Abstract
Intracellular pathogens, including Mycobacterium tuberculosis, obtain iron from the host for their survival. Ferroportin 1 (FPN1; SLC40A1) is the sole iron exporter from mammalian cells and is expressed in the duodenum and macrophages. In the present study, we show that FPN1 mRNA levels in the mouse macrophage cell line RAW264.7 are synergistically induced by treatment with live or γ-irradiated M. tuberculosis and IFN-γ. FPN1 mRNA levels were also induced by Mycobacterium avium and IFN-γ in RAW264.7 cells and the mouse alveolar macrophage cell line AMJ2-C8. Treatment of mouse resident peritoneal macrophages with M. tuberculosis and IFN-γ resulted in a sixfold increase in FPN1 mRNA expression. In contrast, M. tuberculosis and IFN-γ inhibited FPN1 mRNA expression in bone marrow-derived macrophages and lung macrophages, which have high basal levels of FPN1 mRNA expression. Using confocal microscopy, FPN1 protein localized rapidly to M. tuberculosis phagosomes after infection in RAW264.7 macrophages. In RAW264.7 cells expressing wild-type natural resistance-associated macrophage protein 1 (Nramp1Gly169), FPN1 and Nramp1 partially colocalized in late endosomes/lysosomes prior to infection. After 2 h of infection, Nramp1 and FPN1 were present in M. tuberculosis phagosomes. Our studies provide evidence for transcriptional regulation of FPN1 by pathogenic mycobacteria and IFN-γ, which is dependent on the macrophage type. The trafficking of FPN1 to the M. tuberculosis phagosome suggests that it is involved in regulating iron availability to the mycobacteria in this locale.
Keywords: FPN1, Mycobacterium tuberculosis, IFN-γ
INTRODUCTION
Iron is an essential nutrient but in excess, can result in tissue damage as a result of increased oxidative stress [1, 2] and lack of iron results in anemia. Iron homeostasis is maintained by the intestinal uptake of iron from the diet and by recycling of iron from senescent erythrocytes. The reticuloendothelial system recycles 25 mg iron each day from ∼360 billion senescent erythrocytes [3, 4]. Transport of iron is mediated by transport proteins expressed in the duodenum and reticuloendothelial macrophages. Natural resistance-associated macrophage protein 2 (Nramp2; Slcl1a2) is responsible for transport of dietary iron into the duodenal enterocyte [5, 6] and in other cells, transports iron into the cytosol from recycling endosomes [7]. Nramp1 (Slc11a1) is expressed in macrophages and neutrophils [8,9,10]. Upon phagocytosis of a pathogen, Nramp1 is recruited to the phagosome [11, 12] and is responsible for transport of iron and other divalent cations across the phagosomal membrane [13,14,15,16]. Ferroportin 1 (FPN1; also known as iron-regulated transporter 1, metal transporter protein 1, SLC40a1) is the sole iron export protein identified in mammals and is responsible for iron export from enterocytes of the duodenum and macrophages [17,18,19]. In the dudonenal enterocytes, FPN1 is expressed on the basal membrane and exports iron into the portal blood circulation. In macrophages, FPN1 is expressed in intracellular vesicles. Upon phagocytosis of senescent erythrocytes, FPN1 localizes to the cell membrane of macrophages, where it transports iron out of the macrophages [2, 20,21,22].
During infection, a competition for iron occurs between the macrophage of the infected host and the pathogen. Invading pathogens secrete siderophores or use host-derived iron storage/transport proteins to capture iron from the host [23]. The host macrophage attempts to suppress pathogen proliferation by complex iron-withholding mechanisms. One such mechanism involves changes in the levels of iron transport proteins. Our laboratory has shown that infection of macrophages with the intracellular pathogen Mycobacterium avium results in an increase in expression of mRNA for the iron transport proteins Nramp1 (Slc11a1) and Nramp2 (Slc11a2) and a decrease in the expression of transferrin receptor mRNA [24].
Regulation of FPN1 expression in macrophages by infection and inflammatory stimuli has only recently begun to be characterized. Inflammatory stimuli appear to have a negative regulatory effect on FPN1 expression. In vivo, inflammation induced by LPS in C57BL/6J mice was shown to inhibit expression of FPN1 protein in macrophages of the spleen, liver, and bone marrow [25]. In isolated mouse splenic macrophages and bone marrow-derived macrophages (BMDM), LPS stimulation also decreased FPN1 mRNA [26]. Down-regulation of FPN1 mRNA expression has also been reported in the human macrophage-like cell lines THP-1 and U937 stimulated with IFN-γ and LPS [27]. FPN1 expression is also positively regulated by macrophage iron levels. In BMDM and the mouse J774A.1 macrophage-like cell line, it has been shown that iron loading and phagocytosis of senescent erythrocytes increase the expression of FPN1 mRNA and protein [20, 21].
FPN1 protein expression is also regulated in vivo by hepcidin, which is an antimicrobial peptide produced by the liver during an acute-phase inflammatory response and by iron overload [28,29,30]. Hepcidin regulates iron homeostasis by inhibiting iron absorption by the duodenum [31, 32] and iron recycling by macrophages [22]. Nemeth et al. [33] showed that hepcidin blocks iron export by binding to FPN1, present at the cell membrane, resulting in the internalization and degradation of FPN1. Recently, we reported that hepcidin is induced in macrophages following infection with Mycobacterium tuberculosis and IFN-γ stimulation and localized to the phagosome [34]. We also showed that hepcidin in vitro inhibits M. tuberculosis growth and causes structural damage to the mycobacteria.
The purpose of the present study was to determine the effect of an intracellular bacterium on FPN1 mRNA and protein expression. We show that FPN1 mRNA expression is positively and negatively regulated in different macrophage populations following infection with M. avium and M. tuberculosis and stimulation with IFN-γ. Further, we localized FPN1 to multiple intracellular vesicle compartments and show that upon phagocytosis of M. tuberculosis, FPN1 and Nramp1 are present in the phagosome. These studies indicate that FPN1 expression is regulated by pathogenic mycobacteria and the key cytokine IFN-γ and likely plays a role in regulating iron availability in the mycobacterial phagosome.
MATERIALS AND METHODS
Mycobacteria
M. avium strain Mac 101 (ATCC 70998) was obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). Mycobacteria were cultured in Middlebrook 7H9 broth supplemented with oleic acid-albumin-dextrose-catalase (Difco, Detroit, MI, USA) at 37°C until mid-log phase. Bacteria were aliquoted in 1 ml amounts at 2 × 106 CFU/ml and stored frozen in 10% glycerol at −80°C until used. The number of bacteria was confirmed by plate-counting on 7H11 agar plates. γ-Irradiated M. tuberculosis, H37Rv (Colorado State University, Fort Collins, CO, USA; National Institutes of Health Contract NIAID-N01-AI-40091), was resuspended in PBS, briefly sonicated, and centrifuged at 800 rpm for 10 min to eliminate bacterial clumping. The protein concentration of the supernatant was determined via the Bradford protein assay (Bio-Rad, Hercules, CA, USA). Live M. tuberculosis H37Rv (ATCC 27294) was grown for 9–11 days on Middlebrook 7H11 agar. Immediately prior to the infection of macrophages with M. tuberculosis, bacteria were scraped from the plates into a 2.0-ml polypropylene tube containing two 3 mm glass beads and 1 ml DMEM medium. To reduce clumping, the samples were pulse-vortexed six times (∼1 s per pulse), and the resulting suspensions were kept stationary for 30 min to allow for the settling of clumps as described previously [35]. This bacterial suspension contained 1 × 108 bacteria/ml with minimal clumping (<10%). The number of bacteria present in the suspension was confirmed by counting in a Petroff–Hauser chamber.
Mice
Male C57/BL6 mice, 4–6 weeks of age, were purchased from Charles River Laboratories (Wilmington, MA, USA). They were allowed free access to water and fed a standard diet. The mice were treated according to the Institutional Laboratory Animal Care and Use Committee guidelines regarding the use of animals for research at The Ohio State University (OSU; Columbus, OH, USA).
Cell culture
The RAW 264.7 mouse macrophage cell line (ATCC TIB-71) and the AMJ2-C8 mouse alveolar cell line (ATCC CRL-2455) were obtained from ATCC. To generate a RAW264.7 cell line expressing wild-type Nramp1 (Nramp1Gly169), full-length, wild-type Nramp1 cDNA was first cloned into the p3XFlag-CMV-14 expression vector, which creates a C-terminal 3× Flag Nramp1 fusion protein. The insert from this expression vector was then subcloned by PCR into the NotI and EcoRV sites of the pcDNA3.1/Hygro(–) vector (Invitrogen, Carlsbad, CA, USA). The sequence was confirmed by DNA sequencing. RAW264.7 cells were transfected with Nramp1-3× Flag plasmid DNA using Lipopfectamine Plus (Invitrogen), and a stable clone was obtained by hygromycin selection and limiting dilution cloning. Clones were screened for expression of Nramp1-Flag by Western blotting using anti-Flag M2 mAb (Sigma Chemical Co., St. Louis, MO, USA).
