Abstract
Type II isopentenyl diphosphate (IPP) isomerase catalyzes the interconversion of IPP and dimethylallyl diphosphate (DMAPP). Although the reactions catalyzed by the type II enzyme and the well-studied type I IPP isomerase are identical, the type II protein requires reduced flavin for activity. The chemical mechanism, including the role of flavin, has not been established for type II IPP isomerase. Recombinant type II IPP isomerase from Thermus thermophilus HB27 was purified by Ni2+ affinity chromatography. Aerobically purified enzyme was inactive until the flavin cofactor was reduced by NADPH, dithionite, or photochemically. The inactive oxidized flavin-enzyme complex bound IPP in a Mg2+ dependent manner with KD ~ KmIPP, suggesting that the substrate binds to the inactive oxidized and active reduced forms of the protein with similar affinities. N,N-dimethyl-3-amino-1-propyl diphosphate (NIPP), a transition state analog for the type I isomerase, competitively inhibits the type II enzyme, but with much lower affinity. pH dependent spectral changes indicate that the binding of IPP, DMAPP, and a saturated analogue isopentyl diphosphate promotes protonation of anionic reduced flavin. Electron paramagnetic resonance (EPR) and UV-visible spectroscopy show a substrate-dependent accumulation of the neutral flavin semiquinone during both the flavoenzyme reduction and re-oxidation processes in the presence of IPP and related analogues. Redox potentials of IPP-bound enzyme indicate that the neutral semiquinone state of the flavin is stabilized thermodynamically relative to free FMN in solution.
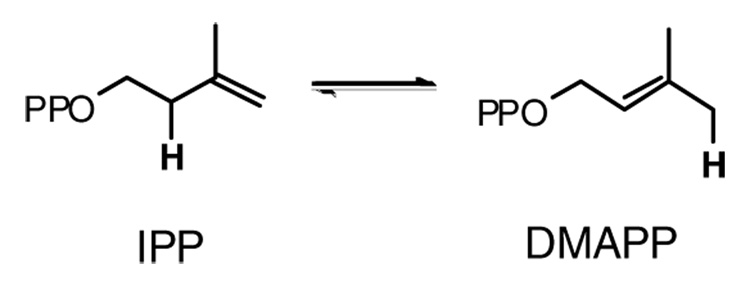
Isopentenyl diphosphate (IPP) isomerase catalyzes the interconversion of isopentenyl diphosphate and dimethylallyl diphosphate (DMAPP), the basic building blocks of isoprenoid molecules (Scheme 1). This family of natural products now consists of over 35,000 compounds (1), which comprise several classes of biologically important molecules, including sterols, carotenoids, prenylated proteins, dolichols, heme A, and ubiquinones. IPP isomerase is essential in organisms that generate IPP through the mevalonate pathway found in eukaryotes, archaea, and some gram-positive eubacteria (2). The enzyme, although not essential, is also found in most organisms that utilize the methylerythritol phosphate pathway, where isomerase activity is probably important for balancing the pools of IPP and DMAPP (3).
Scheme 1.
Interconversion of IPP and DMAPP.
Two structurally unrelated forms of IPP isomerase have been identified. The type I enzyme resides in eukaryotes and in some eubacteria, while the type II protein is found in archaea and other eubacteria (4). Type I IPP isomerase was discovered over 40 years ago and has been extensively characterized (5). The enzyme is a zinc metalloprotein (6) that catalyzes the isomerization of IPP and DMAPP by an antarafacial (7–9) protonation-deprotonation mechanism (10;11). Structural studies and site directed mutagenesis experiments have provided important information about how the substrate binds and have identified several active site residues essential for catalysis (12–14).
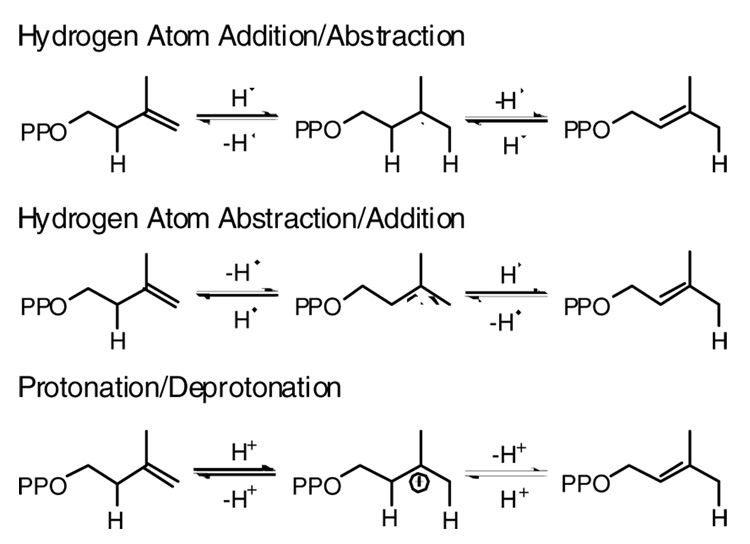
Type II IPP isomerase was first reported in 2001 (15). In contrast to the type I enzyme, type II isomerase requires flavin mononucleotide (FMN), a reducing agent, typically NADPH, and a divalent metal for activity. Presumably NADPH is required to reduce the flavin to generate an active complex. Hemmi and coworkers proposed a radical transfer mechanism for the isomerization reaction with the transient formation of a flavin semiquinone (Scheme 2) (16), while others have suggested a structural role for the cofactor (17;18) in a protonation-deprotonation mechanism similar to the type I enzyme. Type II IPP isomerase is an essential enzyme in Streptococcus pneumoniae and Staphylococcus aureus, both of which utilize the mevalonate route to isoprenoids. Since the type I enzyme is the exclusive IPP isomerase in eukaryotes, the type II protein represents a logical target for antibacterial drugs (15). Our efforts to obtain a type II isomerase suitable for both mechanistic and structure-function studies led to the protein from Thermus thermophilus. We report the results of kinetic and spectroscopic studies that reveal a ligand-induced change in the state of flavin in the resting enzyme·FMNH − complex upon binding IPP, DMAPP, isopentyl diphosphate, or NIPP
Scheme 2.
Proposed mechanisms for the isomerization reaction catalyzed by type II IPP isomerase.
Experimental Procedures
Materials
IPP and isopentyl diphosphate were synthesized from their corresponding alcohols according to the procedure of Davisson et al. (19). NIPP was prepared from 2-dimethylaminoethyl-chloride as described (10). DMAPP was provided by Dr. Wenyun Gao and Nicole Heaps. [14-C] IPP was purchased from G.E. Healthcare (formerly Amersham Biosciences). The gene encoding Thermus thermophilus type II IPP isomerase (locus: TT_P0067) was previously cloned from T. thermophilus HB27 genomic DNA (20). Type II isomerase was expressed in Escherichia coli and purif ied as described (20), with the modification that DTT was excluded from the dialysis buffer. Additional modifications were applied for protein purified for use in spectral assays. Riboflavin (200 µg/mL) was added to the expression media and the purified protein was concentrated to ~1 mM with an Amicon Ultra-15 30K (Millipore) and then dialyzed against 10 mM Tris buffer, pH 8.0, containing 20% glycerol, prior to storage at −80 °C. Protein used for assays in D2O was dialyzed in a corresponding D2O buffer. Glycerol, HEPES, Tris, and guanidinium·HCl were from USB Corporation. D2O was purchased from Cambridge Isotopes. Other reagents, unless specified, were purchased from Sigma.
Mass Spectrometry
Concentrated protein (10 µL) was diluted into 500 µL of water:acetonitrile:formic acid (80:20:0.2) and concentrated via ultrafiltration (Microcon® YM-30, Millipore). The protein was resuspended in the same solution, concentrated, and resuspended in 250 µL of water:acetonitrile:formic acid (50:50:0.2) to give a final concentration of 15 µM protein. Positive ion ESI-MS was performed on a Waters Micromass Quattro II triple quadrupole mass spectrometer.
Protein and Flavin Analysis
The concentration of purified protein was determined by the BCA and Bradford protein assays (Pierce). The concentration of bound flavin was determined as follows: protein (60 µL, ~6 mg/ml in 25 mM HEPES buffer, pH 7.5, 25 °C) was mixed with an equivalent volume of 10% TCA. The mixture was vortexed, placed on ice for ~ 2 min, and then centrifuged (2 min, 14,000 × g, 4 °C). The supernatant was neutralized with an equal volume of 1 M Na2HPO4. Alternatively, protein was diluted to 1–2 mg/mL with 6 M guanidinium·HCl, 50 mM HEPES buffer, pH 7.5, and incubated for 30 min at room temperature. Flavin and protein were separated by ultrafiltration (Microcon® YM-30, Millipore, 12 min, 14,000 × g, 4 °C). As a control, a sample of protein-free FMN was carried through the identical procedure. Absorption spectra were measured with an Agilent 8453 diode array spectrophotometer. LC-MS analyses of guanidine denatured samples were performed with a Waters Alliance 2695 HPLC connected to a Micromass Quattro II triple quadrupole mass spectrometer. Samples were chromatographed on a Phenomenex Prodigy™ ODS column, loaded and eluted with a 5 mM ammonium acetate pH 6.0: methanol (90:10) mobile phase, and analyzed by negative ion ESI-MS.
