Abstract
In some mammalian systems small interfering RNAs (siRNA) targeting homologous sequences in promoter regions of genes induce transcriptional gene silencing (TGS). We have previously reported the induction of TGS by an siRNA (prom-A siRNA) targeting the tandem NF-κB-binding motifs within the human immunodeficiency virus, type 1 (HIV-1), promoter region. Here we report that induction of TGS by prom-A siRNA is accompanied by immediate and sustained local recruitment of Argonaute-1 (Ago1), histone deacetylase-1 (HDAC1), and induction of dimethylation of histone 3 at lysine 9 (H3K9me2), processes known to be associated with transcriptional silencing. Elevated levels of H3K9me2 and HDAC1 spread upstream of the target sequence, and elevated H3K9me2 levels also spread downstream into the coding region. Moreover, this siRNA induces an immediate change in DNA accessibility to restriction enzyme digestion in the region of the transcription initiation site of the HIV-1. This change in accessibility is because of the relocation of a nucleosome known to be associated with this region of the integrated pro-virus. Although there is a theoretical possibility that the observed viral suppression could be mediated by the PTGS mechanism with this siRNA acting at the 3 ®-long term repeat of the virus, we demonstrate that this siRNA, and three other U3 targeted siRNAs, are inefficient inducers of PTGS. These data strongly suggest that siRNA targeting the promoter region acts predominantly at a site within the 5 ®-long term repeat of HIV to induce transcriptional silencing and alterations to chromatin structure of the HIV promoter region that extend well beyond the immediate siRNA target site. These induced changes are consistent with those described in latent HIV-1 infection.
RNA duplexes can induce gene silencing by a number of mechanisms, including post-transcriptional gene silencing (PTGS),4 transcriptional gene silencing (TGS), and translational silencing by microRNA (miRNA) (1-3). It is accepted that PTGS can be induced by small interfering RNA (siRNA) in a wide range of genes in numerous cell types. It has been also demonstrated that TGS can be induced by RNA duplexes homologous to certain sequences within the promoter regions of genes in human cells, in a similar manner to that previously reported in plants, yeasts, and Drosophila (4-23). However, the ability of siRNAs targeting promoter regions to induce TGS in mammalian cells has been controversial, and the mechanisms underlying this process are still being defined. Recently it has been demonstrated that this process is associated with the recruitment of at least one of the argonaute proteins (Ago1 and/or Ago2), which appear to be central components of the RNA-induced initiation of transcriptional gene silencing (RITS) complex in mammalian cells (18, 19, 24, 25), as has recently been demonstrated in plants and fission yeast (26, 27). Furthermore, the process involves changes to the histone code, involving alteration in methylation status of histone tails (e.g. dimethylation of lysine 9 of histone 3 (H3K9me2) and trimethylation of lysine 27 of histone 3), and the recruitment of both histone deacetylase-3 (HDAC3) and the polycomb group protein, enhancer of zeste homolog 2 (10, 17, 19, 21, 22). These changes are consistent with the formation of repressive chromatin structure (17, 19, 28). Recent reports have shown that the antisense strand of siRNA duplexes, acting alone, interacting with an RNA polymerase II-transcribed promoter-associated RNA species, can direct sequence-specific transcriptional gene silencing (17, 22, 29). This process is associated with recruitment of both DNA methyltransferases and trimethylation of lysine 27 of histone 3.
Peptide nucleic acids can also induce TGS and chromatin changes. These directly target the transcription start site and induce repressive changes in chromatin architecture of the promoter region, which extend through this site (13). It is also becoming clear that miRNAs can also induce both changes in the histone code and chromatin remodeling resulting in repressive chromatin structures. This process is initiated by the ability of miRNA to suppress specific genes at the translational level (30-33). There are therefore several mechanisms by which small RNA species, including siRNA and miRNA, induce transcriptional gene silencing. In each case this silencing is associated with biochemical changes in chromatin consistent with heterochromatin formation. In addition, it has also recently been reported that peptides containing a zinc finger motif, fused with DNA methyltransferase can induce DNA cytosine methylation of certain promoter regions in vitro (34). Therefore the mechanisms underlying heterochromatin formation are multiple and complex and can result from the actions of a number of different regulatory molecules.
We have previously demonstrated that certain siRNAs with sequences homologous to sequences within the U3 region of the 5′-LTR of either the HIV-1 or simian immunodeficiency virus can induce suppression of viral replication by TGS (12, 35). In HIV-1, the effectiveness of this silencing correlates with the degree and density of CpG methylation (12). Here we provide evidence that this process involves the recruitment of components of the RITS complex and changes in the histone code. We demonstrate these changes extend beyond the promoter region and result in the rearrangement of the nucleosome structures adjacent to the transcription start site within the integrated HIV-1 genome. Finally, we demonstrate that these U3-targeted siRNAs are poor inducers of the PTGS pathway, adding weight that this process is the result of the siRNA acting at the promoter region of HIV-1 5′-LTR.
EXPERIMENTAL PROCEDURES
RNA Duplexes—Double-stranded RNA duplexes were synthesized by Dharmacon Research Inc. (Lafayette, CO). The sequences of the RNA duplexes used are as follows: prom-A, 5′-GGGACUUUCCGCUGGGGACTT-3′ (sense) and 5′-GUCCCCAGCGGAAAGUCCCTT-3′ (antisense); prom-B, 5′-GGCCCGAGAGCUGCAUCCGGTT-3′ (sense) and 5′-CCGGAUGCAGCUCUCGGGCCTT-3′ (antisense); prom-C, 5′-GACUGCUGACAUCGAGCUUTT-3′ (sense) and 5′-AAGCTCGATGTCAGCAGTCTT-3′ (antisense); prom-D, 5′-CUGGGGAGUGGCGAGCCCUTT-3′ (sense) and 5′-AGGGCUCGCCACUCCCCAGTT-3′ (antisense); and code-R, 5′-GCCUCAAUAAAGCUUGCCTT-3′ (sense) and 5′-GGCAAGCUUUAUUGAGGCTT-3′ (antisense).
HIV Infection, Transfection of siRNA, and Viral Quantification—5 × 106 MAGIC-5 cells (HeLa cells stably transfected with CD4, CCR-5, and CXCR-4) were infected with the HIV-1NL4-3 subtype B strain (infecting dose equivalent to 38 × 106 copies of HIV-1-RNA), and infection was allowed to establish for 2 days. On day 3, cells were detached with 0.25% trypsin, 1 mm EDTA and washed three times with PBS. Transfection of cells was performed using an AMAXA device (Cologne, Germany) with 12 μl of 20 μm solution of siRNA according to the kit insert. After electroporation the cells were resuspended in 2.5 ml of culture medium in standard 6-well culture plates and maintained according to standard protocols (36). Reverse transcriptase (RT) activity in culture supernatants was determined as described previously (37).