For experiments, the macrophage cell lines were plated at 5 × 106 cells per well in six-well culture plates containing DMEM media supplemented with 10% FBS (Atlanta Biologicals, Lawrenceville, GA, USA) and penicillin-streptomycin. The macrophages were allowed to adhere for 4 h at 37°C in a 5% CO2 incubator. The nonadherent cells were then washed away with DMEM media without antibiotics, and the macrophages monolayers were treated with M. tuberculosis, as indicated in each experiment. At the same time, the macrophage monolayers were treated with 200 U/ml mouse IFN-γ alone or M. tuberculosis and 200 U/ml mouse IFN-γ. In some experiments, RAW264.7 cells were treated with polystyrene beads coated with human serum albumin (HSA; control) or Man-LAM from M. tuberculosis as described previously [36].
Bone marrow macrophage isolation
Bone marrow cells were isolated from the marrow of the femurs and tibias of C57/BL6 mice. The cells were plated in complete DMEM supplemented with 10 ng/ml GM-CSF (Peprotech, Rocky Hill, NJ, USA). On the 3rd day of culture, 50% of the medium was removed and replaced with fresh media supplemented with GM-CSF. On the 5th day of culture, 75% of the media was removed and replaced with fresh media supplemented with GM-CSF. Mature, adherent BMDM were obtained after 7 days of culture. The macrophages were then plated in six-well culture plates at a concentration of 4 × 106 macrophages per well. After adhering overnight, the macrophages were stimulated for 8 h with 100 μg/ml γ-irradiated M. tuberculosis, 200 units/ml IFN-γ, or a combination of IFN-γ and γ-irradiated M. tuberculosis.
Lung macrophage isolation
Mice were anesthetized with CO2, and the lungs were perfused via the heart with 0.02% EDTA in PBS. The lungs were removed and sliced into 1–2 mm pieces and then incubated in RPMI with 10% FBS, penicillin-streptomycin, 10 mM HEPES, collagenase (0.70 mg/ml), and DNase (50 U/ml) for 1.5 h at 37°C. A single-cell suspension was obtained by pipetting vigorously and washed twice with HBBS supplemented with 2% FBS. The cells were then resuspended in RPMI with 10% FBS, glutamine, penicillin-streptomycin, 1 mM pyruvate, and 10 mM HEPES. The macrophages were isolated by adherence to plastic by culturing the cells for 1.5 h at 37°C in 100 mm petri dishes at a concentration of 20 million cells/petri dish. The nonadherent cells were removed by washing three times in warm HBSS containing 2% FBS. The adherent cells were then removed by gently scrapping. The cells were then counted and plated at 4 × 106 macrophages per well in six-well culture plates. Following a 24-h incubation, the media were replaced, and the cells were stimulated as indicated.
RNA isolation
RAW264.7 monolayers were lysed using Qiagen lysis buffer containing 2-ME and homogenized by passing the cell lysates through QiaShredders (Qiagen, Valencia, CA, USA). The RNA was then isolated using the RNeasy mini kit (Qiagen). RNA was isolated from BMDM and lung macrophages using the High Pure RNA isolation kit (Roche, Indianapolis, IN, USA). In both procedures, residual DNA was removed during RNA purification by on-column DNase digestion using RNase-free DNase.
Quantitative RT-PCR (RT-PCR)
Total RNA (1 μg) was reversed-transcribed using 100 μM dNTPs, 15 units avian myloblastosis virus reverse transcriptase, and 0.5 μg oligo(dT)15 primer in RT buffer for 1 h at 42°C (Promega, Madison, WI, USA). The expression of mouse GAPDH and FPN1 mRNA was analyzed by real-time RT-PCR using SYBR green PCR master mix (Roche). Reactions were run on the Roche LightCycler 2. The following sense and antisense sequences were used: mouse GAPDH, sense (5′–3′) GTGTGAACGGA-TTTGGCCGTATTGGGCG, antisense (5′–3′) TCGCTCCTGGAAGATGGTGATG-GGC; mouse FPN1, sense (5′–3′) TGGATGGGTCCTTACTGTCTGCTAC, antisense (5′–3′) TGCTAATCTGCTCCTGTTTTCTCC. Primers used for GAPDH and FPN1 were designed using the MacVector primer software (Accelrys, San Diego, CA, USA). The amplification conditions were 95°C for 10 min followed by 40 cycles of 95°C for 15 s, 60°C for 5 s, and 72°C for 20 s. The relative expression of each sample was calculated using mouse GAPDH as a reference and the Δ comparative threshold method as described previously [37].
Immunofluorescence
RAW 264.7 cells were plated on coverslips in 24-well plates at a concentration of 2.5 × 105 cells/well. The cells were then treated with live M. tuberculosis and IFN-γ for 8 h. Cells were fixed with 4% paraformaldeyde for 20 min and permeabilized with 0.10% Triton X-100 for 10 min at room temperature. The cells were washed twice in PBS and blocked for 3 h at room temperature using a blocking solution containing 1% BSA and 10% heat-inactivated goat serum in PBS. Rabbit anti-mouse FPN1 antibody (Alpha Diagnostic, San Antonio, TX, USA) was absorbed twice with γ-irradiated M. tuberculosis overnight at 4°C to remove M. tuberculosis-reactive antibodies. Removal of M. tuberculosis antibodies was confirmed by Western blot of SDS lysates of M. tuberculosis and by immuofluorescence with M. tuberculosis adhered to coverslips. The absorbed antibody was then added to each well at a final concentration of 1:500 overnight. After washing, the secondary antibody, Alexa488-coupled (Fab′)2 goat anti-rabbit IgG antibody (Invitrogen), was added at room temperature for 1 h. To detect M. tuberculosis in the infected cells, coverslips were stained with auramine-rhodamine (Difco Laboratories, Detroit, MI, USA) and counterstained with 5% potassium permanganate as described previously [38]. Expression of FPN1 was then detected as described above. Coverslips were removed from the 24-well plates and mounted on slides with ProLong® Gold antifade reagent (Invitrogen). Fluorescence was visualized with a Zeiss Meta 510 confocal microscope at the OSU Campus Microscopy and Imaging Facility. Green fluorescence intensity was analyzed from images using Sigma ScanPro image analysis software (SPSS Science, Chicago, IL, USA).
For the RAW-Nramp1Gly169 experiments, the cells were plated on coverslips, and the presence of FPN1 and Nramp1 in the M. tuberculosis phagosome was determined as described. To detect Nramp1, anti-Flag M2 mAb was used as the primary antibody and Alexa488-coupled (Fab′)2 goat anti-mouse IgG antibody (Invitrogen) as the secondary antibody. To study the localization of Nramp1 and FPN1 to vesicular compartments, chicken anti-mouse early embryonic antigen 1 (EEA1) antibody (Zymed Laboratories, S. San Francisco, CA, USA; Invitrogen), rat anti-mouse cathepsin D mAb (R&D Systems, Minneapolis, MN, USA), and rat anti-lysosome-associated membrane protein 1 (Lamp1) mAb (Santa Cruz Biotechnology, Santa Cruz, CA, USA) were used followed by Alexa594-coupled secondary antibodies (Invitrogen). The percentage of Lamp1, cathepsin D, and EEA1 intracellular vesicles that express FPN1 or Nramp1 was determined from confocal images by counting a minimum of 300 vesicles per experiment.
Western blot analysis
RAW264.7 cells were lysed in RPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 0.50% sodium deoxycholate, 0.10% SDS) with protease mixture inhibitor tablets (Roche) and centrifuged at 19,000 g for 15 min at 4°C. Proteins (65 μg) were separated on 10% Tris-glycine gels and transferred to Hybond-P membranes (Amersham Biosciences, Piscataway, NJ, USA). Immunostaining was performed by incubation overnight with rabbit anti-FPN1 antibody (Alpha Diagnostic), absorbed 2× with irradiated M. tuberculosis and rabbit anti-GAPDH mAb (Cell Signaling Technology, Beverly, MA, USA). The secondary antibody detection step was performed with HRP-coupled anti-rabbit IgG (Amersham Biosciences). Blots were developed with the femto-Lucent chemiluminescence detection system (G-Biosciences, St. Louis, MO, USA).
Statistics
Results were analyzed by one-way ANOVA with Turkey’s test using SigmaSTAT (SPSS Science).