IPP Isomerase Assays
Isomerase activity was measured via the acid-lability method (21) with modifications similar to those described by Kaneda (15). The conditions for assays are provided in Table 1. Reactions were initiated by the addition of enzyme, diluted in 1 mg/mL of BSA, 10 mM HEPES buffer, pH 7.6, to the assay cocktail. After 10 min, the reaction was quenched with 200 µL of 4:1 methanol:HCl and followed by a 10 min incubation at 37 °C. Organic soluble products were extracted with 1 ml of ligroine (boiling point 90–110°C, Fisher Scientific). Radioactivity in 500 µL of the extract was measured by liquid scintillation (Ultima Gold™ Cocktail, PerkinElmer®). Assays were performed at less than 10% substrate conversion. Activity versus concentration data were fit with the Michealis-Menten equation via non-linear regression with Grafit 5.0 (Erithacus Software).
Table 1.
Steady state kinetic parameters for type II IPP isomerases
IPP Binding assays
Ultrafiltration assays (500 µl) contained 0–20 µM enzyme, 1 µM [14-C]IPP (59 µCi/µmol), 100 mM HEPES buffer, pH 7.0, containing 100 mM KCl, 2 mM MgCl2, 40 µM FMN, and 0.8% glycerol. The mixtures were incubated for 10 min at 37°C and transferred to a Microcon® YM-30 (Millipore) filter unit. Following an additional 5 min at 37 °C, the samples were quickly centrifuged (1.5 min, 7200 × g). Approximately 70 µL of the 500 µL sample passed through the membrane (30 kD cutoff) during a typical spin. Scintillation counting was performed on 50 µL of the flow-through and 50 µL of the original sample. [E·IPP] was calculated from the difference of [IPP]total and [IPP]free. The values for [E·IPP] at different concentrations of enzyme were fit to equation 1 (22) to determine KDIPP. An offset parameter C was included in equation 1 to correct for a small observed adsorption of unbound IPP onto the membrane.
| (1) |
Dependence of IPP binding and isomerization on MgCl2
Concentrated protein was dialyzed for 24 h at 4 °C against 10 mM Tris buffer, pH 7.5, containing 20% glycerol, and 5% Chelex-100 (Bio-Rad Laboratories) to remove divalent metal. Assay buffers and reagents were mixed with 5% Chelex for 1 h at 25 °C and filtered. Plasticware used in assays was washed with H2O that had been treated with Chelex and filtered. Assays were conducted at 0–10 mM MgCl2. The data were fit with the Michaelis-Menten equation to calculate KDMgCl2. Ultrafiltration assays were used to determine the effect of Mg2+ on binding of IPP to E·FMNox. Microcon® YM-30 (Millipore) filtration units were incubated with 200 mM EDTA, centrifuged, and then rinsed twice with Chelex-treated water. Assays were performed as described above with 1 µM [14-C]IPP (59 µCi/µmol), 0 or 12 µM Chelex-treated enzyme, and 0 or 2 mM MgCl2. MgCl2 did not alter the small amount of IPP absorbed on the membrane observed in the absence of protein. In a second experiment, 1 mM EDTA with either 0 or 5 mM MgCl2 was employed instead of chelex treatment. KD in the presence of 1 mM EDTA without MgCl2 was measured as described above.
Spectroscopic Assays with NADPH
Anaerobic assays used 800 µL reactions in 200 mM HEPES buffer, pH 7.0, containing 25 µM enzyme-bound FMN and 10 mM MgCl2 at 37 °C. The mixtures were placed in a cuvette sealed with a silicone septum and purged with nitrogen. NADPH (2 mM) followed by 50 µM IPP were added anaerobically via air-tight syringes.
Photochemical Spectroscopic Assays
Photoreduction was performed in a 3.5 mL gas-tight all- glass cuvette (Starna Cells) with two side arms (one with a stopcock) and a three-way stopcock. Joints and stopcocks were sealed with Apiezon® N high vacuum grease. The cuvette chamber contained 2.1 mL (final concentrations are indicated for 2.2 mL after mixing) of 100 mM HEPES buffer, pH 7.0 or 8.5, containing 100 mM (pH 7.0) or 40 mM (pH 8.5) KCl, 20 µM enzyme-bound FMN (30–40 µM total enzyme), 5 µM added FMN, 2.25 µM riboflavin, 5% glycerol, 2 mM MgCl2, and 1 mM sodium oxalate at 37 °C. The substrate side arm contained 100 µL of 50 mM HEPES buffer, pH 7.0 or 8.5, containing 55 µM (final at pH 7.0) or 200 µM (final at pH 8.5) IPP, 5.5 µM riboflavin, and 1 mM sodium oxalate at 37 °C. The final IPP concentration was saturating at each pH. Related experiments were conducted with NIPP at pH 7.0 (250 µM) and 8.5 (1 mM) and isopentyl diphosphate at pH 7.0 (10 mM). The side arm with the stopcock was filled with 1 mL of 100 mM Tris buffer, pH 8, containing 1 mM methyl viologen, 3 µM proflavine hemisulfate, and 10 mM EDTA. Six to eight cycles of degassing under vacuum and re-pressurization with OxiClearTm (LabClearTm) purified argon were performed before the cuvette was sealed. The samples were photoreduced by irradiation with a 300 W slide projector. To minimize oxygen contamination, the side arm containing methyl viologen was photoreduced and allowed to oxidize over time with shaking (23). This procedure was conducted three times over a 20–30 min period. The side arm with substrate was also photoirradiated briefly. Photoreduction of the main solution, prior to or after mixing with substrate, was achieved within 15 min of photoirradiation as determined from spectra taken at ~1 min intervals. The spectrum of the enzyme-substrate mixture was monitored over time at 37°C with an Agilent 8453 diode array spectrophotometer.
EPR spectroscopy
Reactions for EPR spectroscopy were performed in a Coy Labs anaerobic chamber. Samples and buffers were purged with purified argon before being placed in the glove box. To generate the flavin semiquinone, 5 µL of 50 mM sodium dithionite in 50 mM Tris buffer, pH 8.7, was added to a 250 µL mixture of 200 mM HEPES buffer, pH 7.7 or 7.2, containing 2 mM MgCl2, 3–10% glycerol, 100 µM enzyme-bound FMN, and either 250 µM IPP, 250 µM DMAPP, or 10 mM isopentyl diphosphate. Reactions in D2O were performed in a similar manner at pD 7.7 with buffers and enzyme prepared in or dialyzed against D2O. Following reduction, samples were quickly transferred to an EPR tube with a rubber septum, removed from the glove box, and frozen in liquid nitrogen. In an attempt to detect a substrate radical/flavin radical pair, 6.25 µL of 200 mM sodium dithionite was first added to a mixture containing 400 µM enzyme-bound FMN without substrate. After ~2 min incubation to ensure complete flavin reduction, the substrate was added to mixture and treated as above. Continuous wave EPR spectra were obtained at 77 °K with a Bruker ESP300 spectrophotometer operating at X-band frequencies. Conditions for measurement are provided in Figure 4. 50 µM TEMPO in methanol:H2O (98:2) was employed as a standard to calculate g factors (24).
