, Törnroth S, Byrne B, Iwata B (2002) Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. Science 295: 1863–1868 [DOI] [PubMed] [Google Scholar]
- Mori H, Cline K (2002) A twin arginine signal peptide and the pH gradient trigger reversible assembly of the thylakoid Delta pH/Tat translocase. J Cell Biol 157: 205–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M (2005) Twin-arginine specific export in Escherichia coli. Res Microbiol 156: 131–136 [DOI] [PubMed] [Google Scholar]
- Oates J, Barrett CML, Barnett JP, Byrne KG, Bolhuis A, Robinson C (2005) The Escherichia coli twin-arginine translocation apparatus incorporates a distinct form of TatABC complex, spectrum of modular TatA complexes and minor TatAB complex. J Mol Biol 346: 295–305 [DOI] [PubMed] [Google Scholar]
- Oresnik IJ, Ladner CL, Turner RJ (2001) Identification of a twin-arginine leader-binding protein. Mol Microbiol 40: 323–331 [DOI] [PubMed] [Google Scholar]
- Randall LL, Hardy SLS (1986) Correlation of competence for export with lack of tertiary structure of the mature species: a study in vivo of maltose-binding in E. coli. Cell 46: 921–928 [DOI] [PubMed] [Google Scholar]
- Ray N, Oates J, Turner RJ, Robinson C (2003) DmsD is required for the biogenesis of DMSO reductase in Escherichia colibut not for the interaction of the DmsA signal peptide with the Tat apparatus. FEBS Lett 534: 156–160 [DOI] [PubMed] [Google Scholar]
- Richter S, Lindenstrauss U, Lücke C, Bayliss R, Brüser T (2007) Functional Tat transport of unstructured, small, hydrophilic proteins. J Biol Chem 282: 33257–33264 [DOI] [PubMed] [Google Scholar]
- Robinson C, Bolhuis A (2004) Tat-dependent protein targeting in prokaryotes and chloroplasts. Biochim Biophys Acta 1694: 135–147 [DOI] [PubMed] [Google Scholar]
- Santini CL, Bernadac A, Zhang M, Chanal A, Ize B, Blanco C, Wu LF (2001) Translocation of jellyfish green fluorescent protein via the Tat system of Escherichia coli and change of its periplasmic localization in response to osmotic up-shock. J Biol Chem 276: 8159–8164 [DOI] [PubMed] [Google Scholar]
- Santini CL, Ize B, Chanal A, Müller M, Giordano G, Wu LF (1998) A novel sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J 17: 101–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent F (2007) The twin-arginine transport system: moving folded proteins across membranes. Biochem Soc Trans 35: 835–847 [DOI] [PubMed] [Google Scholar]
- Sargent F, Bogsch EG, Stanley NR, Wexler M, Robinson C, Berks BC, Palmer T (1998) Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J 17: 3640–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent F, Stanley NR, Berks BC, Palmer T (1999) Sec-independent protein translocation in Escherichia coli. A distinct and pivotal role for the TatB protein. J Biol Chem 274: 36073–36082 [DOI] [PubMed] [Google Scholar]
- Settles AM, Yonetani A, Baron A, Bush DR, Cline K, Martienssen R (1997) Sec-independent protein translocation by the maize Hcf106 protein. Science 278: 1467–1470 [DOI] [PubMed] [Google Scholar]
- Stanley NR, Palmer T, Berks BC (2001) The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J Biol Chem 275: 11591–11596 [DOI] [PubMed] [Google Scholar]
- Thomas JD, Daniel RA, Errington J, Robinson C (2001) Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol Microbiol 39: 47–53 [DOI] [PubMed] [Google Scholar]
- Tullman-Ercek D, Delisa MP, Kawarasaki Y, Iranpour P, Ribnicky B, Palmer T, Georgiou G (2007) Export pathway selectivity of Escherichia coli twin arginine translocation signal peptides. J Biol Chem 282: 8309–8316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner JH, Bilous PT, Shaw GM, Lubitz SP, Frost L, Thomas GH, Cole JA, Turner RJ (1998) A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell 93: 93–101 [DOI] [PubMed] [Google Scholar]
- Wexler M, Sargent F, Jack RL, Stanley NR, Bogsch EG, Robinson C, Berks BC, Palmer T (2000) TatD Is a cytoplasmic protein with DNase activity. No requirement for TatD family proteins in Sec-independent protein export. J Biol Chem 275: 16717–16722 [DOI] [PubMed] [Google Scholar]