Abstract
A general approach was developed for the regio- and chemoselective covalent immobilization of soluble proteins on glass surfaces through an unnatural amino acid created by posttranslationally modifying the cysteine residue in a CaaX recognition motif with functional groups suitable for “click” chemistry or a Staudinger ligation. Farnesyl diphosphate analogs bearing ω-azide or ω-alkyne moieties were attached to the cysteine residue in Cys-Val-Ile-Ala motifs at the C-termini of engineered versions of green fluorescent protein (GFP) and glutathione S-transferase (GST) by protein farnesyltransferase. The derivatized proteins were attached to glass slides bearing linkers containing azide (“click” chemistry) or phosphine (Staudinger ligation) groups. “Click” immobilized proteins were detected by fluorescently labeled antibodies and remained attached to the slide through two cycles of stripping under stringent conditions at 80 °C. GFP immobilized by a Staudinger ligation was detected by directly imagining the GFP fluorophore over a period of 6 days. These methods for covalent immobilization of proteins should be generally applicable. CaaX recognition motifs can easily be appended to the C-terminus of a cloned protein by a simple modification of the corresponding gene, and virtually any soluble protein or peptide bearing a CaaX motif is a substrate for protein farnesyltransferase.
Protein “chips” are useful for studying protein-ligand and protein-protein interactions,i including the detection of antibody-antigen interactions,ii and permit high-throughput screening of limited quantities of analytes in a microarray format.iii Devices with covalently attached proteins1,iv are more robust than their non-covalent counterparts.v In addition, substantial enhancements in sensitivity are seen when proteins are attached in a uniform manner.4,5,vi Typically covalent immobilization is accomplished by reactions that rely on nucleophilic moieties found in the side chains of naturally occurring amino acids.i,vii We now report a general approach for the regio- and chemoselective covalent immobilization of soluble proteins on glass surfaces through an unnatural amino acid created by posttranslationally modifying a cysteine residue with functional groups suitable for “click” chemistryviii or a Staudinger ligation.ix
Protein farnesyltransferase (PFTase) catalyzes the alkylation of the sulfhydryl moiety in the cysteine located in C-terminal CaaX motifs, where X = A, S, M, or Q, by farnesyl diphosphate (1) (see Scheme 1).x The reaction is general for any soluble protein bearing a CaaX motif. We synthesized farnesyl analogs 2 and 3, both of which are excellent alternate substrates for yeast PFTase with catalytic efficiencies (1.6 and 0.58 µM−1s−1) similar to that of 1 (0.76 µM−1s−1).xi As model proteins for immobilization, we engineered C-terminal CVIA motifs into green fluorescent protein (GFP) and glutathione S-transferase (GST). Incubation of the proteins with azide analog 2 or alkyne analog 3 gave GFP-N3, GFP-C2, GST-N3, or GST-C2, respectively.
Scheme 1.
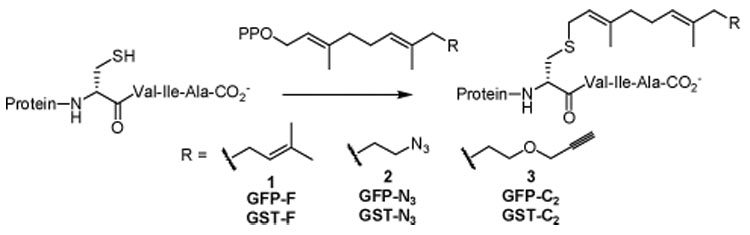
Modification of proteins with CaaX Recognition Motifs
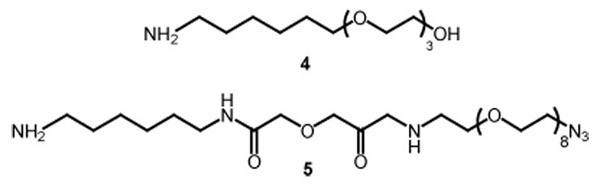
Azide-derivatized glass slides suitable for immobilization of the complementary alkyne-derivitized proteins by “click chemistry” were prepared as outlined in Scheme 2. The surface of amino-activated glass slides was prepared in three steps.xii The amine groups were first converted to the corresponding O-succinimidyl carbamates, and PEG-containing amine linkers were then attached through urea linkages, using a 5:1 molar ratio of 4 to 5.xiii The slides were then treated with ethanolamine to deactivate residual carbamate groups. Phosphine-derivativized glass slides for Staudinger ligations were prepared according to the procedure described by Raines and coworkers.xiv
Scheme 2.