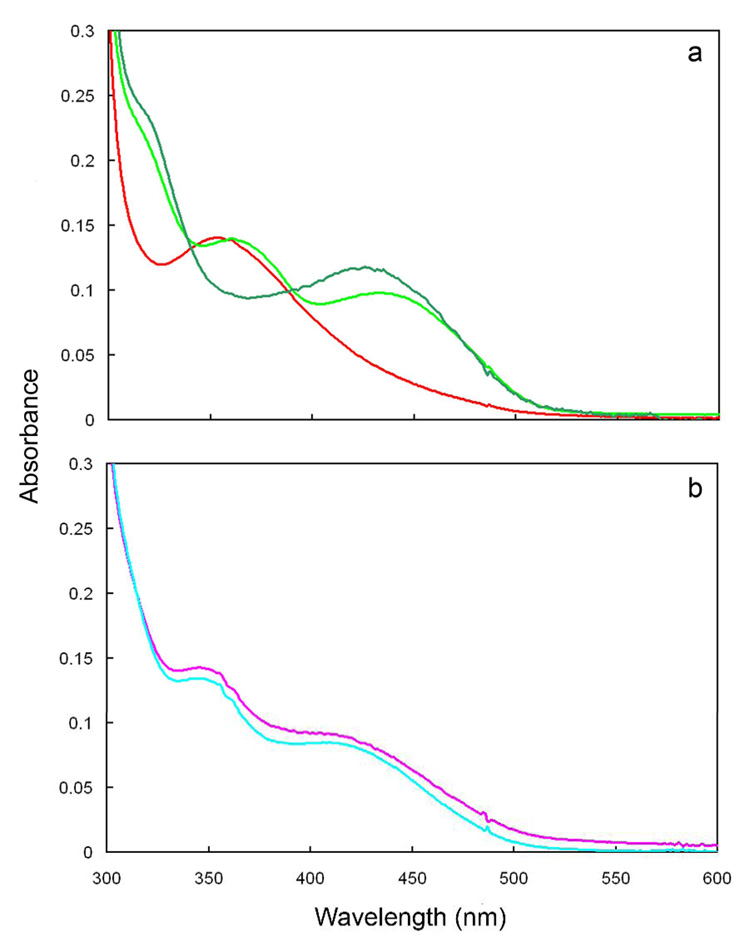
Figure 4.
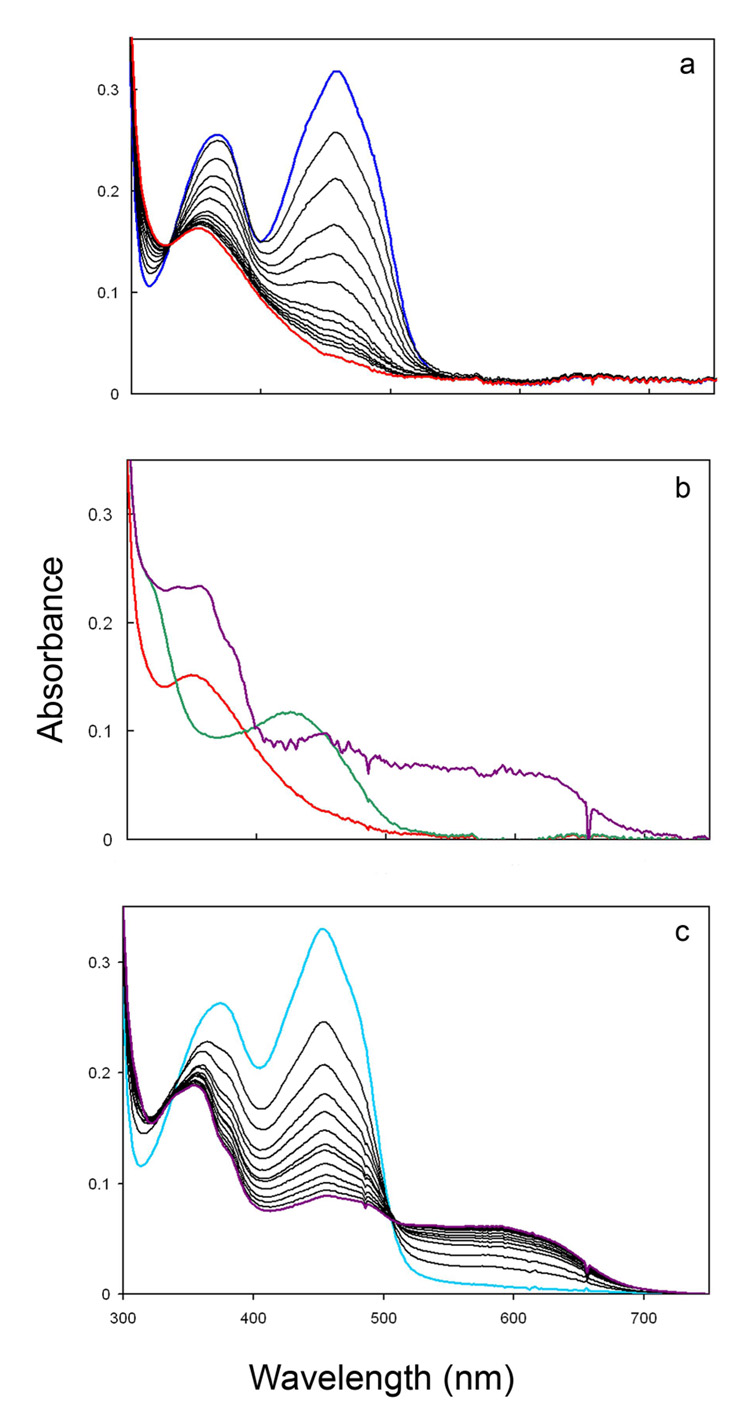
Effect of pH on the spectra of isomerase·flavin complexes at 37°C. Part a. Spectra of enzyme·FMNH− (red) and enzyme·FMNH2·IPP (dark green) at pH 7.0 and enzyme·FMNH2·IPP (light green) at pH 8.5. Part b. Spectra of enzyme·FMNH−·NIPP at pH 7.0 (pink) and 8.5 (cyan).
Redox potentiometry
Redox titrations were carried out in a glass anaerobic cuvette similar to the apparatus described by Dutton (25). This device was fitted with a silicone stopper containing a reference silver/silver chloride electrode in 3 M NaCl and a platinum measuring electrode. The cuvette chamber contained 7 mL of degassed 200 mM HEPES buffer, pH 8.0, containing 2 mM MgCl2, 150 µM IPP, and 30–60 µM redox mediator at 25°C. The redox mediator dyes used for the titrations were: neutral red (Em = −325 mV), anthraquinone 2-sulfonate (Em = −225 mV), 2-hydroxy-1,4-naphthoquinone (Em = −152 mV), 2,5-dihydroxy-p-benzoquinone (Em = −60 mV), menadione (Em = 0 mV), and phenazine ethosulfate (Em = +55 mV). Purified argon was bubbled into the solution for 3–4 hr. Enzyme (25 µM final) was added via syringe, and the solution was further deoxygenated over 1–3 hr. Reductive titrations were performed with 3 mM sodium dithionite, and oxidative titrations, with 24 mM potassium ferricyanide. Once the voltage reading stabilized at each titration point, absorption spectra were monitored with an Agilent 8453 diode array spectrophotometer. Changes in the absorbance at 620 nm, representing the neutral semiquinone, were plotted as a function of the ambient redox potential. Data were fit using a Nernst function describing a one-electron redox process. The calculated midpoint redox potentials were corrected for the potential of Ag/AgCl reference electrode by adding +209 mV.
Results
Mass Spectrometry and Flavin Content
Type II IPP isomerase from T. thermophilus bearing an N-terminal his6 tag was expressed in E. coli and purified in two steps via heat treatment followed by Ni-NTA chromatography (20). The protein maintained a yellow color throughout purification, consistent with retention of bound flavin. A sample of protein prepared for ESI-MS was colorless and gave an observed mass of 38098 ± 0.01% Da. This value agrees with the calculated mass of 38100 Da for the recombinant apoenzyme with the N-terminal his6 tag and a factor Xa protease site.
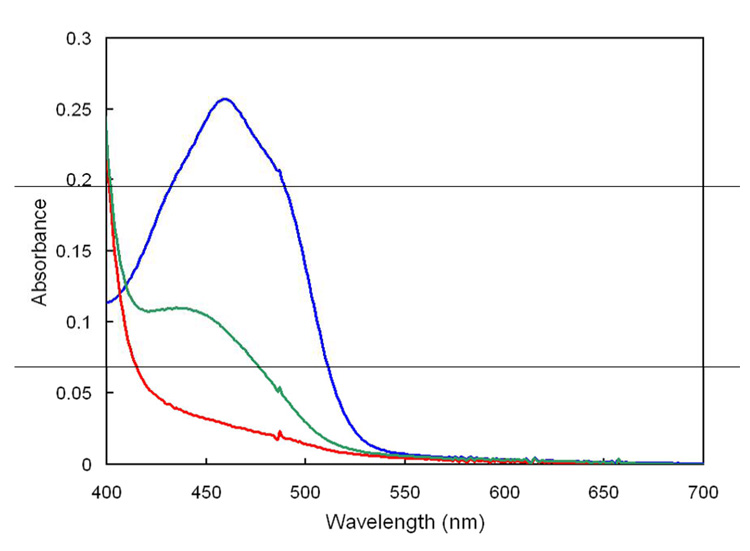
The protein was denatured by treatment with 5% trichloroacetic acid (TCA) followed by centrifugation or by treatment with 6 M guanidinium hydrochloride, followed by ultrafiltration, to release the bound flavin and separate the cofactor from the protein. Both methods yielded similar quantities of free flavin, as determined by UV-vis spectroscopy. Spectra of free and enzyme bound cofactor at equal-molar concentrations are shown in Figure 1. Free flavin has absorbance maxima at 374 nm and 446 nm, consistent with the reported spectrum of FMN (26). LC-MS of the denatured samples gave a peak at m/z of 455 for FMN, while the fluorescence properties of the guanidinium-released cofactor closely resemble free FMN (See Supporting Information). The absorption maxima shift to 368 nm and 460 nm, respectively, with a slight decrease in intensity in the enzyme·FMN complex. Based on a comparison of spectra for bound and released/free FMN (ε445nm= 12,500 M−1 cm−1), an extinction coefficient of 11,300 M−1 cm−1 at 460 nm was calculated for the enzyme·FMN complexwith an FMN occupancy of 32% or 25%, employing the BCA or the Bradford assay, respectively. Supplementation of expression media with 200 µg/mL of riboflavin increased the occupancy up to 70%. The activity of purified enzyme was also enhanced by addition of exogenous FMN. A plot of v/ET versus [FMN] was hyperbolic with a half-maximal increase in rate at ~5 µM FMN (See Supporting Information). Assuming that the maximal rate reflects saturation of IPP isomerase by FMN, the 2.9-fold increase in rate at 37 °C is consistent with a predicted increase of 3.1-fold, corresponding to 32% occupancy determined for the purified protein by the above spectral analyses and the BCA protein assay. The increase in activity upon pre-incubation of the enzyme with 250 nM FMN during time course measurements indicated slow tight binding of the cofactor (See Supporting Information). The data were fit to a first pseudo-order equation to give kobs = (1.0 ± 0.2) × 10−3 s−1, which provides an approximate kon near 104 M−1sec−1 for addition of FMN to the type II protein at 37 °C.