RESULTS
Infection of RAW264.7 cells with mycobacteria and stimulation with IFN-γ induce the FPN1 mRNA and protein expression
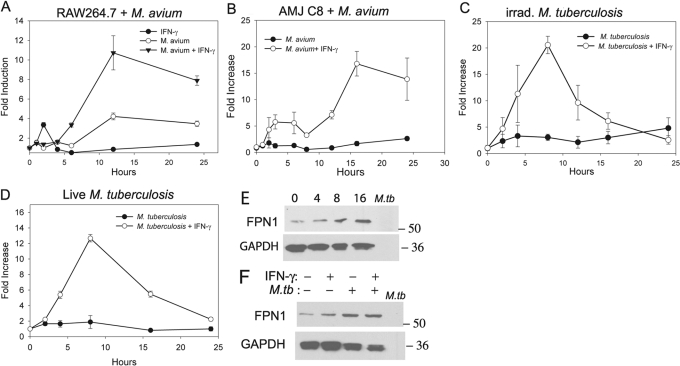
We first examined the expression of FPN1 mRNA in the mouse RAW264.7 macrophage cell line by real-time RT-PCR. The basal expression of FPN1 was low in these cells. The results in Figure 1A show that FPN1 mRNA expression is induced in RAW264.7 cells infected with M. avium and stimulated with IFN-γ, which was chosen based on its importance in activating macrophages. IFN-γ treatment of macrophages has been shown to increase the expression of MHCII as well as enhance the ability of macrophages to control the growth of M. tuberculosis via the induction of inducible NO synthase, which is responsible for the production of NO [39,40,41,42]. A 12-fold increase in the expression of FPN1 mRNA was observed after 12 h of stimulation. Treatment with M. avium alone resulted in a fourfold increase in expression, and stimulation with IFN-γ alone had no effect on FPN1 mRNA expression. M. avium infection and stimulation with IFN-γ also increased FPN1 mRNA expression in AMJ2-C8, a mouse alveolar macrophage cell line (Fig. 1B).
Fig. 1.
Effect of mycobacteria infection on FPN1 mRNA expression in RAW264.7 and AMJ2-C8 cells. (A) RAW264.7 cells were infected with M. avium [multiplicity of infection (MOI) 20:1] and stimulated with 200 units/ml IFN-γ. (B) AMJ2-C8 cells were treated with 20:1 M. avium and stimulated with 200 units/ml IFN-γ. (C) RAW264.7 cells were treated with γ-irradiated (irrad.) M. tuberculosis (100 μg protein/ml) and stimulated with IFN-γ (200 units/ml). (D) RAW264.7 cells were infected with live M. tuberculosis H37Rv (MOI 10:1) and stimulated with 200 units/ml IFN-γ. RNA was isolated at each time-point, and mRNA expression of FPN1 was determined by real-time RT-PCR. The results were normalized to GAPDH and expressed as the fold-increase in FPN1 expression compared with control, unstimulated cells. The results are the mean ± sd of three independent experiments. (E) Western blot of FPN1 expression in RAW264.7 cells stimulated with γ-irradiated M. tuberculosis (100 μg/ml) and IFN-γ (200 units/ml) for 0, 4, 8, and 16 h. (F) Western blot of FPN1 expression in RAW264.7 cells stimulated for 12 h with γ-irradiated M. tuberculosis (M.Tb; 100 μg/ml) and IFN-γ (200 units/ml). (E and F) Also included on the blots are SDS lysates of irradiated M. tuberculosis, which are the negative control for the removal of M. tuberculosis-reactive antibodies, and results are representative blots from three experiments.
Stimulation of RAW264.7 cells with γ-irradiated M. tuberculosis and IFN-γ also synergistically increased FPN1 mRNA expression (Fig. 1C). Expression peaked at 8 h of stimulation with a 30-fold increase and returned to baseline by 24 h. Treatment with γ-irradiated M. tuberculosis alone resulted in only a fivefold increase in FPN1 mRNA levels. Similarly, infection with live M. tuberculosis and stimulation with IFN-γ resulted in a 15-fold induction following 8 h of stimulation and only a fivefold increase when treated with live M. tuberculosis alone (Fig. 1D). Thus, these results show that live and γ-irradiated M. tuberculosis induces a similar level of FPN1 mRNA. We next determined if phagocytosis was sufficient in inducing FPN1 mRNA expression by treating RAW264.7 cells with polysyterene beads coated with HSA or Man-LAM. However, neither HSA beads nor Man-LAM beads altered FPN1 mRNA expression alone or in combination with IFN-γ (data not shown), indicating that the process of phagocytosis itself does not induce FPN1 expression.
FPN1 protein expression was measured by Western blotting using a rabbit anti-FPN1 antibody, which was absorbed twice with γ-irradiated M. tuberculosis to remove mycobacteria-reactive antibodies. Figure 1E shows that FPN1 protein expression in RAW264.7 is up-regulated by M. tuberculosis + IFN-γ at 8 and 16 h after stimulation. FPN1 protein expression was increased by M. tuberculosis alone and by the combination of M. tuberculosis and IFN-γ at 12 h after stimulation (Fig. 1F). However, in contrast to the mRNA levels, M. tuberculosis + IFN-γ did not increase FPN1 protein expression compared with M. tuberculosis alone.
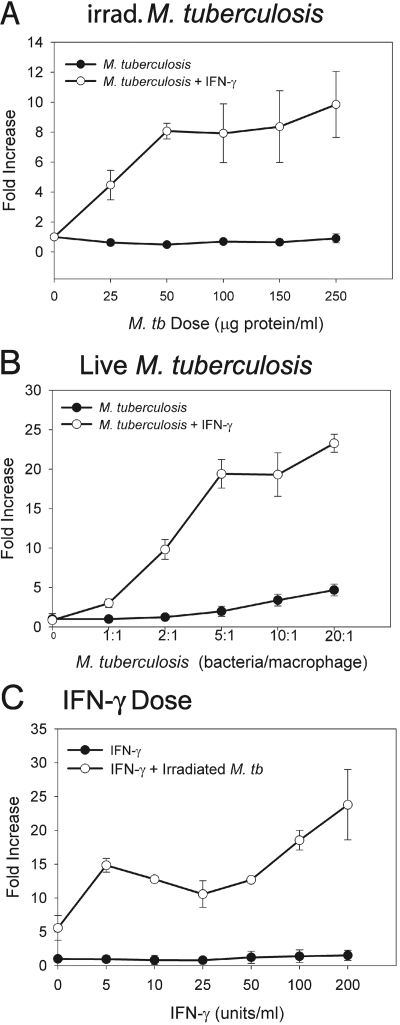
To determine if FPN1 mRNA is induced in a dose-dependent manner, we treated RAW264.7 cells with varying doses of γ-irradiated and live M. tuberculosis and stimulated with IFN-γ. Results in Figure 2A show that FPN1 mRNA expression increased with increasing doses of irradiated M. tuberculosis. Maximal induction of FPN1 mRNA expression was obtained at a concentration of 100 μg protein/ml γ-irradiated M. tuberculosis. FPN1 mRNA expression also increased with increasing doses of live M. tuberculosis (Fig. 2B). Peak levels of FPN1 mRNA were obtained with a MOI of 20 bacteria/macrophage. Results in Figure 2C show that the effect of IFN-γ on FPN1 expression in M. tuberculosis-infected macrophages is also dose-dependent.
Fig. 2.
Effect of varying doses of IFN-γ and mycobacteria on FPN1 mRNA expression. (A) RAW264.7 cells were treated with varying does of γ-irradiated M. tuberculosis and stimulated with 200 units/ml IFN-γ. (B) RAW264.7 cells were infected with a varying MOI of live M. tuberculosis and stimulated with 200 units/ml IFN-γ. (C) RAW264.7 cells were stimulated with varying doses of IFN-γ and treated with 100 μg protein/ml γ-irradiated M. tuberculosis. RNA was isolated from each treatment at 8 h, and expression of FPN1 mRNA was determined by real-time RT-PCR. The results were normalized to GAPDH and expressed as fold-increase in FPN1 expression compared with control, unstimulated cells. The results are the mean ± sd of three independent experiments.