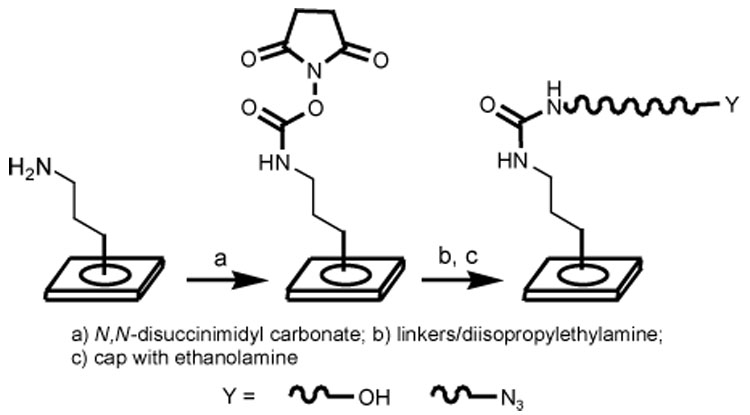
Preparation of glass slides for protein immobilization by “click chemistry”
One microliter samples of solutions of GFP-C2 or GST-C2 in 3:1 (v/v) water:glycerol, containing 1.7 mM Cu+ were spotted into wells on a silicone-masked azido-derivatived glass slide. The slides were maintained at 4 °C in a humidified chamber for 2 h before addition of Block It™ followed by incubation for 5h. The wells were then washed five times with phosphate-buffered saline containing 0.1% Tween 20® (PBST). For slides developed with fluorescent antibodies to GFP and GST, a solution containing 4 µL of the appropriate antibody (4 µg/mL) was added to each well, and the slide was incubated at 4 °C for 16 h. Fluorescence intensities were measured by phosphorimaging.
The results of an immobilization experiment are summarized in Figure 1. The concentrations of GFP-C2 and GST-C2 varied from 1 to 20 µM. No fluorescent signal was seen when the antibodies were added to wells that did not contain GFP or GST proteins (wells 2c, 3c). In order to distinguish between specific and non-specific binding, GFP-F (wells 2a, 2b, 2d, 2e) and GST-F (wells 3a, 3b, 3d, 3e) bearing a farnesyl group instead of the reactive alkyne moiety were used as controls. Background fluorescence was seen for both of the farnesylated proteins. The fluorescence signals for wells containing proteins with covalently attached alkyne groups were easily detected and significantly above background. Over a range of 1–10 µM, the intensity of the signal increased by ~50% for each 2-fold increase in the concentration of the derivatized protein. A maximum intensity was achieved at ~20 µM protein. The intensity of the signals did not increase when the time of the immobilization reaction was extended from 2 to 7 h.
Figure 1.
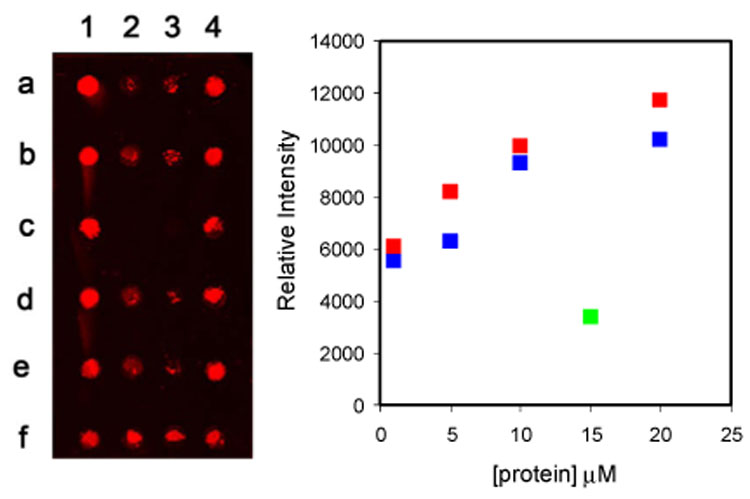
Plate: Fluorescence intensities of immobilized proteins and controls. GFP-C2 – 1 µM (2f), 5 µM (1e, 1f), 10 µM (1c, 1d), 20 µM (1a, 1b); GFP-F – 15 µM (2a, 2b, 2d, 2e); GST-C2 - 1 µM (3f), 5 µM (4e, 4f), 10 µM (4c, 4d), 20 µM (4a, 4b); GST-F – 15 µM (3a, 3b, 3d, 3e); GFP antibody (2c); GST antibody (3c). Plot: Average values of relative fluorescence intensity versus protein concentration. GFP proteins ( ); GST proteins (
); GST proteins ( ); controls (
); controls ( ).