Figure 1.
UV-visible spectra of enzyme bound (dark blue) and 5% TCA released (orange) FMN at equal concentrations.
IPP binding and isomerizaton
The steady state kinetic constants for the conversion of IPP to DMAPP were determined under aerobic conditions at 37 °C (Table 1). Pre-incubation of the enzyme for 1 h at 37 °C prior to the addition of substrate did not result in loss of enzyme activity. While the activity of the enzyme is higher at 60 °C, 37 °C was used in our studies to minimize fluctuations in temperature, minimize changes in concentration due to condensation of water on the sides of the cuvettes, and increase the stability of the enzyme. The affinity of radiolabeled IPP for enzyme·FMN was measured at 37 °C using an ultrafiltration binding assay, KDIPP = 4.4 ± 0.4 µM (See Supporting Information). This value is similar to KMIPP determined under state steady conditions. NIPP, a potent transition state analogue/slow tight-binding inhibitor for type I IPP isomerase (KD < 0.12 nM) (10), was evaluated as an inhibitor of the type II enzyme. Under steady state conditions, NIPP was a competitive inhibitor with KINIPP = 5.1 ± 0.5 µM at 37 °C (See Supporting Information). Time dependent inhibition of NADPH-reduced enzyme by 6 µM NIPP was not observed following 40 min incubation prior to IPP addition (data not shown). Isopentyl diphosphate, a saturated analog of IPP and DMAPP, inhibited turnover of 5 µM IPP, IC50 = 1.2 ± 0.1 mM.
Catalytic Requirement for Reduced FMN
Type II IPP isomerase, when purified and assayed under aerobic conditions, needs a reducing agent for activity (15;18;27–30). The oxidized form of the T. thermophilus enzyme was also inactive. An apparent value KDNADPH = 110 ± 10 µM was measured under aerobic conditions (Table 1). Spectroscopic assays (described in the next section) show that enzyme-bound flavin is rapidly reduced by NADPH. Sodium dithionite also reduces enzyme bound FMN to FMNH− and can substitute for NADPH to produce active enzyme. Under anaerobic conditions, the enzyme is also only active in the presence of reducing agent (J. Johnston, unpublished).
Mg2+ dependence for catalysis and binding
The requirement of Mg2+ for catalysis and for substrate binding was evaluated. When purified IPP isomerase was assayed in a metal-free buffer, the enzyme had ~20% of the activity observed in buffer containing 2 mM MgCl2. The activity decreased to <0.2% when the protein was dialyzed against Chelex and assayed in Chelex-treated metal-free buffer. KD(App)MgCl2 = 130 ± 10 µM (see Table 1). In ultrafiltration experiments, the fraction of radiolabeled IPP (1 µM) bound to excess enzyme (12 µM) increased from 0.08 to 0.7 when MgCl2 was added to the assay buffer. A similar MgCl 2 enhancement in binding was also observed for untreated enzyme (12 µM) with 1 mM EDTA. KD = 84 ± 9 µM was measured for the binding of IPP to oxidized enzyme in the presence of 1 mM EDTA without MgCl2 (data not shown).
Spectroscopic Analysis of Bound FMN and FMNH−
UV-visible spectroscopy was used to characterize the bound flavin species in enzyme·FMN and enzyme·FMNH− complexes (26;31). Under anaerobic conditions the absorbance at 460 nm decreased rapidly upon mixing enzyme·FMN and NADPH (Figure 2). This change was also observed under aerobic conditions. These results are consistent with reduction of enzyme·FMN to enzyme·FMNH−. However, the spectra do not permit FMNH− to be identified unambiguously because of the intense absorption for NADPH at 340 nm. To circumvent this problem, the flavin was reduced photochemically with oxalate under anaerobic conditions (32;33). A time course for photoreduction of FMN in the presence of excess protein is shown in Figure 3a. The reaction proceeds to give a series of spectra with an isosbestic point at 330 nm, indicating that the reaction occurs without the accumulation of an intermediate. The final species has an absorbance maximum at 352 nm (ε ~ 5500 M−1cm−1) characteristic of anionic reduced FMN (Figure 3b), which has a peak at 342 nm when free in solution (26). FMNH− is the typical protonation state for the reduced cofactor in most flavoenzymes (34). The photoreduced enzyme is enzymatically active (Jon Johnston, unpublished results)
Figure 2.
Reduction of enzyme-bound FMN by NADPH under anaerobic conditions, pH 7.0, at 37°C. Shown are spectra of 25 µM FMN in assay buffer (dark blue), FMNH− formed immediately after mixing with 2 mM NADPH (red), and after the subsequent addition of 50 µM IPP (dark green).
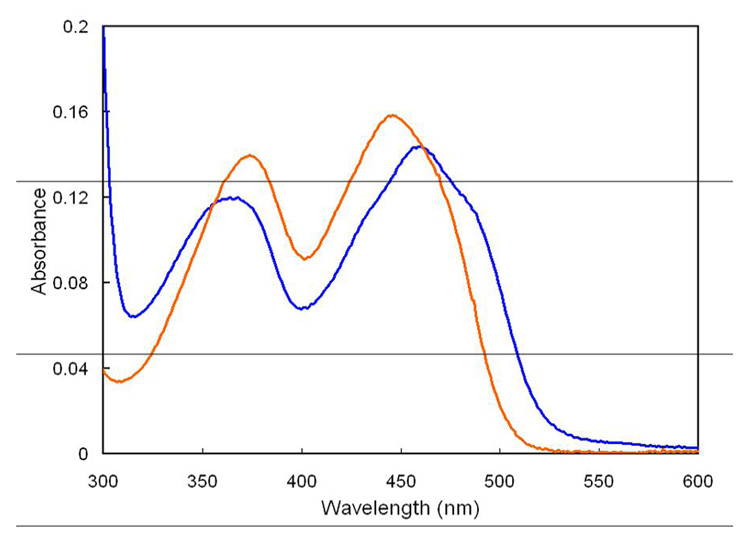
Figure 3.
UV-visible spectra of isomerase·flavin complexes, pH 7.0, at 37°C. Part a. Time course for photoreduction of enzyme·FMN (dark blue) to enzyme·FMNH− (red). Part b. Enzyme·FMNH− (red), enzyme·FMNH2·IPP (dark green), and enzyme·FMNH.·IPP (dark purple) after 14 h of additional incubation. Part c. Time course for photoreduction of oxidized enzyme·FMN·IPP (light blue) to enzyme·FMNH.·IPP (dark purple).
