Abstract
Macrophage-specific Abca1 knock-out (Abca1–M/–M) mice were generated to determine the role of macrophage ABCA1 expression in plasma lipoprotein concentrations and the innate immune response of macrophages. Plasma lipid and lipoprotein concentrations in chow-fed Abca1–M/–M and wild-type (WT) mice were indistinguishable. Compared with WT macrophages, Abca1–M/–M macrophages had a >95% reduction in ABCA1 protein, failed to efflux lipid to apoA-I, and had a significant increase in free cholesterol (FC) and membrane lipid rafts without induction of endoplasmic reticulum stress. Lipopolysaccharide (LPS)-treated Abca1–M/–M macrophages exhibited enhanced expression of pro-inflammatory cytokines and increased activation of the NF-κB and MAPK pathways, which could be diminished by silencing MyD88 or by chemical inhibition of NF-κB or MAPK. In vivo LPS injection also resulted in a higher pro-inflammatory response in Abca1–M/–M mice compared with WT mice. Furthermore, cholesterol depletion of macrophages with methyl-β-cyclodextrin normalized FC content between the two genotypes and their response to LPS; cholesterol repletion of macrophages resulted in increased cellular FC accumulation and enhanced cellular response to LPS. Our results suggest that macrophage ABCA1 expression may protect against atherosclerosis by facilitating the net removal of excess lipid from macrophages and dampening pro-inflammatory MyD88-dependent signaling pathways by reduction of cell membrane FC and lipid raft content.
ABCA1 (ATP-binding cassette transporter A1) is a plasma membrane protein that is widely expressed throughout the body (1, 2) and functions as a primary gatekeeper for eliminating excess free cholesterol (FC)2 from tissues by effluxing cellular FC and phospholipid (PL) to lipid-free apoA-I, resulting in the formation of nascent high density lipoprotein (HDL) particles (3, 4). The nascent discoid-shaped HDL then undergoes a maturation process that involves additional lipid acquisition and conversion of FC to cholesteryl ester (CE) by lecithin:cholesterol acyltransferase to become mature spherical plasma HDL. Mutations that inactivate the human ABCA1 gene result in Tangier disease, which is characterized by extremely low HDL cholesterol concentrations, mildly elevated plasma trigelyceride levels, and accumulation of cholesterol in macrophages (5–10). Targeted deletion of Abca1 in mice and a natural mutation of Abca1 in the Wisconsin hypoalpha mutant chicken recapitulate the Tangier plasma lipid phenotype, supporting the essential role of ABCA1 in HDL formation (11–15). Although ABCA1 is expressed in many cells in the body, recent studies in hepatocyte- and intestinal epithelium-specific Abca1 knock-out mice suggest that the liver contributes 70–80% of the plasma HDL pool, whereas the intestine contributes 20–30% (16, 17). Although mobilization of excess FC from macrophages is dependent on ABCA1 and results in the formation of nascent HDL particles, transplantation of bone marrow from Abca1 knock-out (KO) mice into wild-type (WT) mice or transplantation of WT marrow into Abca1 KO recipients has little effect on plasma HDL concentrations, suggesting that macrophage ABCA1 expression has minimal impact on plasma HDL concentrations (18, 19).
Macrophages are a primary cell type involved in innate immunity. Although macrophage ABCA1 has a minimal impact on plasma lipid levels, there is evidence that its activity modulates the inflammatory response of macrophages to pathogen-associated molecules such as lipopolysaccharide (LPS) (20). Francone et al. (20) reported that LPS-induced septic shock was exacerbated in Abca1–/–/Ldlr–/– mice compared with Ldlr–/– mice, suggesting a potential relationship between ABCA1 function and inflammation. However, macrophages from the Abca1–/–/Ldlr–/– mice were enriched 80-fold in CE content, and plasma HDL concentrations in these mice were extremely low. Thus, it is not clear whether the exacerbated pro-inflammatory response to LPS of macrophages from Abca1–/–/Ldlr–/– mice was due to the massive cellular CE accumulation, the absence of plasma HDL, or some other alteration of macrophages.