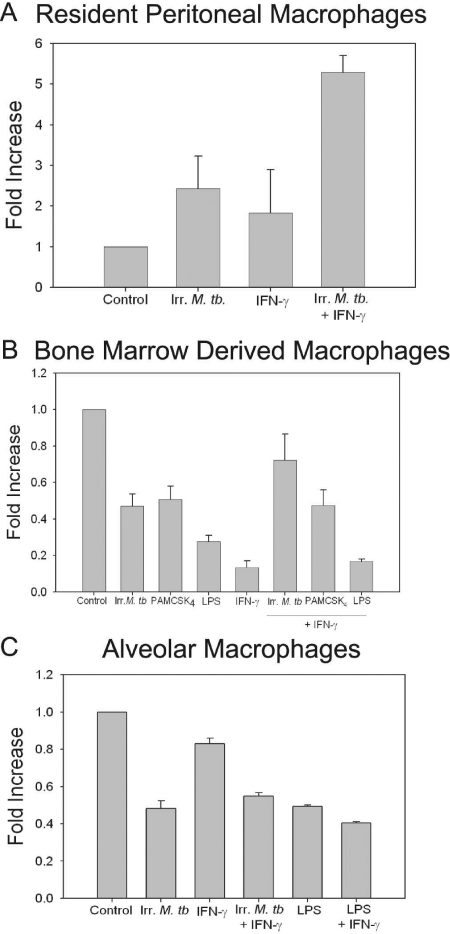
FPN1 mRNA is increased in resident peritoneal macrophages by M. tuberculosis and IFN-γ but not in BMDM or lung macrophages
We further examined the mRNA levels of FPN1 in three different macrophage subpopulations from C57BL/6J mice. Constitutive expression of FPN1 mRNA in unstimulated resident peritoneal macrophages was low. Treatment with γ-irradiated M. tuberculosis and stimulation with IFN-γ resulted in a fivefold increase in FPN1 mRNA compared with a twofold increase with γ-irradiated M. tuberculosis alone (Fig. 3A). In contrast, expression of FPN1 mRNA was constitutively high in BMDM and lung macrophages. Expression of FPN1 mRNA in these cells was inhibited following treatment with γ-irradiated M. tuberculosis or IFN-γ (Fig. 3B). Expression was also inhibited by the TLR2 ligand PAM3CSK4 and the TLR4 ligand LPS, consistent with previous reports in the literature [25, 26]. When the cells were treated with the combination of IFN-γ and M. tuberculosis or the TLR ligands, IFN-γ did not change the down-regulation of FPN1 expression (Fig. 3B). Stimulation of lung macrophages with γ-irradiated M. tuberculosis or LPS also resulted in a down-regulation of FPN1 mRNA expression (Fig. 3C). Addition of IFN-γ did not alter the down-regulation by M. tuberculosis or LPS.
Fig. 3.
Expression of FPN1 mRNA in primary mouse macrophages. (A) Resident peritoneal macrophages from C57/BL6 mice were treated with γ-irradiated (Irr.) M. tuberculosis (100 μg/ml) and stimulated with 200 units/ml IFN-γ. (B) BMDM from C57/BL6 mice were treated with γ-irradiated M. tuberculosis (100 μg/ml), palmitoyl-3-cysteine-serine-lysine-4 (Pam3CSK4; 500 ng/ml), or LPS (100 ng/ml) and stimulated with 200 units/ml IFN-γ. (C) Alveolar macrophages from C57/BL6 mice were treated with γ-irradiated M. tuberculosis (100 μg/ml) or LPS (100 ng/ml) and stimulated with 200 units/ml IFN-γ. RNA was isolated from each treatment, and expression of FPN1 mRNA was determined by real-time RT-PCR. The results were normalized to GAPDH and expressed as fold-increase in FPN1 mRNA expression compared with control, unstimulated cells. The results are the mean ± sd of three independent experiments.
FPN1 localizes to the mycobacteria-containing phagosomes
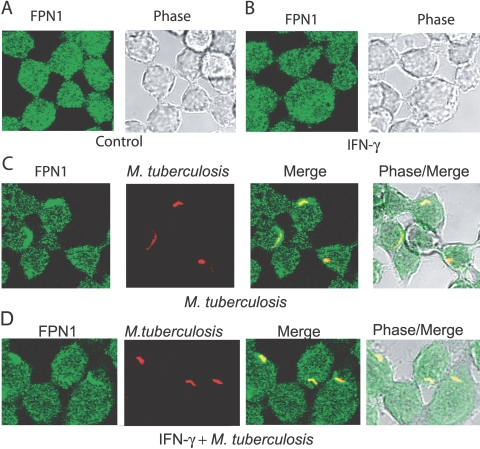
Previous studies [20, 21] have shown that FPN1 in macrophages is present in intracellular vesicles and upon phagocytosis of senescent erythrocytes, traffics to the plasma membrane. As trafficking of FPN1 during phagocytosis of a pathogen has not been studied, we examined FPN1 expression by confocal microscopy in RAW264.7 cells infected with live M. tuberculosis and stimulated with IFN-γ. We observed a punctuate pattern of fluorescence in unstimulated RAW264.7 cells, consistent with localization of FPN1 within intracellular vesicles (Fig. 4A). When the RAW264.7 cells were treated with IFN-γ, the intensity of FPN1 immunfluorescence was comparable with control, unstimulated RAW264.7 cells (Fig. 4B). Pretreatment of the rabbit anti-FPN1 antibody with a blocking peptide abolished activity (no fluorescence), indicating that the immunofluorescence was specific (not shown). Also, controls without primary antibody showed no fluorescence (not shown).
Fig. 4.
Localization of FPN1 to the mycobacteria-containing phagosomes. RAW264.7 cells were stimulated with IFN-γ (200 u/ml), live M. tuberculosis (MOI 5:1), and IFN-γ + live M. tuberculosis for 8 h. M. tuberculosis was detected by staining with auramine-rhodamine. FPN1 was detected by cross-sectional confocal microscopy using rabbit anti-mouse FPN1 antibody. The secondary antibody was Alexa488-coupled (Fab′)2 goat anti-rabbit IgG. Representative confocal images of unstimulated control RAW264.7 cells (A), RAW264.7 cells stimulated with IFN-γ (B), RAW264.7 cells infected with live M. tuberculosis (C), and RAW264.7 cells infected with live M. tuberculosis and stimulated with IFN-γ (D). (A and B) RAW264.7 cells with FPN1 contained within small, intracellular vesicles. (C and D) FPN1 localized to the M. tuberculosis containing phagosomes. Data are representative of three experiments.
In RAW264.7 cells infected with M. tuberculosis, FPN1 was highly localized to the M. tuberculosis phagosome (Fig. 4C). FPN1 also localized to the M. tuberculosis phagosome in RAW264.7 cells infected with M. tuberculosis and stimulated with IFN-γ (Fig. 4D). Again, pretreatment of the rabbit anti-FPN1 antibody with a blocking peptide abolished activity (no fluorescence), indicating that the immunofluorescence was specific (not shown). Quantitative analysis showed that 80–90% of the phagosomes were positive for FPN1 in the cells infected with M. tuberculosis alone and those infected with M. tuberculosis and stimulated with IFN-γ. The fluorescence intensity of FPN1 in phagosomes stimulated with M. tuberculosis + IFN-γ was 1.23 ± 0.13 × 105 green fluorescence intensity units/phagosome compared with 0.742 ± 0.059 × 105 for phagosomes in macrophages infected with M. tuberculosis alone (P<.005). Although the fluorescence intensity is significantly higher in the cells stimulated with M. tuberculosis + IFN-γ, the magnitude of the increase (1.7×) was much less than we observed in FPN1 mRNA expression. These results provide evidence that FPN1 localizes to the phagosome following phagocytosis of M. tuberculosis, a process that is enhanced by simultaneous stimulation with IFN-γ.
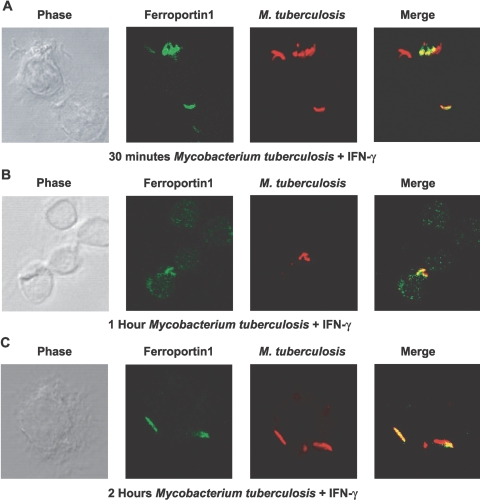
To examine the trafficking of FPN1 to the mycobacteria-containing phagosomes, we performed a time-course experiment following infection of RAW264.7 cells with M. tuberculosis (Fig. 5). FPN1 rapidly localized to M. tuberculosis-containing phagosomes with partial colocalization by 30 min of infection and stimulation with IFN-γ (Fig. 5A). As shown in Figure 5A, mycobacteria not phagocytosed by the macrophage stained with auramine-rhodamine but were not positive for FPN1, which was almost completely associated with the M. tuberculosis-containing phagosomes by 1 h (Fig. 5B) and completely associated following 2 h of infection (Fig. 5C). By 2 h, ∼90% of the phagosomes were positive for FPN1.
Fig. 5.