Chromatin Immunoprecipitation (ChIP) Assay—ChIP assays were conducted according to the standard kit procedure (17-295, Upstate). In short, 2-3 million cells were fixed with 1% formaldehyde for 10 min at room temperature and incubated for 5 min with 0.125 m glycine, and after three washes with cold PBS, the pellets were resuspended into 400 μl of SDS lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris, pH 8.0), and the lysate was sonicated for 2 min using a 130-watt ultrasonic processor (Cole-Parmer Inc.) to shear DNA. The lysate was then pelleted by centrifugation at 10,000 × g for 5 min, and the supernatant was kept for immunoprecipitation. Various immunoprecipitating antibodies were added to aliquots of lysate. Antibody was then recovered with salmon sperm DNA/protein-A-agarose bead slurry. The beads were washed and resuspended into TE buffer. DNA was extracted by the standard phenol/chloroform procedure. Real time PCR analysis was then conducted. Antibodies used for ChIP assay were as follows: HDAC1 (SC-782, Santa Cruz Biotechnology), Ago1 (07-599, Upstate), H3K9me2 (07-449, Upstate), histone H3 acetylation (H3Ac, 06-599, Upstate), and RNA polymerase II (pol II, SC-900, Santa Cruz Biotechnology). PCR primers used for real time PCR amplification of DNA isolated by the ChIP process were as follows: LTR-forward, 5′-TACAAGGGACTTTCCGCTGG-3′, and LTR-reverse, 5′-AGCTTTATTGAGGCTTAAGC-3′ for the Nucleosome-1 (Nuc-1) region of the LTR; LTRup-forward, 5′-CCAAAGAAGACAAGATATCCTTGA-3′, and LTRup-reverse, 5′-TCATCCATTCCATGCAGGC-3′ for Nuc-0 region of the LTR; SK145, 5′-AGTGGGGGGACATCAAGCAGCCATGCAAAT-3′; SKCC1B, 5′-TACTAGTAGTTCCTGCTATGTCACTTCC-3′ for gag region. PCR conditions were 94 °C for 3 min, followed by 50 cycles of 94 °C for 15 s, 56 °C for 15 s, and 68 °C for 35 s with AcuuPrime Taq polymerase with 1 mm SYTO-9 (Invitrogen) with Rotor-Gene 4000 (Corbett, Sydney, Australia). HIV copy number was measured with a standard curve generated with serial dilutions of HIV-1 plasmid (pNL4-3) from 10 to 107 copies per reaction.
Chromatin Accessibility Real Time PCR (CHART) Assay—CHART assays were conducted as reported previously (38) with some modifications to specifically target the HIV-1 LTR region. MAGIC-5 cells were washed with cold PBS two times followed by treatment with a lysis buffer (3 mm MgCl2, 10 mm NaCl, 0.5% Nonidet P-40, 10 mm Tris, pH 7.5) for 5 min. Cells were washed with the lysis buffer once followed by another wash with RE-2 buffer (10 mm MgCl2, 50 mm NaCl, 0.03 mm spermine, 0.1 mm spermidine, 10 mm Tris, pH 7.5). The nuclei were suspended into NE Buffer 2 (New England Biolabs, Ipswich, MA) and incubated with 10 units of BglII. DNA was extracted by standard phenol/chloroform procedure. LTR-specific PCR primers used for CHART assay were as follows: LTRkBprimerFOR-1, 5′-AGGTTTGACAGCCGCCTAGCA-3′ and HIVTarREV-1, 5′-TCTGAGGGATCTCTAGTTACCAGAGTC-3′. The cycling conditions employed for real time PCR analysis were 94 °C for 3 min, followed by 50 cycles of 94 °C for 15 s, 60 °C for 30 s, and 68 °C for 45 s with AcuuPrime Taq polymerase with SyBr Green diluted by a factor of 5× 104 (Invitrogen). HIV copy number was determined against serial dilutions of HIV-1 plasmid (pNL4-3).
Assessment of Extent of PTGS Induced by U3 Region-targeted siRNAs—The HIV-1 3′-LTR region was amplified by PCR from HIV-1 plasmid (pNL4-3) with primer set of Nef3′-LTR-Fw1, 5′-CGCTCTAGAGTAGTGTGATTGGATGGCCTGCTGT-3′ (XbaI site underlined), and Nef3′-LTR-Rv1, 5′-CGCGGATCCTGCTAGAGATTTTCCACACTGACTA-3′ (BamHI site underlined). The resulting 3′-LTR amplicon was cloned into an expression vector (pcDNA3.1/Hygro; Invitrogen) using XbaI and BamHI restriction enzyme sites so that its expression was controlled by the CMV immediate early promoter. Resultant purified plasmids were screened by restriction enzyme digest, and correct insertion was confirmed by sequence analysis. HeLa cells were transfected with a clone of the resulting HIV-1 3′-LTR plasmid using Oligofectamine-2000 (Invitrogen) according to the manufacturer's instructions. Hygromycin selection (300 μg/ml) was carried out 2 days after transfection. After 10 days, cloning by limiting dilution was performed. After expansion of these cultures for 50 days, expression of the 3′-LTR was screened for by reverse transcriptase-PCR using the primer set of NUAf, 5′-CCAAAGAAGACAAGATATCCTTGA-3′, and Chips2r, 5′-GCAGCTGCTTATATGCAGCATCTG-3′. A stably transfected clone with high levels of 3′-LTR expression, designated CMV-3LTR1-4, was selected for further study.
RESULTS
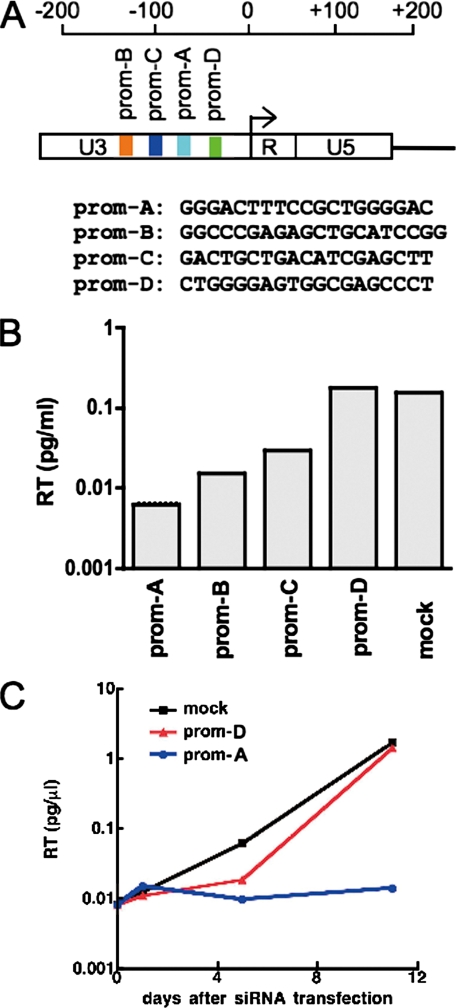
siRNA Targeting HIV-1 Promoter Induces Gene Silencing—Four siRNAs were designed with sequences homologous to target sequences within the U3 promoter region of HIV-1 LTR (Fig. 1A). MAGIC-5 cells were infected with the HIV-1NL4-3 strain. Two days later separate cultures were transfected with each of the four synthetic siRNAs. New virus production was substantially reduced following transfection of prom-A and prom-B siRNAs. Prom-C siRNA had a less marked effect on viral production, whereas prom-D siRNA had a transient effect at day 6 post-transfection that was lost by day 11 (Fig. 1B). We employed two of four siRNAs for further analysis on virus production as follows: prom-A, which induced the most efficient suppression, and prom-D, which induced the least efficient suppression. Furthermore, we confirmed that prom-A siRNA substantially reduced both cell-associated viral RNA levels and syncytia formation compared with mock-transfected cultures, whereas prom-D siRNA had only a marginal effect on either of these read-outs (Fig. 1C and supplemental Fig. 1, A and C). Proviral DNA was detected in all cultures (supplemental Fig. 1B), providing evidence that in this system productive viral infection is suppressed at a stage of the viral life cycle after reverse transcription. These data are similar to our previously reported results, where we demonstrated by nuclear run-on assays that inhibition of viral replication by prom-A siRNA was because of silencing of HIV-1 transcription (12).
FIGURE 1.