).
The antibodies could be removed by treatment with an acidic saline solution for 2 h at 80 °C. Incubation of the stripped slides with anti-GFP for 16 h at 4 °C, only gave signals corresponding to locations of immobilized GFP. A second cycle of stripping, followed by incubation with anti-GST only gave signals at the locations of immobilized GST. Thus, GFP and GST remain attached to the slides under conditions sufficiently stringent to disrupt interactions between the immobilized proteins and their respective antibodies.
In another experiment, GFP-C2 (25 – 100 µM) was immobilized by the “click” procedure, and the slide was analyzed directly by phosphorimaging without conjugation with fluorescent antiGFP. A fluorescent signal was detected after the slide had been thoroughly washed with PBST. The slide was then allowed to stand in phosphate buffer (pH 7.0) at 4 °C for two days. After two days, the signal had only diminished by 22%. Thus, GFP retained its native fold during the immobilization and subsequent storage in buffer.
Staudinger ligations were performed with GFP-N3 and GST-N3 using slides derivatized with diphenylphosphine groups.14 Preliminary experiments using an aqueous buffer for the immobilization reactions gave slides with high backgrounds for GFP-F and GST-F visualized by fluorescent antibodies. Lower backgrounds were achieved when the ligation was carried out in 50:1 DMF/water. We were concerned that these conditions were not compatible with preserving the native fold of proteins. GFP-N3 was immobilized by the Staudinger ligation. The slides were stored in buffer and visualized by direct imaging of the GFP fluorophore over a period of 6 days. The fluorescence intensity of spots corresponding to immobilized GFP-N3 and the GFP-F control decreased during the first 4 days; however, the difference between the intensities for immobilized GFP and the control remained constant (see Figure 2). After 4 days signals for the control samples were reduced to background levels; whereas, the signal for covalently bound GFP remained constant.
Figure 2.
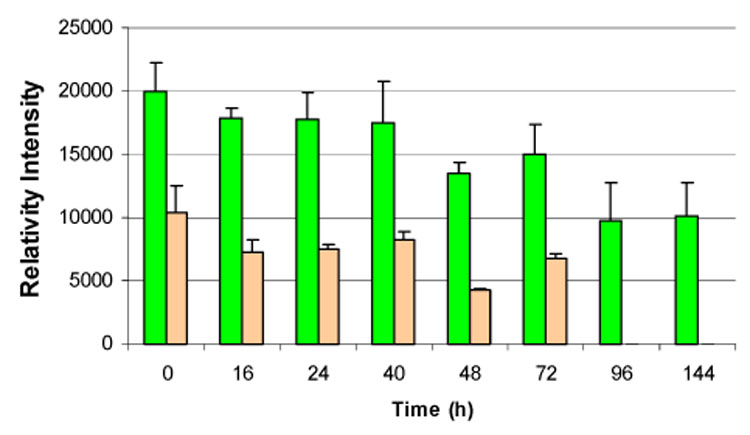
Fluorescence intensities of GFP-N3 ( ) immobilized by Staudinger ligation and the GFP-F (
) immobilized by Staudinger ligation and the GFP-F ( ) control after storage in buffer.
) control after storage in buffer.
In summary, we have developed a new strategy of the regio- and chemoselective immobilization of proteins on surfaces through an unnatural amino acid created posttranslationally at the cysteine residue in a C-terminal CaaX motif. The CaaX recognition motif can be easily appended to the C-terminus of a cloned protein by a simple modification of the corresponding gene. Virtually any soluble protein or peptide bearing the CaaX motif is a substrate for PFTase. The cysteine residue can then be modified with farnesyl analogs bearing either an ω-alkyne moiety useful for immobilization of the protein to a surface by “click” chemistry or an ω-azide for Staudinger ligations. The triazole and amide linkages produced by the respective reactions are chemically robust.