In macrophages, excess FC accumulation induces cytotoxicity and apoptosis, which is thought to be triggered through the endoplasmic reticulum (ER) stress pathway (21–23). In addition, mounting evidence suggests an association between the presence of lipid rafts in the plasma membrane and the pro-inflammatory activation of macrophages by LPS (24–26). Lipid rafts consist of an ordered membrane microdomain enriched in FC, glycosphingolipid, and glycosylphosphatidylinositol-anchored proteins and are platforms for signal transduction (27, 28). Recently, Landry et al. (29) reported that ABCA1 overexpression in baby hamster kidney cells disrupted membrane lipid rafts, enhanced FC efflux to apoA-I, and resulted in a significant redistribution of FC and sphingomyelin from lipid rafts to non-raft regions of the membrane. Conversely, Koseki et al. (30) reported an increase in lipid raft content and an accelerated tumor necrosis factor-α (TNF-α) secretion induced by LPS in Abca1 KO macrophages and suggested an association between macrophage membrane lipid raft content and cellular sensitivity to LPS.
To determine the role of macrophage ABCA1 in innate immunity with minimal alteration in lipid and lipoprotein metabolism, we developed the macrophage-specific Abca1 KO (Abca1–M/–M) mouse by crossing the conditionally targeted (“floxed”) Abca1 mouse with mice expressing Cre recombinase “knocked in” to the lysozyme M locus. These mice were then used to determine the effect of macrophage Abca1 gene deletion on the concentration of plasma lipids and lipoproteins, macrophage sterol accumulation, and the response of macrophages to a pro-inflammatory stimulus, LPS. Our results indicate that a small increase in membrane FC and lipid raft content causes macrophage hypersensitivity to LPS in the absence of significant changes in plasma lipid and lipoprotein concentrations. Thus, macrophage ABCA1 expression plays a critical regulatory role in plasma membrane cholesterol/lipid raft homeostasis and macrophage innate immune response.
EXPERIMENTAL PROCEDURES
Animals—Abca1 floxed mice were generated by insertion of LoxP sites into introns 44 and 46 of the murine Abca1 gene using homologous recombination as described previously (17). Mice with macrophage-specific deletion of Abca1 were generated by crossing Abca1 floxed (Abca1flox/flox) mice with C57BL/6 mice expressing the Cre recombinase transgene under control of the lysozyme M promoter (B6.129-Lyzstm1(cre)Ifo/J, The Jackson Laboratory). Progeny from the cross that were heterozygous at the Abca1 locus and inherited the LysCre allele were identified by PCR (i.e. Abca1+/–M). Intercrossing Abca1+/–M mice resulted in the production of the Abca1+/+ (WT), Abca1+/–M (heterozygous), and Abca1–M/–M (homozygous) mice used in this study. Abca1 total KO (Abca1–/–) mice were generated as reported previously (15). Animals were housed in a specific pathogen-free facility with 12-h light/dark cycles and received a standard laboratory chow diet. All animal procedures were approved by the Wake Forest University School of Medicine Animal Care and Use Committee.
Peritoneal Macrophage (PM) and Bone Marrow-derived Macrophage (BMDM) Isolation and Culture—Elicited PMs were collected 4 days after injection of 1 ml of 10% thioglycolate into the peritoneal cavity of 9–15-week-old mice as described previously (17). PMs were suspended in RPMI 1640 medium or Dulbecco's modified Eagle's medium containing 10% heat-in-activated fetal bovine serum (FBS) or 1% Nutridoma-SP (Roche Applied Science), 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mm l-glutamine and then plated in tissue culture plates and cultured at 37 °C under a 5% CO2 atmosphere. Two hours later, non-adherent cells were removed by washing with phosphate-buffered saline (PBS), and adherent macrophages were used for studies. BMDMs were obtained as described previously with minor modification (31). Briefly, cleaned femurs and tibias from mice were cut at one end, and the marrow was collected by centrifugation into Eppendorf tubes and then plated in Petri dishes (Fisher) in low glucose Dulbecco's modified Eagle's medium containing 30% L929 cell-conditioned medium, 20% FBS, 2 mm l-glutamine, 1 mm sodium pyruvate, 100 units/ml penicillin, and 100 μg/ml streptomycin for ∼7 days until the cells reached confluence. BMDMs were then washed once with PBS and removed from the dishes with 0.53 mm EDTA (pH 7.4) and incubated overnight in tissue culture plates with RPMI 1640 medium containing 1% Nutridoma-SP before experiments were initiated.
Southern Blot Analysis—Southern blotting was performed as described previously (17).