HIV replication is inhibited by siRNA targeting the HIV-1 promoter. A, map of the HIV-1 5′-LTR, with the location of sequences targeted by the four siRNAs indicated, prom-A, -B, -C, and -D. The arrow marks the HIV-1 transcription start site. The number indicates the relative location from the transcription start site. B, various levels of inhibition are induced by the four siRNAs targeting the HIV promoter. Reverse transcriptase (RT) levels at day 6 post-transfection of siRNA in supernatant of viral cultures are shown. C, effect of prom-A and -D siRNAs on the time course of HIV-1NL4-3 production in MAGIC-5 cells. Prom-A siRNA reduces viral production 200-fold by day 11 post-transfection compared with prom-D siRNA or mock transfection. The zero time point, when siRNAs are transfected into the cells, is 2 days after infection of the cells with HIV-1NL4-3. Reverse transcriptase (RT) levels are shown for cultures transfected with prom-A siRNA (blue circle), prom-D siRNA (red triangle), and mock-transfected cells (black square).
Ago1, H3K9me2, and HDAC1 Are Recruited to the Promoter Region—Having confirmed our previous observations, we set out to demonstrate that siRNA-induced TGS of HIV-1 was associated with recruitment of important components of RITS and alterations in histone methylation status. We performed chromatin immunoprecipitation (ChIP) analyses using antibodies to Ago1, an integral component of the RITS complex (18, 19), and to dimethylated histone 3 at lysine 9 (H3K9me2), a marker of heterochromatin formation (39, 40). Transfection of prom-A siRNA into productively infected cultures was associated with recruitment of Ago1 (Fig. 2A) and induction of H3K9me2 (Fig. 2B) in the U3 region of the viral LTR by day 1 post-transfection. These changes were maintained until at least day 11 post-transfection. Transfection of prom-D siRNA was not associated with recruitment of Ago1 and induced less marked enrichment of H3K9me2 (Fig. 2, A and B).
FIGURE 2.
Enrichment of Ago1, H3K9me2, and HDAC1 associated with HIV-LTR in prom-A siRNA-transfected cells. ChIP assays were conducted at days 1 and 11 post-transfection on extracts from formaldehyde-fixed MAGIC-5 cells treated with prom-A, prom-D, or mock transfection. DNA fragments from whole-cell extracts were co-precipitated with antibodies against the following: A, Ago1; B, H3K9me2, and C, HDAC1, and then amplified by PCR using the LTR-forward and LTR-reverse primer pair (the position of this amplicon is between -110 and +80 relative to transcription start site). HIV-1-LTR copy numbers obtained from each immunoprecipitation were normalized against that obtained from whole-cell extracts. Each value shown is the relative enrichment of three separate experiments (mean ± S.E.) normalized to the value obtained from the mock transfection culture.
Furthermore, we used ChIP to investigate the recruitment of histone deacetylase-1 (HDAC1) to this region. Treatment with prom-A siRNA was followed by an enrichment of HDAC1 bound to HIV-1 LTR (Fig. 2C). This change occurred within 24 h and was sustained for 11 days. This is particularly significant because previous studies have demonstrated that elevated levels of HDAC1 are associated with the 5′-LTR of latent forms of HIV-1 and repressive changes in chromatin structure of the HIV-1 promoter region (41-48). In contrast, prom-D induced a less marked recruitment of HDAC1 at day 1. However, consistent with the viral production profile in this culture, this recruitment was not sustained, and by day 11 there appeared to be less HDAC1 associated with HIV-1 LTR in the prom D-treated cultures than the mock-transfected cultures. In addition we assessed H3 acetylation status and recruitment of pol II at the Nuc-1 region at day 5 (supplemental Fig. 2). The data showed a failure of enrichment of H3Ac and an inhibition of recruitment of pol II in prom-A siRNA-treated cultures compared with mock-treated cultures. In contrast, there was substantial maintenance of levels of H3Ac, but only partial recruitment of pol II, to this region in prom-D siRNA-treated cultures. These data suggest the induction of stable heterochromatin in prom-A siRNA-treated cultures, whereas in the prom-D siRNA-treated cultures there are partial changes in this region of chromatin, resulting in partial and transient inhibition of transcription.
siRNA Induces Recruitment of a Nucleosome at the Transcription Initiation Site—The above data suggested that prom-A siRNA treatment may have induced sustained alterations in the structure of the chromatin associated with the LTR of HIV-1, although prom-D did not. The architecture of the nucleosomes associated with HIV-1 5′-LTR after integration has been well described (41-43) and is summarized in Fig. 3A. Previous reports have suggested that during active HIV-1 transcription, Nuc-1 is shifted or disrupted from its position close to the transcription initiation site in latent infection to a more downstream position (41-46, 48). We designed a CHART assay (38) based on the hypothesis that during active transcription, the BglII site 20 bases downstream of the transcription initiation site would be accessible to digestion, whereas during transcriptional silencing, re-positioning of Nuc-1 would induce relative protection of this site. The accessibility of HIV-1 LTR region to BglII digestion was quantified before and after restriction enzyme digestion by real time PCR using primers flanking this region (Fig. 3A). First, we demonstrated that the BglII site, in nuclei isolated from productively infected cells, was susceptible to digestion, with the extent of digestion increasing with incubation time. Digestion of the same substrate by EcoRI did not result in any cutting of this region, supporting the specificity of this assay (Fig. 3, B and C). Furthermore, we confirmed that the gag copy number was similar across each sample confirming similar levels of pro-virus in each culture (Fig. 3D).
FIGURE 3.
The BglII site in HIV-1 promoter is protected after prom-A siRNA transfection as determined by CHART assay. A, design of CHART assay. Schematic figures show the relative positions of nucleosomes within the 5′-LTR of HIV-1. The nucleosomes (Nuc-0, -1, and -2) are precisely positioned in the HIV-1 LTR after integration of viral DNA into host genome (diagram modified from Ref. 41). Nuc-1 is tightly associated with the transcription initiation site, indicated by the arrow, in the silenced template. The position of Nuc-1 is shifted or disrupted following activation of viral transcription. A BglII site is located 20 bases from transcription initiation position close to the putative location of Nuc-1 in latent virus. The NF-Tar PCR primer pair was used to amplify an amplicon spanning the BglII site (indicated by double arrows) from nuclear DNA. DNA accessibility was assessed by the percentage decrease in HIV copy number measured by real time PCR with and without BglII digestion. B, productive infection is associated with an accessible BglII site. The nuclear fraction of MAGIC-5 cells productively infected with HIV-1 was subjected to BglII digestion at 37 °C for periods up to 60 min, and the extent of digestion was assessed by real time PCR. C, EcoRI digestion of the same DNA did not result in any change in real time PCR readout, supporting the specificity of the BglII CHART assay. D, real time PCR of the gag region confirms similar levels of cell-associated virus were present in all samples. In the following experiment, results are reported as the extent of digestion after 60 min of incubation of nuclei with BglII. E, CHART assay was applied to productively infected MAGIC-5 cells at days 1 and 11 after prom-A siRNA, prom-D siRNA, or mock transfection. In productively infected cells, the BglII site is highly susceptible to digestion with close to 100% of LTR copies cut. Within 24 h of transfection of prom-A siRNA, the BglII site is protected, and this protection is maintained for at least 11 days. Transfection with prom-D siRNA is associated with transient partial protection of this site. Protection of the BglII site suggests an alteration in chromatin structure that reduces DNA accessibility because of the re-positioning of Nuc-1 as proposed in A.
Next we asked if siRNA transfection induced a change in the accessibility of this BglII site. Nuclei isolated from MAGIC-5 cells, productively infected with HIV-1 for 2 days, had a highly accessible BglII site prior to siRNA transfection, suggesting open chromatin at the HIV transcription start site (Fig. 3E). In the same cultures, 24 h after transfection with prom-A siRNA, there was a marked change in the accessibility of this site, such that <3% of sites within the treated culture were susceptible to BglII digestion compared with >75% in the mock-transfected control. This difference was maintained and became more marked by day 11 post-transfection (Fig. 3E). In prom-D-treated cultures, there is a transient and less marked protection of the BglII site at day 1 post-transfection, which is lost by day 11. These changes are consistent with prom-A siRNA, but not prom-D siRNA, inducing sustained changes in chromatin architecture in the region of the transcription initiation site and are consistent with the changes in Nuc-1 positioning predicted by the model presented in Fig. 3A. These data are again consistent with the changes seen over time in viral production in cultures treated with these siRNAs. These data are both compatible with and extend our ChIP-based observations regarding siRNA-induced changes in chromatin of the HIV-1 promoter region.