Supplementary Material
Supporting Information Available Procedures for synthesis of linkers, preparation of slides, cloning, protein expression and purification, derivatization and microarray-related experiments. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgements
The authors are grateful to Prof. R. T. Raines for a sample of protected phosphinothiol and helpful discussions. This project was supported by NIH grant GM 21328.
References
- (1).MacBeath G, Schreiber SL. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- (ii).(a) Nielsen UB, Cardone MH, Sinskey AJ, MacBeath G, Sorger PK. Proc. Natl. Acad. Sci. USA. 2003;100:9330–9335. doi: 10.1073/pnas.1633513100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) de Wildt RM, Mundy CR, Gorick BD, Tomlinson IM. Nat. Biotechnol. 2000;18:989–994. doi: 10.1038/79494. [DOI] [PubMed] [Google Scholar]
- (iii).For recent reviews, see:Zhu H, Snyder M. Curr. Opin. Chem. Biol. 2003;7:55–63. doi: 10.1016/s1367-5931(02)00005-4. Wilson DS, Nock S. Angew. Chem. Int. Ed. 2003;42:494–500. doi: 10.1002/anie.200390150. Ramachandran N. Curr. Opin. Chem. Biol. 2005;9:14–19. doi: 10.1016/j.cbpa.2004.12.006.
- (iv).(a) Houseman BT, Huh JH, Kron SJ, Mrksich M. Nat. Biotechnol. 2002;20:270–274. doi: 10.1038/nbt0302-270. [DOI] [PubMed] [Google Scholar]; (b) Toepert F, Knaute T, Guffler S, Pirés JR, Matzdorf T, Oschkinat H, Schneider-Mergener J. Angew. Chem. Int. Ed. 2003;42:1136–1140. doi: 10.1002/anie.200390298. [DOI] [PubMed] [Google Scholar]
- (v).Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, Mitchell T, Miller P, Dean RA, Gerstein M, Snyder M. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- (vi).Sigal GB, Mrksich M, Whitesides GM. J. Am. Chem. Soc. 1998;120:3404–3473. [Google Scholar]
- (vii).Zheng T, Peelen D, Smith LM. J. Am. Chem. Soc. 2005;127:9982–9983. doi: 10.1021/ja0505550. [DOI] [PubMed] [Google Scholar]
- (viii).Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew. Chem. Int. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- (ix).(a) Köln M, Breinbauer R. Angew. Chem. Int. Ed. 2004;43:3106–3116. doi: 10.1002/anie.200401744. [DOI] [PubMed] [Google Scholar]; (b) Watzke A, Kohn M, Gutierrex-Rodrigues M, Wacker R, Schroder H, Brienbauer JK, Alexandrov K, Niemeyer CM, Goody RS, Waldmann H. Angew. Chem. Int. Ed. 2006;45:1–5. doi: 10.1002/anie.200502057. [DOI] [PubMed] [Google Scholar]
- (x).Spence RA, Casey PJ. Enzymes. 2001;21:1–18. [Google Scholar]
- (xi).For other exemples of farnesyl azide derivatives, see:Rose MW, Xu J, Kale TA, O’Doherty G, Barany G, Distefano MD. Biopolymers. 2005;80:164–171. doi: 10.1002/bip.20239.
- (xii).Kanoh N, Kumashiro S, Simizu S, Kondoh Y, Hatakeyama S, Tashiro H, Osada H. Angew. Chem. Int. Ed. 2003;42:5584–5587. doi: 10.1002/anie.200352164. [DOI] [PubMed] [Google Scholar]
- (xiii).Camarero JA, Kwon Y, Coleman MA. J. Am. Chem. Soc. 2004;126:14730–14731. doi: 10.1021/ja0456611. [DOI] [PubMed] [Google Scholar]
- (xiv).(a) Soellner MB, Dickson KA, Nilsson BL, Raines RT. J. Am Chem. Soc. 2003;125:11790–11791. doi: 10.1021/ja036712h. [DOI] [PubMed] [Google Scholar]; (b) Soellner MB, Nilsson BL, Raines RT. J. Org. Chem. 2002;67:4993–4996. doi: 10.1021/jo025631l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Available Procedures for synthesis of linkers, preparation of slides, cloning, protein expression and purification, derivatization and microarray-related experiments. This material is available free of charge via the Internet at http://pubs.acs.org.