Trafficking of FPN1 to the mycobacteria-containing phagosomes. RAW264.7 cells were infected with live M. tuberculosis for 30 min, 1 h, and 2 h. Detection of M. tuberculosis was carried out with auramine-rhodamine and counterstained with 5% potassium permanganate. FPN1 was detected by immunofluorescence using rabbit anti-mouse FPN1 primary antibody. The secondary antibody was an Alexa488-coupled (Fab′)2 goat anti-rabbit IgG antibody. Confocal microscopy images shown are representative of three independent experiments.
FPN1 and Nramp1 localize to the mycobacteria-containing phagosomes in RAW264.7 cells expressing wild-type Nramp1
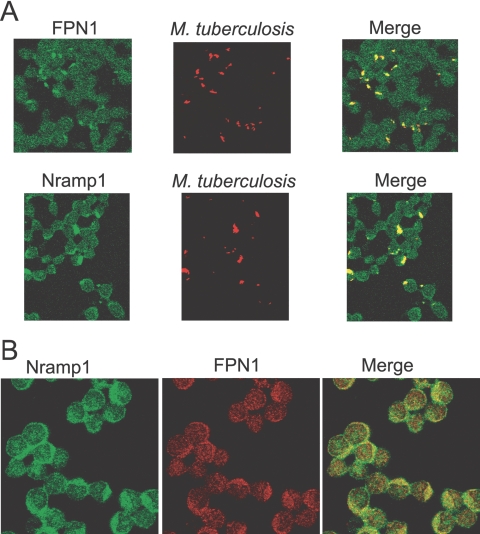
As previous studies [11, 12] have shown that the iron transport protein Nramp1 also localizes to phagosomes, we determined whether FPN1 and Nramp1 are present in the phagosome following a 2-h infection with M. tuberculosis. As RAW264.7 cells express mutant Nramp1Asp169, which does not leave the endoplasmic reticulum (ER) [43, 44], we used a RAW264.7 cell line that constitutively expresses wild-type Nramp1Gly169 tagged with Flag at the C terminus. As shown in Figure 6A, Nramp1 and FPN1 are present in the M. tuberculosis phagosome.
Fig. 6.
Trafficking of Nramp1 and FPN1 to mycobacteria-containing phagosomes in RAW264.7 cells that express wild-type Nramp1Gly169. (A) RAW.264.7-Nramp1Gly169 cells were infected with live M. tuberculosis for 2 h. Detection of M. tuberculosis was carried out with auramine-rhodamine and counterstained with 5% potassium permanganate. FPN1 was detected by immunofluorescence using rabbit anti-mouse FPN1 as a primary antibody and Alexa488-coupled (Fab′)2 goat anti-rabbit IgG antibody as a secondary antibody. Nramp1Gly169 was detected using mouse anti-Flag mAb as a primary antibody and Alexa488-coupled (Fab′)2 goat anti-mouse IgG antibody as a secondary antibody. (B) Colocalization of Nramp1 and FPN1 in noninfected RAW.264.7-Nramp1Gly169 cells. Nramp1Gly169 was detected by immunofluorescence using mouse anti-Flag mAb as a primary antibody and Alexa488-coupled (Fab′)2 goat anti-mouse IgG antibody as a secondary antibody. FPN1 was detected using rabbit anti-mouse FPN1 as a primary antibody and Alexa594-coupled (Fab′)2 goat anti-rabbit Ig antibody as a secondary antibody. Confocal microscopy images shown are representative of three independent experiments.
FPN1 and Nramp1 are present in the late endosome and lysosomal compartments
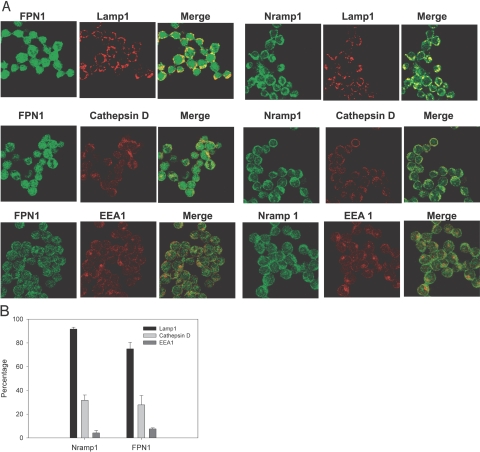
We first determined if Nramp1 and FPN1 are expressed in the same vesicular compartments in the RAW264.7-Nramp1Gly169 cells. Nramp1 highly localized with FPN1 in vesicular compartments in noninfected cells (Fig. 6B); however, FPN1 is also present in vesicular compartments that do not contain Nramp1. Likewise, Nramp1 is also present in vesicles that do not contain FPN1. We next determined whether FPN1 and Nramp1 are present in early endosomes, late endosomes, and lysosomes in the RAW264.7-Nramp1Gly169 cells. Nramp1 highly localized with Lamp1, a marker of late endosomes and lysosomes, and partially localized with cathepsin D, a lysosomal marker (Fig. 7A). Quantitative analysis showed that ∼90% of the Lamp1-positive vesicles and 30% of the cathepsin D-positive vesicles contained Nramp1. This localization of Nramp1 to late endosomes and lysosomes is consistent with previous studies [11, 12]. FPN1 also localized to the Lamp1-positive vesicles and cathepsin D vesicles (Fig. 7, A and B). To determine localization to early endosomes, we used antisera to EEA1. Nramp1 and FPN1 showed little colocalization with EEA1 (Fig. 7A). Less than 10% of the EEA1-positive vesicles contained Nramp1 or FPN1 (Fig. 7B). The above data suggest that although Nramp1 and FPN1 are present in the late endosomal compartments, FPN1 is also expressed highly in intracellular vesicular compartments that are not positive for the early endosome marker EEA1 or the late endosome markers Lamp1 and cathepsin D.
Fig. 7.
Localization of FPN1 and Nramp1 in intracellular vesicular compartments. (A) RAW-264.7-Nramp1Gly169 cells were cultured overnight on coverslips and then fixed and permeablized. FPN1 was detected with a rabbit anti-FPN1 primary antibody and Alexa488-coupled (Fab′)2 goat anti-rabbit Ig secondary antibody. Nramp1Gly169 was detected with a mouse anti-Flag mAb and Alexa488-coupled (Fab′)2 goat anti-mouse IgG secondary antibody. Late endosomes and lysosomes were detected using rat anti-Lamp1 and rat anticathepsin D antibodies as primary antibodies and an Alexa594-coupled (Fab′)2 goat anti-rat IgG secondary antibody. Early endosomes were detected with a chicken anti-EEA1 antibody as a primary antibody and Alexa594-coupled goat anti-chicken IgG secondary antibody. Controls stained with secondary antibodies alone were negative (not shown). Results are representative confocal images of three to four independent experiments. (B) Quantitative analysis of confocal images. Results shown are the percentage of Lamp1, cathepsin D, and EEA1-positive vesicles that are also positive for Nramp1 or FPN1. Results are mean ± sd of three independent experiments.
DISCUSSION
Previous studies have shown that FPN1 expression in mouse macrophages is negatively regulated by LPS and the gram-negative bacteria Pseudomonas aeruginosa [45]. In this report, we demonstrate that FPN1 mRNA expression is differentially expressed in macrophage subpopulations and macrophage cell lines following infection with mycobacteria and stimulation with IFN-γ. Two patterns of expression were observed. In BMDM and lung macrophages, FPN1 mRNA expression was down-regulated. In resident peritoneal macrophages and macrophage cell lines, RAW264.7 and AMJ2-C8, infection with mycobacteria, and stimulation with IFN-γ synergistically increased FPN1 mRNA. Thus, in these cells, FPN1 expression is increased by the classical, two-signal pattern of macrophage activation, one signal being initiated by the interaction of the mycobacteria with the macrophage, and the second signal being initiated by IFN-γ. One possible explanation for the different results is that the different macrophage populations are engaging different TLRs. This does not appear likely, as a TLR2 ligand (Pam3CSK4) and a TLR4 ligand (LPS) also inhibited FPN1 expression in the BMDM. This differential expression may relate to the basal level of FPN1 mRNA in unstimulated macrophages. Resident peritoneal macrophages, RAW264.7 cells, and AMJ2-C8 cells have low levels of constitutive mRNA expression of FPN1. BMDM and lung macrophages have high constitutive mRNA expression. Another possible explanation for differential expression of FPN1 mRNA in macrophage populations results from differences in transcriptional activation of the FPN1 gene in unstimulated and activated macrophages. Although the 5′ upstream region of the mouse FPN1 gene has been shown to have promoter activity [46], the signaling pathways and transcription factors involved in regulating expression are unknown.