Closed Chromatin Structure Is Induced in the Upstream Promoter Region and the Coding Region of the HIV-1 Genome by siRNA—Recent reports regarding siRNA-induced TGS in yeast revealed that heterochromatin-specific chromatin modifications, such as H3K9me2, spread beyond the nucleation site provided by the siRNA-induced RITS complex, resulting in global heterochromatin formation (8, 49, 50). To explore the extent of heterochromatin formation, we performed further ChIP analyses for enrichment of H3K9me2 and HDAC1 at sites within both the upstream promoter and down-stream coding regions of HIV-1. We focused on the Nuc-0 region ∼300 bp upstream of the siRNA target sites and a region of gag about 800 bp downstream. At day 11 we found enrichment of both HDAC1 and H3K9me2 in the Nuc-0 region, in cultures silenced by prom-A siRNA treatment, but not in cultures treated with prom-D siRNA or mock transfection (Fig. 4A). ChIP analyses of the gag region showed elevated H3K9me2 levels in the silenced cultures but no enrichment of HDAC1 (Fig. 4B). These data suggest siRNA-induced TGS of HIV-1 is associated with extended regional heterochromatin formation, as indicated by H3K9me2, which spreads across the adjacent HIV promoter and well into the coding regions of the genome. HDAC1 recruitment is, however, tethered to regions closer to the siRNA target site (Fig. 4C).
FIGURE 4.
Enrichment of H3K9me2 associated with HIV-LTR and HIV-gag in prom-A siRNA-transfected cells indicates extensive regional heterochromatin formation. ChIP assays were conducted as in Fig. 2 at day 11 post-transfection with antibodies against HDAC1 and H3K9me2 for the analysis of the following: A, upstream Nuc-0 region; B, downstream site within the coding region of gag. The Nuc-0 region was amplified using the LTRup-forward and LTRup-reverse primer pair for quantitative analysis by real time PCR (the position of this amplicon is between positions -435 and -299 relative to the transcription start site). The gag region was amplified with the primer pair SK145 and SCCIB (the position of this amplicon is between +1359 and +1513 relative to transcription start site). Values shown are the relative enrichment of three independent experiments (mean ± S.E.) normalized to the value obtained from the mock-transfected culture. C, model of heterochromatin formation induced by siRNA targeting HIV-1 promoter region. siRNA acts as a nucleation center for recruitment of the RNA-induced transcriptional silencing (RITS) complex and closed chromatin formation extends both upstream and downstream to include adjacent promoter and mRNA coding regions. siRNA (purple line) is loaded into Ago1. The RITS machinery, including histone deacetylase (HDAC1) and histone methyltransferase (HMT), induces H3K9me2 (blue flag) and recruits heterochromatin protein 1 (HP1) to the area surrounding the siRNA target site. Complexes including HP1 and HMT spread beyond the initial site of recruitment, creating broad domains of heterochromatin structure as indicated by H3K9me2 status of both the upstream promoter and downstream gag regions.
siRNA Targeting HIV-1 U3 Region Induces Limited Gene Silencing by the PTGS Pathway—During the process of reverse transcription, HIV-1 creates two identical LTR regions as follows: the 5′-LTR, which acts as a promoter, and the 3′-LTR, which plays a role in the termination of transcription of mRNA, providing the machinery for polyadenylation (51). Therefore, siRNAs with homology for the U3 region have the potential to act as inducers of PTGS-mediated degradation of all HIV mRNA species, both spliced and unspliced. To evaluate the contribution of these 3′-LTR sites to the observed reduction in viral mRNA and viral replication, siRNAs were tested on a HeLa cell clone stably transfected with the HIV-3′-LTR expressed under the immediate early CMV promoter, (CMV-3LTR1-4, Fig. 5, A and B). CMV-3LTR1-4 HeLa cells were transfected with each of the four siRNAs targeting the U3 region and an siRNA targeting a sequence in the R region of the LTR, which is an effective inducer of PTGS as a positive control (Fig. 5, A and C). In this system, no significant reduction in LTR mRNA was observed with any of four siRNAs targeting U3 regions, whereas the siRNA targeting the R region reduced these levels by ∼70%. These data suggest that siRNAs targeting the HIV U3 region, including prom-A siRNA, show limited capacity to induce PTGS. These data, along with the other data presented here and previously (12), suggest that prom-A siRNA inhibits productive HIV-1 infection primarily through TGS.
FIGURE 5.
Short siRNAs targeting the U3 region of the HIV-1 promoter have limited PTGS activity. A, map of the HIV-1 3′-LTR under the control of the immediate early CMV promoter, with the location of sequences targeted by four previously described siRNAs and code-R siRNA. Code-R siRNA has sequence homologous to part of the R region of HIV-1 3′-LTR upstream of the polyadenylation sites. The position of PCR primers used for the detection of mRNA of HIV-LTR is indicated with arrows. B, identification of two clones stably transfected with high expression of HIV-3′-LTR under the immediate early CMV promoter. RT-PCR was used to detect 3′-LTR expression, and two expressing clones are shown. One clone, CMV-3LTR1-4, with the highest level of expression of 3′-LTR messenger RNA, was chosen for further study. C, assessment of the extent of gene silencing by PTGS following transfection of clone CMV-3LTR1-4, with each of four U3 region-targeted siRNAs or an R region-targeted siRNA. LTR mRNA levels were assessed by real time RT-PCR 48 h after transfection. Real time PCR data are shown as a relative reduction in HIV-mRNA levels normalized to the value obtained from mock transfection experiments. Results shown are from three independent experiments (mean ± S.E.).
DISCUSSION
ChIP analyses of viral cultures suppressed by prom-A siRNA revealed that transcriptional silencing of HIV-1 is accompanied by the recruitment of Ago1, induction of dimethylation of H3K9, and recruitment of HDAC1 within the chromatin of the HIV-LTR in the immediate vicinity of the siRNA target site. Furthermore, we demonstrate that these siRNA-induced epigenetic changes extend well beyond the U3 region, into both the adjacent promoter sequences upstream and into the coding regions of the HIV-1 genome downstream of the target site. In addition, these induced biochemical changes are accompanied by structural changes in the chromatin associated with the transcription start site of HIV-1. Specifically, we demonstrate that there is an associated alteration in the position of Nuc-1. These changes are initiated within 24 h of transfection of the siRNA and are maintained over a period of 11 days. These findings are consistent with, and extend, current models of siRNA-induced TGS, which suggest RNA duplexes act as a nucleation site for recruitment of the RITS complex (8, 49, 50). Our data are consistent with that found in other mammalian systems, where it has been shown that recruitment of Ago1 is rapid and is followed by a range of biochemical changes in the promoter-associated chromatin (19).
In our system, the maintenance of the changes in chromatin, despite considerable cell division, suggests that the changes must be maintained or sustained by an active process during cell division, allowing the induced changes to be passed onto daughter cells. This process is unlikely to be maintained by the direct ongoing action of the transfected siRNAs as there would inevitably be considerable reduction in siRNA concentration because of both cell division and the degradation of siRNA species.
The two siRNAs studied here, prom-A and prom-D, both target transcription factor-binding sites within the HIV-1-LTR (NF-κB and Sp-1, respectively) separated by only 50 bp. However, their efficacy differs markedly, both in terms of viral suppression and induction of changes in chromatin biochemistry and structure. Prom-D siRNA induces reproducible but modest and transient reductions in reverse transcriptase activity in culture supernatant (Fig. 1C). It induces some initial local changes in chromatin structure and biochemistry, such as partial protection of the BglII site (Fig. 4E), and slight enrichment of H3K9me2 and HDAC1 (Fig. 2, B and C). Unlike the changes observed after prom-A siRNA transfection, these changes are transient and do not spread to adjacent regions of the genome (Fig. 4). Therefore, the extent and duration of viral suppression correlates with the extent of changes in chromatin biochemistry and structure, as well as the extent and density of CpG methylation, of the HIV-1 promoter region (12).