When IPP is added to enzyme·FMNH− obtained by photoreduction at pH 7.0, the UV-visible absorbance spectrum of the flavin gives a new spectrum with a peak at 426 nm (ε ~ 4300 M−1cm−1) and a shoulder ~320 nm (Figure 3b). A similar change is seen in the 400–450 nm region when IPP is added to enzyme·FMNH − generated by reduction with NADPH (Figure 2). The absorption features for the isomerase·FMNH2·IPP complex resemble those reported for FMNH2 bound to E. coli thioredoxin reductase, where the N1 position of the flavin is protonated at a physiological pH (26;35). In addition, the UV-visible absorbance spectrum of neutral reduced flavin in a polar organic solvent is similar to those of the two flavoenzymes (26). These similarities suggest that the binding of IPP induces concomitant protonation of the flavin in the enzyme·FMNH− complex at pH 7.0. When the pH of the buffer is raised from 7.0 to 8.5, the UV-visible spectrum of the enzyme·FMNH− complex is essentially unaltered. In contrast, the spectrum of the isomerase·FMNH2·IPP complex changed to one consistent with an isomerase·FMNH−·IPP species (Figure 4a). Reduced enzyme bound to NIPP at either pH 7.0 or 8.5 exhibits a UV-spectrum similar to the IPP complex at 8.5 (Figure 4b), consistent with stabilization of the anionic form of the reduced cofactor by the positive charge at N3 of NIPP. Isopentyl diphosphate elicited spectral changes consistent with flavin protonation (data not shown). The retention of a peak near 430 nm for each of the complexes at pH 8.5 may indicate substrate-stabilization of a planar conformation for bound FMN (36).
Upon prolonged incubation in the presence of small amounts of residual oxygen, the enzyme·FMNH− complex slowly oxidizes to enzyme·FMN without the formation of a detectable intermediate. In contrast, under similar conditions enzyme·FMNH2·IPP oxidizes to an intermediate with an absorption at 550–650 nm (green, Figure 3b). Although the absorption at 550–650 nm could represent a charge-transfer complex, the accompanying intense peak near 350 nm indicates formation of the neutral flavin semiquinone (31;37). The enzyme·FMNH.·IPP complex was stable to extended incubation in the sealed glass cuvette. The spectrum shifted to one characteristic of the enzyme·FMN·IPP complex only after the sample was fully exposed to atmospheric oxygen.
Oxidized enzyme·FMN and enzyme·FMN·IPP give similar spectra (Figures 3a and 3c). Photoreduction of enzyme·FMN cleanly generates enzyme·FMNH− (Figure 3a). In contrast, photoreduction of enzyme·FMN·IPP cleanly produces enzyme·FMNH.·IPP (Figure 3c). An extended 40 min illumination failed to generate the spectrum for the enzyme·FMNH2·IPP that was observed when IPP was added to photoreduced enzyme·FMNH−.
EPR spectroscopy of the flavin semiquinone
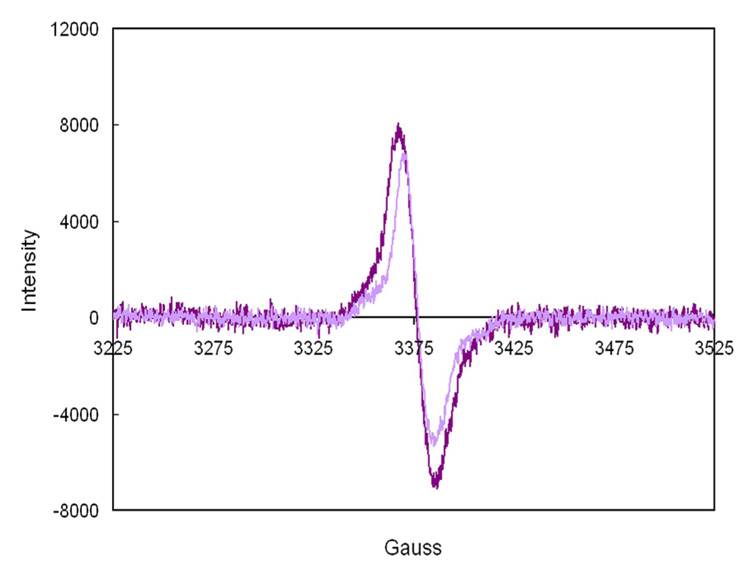
EPR spectroscopy was employed to evaluate the nature of the putative substrate-dependent flavin radical. Dithionite reduction of the enzyme·FMN·IPP complex initially produced the neutral semiquinone species observed during photoreduction (unpublished results). EPR spectra for the partially reduced complex at pH (or pD) 7.7 are shown in Figure 5. A strong signal was seen with a g factor of 2.0046 and a linewidth of 19 G, which narrows to 15 G in D2O. These features are consistent with the presence of a neutral flavin radical (38). Similar spectra were observed upon dithionite reduction of the IPP (pH 7.2 and pH 7.7) and isopentyl diphosphate complexes, but not for enzyme·FMN. A signal for the flavin radical signal was also observed when NADPH reduced enzyme·FMNH2·IPP was briefly exposed to oxygen. In an attempt to detect a substrate/flavin radical pair, 500 µM IPP was added to fully reduced 400 µM enzyme·FMNH −. Only faint EPR signals, similar to those seen for the enzyme·FMNH.·IPP complex generated by reduction of the flavin in the presence of IPP were observed (data not shown). At this point we cannot rule out the possibility of contamination of the sample by trace quantities of oxygen. In addition, a semiquinone signal was not observed via absorption spectroscopy when 250 µM IPP was added anaerobically to 200 µM enzyme·FMNH−.
Figure 5.
EPR spectra of enzyme·FMNH.·DMAPP at 77 °K produced by reduction ( pH 7.7, 25°C) of enzyme·FMN·DMAPP in H2O (dark purple) and D2O (light purple). Spectrometer settings were as follows: microwave frequency, 9.48 GHz; microwave power, 20 µW; modulation frequency, 100 kHz; modulation amplitude, 1 G. The linewidths are 19 G in H2O and 15 G in D2O. The g factor was 2.0046 based on a TEMPO standard.
Redox potentiometry
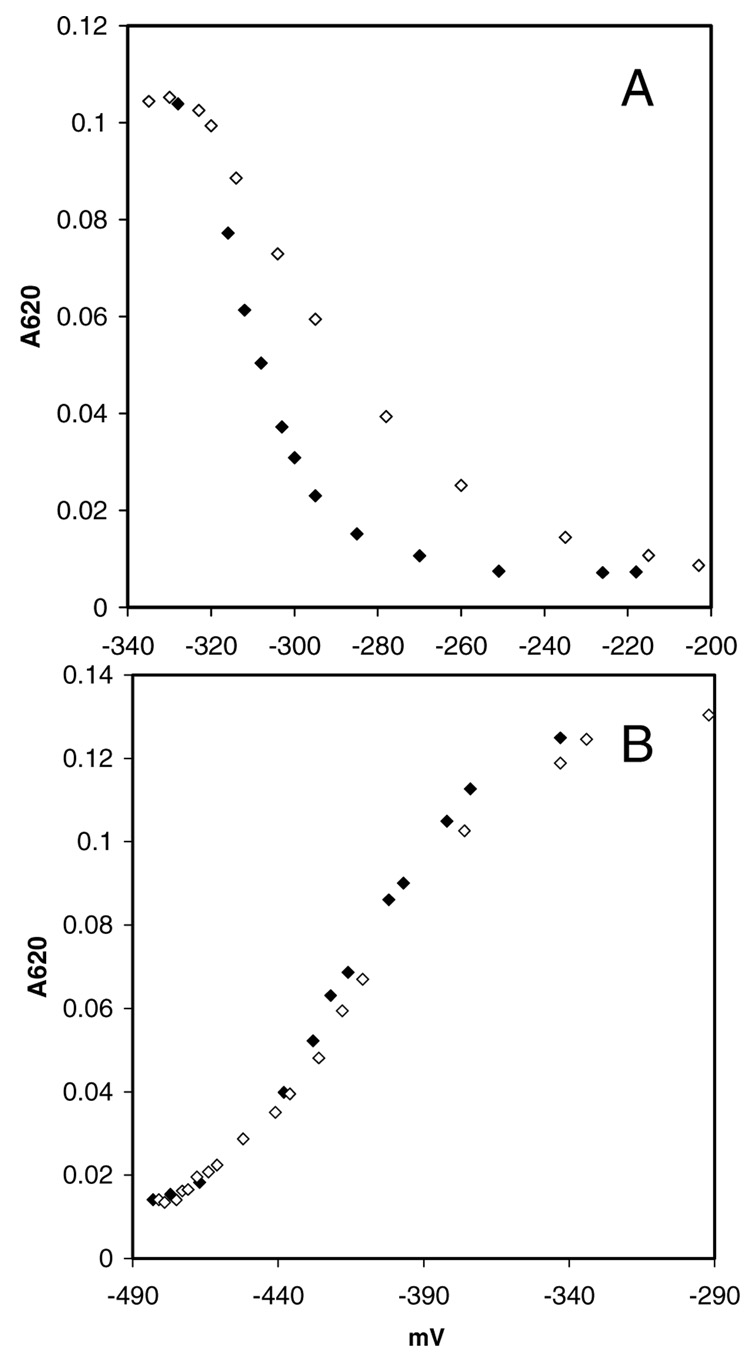
Anaerobic redox titrations were performed to determine the midpoint redox potentials for enzyme-bound flavin in the presence of IPP. Oxidative and reductive titrations employing dye mediators were carried out at pH 8.0 and 25°C. The spectra at each titration point were recorded (See Supporting Information). The formation or loss the neutral semiquinone, based on absorbance at 620 nm, was plotted as function of the ambient redox potential (Figure 6). A small 18 mV difference was observed for the oxidized flavin/semiquinone couple midpoint redox potentials in the two directions (part A), which probably results from incomplete equilibration before the measurements. There was no difference for the semiquinone/reduced flavin couple (part B). The measured midpoint redox potentials (n = 1) for the oxidized/neutral semiquinone (oxidative titration) and neutral semiquinone/reduced couples are −83 ± 1 mV and −196 ± 1 mV, respectively. These contrast with the redox potentials for free flavin at pH 8 of −364 mV for the oxidized/ semiquinone couple and −119 mV for semiquinone/reduced couple, calculated from the reported values (−301 mV and −101 mV) at pH 7 and 20°C (39).