In this study, we also examined FPN1 protein expression in RAW264.7 cells by confocal microscopy and Western blotting. In unstimulated cells, a constitutive level of FPN1 expression was observed that localized to intracellular vesicles. In cells infected with M. tuberculosis and stimulated with IFN-γ, a small (1.7×) increase in FPN1 immunofluorescence in the phagosome was observed when compared with cells that were only infected with M. tuberculosis. However, there was no increase in protein expression detected by Western blotting. Thus, this increase in FPN1 immunofluorescence is not a result of an overall increase in protein expression. One possible explanation is that more FPN1-positive vesicles have fused to phagosomes in the IFN-γ-stimulated cells. The Western blots and confocal microscopy show that although there is an increase in FPN1 mRNA expression in RAW264.7, there is no corresponding increase in protein expression. This suggests that FPN1 is also post-transcriptionally regulated in these cells. Previous studies have shown that FPN1 protein expression is regulated post-transcriptionally by intracellular iron levels. The mRNA of FPN1 contains an iron responsive element (IRE) in the 5′ untranslated region (UTR) that has been shown to confer translational regulation by iron through the iron regulatory proteins in a manner similar to ferritin and other 5′UTR-IRE-regulated genes [17, 19, 46]. IFN-γ has been shown to down-regulate macrophage iron levels by decreasing expression of the transferrin receptor and ferritin synthesis [12, 47,48,49]. Thus, the difference in FPN1 mRNA and protein expression in the RAW264.7 cells following infection and stimulation with IFN-γ is most likely a result of the activity of the IRE in the 5′UTR of the FPN1 mRNA, which under low iron, represses translation of the protein.
Previous studies have shown that FPN1 is expressed in intracellular vesicles, and upon phagocytosis of senescent erythrocytes, it localizes to the cell membrane [2, 20, 22]. However, the intracellular vesicular compartments containing FPN1 have not been identified Here, we show that FPN1 is expressed in several vesicular compartments. It was found to colocalize with Nramp1 in late endosomes/lysosomes but is also present in other intracellular vesicles. After phagocytosis of M. tuberculosis, we also show that FPN1 is more highly localized to the phagosome than to the cell membrane of infected cells. The low level of trafficking of FPN1 to the cell membrane is possibly a result of the low levels of iron in these cells. Previous studies [20, 21] have shown that iron loading of macrophages induces trafficking of FPN1 to the plasma membrane, which suggests that the trafficking of FPN1 to the plasma membrane is regulated by a signal pathway that is activated by intracellular iron levels. Thus, we speculate that there are two pathways by which FPN1 translocates in macrophages. One pathway delivers FPN1 from intracellular vesicles to the plasma membrane when intracellular iron levels are high. The second pathway involves late endosomes/lysosomes that fuse with the phagososome. As intracellular iron levels decrease following phagocytosis of M. tuberculosis and stimulation with IFN-γ, FPN1 traffics primarily to the phagosome, and there is little trafficking of FPN1 to the plasma membrane
Previous studies have shown that the iron transporter, Nramp1 (SLC11a1), is also present in late endosomal vesicles [11, 12] that fuse with the phagosome. The Nramp1 gene in mice has two alleles that determine resistance to a range of intracellular pathogens: a wild-type-resistant Nramp1Gly169 allele and a mutant-susceptible Nramp1Asp169 allele [8, 9]. Previous studies [12, 14] have shown Nramp1 to localize to phagosomes containing M. avium in macrophages that express the wild-type Nramp1Gly169 allele, whereas Nramp1 is retained within the ER in macrophages that express the mutant Nramp1Asp169 allele [43, 44]. In the current study, we showed that Nramp1 colocalizes with FPN1 in late endosomes/lysosome vesicles in RAW264.7 cells that express the wild-type Nramp1Gly169. We also report that FPN1 and Nramp1 are present in the M. tuberculosis phagosome within 2 h after infection.
The presence of both iron transport proteins in the mycobacteria-containing phagosome raises the question of what their functions are during iron transport. Our lab was the first to show that Nramp1 transports iron into phagosomes and mediates resistance to pathogens by catalyzing the production of hydroxyl radicals by the Fenton reaction [13, 14, 50]. An opposite conclusion was reached by Jabado et al. [15], suggesting that Nramp1 transports iron out of the phagosome and mediates resistance to pathogens by removing iron required for growth. In this model, Nramp1 functions in a similar manner as Nramp2, which transports iron out of recycling endosomes. However, expression of Nramp1 in frog oocytes suggests that Nramp1 is a pH-dependent antiporter that fluxes divalent metals (Fe+2, Mn+2, Zn+2) against a proton gradient and can transport divalent cations in either direction dependent on proton and divalent cation concentrations [16]. In contrast, Nramp2 is a pH-dependent symporter that transports bivalent metals with a proton gradient [5]. In a recent yeast complementation study, Techau et al. [51] showed that SLC11a1 (Nramp1) but not SLC11a2 (Nramp2) expression rescues a divalent cation stress phenotype in mutant bsd2A/rer1Δ yeast. In contrast, SLC11a2 but not SLC11a1 can complement EGTA sensitivity in mutant smf1Δ/smf2Δ/smf3 yeast. Interpretation of these studies is that SLC11a1 rescues the divalent cation stress by transporting divalent cations out of the yeast cells, and SLC11a2 rescues EGTA sensitivity by transporting divalent cations into the yeast cells. The discrepancy in the Nramp1 transport studies can best be explained by the ability of Nramp1 to transport divalent cations in either direction, dependent on the divalent cation concentration. Thus, Nramp1 would be expected to transport iron into phagosomes with low iron content, such as phagosomes containing pathogens, and transport iron out of phagosomes with high iron content, such as phagosomes containing senescent erythrocytes. This bidirectional transport of divalent cations would explain why we were able to measure iron transport into mycobacteria-containing phagosomes [13, 14], and Jabado et al. [15] was able to measure Mn+2 transport out of phagosomes containing zymosan particles loaded with bound Mn+2.
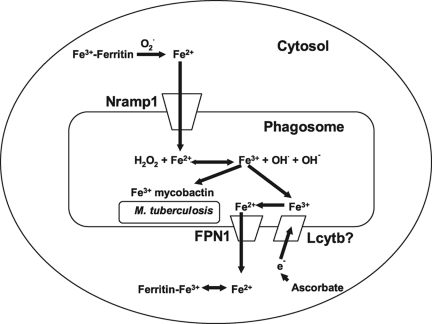
Expression of FPN1 in oocytes results in iron export out of the oocyte, confirming that FPN1 is an iron export protein [17, 18]. Thus, the presence of FPN1 in the phagosomal membrane suggests that FPN1 may be involved in the export of iron from the phagosome into the cytosol. We favor a model (Fig. 8) in which Nramp1 and FPN1 act together to regulate phagosomal iron levels. In this model, Fe+2 is transported into the mycobacteria- containing phagosome, where the iron catalyzes the production of highly toxic hydroxyl radicals. As the result of the Fenton reaction, the iron is oxidized to Fe+3. M. tuberculosis expresses mycobactin, a siderophore that captures Fe+3 [52, 53]. Thus, it is critical that the oxidized iron be removed from the phagosome to prevent growth of surviving mycobacteria. We propose that FPN1 is the principal iron exporter from the phagosome. If the iron concentrations are high enough, Nramp1 may also be involved in iron export. Nramp1 and FPN1 transport Fe+2. Thus, we would expect that the phagosomal membrane also contains a ferrireductase that is closely associated with the iron transport proteins. This ferrireductase remains to be identified. A possible candidate is the recently characterized ferrireductase Lcytb, which is expressed in lysosomal vesicles and is a member of the cytochrome b561 protein family of ascorbate-dependent ferrireductases [54, 55].
Fig. 8.
Proposed model for action of FPN1 and Nramp1 in the mycobacteria-containing phagosome. Iron (Fe+2) is transported from the intracellular iron pool into the phagosome by Nramp1, possibly facilitated by the release of Fe+2 from ferritin through the action of reactive oxygen species. In the phagosome, iron catalyzes the production of hydroxyl radicals via the Fenton reaction, resulting in the oxidation of Fe+2 to Fe+3. As Fe+3 can be acquired by M. tuberculosis via the mycobacterial mycobactins, Fe+3 needs to be recycled out of the phagosome quickly. In this model, Fe+3 is reduced by a phaogsomal membrane-associated ferrireductase, possibly Lcytb, and exported out of the phagosome by FPN1 into the cytoplasm, where it can reassociate with ferritin.
The expectation is that the ability of Nramp1 and FPN1 to cycle iron through the phagosome would be most efficient in activated macrophages. Not only is expression of Nramp1 and FPN1 up-regulated by IFN-γ and infection with mycobacteria (this paper and ref. [24]), but also, IFN-γ activation results in the acidification of the phagosome [56, 57] and increased NADPH oxidase expression [58, 59]. Recently, we also showed that hepcidin is induced by mycobacteria and IFN-γ and also locates to mycobacteria-containing phagosomes [34]. However, the appearance of hepcidin in the phagosome is much later than FPN1 and Nramp1 expression, peaking at 24 h after infection of the macrophage. We reason that hepcidin in the phagosome primarily acts as an antimicrobial peptide. However, we showed that hepcidin can also be secreted by macrophages and thus, could affect the iron homeostasis of nearby cells by interacting with and causing the degradation of cell surface-expressed FPN1 [33].