The recruitment of Ago1 by prom-A, but not prom-D, is consistent with the relative efficacy of the two siRNAs. The time course of its recruitment is consistent with other reports regarding siRNA-induced TGS in mammalian models. Others have observed that peak enrichment of Ago1, but not Ago2, occurs 12 h after transfection. Thereafter, enrichment of Ago1 was maintained for at least 30 h but at substantially lower levels (19). It may be that the relative enrichment of Ago1 and Ago2 varies with cell type and/or the targeted promoter. This has been demonstrated in an alternative mammalian system, where siRNA-induced TGS is associated with sustained enrichment of both Ago proteins (18). Although our data and that of others suggest that argonaute proteins can play a central role in both miRNA- and siRNA-directed TGS in mammals by forming the large protein complexes of RITS, as has been suggested in fission yeast and plant systems (18, 26, 27, 52), the exact role of each of the four argonaute proteins in this process is still unclear, especially in mammalian systems. Furthermore, in currently published data, there is no evidence that directly shows that any argonaute protein loaded with siRNA interacts with the target promoter DNA, or that these proteins direct the induction of chromatin modification in mammalian cells. Although Ago2 has enzymatic activity, which plays an important role in PTGS, the importance of this protein relative to Ago1 in TGS is unclear. Our data, however, indicate that Ago1 appears important as it is recruited to the HIV-1 LTR in cultures treated with the effective prom-A siRNA but not in cultures treated with the relatively ineffective prom-D siRNA. The elevated H3K9me2 in HIV-1-LTR in prom-D siRNA-transfected cultures, despite substantial HIV RT activity and therefore viral transcription (Fig. 1C and Fig. 2B), needs further explanation. Given the observations regarding the time course of Ago1 recruitment mentioned above (19), an initial weak enrichment of Ago1 following prom-D siRNA treatment may have been missed here because of the time points studied. This weak recruitment of Ago1 and the RITS complex may have induced the transient suppression of viral production observed and the associated weak induction of H3K9me2, the failure of maintenance of HDAC1 enrichment in the Nuc-1 region, and the failure of recruitment of either HDAC1 and H3K9me2 to the upstream promoter region (Fig. 1C, Fig. 2, and Fig. 4A). This apparent disconnect with a lack of detectable Ago1 recruitment but enrichment of H3K9me2 status has been observed previously in a mammalian model at the 24-h time point after siRNA treatment (19). Another possible interpretation is that one of the other Ago family proteins is critical for initiation of recruitment of RITS in this system, and the differential effectiveness of these two siRNAs is related to the relative ability of these two siRNAs to recruit this protein. The dissection of this observation requires further experimental work. However, it should be noted that at day 5 the partially effective prom-D siRNA also resulted in a partial reduction in the recruitment of pol II to HIV-1 DNA in this region and a simultaneous failure to reduce the acetylation status of the H3 associated with the Nuc-1 region (supplemental Fig. 2). Taken together these data suggest that prom-D is acting as a partially effective repressor, the actions of which result in some changes consistent with heterochromatin in the Nuc-1 region; however, overall, viral transcription is in an active mode. These observations indicate that the relative potency of an siRNA in terms of induction of TGS may not only relate to the sequences targeted but their ability to effectively bind to and recruit Ago proteins; however, confirmation of this requires further experimental work.
As noted above, the time course of H3K9me2 induction reflects the time course of viral production following transfection with both siRNAs. Prom-D siRNA results in a short term reduction in viral load production to day 5, which is not maintained at day 11. Similarly, there is some early induction of H3K9me2 by prom-D. This weak induction is maintained to day 11. By contrast, the sustained reduction in viral production following treatment with prom-A siRNA is mirrored by more profound and sustained induction of H3K9me2. The partial induction of heterochromatin status induced by prom-D siRNA was also observed in the chromatin accessibility assay. These differences may reflect the effectiveness of the two siRNAs. Accumulating evidence suggests that H3K9me2 as well as another protein that is essential for heterochromatin formation, heterochromatin protein-1 (HP1), can counterintuitively be associated with certain genes undergoing active transcription (53, 54, 56-60). The association of these heterochromatin proteins with transcribed genes might reflect regulatory chromatin-modifying activities that are essential for the re-establishment of chromatin structure following transcription. The balance between the silencing and anti-silencing factors is crucial for determining the transcriptional status of a locus (8, 61-63). These observations strongly suggest that the transcriptional activity of the integrated HIV-1 genome cannot be determined by one single marker of heterochromatin formation, such as the induction of H3K9me2. Rather, sustained transcriptional silencing appears to require the induction of a complex of changes in chromatin, which include as a minimum the sustained recruitment of Ago1, the induction of H3K9me2, and the recruitment and maintenance of HDAC1 (Fig. 2). Other processes are likely to be involved such as inhibition of pol II recruitment and the acetylation status of H3 (supplemental Fig. 2). The exact components of RITS involved in this process require further investigation.
Although there are previous reports suggesting that increased levels of HDAC1 are associated with the promoter regions of both latent forms of HIV-1 (41-48), and of integrated viral forms silenced by miRNAs targeting the TAR region downstream of the promoter region of HIV-1 (33), there are no previous reports demonstrating the recruitment of HDAC1 to chromatin in association with siRNA-induced TGS. The recruitment of HDAC1 is associated with the presence of DNA methyltransferases (DNMT), particularly DNMT3A (17). Because we had previously shown that prom-A siRNA induces CpG methylation of the HIV-1-LTR (12), and others have shown that TGS of the EF1A gene is associated with the recruitment of DNMT3A (17), we reasoned that it was likely that HDAC1 would be recruited as part of the process of siRNA-induced TGS and the associated histone and DNA modifications. Our ChIP analyses show that HDAC1 is recruited to the HIV promoter following treatment with prom-A siRNA. Others have shown that the antisense strand of RNA duplexes associate with a complex that contains HDAC3 by protein pulldown assays (17). HDAC3 has been shown to associate with Suv39, histone methyltransferase (HMT), and HDAC1 in repressive chromatin structures (64). These observations, along with our previous observations (12), strongly suggest that the process of viral suppression by siRNA-induced TGS involves recruitment of complexes that induce both DNA and histone methylation.
Treatment with prom-A siRNA induced enrichment of H3K9me2 not only at the targeted site within the promoter region but also well upstream in the U3 region and well downstream into the coding region of the genome. HDAC1 is enriched throughout the promoter region but not in the coding region. These observations suggest the siRNA acts as a nucleation site from which changes in chromatin structure extend upon induction and maintenance of effective TGS. This observation is consistent with those made in recent publications that showed, using genome-wide ChIP-on-chip analysis, that histone methylation is observed in both promoter and coding regions of repressed genes (8, 65). Furthermore, it has been shown that H3K9me2 serves as a binding region for HP1, initiating the formation of heterochromatin (64, 66). In the fission yeast Schizosaccharomyces pombe, the mechanism of siRNA-mediated induction of heterochromatin is well studied (25). In that system RITS contains Ago1 and siRNAs that appear to form a nidus at the targeted site for heterochromatin formation with recruitment of the Clr4 histone methyltransferase (HMT), which induces H3K9 methylation and subsequent recruitment of histone deacetylases (HDACs). H3K9 methylation can spread through the activity of an RNA-dependent RNA polymerase-2 (RdR2)-containing complex, which interacts with siRNA loaded into Ago1-RITS complex to synthesize a double-stranded RNA using the cleaved transcript as template. The synthesized double-stranded RNA is subsequently processed by Dicer producing secondary siRNAs, which is subsequently loaded to form a secondary Ago1-RITS complex. Then HMT is recruited to modify histone tails, allowing heterochromatin to spread to adjacent regions (49, 50). However, the precise mechanism as to how siRNA induces both local heterochromatin formation and its spread to adjacent regions in mammalian cells requires further investigation as the mechanism that has been invoked in plants, which appears to be dependent on RdR2, is not present in genomes of mammalian cells.