Figure 6.
Changes in the enzyme·FMN·IPP oxidation state as a function of the ambient redox potential (pH 8.0, 25°C). Shown are the spectral changes associated with the interconversion of the oxidized/semiquinone states (A) and semiquinone/reduced states (B). Titrations were performed reductively with sodium dithionite (◆) and oxidatively with potassium ferricyanide (◇).
Discussion
As a prelude to work with type II IPP isomerase, we cloned and conducted preliminary experiments with the enzymes from Synechocystis sp. strain PCC 6803 (28), Methanothermobacter thermautotrophicus(27), Streptococcus pneumoniae, Thermoplasma acidophilum, and Thermus thermophilus HB27. Of these, the enzyme·FMN complex from T. thermophilus HB27 was soluble and stable, even when studied at higher temperatures, and that protein was selected for additional studies. As reported for other type II isomerases, the T. thermophilus enzyme catalyzed the isomerization of IPP to DMAPP in the presence of bound FMN, a reducing agent, and MgCl2. The value for kcat at 37 °C (Table 1) was similar to those for mesophilic enzymes from Bacillus subtilis (29;40) and Synechocystis sp. strain PCC 6803 (28) and increased ~10-fold at the optimal temperature near 60 °C (unpublished results). Freshly purified T. thermophilus type II IPP isomerase contained 0.3 to 0.7 equiv of FMN, depending on the conditions employed for expression. Upon incubation with FMN, the protein binds additional cofactor in a slow-tight binding process similar to that proposed for the type II isomerase from Sulfolobus shibatae (30).
Type II isomerase, typically purified in the presence of oxygen, gave inactive enzyme containing oxidized flavin. Reduction of the cofactor with NADPH or dithionite was required to generate an active enzyme·reduced flavin complex (15;16;27;29). Laupitz and coworkers (18) reported that B. subtilis type II isomerase purified from an recombinant E. coli strain under anaerobic conditions was active without addition of NADPH to the assay buffer. It is curious to note, that they also reported that added FMN was required for activity in the anaerobic assay and that FMN was not reduced by NADPH in the presence of enzyme. We found that FMN in the T. thermophilus enzyme was rapidly reduced by NADPH or dithionite, and was reduced photochemically to the fully reduced anion. The enzyme was active regardless of the method of reduction. Thus, FMNH− is the form of the flavin cofactor in the resting state of the catalytically active enzyme.
Type II IPP isomerase requires a divalent metal, Mg2+, Mn2+, or Ca2+ for activity (15;18;29). A specific role for the metal has not been established. The type I enzyme requires two divalent metals (13). Zn2+ is located in a hexacoordinate His3Glu2 binding site that is to be part of the catalytic machinery for protonation of the carbon-carbon double bond in IPP. The second metal, Mg2+, helps anchor IPP by coordinating to non-bridging oxygens in the diphosphate moiety and residues in the protein. Our data support the latter role for the divalent metal required by the type II enzyme. A comparison of KDIPP for type II isomerase in the presence (4.4 ± 0.4 µM) and absence (84 ± 9 µM) of MgCl2 demonstrates that the metal enhances substrate binding. The dissociation constant for IPP binding to enzyme·FMN in buffer containing Mg2+ is similar to KMIPP determined from steady state kinetic assays. Thus, it appears that oxidation state of the cofactor is not important for IPP binding.
Upon binding IPP, the UV-visible absorbance spectrum of the cofactor changes to one characteristic of neutral reduced flavin, indicating that the reduced flavin anion in the enzyme·FMNH− complex is protonated. Thus, the catalytically active form of the type II isomerase appears to be an enzyme·FMNH2·IPP complex. The pKa of reduced FMN in solution is 6.7 (41). Spectral changes associated with protonation of enzyme·FMNH− were not observed in the pH range of 6–10. However, observed changes in the presence of IPP over this range suggest a pKa near 8 for the enzyme·FMNH2·IPP complex.
A second consequence of IPP binding is the accumulation of a neutral semiquinone during reduction and re-oxidation of the enzyme-bound flavin, whose nature was established by UV and EPR experiments. Photoreduction of flavoenzymes with free flavin as a catalyst is thought to occur by both 1 e− and 2 e− transfers from reduced free flavin to enzyme-bound cofactor (32). Our inability to photoreduce flavin in the enzyme·FMN·IPP complex beyond the neutral semiquinone state suggests that the FMNH−/FMNH. redox potential in the enzyme·FMN·IPP complex is substantially altered relative to free flavin. The neutral semiquinone also forms during re-oxidation of enzyme·FMNH2·IPP by molecular oxygen, a process that proceeds through sequential 1 e− transfers (31;42). Accumulation of semiquinone during photoreduction/re-oxidation as the result of a shift in the FMNH− (or FMNH2)/FMNH. redox potential has also been observed for flavodoxin (23;32). Direct participation of IPP in the flavin reduction and oxidation processes is unlikely, given the similar results seen for IPP and the fully saturated analogue isopentyl diphosphate. The measured midpoint redox potentials in the presence of IPP indicate that the accumulation of the neutral semiquinone derives at least in part from thermodynamic stabilization of the flavin radical state. We have also observed the accumulation of the anionic semiquinone during dithionite titrations without substrate in the presence of either benzyl viologen or indigo carmine as mediators (unpublished results). Thus, the enzyme binds flavin in a manner that thermodynamically stabilizes the semiquinone state.
Three different mechanisms, with different roles for the flavin cofactor, have been suggested for the reaction catalyzed by type II IPP isomerase (Scheme 2). A hydrogen atom addition/abstraction was originally suggested by Bornemann (17) and later by Hemmi and coworkers (16), based on the observation that reduced flavin was required for catalysis and that apoenzyme reconstituted with 5-deazaFMN, an analog that only participates in 2 e− transfer reactions, was inactive. The initial step would generate a flavin semiquinone/IPP radical pair, followed by hydrogen atom abstraction to give DMAPP and regenerate FMNH2. This mechanism requires the transient 1 e− oxidation of the flavin cofactor. We find that the semiquione state is stabilized by the enzyme. However, related behaviour has been reported for flavoenzymes that do not employ a radical mechanism(43;44). The second possibility, a hydrogen atom abstraction/addition mechanism with an intermediate allylic radical, seems unlikely. This mechanism involves a transient 1 e− reduction of the cofactor, which is not consistent with isomerase·FMNH2·IPP as the catalytically active complex. A third possibility is a protonation/deprotonation mechanism similar to the isomerization catalyzed by the type I enzyme. In this case, FMNH2 could passively serve in a structural role (17) or could actively participate by protonating the double bond in IPP with a concomitant transient 2 e− oxidation of the cofactor. A protonation/deprotonation mechanism was recently suggested by Hoshino and coworkers (45), following the discovery that an epoxide analogue of IPP, a potent mechanism-based inhibitor of type I IPP isomerase (46), covalently inactivated the type II enzyme. However, it is not clear that the mechanisms for inactivation of the two proteins by the epoxide are similar. We found that NIPP, a potent transition state analogue for the type I enzyme, is a modest inhibitor for type II isomerase.
A protonation/deprotonation mechanism that actively involves the flavin cofactor might use the zwitterionic 5,5-tautomer of FMNH2 to protonate the double bond in IPP. The 5,5-zwitterion was recently implicated in acid/base chemistry during catalysis (47), and its involvement is consistent with the inability of 5-deazaFMN to substitute for FMN (16). The UV-visible spectrum of a 5,5-dimethyl analog of the 5,5-FMN tautomer has a peak at ~310–320 nm (48). We see a shoulder near 320 nm in the spectrum of the isomerase·FMNH2·IPP complex, although a peak at this wavelength was also reported for the 1,5-tautomer bound to thioredoxin reducase (26;35). It is interesting to note that the reduction potential for the t-butyl cation in a polar solvent is + 90 mV (49). Thus, the redox potential (−196 mV) we measured for oxidation of enzyme·FMNH2/FMNH−·IPP to enzyme·FMNH.·IPP suggests that an FMNH−·IPPH+ ion pair produced by protonation of the double bond in IPP might be unstable with respect to a FMNH.·IPPH. radical pair, unless selectively stabilized by the enzyme. Otherwise, one would anticipate that at some point along a putative protonation reaction coordinate an electron transfer would generate the radical pair. At this point additional work is needed to convincingly resolve the mechanism of reaction catalyzed by type II IPP isomerase.