In conclusion, we demonstrate that FPN1 expression is differentially expressed in macrophage populations, and expression is regulated at multiple levels, including mRNA expression, protein expression, and trafficking. We also show that FPN1 localizes to the M. tuberculosis phagosome, where it may have antimycobacterial activity by limiting iron available to the mycobacteria. Although the antimycobacterial activity of FPN1 needs to be determined experimentally, FPN1 has recently been shown to have antibacterial activity against the intracellular growth of Salmonella enterica [60]. Further studies to determine if FPN1 is expressed in macrophages during tuberculosis may lead to development of a strategy to up-regulate expression of FPN1, which may contribute to the treatment of diseases caused by M. tuberculosis.
Acknowledgments
This work was supported by National Institutes of Health Grants AI45673 and DK57667. We are grateful to Gail Alvarez for technical assistance.
References
- Ratledge C. Iron, mycobacteria and tuberculosis. Tuberculosis (Edinb) 2004;84:110–130. doi: 10.1016/j.tube.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Chung J, Haile D J, Wessling-Resnick M. Copper-induced ferroportin-1 expression in J774 macrophages is associated with increased iron efflux. Proc Natl Acad Sci USA. 2004;101:2700–2705. doi: 10.1073/pnas.0306622101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratosin D, Mazurier J, Tissier J P, Slomianny C, Estaquier J, Russo-Marie F, Huart J J, Freyssinet J M, Aminoff D, Amesisen J C, Montreuil J. Molecular mechanisms of erythrophagocytosis. Characterization of the senescent erythrocytes that are phagocytized by macrophages. C R Acad Sci III. 1997;320:811–818. doi: 10.1016/s0764-4469(97)85017-2. [DOI] [PubMed] [Google Scholar]
- Bothwell T H. Overview and mechanisms of iron regulation. Nutr Rev. 1995;53:237–245. doi: 10.1111/j.1753-4887.1995.tb05480.x. [DOI] [PubMed] [Google Scholar]
- Gunshin H, Mackenzie B, Berger U V, Gunshin Y, Romero M F, Boron W F, Nussberger S, Golian J L, Hediger M A. Coning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- Crichton R R, Wilmet S, Legssyer R, Ward R J. Molecular and cellular mechanisms of iron homeostasis and toxicity in mammalian cells. J Inorg Biochem. 2002;91:9–18. doi: 10.1016/s0162-0134(02)00461-0. [DOI] [PubMed] [Google Scholar]
- Gruenheid S, Cannonne-Hergaux F, Gauthier S, Hackman D J, Grinstein S, Gros P. The iron transport protein NRAMP2 is an integral membrane glycoprotein that colocalizes with transferrin in recycling endosomes. J Exp Med. 1999;189:831–841. doi: 10.1084/jem.189.5.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal S M, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell. 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- Vidal S, Tremblay M L, Govoni G, Gauthier S, Sebastini G, Malo D, Skamene E, Oliver S, Jothy S, Gros P. The Ity/Lsh/Bcg locus: natural resistance to infection with intracellular parasites is abrogated by disruption of the Nramp1 gene. J Exp Med. 1995;182:655–666. doi: 10.1084/jem.182.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govoni G, Gauthier S, Billia F, Iscone N N, Gros P. Cell specific and inducible Nramp1 gene expression in mouse macrophages in vitro and in vivo. J Leukoc Biol. 1997;62:277–286. doi: 10.1002/jlb.62.2.277. [DOI] [PubMed] [Google Scholar]
- Gruenheid S, Pinner E, Desjardins M, Gros P. Natural resistance to infection with intracellular pathogens: the Nramp1 protein recruited to the membrane of the phagosome. J Exp Med. 1997;185:717–730. doi: 10.1084/jem.185.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle S, Bright N A, Roach T I A, Atkinson P G P, Barton C H, Melon R H, Blackwell J M. Localization of Nramp1 in macrophages: modulation with activation and infection. J Cell Sci. 1998;111:2855–2866. doi: 10.1242/jcs.111.19.2855. [DOI] [PubMed] [Google Scholar]
- Kuhn D E, Baker B D, Lafuse W P, Zwilling B S. Differential iron transport into phagosomes isolated from the RAW264.7 macrophage cell lines transfected with Nramp1Gly169 or Nramp1Asp169. J Leukoc Biol. 1999;66:113–119. doi: 10.1002/jlb.66.1.113. [DOI] [PubMed] [Google Scholar]
- Kuhn D E, Lafuse W P, Zwilling B S. Iron transport into Mycobacterium avium containing phagosomes from an Nramp1Gly169 transfected RAW264.7 macrophage cell line. J Leukoc Biol. 2001;69:43–49. [PubMed] [Google Scholar]
- Jabado N, Jankowski A, Dougaparsad S, Picard V, Grinstein S, Gros P. Natural resistance to intracellular infections: natural resistance-associated macrophage protein (NRAMP1) functions as a pH-dependent manganese transporter at the phagosomal membrane. J Exp Med. 2000;192:1237–1248. doi: 10.1084/jem.192.9.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami T, Bhattachargee A, Bapal P, Searle S, Moore E, Li M, Blackwell J M. Natural resistance associated macrophage protein 1 is a H+/bivalent cation antiporter. Biochem J. 2001;354:511–519. doi: 10.1042/0264-6021:3540511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKie A T, Marciani P, Rolfs A, Brennan K, Wehr K, Barrow D, Miret S, Bomford A, Peters T J, Farzaneh F, Hediger M A, Hentze M W, Simpson R J. A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron into the circulation. Mol Cell. 2000;5:299–309. doi: 10.1016/s1097-2765(00)80425-6. [DOI] [PubMed] [Google Scholar]
- Donovan A, Brownlie A, Zhou Y, Shepard J, Pratt S J, Moynihan J, Paw B H, Drejer A, Barut B, Zapata A, Law T C, Brugnara C, Lux S E, Pinkus G S, Pinkus J L, Kingsley P D, Palis J, Fleming M D, Andrews N C, Zon L I. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature. 2000;403:776–781. doi: 10.1038/35001596. [DOI] [PubMed] [Google Scholar]
- Abboud S, Haile D J. A novel mammalian iron regulated protein involved in intracellular iron metabolism. J Biol Chem. 2000;275:19906–19912. doi: 10.1074/jbc.M000713200. [DOI] [PubMed] [Google Scholar]
- Delaby C, Pilard N, Gonclaves A S, Beaumont C, Cannone-Hergaux F. Presence of the iron exporter ferroportin at the plasma membrane of macrophages is enhanced by iron loading and down-regulated by hapcidin. Blood. 2005;106:3979–3984. doi: 10.1182/blood-2005-06-2398. [DOI] [PubMed] [Google Scholar]
- Knutson M D, Vafa M R, Haile D J, Wessling-Resnick M. Iron loading and erythrophagocytosis increases ferroportin 1 (FPN1) expression in J774 macrophages. Blood. 2003;102:4191–4197. doi: 10.1182/blood-2003-04-1250. [DOI] [PubMed] [Google Scholar]
- Knutson M D, Ukka M, Koss L M, Aydemir F, Wessling-Resnick M. Iron release from macrophages after erythrophagocytosis is upregulated by ferroportin1 overexpression and down-regulated by hepcidin. Proc Natl Acad Sci USA. 2005;102:1324–1328. doi: 10.1073/pnas.0409409102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miethke M, Marahiel M A. Siderophore-based iron acquisition and pathogen control. Microbio Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Lafuse W P, Zwilling B S. Infection with Mycobacterium avium differentially regulates the expression of iron transport protein mRNA in murine peritoneal macrophages. Infect Immun. 2001;69:6618–6624. doi: 10.1128/IAI.69.11.6618-6624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Liu X B, Quinones M, Melby P, Ghio A, Haile D J. Regulation of reticuloendothelial iron transporter MTP1 (Slc11a3) by inflammation. J Biol Chem. 2002;277:39786–39791. doi: 10.1074/jbc.M201485200. [DOI] [PubMed] [Google Scholar]
- Liu X B, Yang F, Haile D J. Functional consequences of ferroportin1 mutations. Blood Cells Mol Dis. 2005;35:33–46. doi: 10.1016/j.bcmd.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Ludwiczek S, Aigner E, Theurl I, Weiss G. Cytokine-mediated regulation of iron transport in human monocytic cells. Blood. 2003;101:4148–4154. doi: 10.1182/blood-2002-08-2459. [DOI] [PubMed] [Google Scholar]