An siRNA targeting the HIV-1 U3 region has two potential target sites as follows: a TGS target site in the U3 region of 5′-LTR, the HIV-1 promoter region, and a PTGS target site in the U3 region of 3′-LTR, which is transcribed prior to termination of transcription of all HIV mRNA species, spliced or un-spliced (51). To evaluate the contribution of mRNA degradation induced by the siRNA-targeting HIV-1-LTR region, we used a HeLa cell clone stably expressing the HIV-1 3′-LTR. Based on our data, both prom-A and prom-D siRNAs were poor mediators of PTGS (Fig. 5C). It should be noted that all four U3 targeted siRNAs we tested, prom-A, -B, -C, and -D, scored poorly against criteria for prediction of the effectiveness of siRNAs as mediators of PTGS (67). These observations, along with the data presented in our previous report (12), strongly suggest that the observed viral suppression, mediated by transfection of prom-A siRNA, is mediated predominantly through TGS. Recent reviews suggest that certain siRNAs may mimic miRNAs and act at a translational level (68, 69). However, given that we have previously demonstrated that prom-A abrogates transcriptional activity of HIV-1 in a run-on assay (12), any action at a translational level would be a minor component of the observed inhibition of viral production. Given this earlier observation, we have not tried to entirely rule out the possibility of the contribution of an miRNA-like activity but argue that if there is such a contribution it is minor.
The reasons for the observed differences in efficacy to these two siRNAs (prom-A and -D) are not as yet clear. A cogent explanation requires a greater understanding of the underlying mechanisms by which siRNAs induce TGS. This is emphasized by the recent observations that certain dsRNAs targeting promoter regions have been shown to induce activation of the target gene expression. This activation may be target gene-specific, or nonspecific, off-target gene activation (22, 31, 70). Specific target gene activation has been demonstrated using promoter-targeted dsRNAs to several mammalian genes (31, 70). As yet the characteristics differentiating activating from inhibitory promoter-targeted dsRNAs have not been defined; however, these are likely to be subtle and complex (13, 16, 70). Furthermore, although nonspecific “off target” activation has been described by certain antisense and dsRNAs targeting the 5′-LTR of HIV-1 at sites over 100 bases upstream of the prom-A siRNA target site (22), the determinants of these effects are unclear at this point. In that system, other antisense oligomers overlapping the same target sequence or longer constructs, including the same target sequence, do not induce the same nonspecific effects (22). We have previously demonstrated that the effects of prom-A siRNA are virus-specific, having no effect on cell growth characteristics, expression of CD4 or chemokine co-receptors for HIV, or on other genes regulated by NF-κB (12).
Clearly, further work is required before there is a clear understanding of the properties that differentiate effective promoter-targeted siRNAs from ineffective siRNAs and activating RNAs. One testable hypothesis, however, is related to the fact that prom-A and prom-D target sequences that include the binding motifs of the transcription factors NF-κB and Sp-1 respectively. The effectiveness of these siRNAs may relate either to the extent to which the chromatin modifications they induce interfere with the ability of these transcription factors to access their target sites. Alternatively the chromatin changes induced by both siRNAs may have similar inhibitory effects on the access of these transcription factors to their respective binding sites, but a differential in the relative strength of their effects of these two on HIV-1 transcription results in the observed differences in effectiveness. This hypothesis is currently being explored in the laboratory. As mentioned above another possible explanation is that there is a difference in the ability of siRNAs to bind to Ago proteins and therefore in their ability to recruit components of the RITS complexes. The differential effects of these two siRNAs on Ago1 recruitment suggest that this hypothesis is worthy of exploration.
In summary, we show that effective induction and maintenance of siRNA-directed TGS correlates with the recruitment of Ago1, HDAC1, and factors governing repressive histone modifications such as of H3K9me2 as well as the methylation of DNA (12). Hence, chromatin-modifying repressor complexes appear to play a crucial role in TGS-induced heterochromatin formation. Based on our findings we propose a preliminary model as shown in Fig. 4C. Following the targeting of the Ago1-containing siRNA-RITS complex, repressors are recruited to the vicinity of Nuc-1. Such recruitment could either be an active process through interaction with protein domains within the RITS complex or could be a consequence of interference with an activator binding to this region, preventing co-activator complexes from keeping this region in an open chromatin formation. Subsequently, the action of co-repressor complexes leads to methylation of histones and DNA, by way of enlisting heterochromatin protein complexes, including HP1 and polycomb protein complexes (8, 49, 50), resulting in the establishment of a heterochromatic configuration within this region. Recent reports that suggest the formation of H3K9me2 are critical for recruitment of HP1 in Drosophila, and yeast support this model (53, 54, 59, 60, 72, 73). Further support for this model comes from the observation that HMT and HP1 are recruited to the 5′-LTR in latent HIV infection (48). Future experiments should provide evidence for the direct involvement of specific activators, such as NF-κB, in this process and the precise time frame and order in which HMTs and DNMTs are recruited.
The changes induced during siRNA-induced TGS are consistent with those previously reported in in vitro models of latent HIV-1 infection (41-46, 48). Furthermore, the related retrovirus HTLV-1 has naturally occurring latent states of infection, which is associated with changes in DNA methylation and chromatin similar to those described here (74-78). The regulation of these changes requires further study not only for our understanding of the mechanism of action of these siRNAs but also because one of the roadblocks to effective therapy of HIV-1 is a reservoir of transcriptionally silent provirus in long lived lymphocytes (71, 79, 80). Understanding the precise molecular mechanisms of heterochromatin establishment in effective induction of siRNA-directed TGS will have important implications for future therapeutic approaches for HIV-1 infection.
Supplementary Material
Acknowledgments
We thank Drs. Oka and Tastumi for kindly providing MAGIC-5 cells. HIV infectious molecular clone of pNL4-3 was obtained from AIDS Research (catalog number 114), National Institutes of Health. The National Centre in HIV Epidemiology and Clinical Research is supported by the Commonwealth Department of Health and Aging through the Australian National Council on AIDS, Hepatitis C, and Related Diseases.
Author's Choice—Final version full access.
This work was supported in part by a Project Grant from the National Health and Medical Research Council of Australia. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: PTGS, post-transcriptional gene silencing; Ago, Argonaute; CHART, chromatin accessibility real time PCR; ChIP, chromatin immunoprecipitation; EZH2, enhancer of zeste homolog 2; H3K9me2, dimethylation of histone 3 at lysine 9; H3Ac, histone 3 acetylation; HDAC, histone deacetylase; HMT, histone methyltransferase; HP1, heterochromatin protein-1; pol II, RNA polymerase II; RITS, RNA-induced initiation of transcriptional gene silencing; siRNA, small interfering RNA; TGS, transcriptional gene silencing; LTR, long terminal repeat; HIV, human immunodeficiency virus; DNMT, DNA methyltransferase; CMV, cytomegalovirus; RT, reverse transcriptase; dsRNA, double-stranded RNA; miRNA, microRNA; PBS, phosphate-buffered saline.