Supplementary Material
Supporting Information Available Figures for fluorescence of the guanidinium-released flavin and for binding of oxidized enzyme-FMN to IPP as measured via ultrafiltration assays. Figures and experimental details for dependence of isomerase activity on exogenous FMN and for enzyme inhibition by NIPP and isopentyl diphosphate. Anaerobic spectra of dithionite reduced enzyme, spectra for aerobic reduction of enzyme by NADPH, and re-oxidation of photoreduced enzyme. EPR spectra of enzyme·FMNH. complexes. UV-visible spectra of reductive potentiometric titrations. This material is available free of charge via the Internet at http://pubs.acs.org
Acknowledgement
We thank Dr. Wenyun Gao and Nicole Heaps for samples of DMAPP. We also thank Professor Ann Walker and Dr. Andrie Astashkin for assistance with EPR spectroscopy, Dr. Elliot Rachlin for assistance with mass spectrometry, and Professor Vahe Bandarian for assistance with anaerobic experiments and helpful discussions.
ABBREVIATIONS
- DMAPP
dimethylallyl diphosphate
- ESI-MS
electrospray ionization mass spectrometry
- FMN
flavin mononucleotide
- FMNH−
anionic reduced FMN
- FMNH2
reduced FMN
- FMNH.
FMN semiquinone
- IPP
isopentenyl diphosphate
- NIPP
N,N-dimethyl-3-amino-1-propyl diphosphate
- TCA
trichloroacetic acid
- TEMPO
2,2,6,6-tetramethyl-piperidine-1-oxyl.
Footnotes
This work was supported by NIH Grant GM25521. SCR was supported by NIH Postdoctoral Fellowship GM071114.
References
- 1.Rohdich F, Bacher A, Eisenreich W. Perspectives in anti-infective drug design. The late steps in the biosynthesis of the universal terpenoid precursors, isopentenyl diphosphate and dimethylallyl diphosphate. Bioorganic Chemistry. 2004;32:292–308. doi: 10.1016/j.bioorg.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 2.Kuzuyama T, Seto H. Diversity of the biosynthesis of the isoprene units. Nat. Prod. Rep. 2003;20:171–183. doi: 10.1039/b109860h. [DOI] [PubMed] [Google Scholar]
- 3.Rohmer M. A mevalonate-independent rout to isopentenyl diphosphate. In: Cane D, editor. Comprehensive Natural Products Chemistry. Pergamon Press; 1999. pp. 45–67. [Google Scholar]
- 4.Laupitz R, Hecht S, Amslinger S, Zepeck F, Kaiser J, Richter G, Schramek N, Steinbacher S, Huber R, Arigoni D, Bacher A, Eisenreich W, Rohdich F. Biochemical characterization of Bacillus subtilis type II isopentenyl diphosphate isomerase, and phylogenetic distribution of isoprenoid biosynthesis pathways. Eur. J. Biochem. 2004;271:2658–2669. doi: 10.1111/j.1432-1033.2004.04194.x. [DOI] [PubMed] [Google Scholar]
- 5.Agranoff BW, Eggerer H, Henning U, Lynen F. Isopentenyl pyrophosphate isomerase. Journal of the American Chemical Society. 1959;81:1254–1255. [Google Scholar]
- 6.Lee S, Poulter CD. Escherichia coli Type I Isopentenyl Diphosphate Isomerase: Structural and Catalytic Roles for Divalent Metals. Journal of the American Chemical Society Accepted for Publication. 2006 doi: 10.1021/ja063073c. [DOI] [PubMed] [Google Scholar]
- 7.Cornforth JW, Cornforth RH, Popjak G, Yengoyan L. Studies on the biosynthesis of cholesterol. XX. Steric course of decarboxylation of 5-pyrophosphomevalonate and of the carbon to carbon bond formation in the biosynthesis of farnesyl pyrophosphate. J. Biol. Chem. 1966;241:3970–3987. [PubMed] [Google Scholar]
- 8.Cornforth JW, Popjak G. Chemical syntheses of substrates of sterol biosynthesis. Methods Enzymol. 1969:359–371. [Google Scholar]
- 9.Clifford KH, Cornforth JW, Mallaby R, Phillips GT. Stereochemistry of isopentenyl pyrophosphate isomerase. Journal of the Chemical Society D: Chemical Communications. 1971:1599–1600. [Google Scholar]
- 10.Muehlbacher M, Poulter CD. Isopentenyl-diphosphate isomerase: inactivation of the enzyme with active-site-directed irreversible inhibitors and transition-state analogues. Biochemistry. 1988;27:7315–7328. doi: 10.1021/bi00419a021. [DOI] [PubMed] [Google Scholar]
- 11.Reardon JE, Abeles RH. Mechanism of action of isopentenyl pyrophosphate isomerase: evidence for a carbonium ion intermediate. Biochemistry. 1986;25:5609–5616. doi: 10.1021/bi00367a040. [DOI] [PubMed] [Google Scholar]
- 12.Durbecq V, Sainz G, Oudjama Y, Clantin B, Bompard-Gilles C, Tricot C, Caillet J, Stalon V, Droogmans L, Villeret V. Crystal structure of isopentenyl diphosphate : dimethylallyl diphosphate isomerase. Embo Journal. 2001;20:1530–1537. doi: 10.1093/emboj/20.7.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wouters J, Oudjama Y, Ghosh S, Stalon V, Droogmans L, Oldfield E. Structure and mechanism of action of isopentenylpyrophosphate-dimethylallylpyrophosphate isomerase. Journal of the American Chemical Society. 2003;125:3198–3199. doi: 10.1021/ja029171p. [DOI] [PubMed] [Google Scholar]
- 14.Wouters J, Oudjama Y, Barkley SJ, Tricot C, Stalon V, Droogmans L, Poulter CD. Catalytic mechanism of Escherichia coli isopentenyl diphosphate isomerase involves Cys-67, Glu- 116, and Tyr-104 as suggested by crystal structures of complexes with transition state analogues and irreversible inhibitors. Journal of Biological Chemistry. 2003;278:11903–11908. doi: 10.1074/jbc.M212823200. [DOI] [PubMed] [Google Scholar]
- 15.Kaneda K, Kuzuyama T, Takagi M, Hayakawa Y, Seto H. An unusual isopentenyl diphosphate isomerase found in the mevalonate pathway gene cluster from Streptomyces sp strain CL190. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:932–937. doi: 10.1073/pnas.020472198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hemmi H, Ikeda Y, Yamashita S, Nakayama T, Nishino T. Catalytic mechanism of type 2 isopentenyl diphosphate : dimethylailyl diphosphate isomerase: verification of a redox role of the flavin cofactor in a reaction with no net redox change. Biochemical and Biophysical Research Communications. 2004;322:905–910. doi: 10.1016/j.bbrc.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 17.Bornemann S. Flavoenzymes that catalyse reactions with no net redox change. Nat Prod Rep. 2002;19:761–772. doi: 10.1039/b108916c. [DOI] [PubMed] [Google Scholar]
- 18.Laupitz R, Hecht S, Amslinger S, Zepeck F, Kaiser J, Richter G, Schramek N, Steinbacher S, Huber R, Arigoni D, Bacher A, Eisenreich W, Rohdich F. Biochemical characterization of Bacillus subtilis type II isopentenyl diphosphate isomerase, and phylogenetic distribution of isoprenoid biosynthesis pathways. European Journal of Biochemistry. 2004;271:2658–2669. doi: 10.1111/j.1432-1033.2004.04194.x. [DOI] [PubMed] [Google Scholar]
- 19.Davisson VJ, Woodside AB, Poulter CD. Synthesis of allylic and homoallylic isoprenoid pyrophosphates. Methods Enzymol. 1985;110:130–144. doi: 10.1016/s0076-6879(85)10068-6. [DOI] [PubMed] [Google Scholar]
- 20.de Ruyck J, Rothman SC, Poulter CD, Wouters J. Structure of Thermus thermophilus type 2 isopentenyl diphosphate isomerase inferred from crystallography and molecular dynamics. Biochem Biophys Res Commun. 2005;338:1515–1518. doi: 10.1016/j.bbrc.2005.10.114. [DOI] [PubMed] [Google Scholar]
- 21.Satterwhite DM. Isopentenyldiphosphate delta-isomerase. Methods Enzymol. 1985;110:92–99. doi: 10.1016/s0076-6879(85)10064-9. [DOI] [PubMed] [Google Scholar]