- Anderson G J, Frazer D M, Wilkins S J, Becker E M, Millard K N, Murphy T L, McKie A T, Vulpe C D. Relationship between intestinal iron-transporter expression, hepatic hepcidin levels and the control of iron absorption. Biochem Soc Trans. 2002;30:724–726. doi: 10.1042/bst0300724. [DOI] [PubMed] [Google Scholar]
- Nicolas G, Ghauvet C, Viatte L, Danan J L, Biggard X, Devaux I, Beaumont C, Kahn A, Vaulont S. The gene that codes for hepcidin, an iron regulatory hormone, is regulated by anemia, hypoxia, and inflammation. J Clin Invest. 2002;110:1037–1044. doi: 10.1172/JCI15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigeon C, Ilyin G, Courselaud B, Leroyer P, Turlin B, Brissot P, Loreal O. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem. 2001;276:7811–7819. doi: 10.1074/jbc.M008923200. [DOI] [PubMed] [Google Scholar]
- Laftah A H, Ramesh B, Simpson R J, Solansky N, Bahram S, Schumann K, Debnam E S, Srai S K S. Effect of hepcidin on intestinal iron absorption in mice. Blood. 2004;103:3940–3944. doi: 10.1182/blood-2003-03-0953. [DOI] [PubMed] [Google Scholar]
- Yamaji S, Sharp P, Ramesh B, Srai S K S. Inhibition of iron transport across human intestinal epithelial cells by hepcidin. Blood. 2004;104:2178–2180. doi: 10.1182/blood-2004-03-0829. [DOI] [PubMed] [Google Scholar]
- Nemeth E, Tuttle M S, Powelson J, Vaughn M B, Donovan A, Ward D M, Gantz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- Sow F B, Florence W C, Satoskar A R, Schlesinger L S, Zwilling B S, Lafuse W P. Expression and localization of hepcidin in macrophages: a role in host defense against tuberculosis. J Leukoc Biol. 2007;82:934–945. doi: 10.1189/jlb.0407216. [DOI] [PubMed] [Google Scholar]
- Schlesinger L S, Bellinger-Kawahara C G, Payne N R, Horwitz M A. Phagocytosis of Mycobacterium tuberculosis is mediated by human monocyte complement receptors and complement component C3. J Immunol. 1990;144:2771–2780. [PubMed] [Google Scholar]
- Schlesinger L S, Hull S R, Kaufman T M. Binding of the terminal mannosyl units of lipoarabinomannan from a virulent strain of Mycobacterium tuberculosis to human macrophages. J Immunol. 1994;152:4070–4079. [PubMed] [Google Scholar]
- Livak K J, Schmittgen D T. Analysis of relative gene expression data real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ferguson J S, Voelker D R, Ufnar J A, Dawson A J, Schlesinger L S. Surfactant protein D inhibition of human macrophage uptake of Mycobacterium tuberculosis is independent of bacterial agglutination. J Immunol. 2002;168:1309–1314. doi: 10.4049/jimmunol.168.3.1309. [DOI] [PubMed] [Google Scholar]
- Flesch I E, Kaufmann S H E. Mechanisms involved in mycobacterial growth inhibition by γ-interferon-activated bone marrow macrophages: role of reactive nitrogen intermediates. Infect Immun. 1991;59:3213–3218. doi: 10.1128/iai.59.9.3213-3218.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Tanaka K E, Carroll D, Flynn J, Bloom B R. Effects of nitric oxide inhibitors on murine infection with Mycobacterium tuberculosis. Infect Immun. 1995;63:736–740. doi: 10.1128/iai.63.2.736-740.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking J D, North R J, LaCourse R, Mudgett J S, Shah S K, Nathan C F. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci USA. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J L, Scanga C A, Tanaka K E, Chan J. Effects of aminoguanadine on latent murine tuberculosis. J Immunol. 1998;160:1796–1803. [PubMed] [Google Scholar]
- Vidal S M, Pinner E, Lepage P, Gauthier S, Gros P. Nramp1 encodes a membrane phosphoglycoprotein absent in macrophages from susceptible (NRAMP1D169) mouse strains. J Immunol. 1996;157:3559–3568. [PubMed] [Google Scholar]
- White J K, Stewart A, Popoff J F, Wilson S, Blackwell J M. Incomplete glycosylation and defective intracellular targeting of mutant solute carrier family 11 member 1 (Slc11a1) Biochem J. 2004;382:811–819. doi: 10.1042/BJ20040808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyssonnaux C, Zinkernagal A S, Datta V, Lauth X, Johnson R S, Nizet V. TLR4-dependent hepcidin expression by myeloid cells in response to bacterial pathogens. Blood. 2006;107:3727–3732. doi: 10.1182/blood-2005-06-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X B, Hill P, Haile D J. Role of ferroportin iron-responsive element in iron and nitric oxide dependent gene regulation. Blood Cells Mol Dis. 2002;29:315–326. doi: 10.1006/bcmd.2002.0572. [DOI] [PubMed] [Google Scholar]
- Taetle R, Honeysett J M. γ-Interferon modulates human monocyte/macrophage transferrin receptor expression. Blood. 1988;71:1590–1595. [PubMed] [Google Scholar]
- Byrd T F, Horwitz M A. Regulation of transferrin receptor expression and ferritin content in human mononuclear phagocytes. Coordinate upregulation by iron transferrin and downregulation by interferon-γ. J Clin Invest. 1993;91:969–976. doi: 10.1172/JCI116318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero V, Brock J H. Regulation of iron metabolism in murine J774 macrophages: role of nitric oxide-dependent and -independent pathways following activation with γ interferon and lipopolysaccharide. Blood. 1999;94:2383–2389. [PubMed] [Google Scholar]
- Zwilling B S, Kuhn D E, Wikoff L, Brown D, Lafuse W P. Role of iron in Nramp1-mediated inhibition of mycobacterial growth. Infect Immun. 1999;67:1386–1392. doi: 10.1128/iai.67.3.1386-1392.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Techau M E, Valdez-Taubas J, Popoff J-F, Francis R, Seaman R, Blackwell J M. Evolution of differences in transport function in SLC11A family members. J Biol Chem. 2007;282:35646–35656. doi: 10.1074/jbc.M707057200. [DOI] [PubMed] [Google Scholar]
- De Voss J J, Rutter K, Schroeder B G, Barry C E. Iron acquisition and metabolism by mycobacteria. J Bacteriol. 1999;181:4443–4451. doi: 10.1128/jb.181.15.4443-4451.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobin J, Moore C H, Reeve J R, Jr, Wong D K, Gibson B W, Horwitz M A. Iron acquisition by Mycobacterium tuberculosis: isolation and characterization of a family of iron-binding exochelins. Proc Natl Acad Sci USA. 1995;92:5189–5193. doi: 10.1073/pnas.92.11.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D, Asard H. Three mammalian cytochromes b561 are ascorbate-dependent ferrireductases. FEBS J. 2006;273:3722–3734. doi: 10.1111/j.1742-4658.2006.05381.x. [DOI] [PubMed] [Google Scholar]
- Zhang D L, Su D, Bérczi A, Varga A, Asard H. An ascorbate-reducible cytochrome b561 is localized in macrophage lysosomes. Biochem Biophys Acta. 2006;1760:1903–1913. doi: 10.1016/j.bbagen.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Schaible U E, Strugill-Koszycki S, Schlesinger P H, Russell D G. Cytokine activation leads to acidification and increases maturation of Mycobacterium avium-containing phagosomes in murine macrophages. J Immunol. 1998;160:1290–1296. [PubMed] [Google Scholar]
- Via L E, Fratti R A, McFalone M, Pagan-Ramos E, Deretic D, Deretic V. Effects of cytokines on mycobacterial phagosome maturation. J Cell Sci. 1998;111:897–905. doi: 10.1242/jcs.111.7.897. [DOI] [PubMed] [Google Scholar]
- Mazzi P, Donini M, Margotto D, Wientjes F, Dusi S. IFN-γ induces gp91phox expression in human monocytes via protein C-dependent phosphorylation. J Immunol. 2004;172:4941–4947. doi: 10.4049/jimmunol.172.8.4941. [DOI] [PubMed] [Google Scholar]
- Kakar R, Kautz B, Eklund E A. JAK2 is necessary and sufficient for interferon-γ-induced transcription of the gene encoding gp91phox. J Leukoc Biol. 2005;77:120–127. doi: 10.1189/jlb.0704429. [DOI] [PubMed] [Google Scholar]
- Chlosta S, Fishman D S, Harrington L, Johnson E E, Knutson M D, Wessling-Resnick M, Cherayil B J. The iron efflux protein ferroportin regulates the intracellular growth of Salmonella enterica. Infect Immun. 2006;74:3065–3067. doi: 10.1128/IAI.74.5.3065-3067.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]