References
- 1.Lippman, Z., and Martienssen, R. (2004) Nature 431 364-370 [DOI] [PubMed] [Google Scholar]
- 2.de Fougerolles, A., Vornlocher, H. P., Maraganore, J., and Lieberman, J. (2007) Nat. Rev. Drug Discov. 6 443-453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soifer, H. S., Rossi, J. J., and Saetrom, P. (2007) Mol. Ther. 15 2070-2079 [DOI] [PubMed] [Google Scholar]
- 4.Mette, M. F., Aufsatz, W., van der Winden, J., Matzke, M. A., and Matzke, A. J. (2000) EMBO J. 19 5194-5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matzke, M., Matzke, A. J., and Kooter, J. M. (2001) Science 293 1080-1083 [DOI] [PubMed] [Google Scholar]
- 6.Xie, Z., Johansen, L. K., Gustafson, A. M., Kasschau, K. D., Lellis, A. D., Zilberman, D., Jacobsen, S. E., and Carrington, J. C. (2004) Plos Biol. 2 642-652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matzke, M. A., and Birchler, J. A. (2005) Nat. Rev. Genet. 6 24-35 [DOI] [PubMed] [Google Scholar]
- 8.Grewal, S. I., and Jia, S. (2007) Nat. Rev. Genet. 8 35-46 [DOI] [PubMed] [Google Scholar]
- 9.Morris, K. V., Chan, S. W., Jacobsen, S. E., and Looney, D. J. (2004) Science 305 1289-1292 [DOI] [PubMed] [Google Scholar]
- 10.Buhler, M., Mohn, F., Stalder, L., and Muhlemann, O. (2005) Mol. Cell 18 307-317 [DOI] [PubMed] [Google Scholar]
- 11.Castanotto, D., Tommasi, S., Li, M., Li, H., Yanow, S., Pfeifer, G. P., and Rossi, J. J. (2005) Mol. Ther. 12 179-183 [DOI] [PubMed] [Google Scholar]
- 12.Suzuki, K., Shijuuku, T., Fukamachi, T., Zaunders, J., Guillemin, G., Cooper, D., and Kelleher, A. (2005) J. RNAi Gene Silencing 1 66-78 [PMC free article] [PubMed] [Google Scholar]
- 13.Janowski, B. A., Kaihatsu, K., Huffman, K. E., Schwartz, J. C., Ram, R., Hardy, D., Mendelson, C. R., and Corey, D. R. (2005) Nat. Chem. Biol. 1 210-215 [DOI] [PubMed] [Google Scholar]
- 14.Ting, A. H., Schuebel, K. E., Herman, J. G., and Baylin, S. B. (2005) Nat. Genet. 37 906-910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang, M. X., Ou, H., Shen, Y. H., Wang, J., Wang, J., Coselli, J., and Wang, X. L. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 16967-16972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janowski, B. A., Huffman, K. E., Schwartz, J. C., Ram, R., Hardy, D., Shames, D. S., Minna, J. D., and Corey, D. R. (2005) Nat. Chem. Biol. 1 216-222 [DOI] [PubMed] [Google Scholar]
- 17.Weinberg, M. S., Villeneuve, L. M., Ehsani, A., Amarzguioui, M., Aagaard, L., Chen, Z. X., Riggs, A. D., Rossi, J. J., and Morris, K. V. (2006) RNA (N. Y.) 12 256-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janowski, B. A., Huffman, K. E., Schwartz, J. C., Ram, R., Nordsell, R., Shames, D. S., Minna, J. D., and Corey, D. R. (2006) Nat. Struct. Mol. Biol. 13 787-792 [DOI] [PubMed] [Google Scholar]
- 19.Kim, D. H., Villeneuve, L. M., Morris, K. V., and Rossi, J. J. (2006) Nat. Struct. Mol. Biol. 13 793-797 [DOI] [PubMed] [Google Scholar]
- 20.Pulukuri, S. M., and Rao, J. S. (2007) Cancer Res. 67 6637-6646 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Wang, X., Feng, Y., Pan, L., Wang, Y., Xu, X., Lu, J., and Huang, B. (2007) Mol. Cell. Biochem. 301 259-266 [DOI] [PubMed] [Google Scholar]
- 22.Weinberg, M. S., Barichievy, S., Schaffer, L., Han, J., and Morris, K. V. (2007) Nucleic Acids Res. 35 1-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, J. W., Zhang, Y. H., Zern, M. A., Rossi, J. J., and Wu, J. (2007) Biochem. Biophys. Res. Commun. 359 292-297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noma, K., Sugiyama, T., Cam, H., Verdel, A., Zofall, M., Jia, S., Moazed, D., and Grewal, S. I. (2004) Nat. Genet. 36 1174-1180 [DOI] [PubMed] [Google Scholar]
- 25.Verdel, A., Jia, S., Gerber, S., Sugiyama, T., Gygi, S., Grewal, S. I., and Moazed, D. (2004) Science 303 672-676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutvagner, G., and Simard, M. J. (2008) Nat. Rev. Mol. Cell Biol. 105 512-517 [DOI] [PubMed] [Google Scholar]
- 27.Peters, L., and Meister, G. (2007) Mol. Cell 26 611-623 [DOI] [PubMed] [Google Scholar]
- 28.Vire, E., Brenner, C., Deplus, R., Blanchon, L., Fraga, M., Didelot, C., Morey, L., Van Eynde, A., Bernard, D., Vanderwinden, J. M., Bollen, M., Esteller, M., Di Croce, L., de Launoit, Y., and Fuks, F. (2006) Nature 439 871-874 [DOI] [PubMed] [Google Scholar]
- 29.Han, J., Kim, D., and Morris, K. V. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 12422-12427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennasser, Y., Le, S. Y., Yeung, M. L., and Jeang, K. T. (2004) Retrovirology 1 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, L. C., Okino, S. T., Zhao, H., Pookot, D., Place, R. F., Urakami, S., Enokida, H., and Dahiya, R. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 17337-17342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Triboulet, R., Mari, B., Lin, Y. L., Chable-Bessia, C., Bennasser, Y., Lebrig- and, K., Cardinaud, B., Maurin, T., Barbry, P., Baillat, V., Reynes, J., Corbeau, P., Jeang, K. T., and Benkirane, M. (2007) Science 315 1579-1582 [DOI] [PubMed] [Google Scholar]
- 33.Klase, Z., Kale, P., Winograd, R., Gupta, M. V., Heydarian, M., Berro, R., McCaffrey, T., and Kashanchi, F. (2007) BMC Mol. Biol. 8 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, A. E., Hurd, P. J., Bannister, A. J., Kouzarides, T., and Ford, K. G. (2008) J. Biol. Chem. 283 9878-9885 [DOI] [PubMed] [Google Scholar]
- 35.Lim, H. G., Suzuki, K., Cooper, D. A., and Kelleher, A. D. (2008) Mol. Ther. 16 565-570 [DOI] [PubMed] [Google Scholar]
- 36.Aldovini, A., and Walker, B. D. (1990) Techniques in HIV Research: HIV-Laboratory Manuals, pp. 51-128, Macmillan Press, Ltd., Hants, UK
- 37.Suzuki, K., Craddock, B. P., Okamoto, N., Kano, T., and Steigbigel, R. T. (1993) J. Virol. Methods 44 189-198 [DOI] [PubMed] [Google Scholar]
- 38.Rao, S., Procko, E., and Shannon, M. F. (2001) J. Immunol. 167 4494-4503 [DOI] [PubMed] [Google Scholar]
- 39.Nakayama, J., Rice, J. C., Strahl, B. D., Allis, C. D., and Grewal, S. I. (2001) Science 292 110-113 [DOI] [PubMed] [Google Scholar]
- 40.Peters, A. H., Mermoud, J. E., O'Carroll, D., Pagani, M., Schweizer, D., Brockdorff, N., and Jenuwein, T. (2002) Nat. Genet. 30 77-80 [DOI] [PubMed] [Google Scholar]