- 22.Copeland RA. Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis. New York City, NY: Wiley-VCH; 2000. [Google Scholar]
- 23.Yalloway GN, Mayhew SG, Malthouse JP, Gallagher ME, Curley GP. pH-dependent spectroscopic changes associated with the hydroquinone of FMN in flavodoxins. Biochemistry. 1999;38:3753–3762. doi: 10.1021/bi982476p. [DOI] [PubMed] [Google Scholar]
- 24.Murataliev MB. Application of electron spin resonance (ESR) for detection and characterization of flavoprotein semiquinones. Methods Mol. Biol. 1999;131:97–110. doi: 10.1385/1-59259-266-X:97. [DOI] [PubMed] [Google Scholar]
- 25.Dutton PL. Redox potentiometry: determination of midpoint potentials of oxidation-reduction components of biological electron-transfer systems. Methods Enzymol. 1978;54:411–435. doi: 10.1016/s0076-6879(78)54026-3. [DOI] [PubMed] [Google Scholar]
- 26.Ghisla S, Massey V, Lhoste JM, Mayhew SG. Fluorescence and optical characteristics of reduced flavines and flavoproteins. Biochemistry. 1974;13:589–597. doi: 10.1021/bi00700a029. [DOI] [PubMed] [Google Scholar]
- 27.Barkley SJ, Cornish RM, Poulter CD. Identification of an Archaeal type II isopentenyl diphosphate isomerase in methanothermobacter thermautotrophicus. J Bacteriol. 2004;186:1811–1817. doi: 10.1128/JB.186.6.1811-1817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barkley SJ, Desai SB, Poulter CD. Type II isopentenyl diphosphate isomerase from Synechocystis sp. strain PCC 6803. J Bacteriol. 2004;186:8156–8158. doi: 10.1128/JB.186.23.8156-8158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takagi M, Kaneda K, Shimizu T, Hayakawa Y, Seto H, Kuzuyama T. Bacillus subtilis ypgA gene is fni, a nonessential gene encoding type 2 isopentenyl diphosphate isomerase. Biosci Biotechnol Biochem. 2004;68:132–137. doi: 10.1271/bbb.68.132. [DOI] [PubMed] [Google Scholar]
- 30.Yamashita S, Hemmi H, Ikeda Y, Nakayama T, Nishino T. Type 2 isopentenyl diphosphate isomerase from a thermoacidophilic archaeon Sulfolobus shibatae. Eur J Biochem. 2004;271:1087–1093. doi: 10.1111/j.1432-1033.2004.04010.x. [DOI] [PubMed] [Google Scholar]
- 31.Massey V. The chemical and biological versatility of riboflavin. Biochem Soc Trans. 2000;28:283–296. [PubMed] [Google Scholar]
- 32.Massey V, Stankovich M, Hemmerich P. Light-mediated reduction of flavoproteins with flavins as catalysts. Biochemistry. 1978;17:1–8. doi: 10.1021/bi00594a001. [DOI] [PubMed] [Google Scholar]
- 33.Massey V, Hemmerich P. Photoreduction of flavoproteins and other biological compounds catalyzed by deazaflavins. Biochemistry. 1978;17:9–16. doi: 10.1021/bi00594a002. [DOI] [PubMed] [Google Scholar]
- 34.Müller F. Nuclear magnetic resonance studies on flavoproteins. In: Müller F, editor. Chemistry and Biochemistry of Flavoenzymes. Boca Raton, Florida: CRC Press; 1992. pp. 557–595. [Google Scholar]
- 35.Eisenreich W, Kemter K, Bacher A, Mulrooney SB, Williams CH, Jr, Muller F. 13C-, 15N- and 31P-NMR studies of oxidized and reduced low molecular mass thioredoxin reductase and some mutant proteins. Eur J Biochem. 2004;271:1437–1452. doi: 10.1111/j.1432-1033.2004.04043.x. [DOI] [PubMed] [Google Scholar]
- 36.Ghisla S. Fluorescence and optical characteristics of reduced flavins and flavoproteins. Methods Enzymol. 1980;66:360–373. doi: 10.1016/0076-6879(80)66481-7. [DOI] [PubMed] [Google Scholar]
- 37.Massey V, Palmer G. On the existence of spectrally distinct classes of flavoprotein semiquinones. A new method for the quantitative production of flavoprotein semiquinones. Biochemistry. 1966;5:3181–3189. doi: 10.1021/bi00874a016. [DOI] [PubMed] [Google Scholar]
- 38.Kay CMW, Weber S. EPR of radical intermediates in flavoenzymes. In: Gilbert BC, Davies MJ, Murphy DM, editors. Electron Paramagnetic Resonance. London: Royal Society of Chemistry; 2002. pp. 222–253. [Google Scholar]
- 39.Mayhew SG. The effects of pH and semiquinone formation on the oxidation-reduction potentials of flavin mononucleotide. A reappraisal. Eur. J. Biochem. 1999;265:698–702. doi: 10.1046/j.1432-1327.1999.00767.x. [DOI] [PubMed] [Google Scholar]
- 40.Steinbacher S, Kaiser J, Gerhardt S, Eisenreich W, Huber R, Bacher A, Rohdich F. Crystal structure of the type II isopentenyl diphosphate: Dimethylallyl diphosphate isomerase from Bacillus subtilis. Journal of Molecular Biology. 2003;329:973–982. doi: 10.1016/s0022-2836(03)00527-8. [DOI] [PubMed] [Google Scholar]
- 41.Draper RD, Ingraham LL. A potentiometric study of the flavin semiquinone equilibrium. Arch. Biochem. Biophys. 1968;125:802–808. doi: 10.1016/0003-9861(68)90517-1. [DOI] [PubMed] [Google Scholar]
- 42.Eberlein G, Bruice TC. One and two electron reduction of oxygen by 1,5-dihydroflavins. Journal of the American Chemical Society. 1982;104:1449–1452. [Google Scholar]
- 43.Gadda G, Fitzpatrick PF. Biochemical and physical characterization of the active FAD-containing form of nitroalkane oxidase from Fusarium oxysporum. Biochemistry. 1998;37:6154–6164. doi: 10.1021/bi973085y. [DOI] [PubMed] [Google Scholar]
- 44.Fullerton SW, Daff S, Sanders DA, Ingledew WJ, Whitfield C, Chapman SK, Naismith JH. Potentiometric analysis of UDP- galactopyranose mutase: stabilization of the flavosemiquinone by substrate. Biochemistry. 2003;42:2104–2109. doi: 10.1021/bi027077f. [DOI] [PubMed] [Google Scholar]
- 45.Hoshino T, Tamegai H, Kakinuma K, Eguchi T. Inhibition of type 2 isopentenyl diphosphate isomerase from Methanocaldococcus jannaschii by a mechanism-based inhibitor of type 1 isopentenyl diphosphate isomerase. Bioorg. Med. Chem. 2006 doi: 10.1016/j.bmc.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 46.Lu XJ, Christensen DJ, Poulter CD. Isopentenyl-diphosphate isomerase: irreversible inhibition by 3-methyl-3,4-epoxybutyl diphosphate. Biochemistry. 1992;31:9955–9960. doi: 10.1021/bi00156a014. [DOI] [PubMed] [Google Scholar]
- 47.Macheroux P, Ghisla S, Sanner C, Ruterjans H, Muller F. Reduced flavin: NMR investigation of N5-H exchange mechanism, estimation of ionization constants and assessment of properties as biological catalyst. BMC. Biochem. 2005;6:26. doi: 10.1186/1471-2091-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghisla S, Hartmann U, Hemmerich P, Müller F. Die reductive Alkylierung des Flavinkerns 2); Struktur und Reaktivitat von Dihydroflavinen. Liebigs Annalen der Chemie. 1973;8:1388–1415. [Google Scholar]
- 49.Wayner DDM, McPhee DJ, Griller D. Oxidation and reduction potentials of transient free radicals. Journal of the American Chemical Society. 1988;110:132–137. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Available Figures for fluorescence of the guanidinium-released flavin and for binding of oxidized enzyme-FMN to IPP as measured via ultrafiltration assays. Figures and experimental details for dependence of isomerase activity on exogenous FMN and for enzyme inhibition by NIPP and isopentyl diphosphate. Anaerobic spectra of dithionite reduced enzyme, spectra for aerobic reduction of enzyme by NADPH, and re-oxidation of photoreduced enzyme. EPR spectra of enzyme·FMNH. complexes. UV-visible spectra of reductive potentiometric titrations. This material is available free of charge via the Internet at http://pubs.acs.org