- 41.Verdin, E., Paras, P., Jr., and Van Lint, C. (1993) EMBO J. 12 3249-3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Lint, C., Emiliani, S., Ott, M., and Verdin, E. (1996) EMBO J. 15 1112-1120 [PMC free article] [PubMed] [Google Scholar]
- 43.Pazin, M. J., Sheridan, P. L., Cannon, K., Cao, Z., Keck, J. G., Kadonaga, J. T., and Jones, K. A. (1996) Genes Dev. 10 37-49 [DOI] [PubMed] [Google Scholar]
- 44.Jordan, A., Bisgrove, D., and Verdin, E. (2003) EMBO J. 22 1868-1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams, S. A., Chen, L. F., Kwon, H., Fenard, D., Bisgrove, D., Verdin, E., and Greene, W. C. (2004) J. Biol. Chem. 279 42008-42017 [DOI] [PubMed] [Google Scholar]
- 46.Williams, S. A., Chen, L. F., Kwon, H., Ruiz-Jarabo, C. M., Verdin, E., and Greene, W. C. (2006) EMBO J. 25 139-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marban, C., Suzanne, S., Dequiedt, F., de Walque, S., Redel, L., Van Lint, C., Aunis, D., and Rohr, O. (2007) EMBO J. 26 412-423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.du Chene, I., Basyuk, E., Lin, Y. L., Triboulet, R., Knezevich, A., Chable-Bessia, C., Mettling, C., Baillat, V., Reynes, J., Corbeau, P., Bertrand, E., Marcello, A., Emiliani, S., Kiernan, R., and Benkirane, M. (2007) EMBO J. 26 424-435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hall, I. M., Shankaranarayana, G. D., Noma, K., Ayoub, N., Cohen, A., and Grewal, S. I. (2002) Science 297 2232-2237 [DOI] [PubMed] [Google Scholar]
- 50.Buhler, M., Verdel, A., and Moazed, D. (2006) Cell 125 873-886 [DOI] [PubMed] [Google Scholar]
- 51.Coffin, J. M., Hughes, S. H., and Varmus, H. E. (1997) Retroviruses, pp. 205-261, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [PubMed]
- 52.Kim, D. H., and Rossi, J. J. (2007) Nat. Rev. Genet. 8 173-184 [DOI] [PubMed] [Google Scholar]
- 53.Piacentini, L., Fanti, L., Berloco, M., Perrini, B., and Pimpinelli, S. (2003) J. Cell Biol. 161 707-714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vakoc, C. R., Mandat, S. A., Olenchock, B. A., and Blobel, G. A. (2005) Mol. Cell 19 381-391 [DOI] [PubMed] [Google Scholar]
- 55.Deleted in proof
- 56.Weiler, K. S., and Wakimoto, B. T. (1995) Annu. Rev. Genet. 29 577-605 [DOI] [PubMed] [Google Scholar]
- 57.Lu, B. Y., Emtage, P. C., Duyf, B. J., Hilliker, A. J., and Eissenberg, J. C. (2000) Genetics 155 699-708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yasuhara, J. C., and Wakimoto, B. T. (2006) Trends Genet. 22 330-338 [DOI] [PubMed] [Google Scholar]
- 59.Greil, F., van der Kraan, I., Delrow, J., Smothers, J. F., de Wit, E., Busse-maker, H. J., van Driel, R., Henikoff, S., and van Steensel, B. (2003) Genes Dev. 17 2825-2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cryderman, D. E., Grade, S. K., Li, Y., Fanti, L., Pimpinelli, S., and Wallrath, L. L. (2005) Dev. Dyn. 232 767-774 [DOI] [PubMed] [Google Scholar]
- 61.Carrozza, M. J., Li, B., Florens, L., Suganuma, T., Swanson, S. K., Lee, K. K., Shia, W. J., Anderson, S., Yates, J., Washburn, M. P., and Workman, J. L. (2005) Cell 123 581-592 [DOI] [PubMed] [Google Scholar]
- 62.Joshi, A. A., and Struhl, K. (2005) Mol. Cell 20 971-978 [DOI] [PubMed] [Google Scholar]
- 63.Keogh, M. C., Kurdistani, S. K., Morris, S. A., Ahn, S. H., Podolny, V., Collins, S. R., Schuldiner, M., Chin, K., Punna, T., Thompson, N. J., Boone, C., Emili, A., Weissman, J. S., Hughes, T. R., Strahl, B. D., Grunstein, M., Greenblatt, J. F., Buratowski, S., and Krogan, N. J. (2005) Cell 123 593-605 [DOI] [PubMed] [Google Scholar]
- 64.Fuks, F., Hurd, P. J., Deplus, R., and Kouzarides, T. (2003) Nucleic Acids Res. 31 2305-2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morris, S. A., Rao, B., Garcia, B. A., Hake, S. B., Diaz, R. L., Shabanowitz, J., Hunt, D. F., Allis, C. D., Lieb, J. D., and Strahl, B. D. (2007) J. Biol. Chem. 282 7632-7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fuks, F., Hurd, P. J., Wolf, D., Nan, X., Bird, A. P., and Kouzarides, T. (2003) J. Biol. Chem. 278 4035-4040 [DOI] [PubMed] [Google Scholar]
- 67.Reynolds, A., Leake, D., Boese, Q., Scaringe, S., Marshall, W. S., and Khvorova, A. (2004) Nat. Biotechnol. 22 326-330 [DOI] [PubMed] [Google Scholar]
- 68.Eulalio, A., Behm-Ansmant, I., and Izaurralde, E. (2007) Nat. Rev. Mol. Cell Biol. 8 9-22 [DOI] [PubMed] [Google Scholar]
- 69.Parker, R., and Sheth, U. (2007) Mol. Cell 25 635-646 [DOI] [PubMed] [Google Scholar]
- 70.Janowski, B. A., Younger, S. T., Hardy, D. B., Ram, R., Huffman, K. E., and Corey, D. R. (2007) Nat. Chem. Biol. 3 166-173 [DOI] [PubMed] [Google Scholar]
- 71.Han, Y., Wind-Rotolo, M., Yang, H. C., Siliciano, J. D., and Siliciano, R. F. (2007) Nat. Rev. Microbiol. 5 95-106 [DOI] [PubMed] [Google Scholar]
- 72.Haynes, K. A., Caudy, A. A., Collins, L., and Elgin, S. C. (2006) Curr. Biol. 16 2222-2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sugiyama, T., Cam, H. P., Sugiyama, R., Noma, K., Zofall, M., Kobayashi, R., and Grewal, S. I. (2007) Cell 128 491-504 [DOI] [PubMed] [Google Scholar]
- 74.Harbers, K., Schnieke, A., Stuhlmann, H., Jahner, D., and Jaenisch, R. (1981) Proc. Natl. Acad. Sci. U. S. A. 78 7609-7613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu, W. S., Fanning, T. G., and Cardiff, R. D. (1984) J. Virol. 49 66-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clarke, M. F., Trainor, C. D., Mann, D. L., Gallo, R. C., and Reitz, M. S. (1984) Virology 135 97-104 [DOI] [PubMed] [Google Scholar]
- 77.Adachi, A., Gendelman, H. E., Koenig, S., Folks, T., Willey, R., Rabson, A., and Martin, M. A. (1986) J. Virol. 59 284-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koiwa, T., Hamano-Usami, A., Ishida, T., Okayama, A., Yamaguchi, K., Kamihira, S., and Watanabe, T. (2002) J. Virol. 76 9389-9397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Siliciano, J. D., Kajdas, J., Finzi, D., Quinn, T. C., Chadwick, K., Margolick, J. B., Kovacs, C., Gange, S. J., and Siliciano, R. F. (2003) Nat. Med. 9 727-728 [DOI] [PubMed] [Google Scholar]
- 80.Lassen, K., Han, Y., Zhou, Y., Siliciano, J., and Siliciano, R. F. (2004) Trends Mol. Med. 10 525-531 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.