Western Blot Analysis of Macrophages—Protein expression was determined by Western blot analysis of total cell protein harvested from macrophages using radioimmune precipitation assay lysis buffer (50 mm Tris, 150 mm NaCl, 1% Nonidet P-40, 10% sodium cholate, and 1 mm EDTA) containing protease inhibitors (1 mm phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin, 1 μg/ml leupeptin, and 1 μg/ml aprotinin). Cell proteins (50 μg) were separated on 4–16% SDS-polyacrylamide gels, transferred to nitrocellulose membranes (Schleicher & Schüll), and incubated overnight at 4 °C with the following antibodies depending on the experiment: rabbit anti-human/mouse ABCA1 (17), rabbit anti-mouse CD14 (Abcam), rabbit anti-mouse TLR4 (Cell Signaling Technology, Inc.), and rabbit anti-mouse MyD88 and mouse anti-human/rat/mouse β-actin (Sigma). The blots were then incubated with horseradish peroxidase-linked anti-rabbit IgG or anti-mouse IgG (Amersham Biosciences) at room temperature for 1 h. Immunoblots were visualized with a chemiluminescent reagent (Pierce), and the chemiluminescence was captured with an LSA-3000 imaging system (Fujifilm Life Science) or Kodak X-Omat XLS-1 film.
Lipid Efflux Assay—FC and PL efflux assays from elicited PMs were performed as described previously (17).
Plasma Analyses—Plasma was isolated from blood collected through the tail veins of mice fasted for 4 h. Plasma lipid concentrations were determined by enzymatic assays (32). Plasma HDL cholesterol concentrations were measured as described previously (17, 32). ApoA-I levels were measured by a sandwich enzyme-linked immunosorbent assay (ELISA) (17, 33). Fast protein liquid chromatography was performed using two Superose 6 columns (1 × 30 cm; Amersham Biosciences) in series, followed by total cholesterol enzymatic assay of each eluted fraction.
Cholesterol Content in Macrophages—Cellular lipids were extracted with isopropyl alcohol (including 5α-cholestane as an internal standard) at room temperature overnight and analyzed for cholesterol content by gas-liquid chromatography as described previously (34).
Inflammatory Mediator Expression in LPS-treated Macrophages—After overnight incubation, PMs were switched to serum-free RPMI 1640 medium for 2 h before LPS stimulation. BMDMs were incubated overnight in 1% Nutridoma-SP medium before LPS stimulation. Macrophages were treated with 100 ng/ml LPS from Salmonella typhimurium (Sigma) in serum-free RPMI 1640 medium for 0–6 h, after which the culture supernatants were collected and stored at –80 °C for cytokine ELISA (BD Biosciences). Total RNA was isolated from macrophages using TRIzol reagent as described previously (17). Relative mRNA expression of each target gene was analyzed in triplicate, normalized to glyceraldehyde-3-phosphate dehydrogenase, and expressed relative to WT macrophages without LPS treatment using the 2–ΔΔCT method (35). Primers were designed using the default settings of the manufacturer for the Primer Express 2.0 program (Applied Biosystems) or from published sequences and are listed in supplemental Table S1.
Plasma Concentration of Cytokines after LPS Injection into Mice—Abca1+/+ or Abca1–M/–M mice (8–12 weeks old) were injected in the peritoneal cavity with LPS (3 mg/kg) or pyrogen-free PBS (vehicle). Three hours after injection, mice were killed and bled by cardiac puncture, and plasma was isolated by centrifugation at 12,000 × g for 10 min and stored at –80 °C until analyses were performed. The plasma levels of interleukin (IL)-6 and IL-12p40 were measured by ELISA kits (BD Biosciences) according to the manufacturer's instructions. The lower limit of detection for the cytokine assays was 15.6 pg/ml.
Analysis of Signal Pathways in the LPS-treated Macrophage—Analysis of macrophage signaling pathways before and after stimulation with LPS was performed using Western blot analysis and specific inhibitors. Macrophages were serum-starved for 2 h prior to LPS stimulation and then treated with or without 100 ng/ml LPS for 0–6 h, after which cells were processed for Western blot analysis. Antibodies to access activation of signaling pathways were purchased from Cell Signaling Technology, Inc. Immunoblots were visualized as described above.
In separate experiments, 2-h serum-starved macrophages were pretreated for 1 h with IκBα inhibitor Bay 11-7082 (5.0 μm; Calbiochem), MEK/ERK inhibitor U0126 (25 μm; Calbiochem), or p38 inhibitor (p38 MAPK inhibitor III; 2 μm; Calbiochem) and subsequently treated with 100 ng/ml LPS for an additional 3 or 6 h before RNA isolation for reverse transcription (RT)-PCR quantification of pro-inflammatory cytokine expression.
Small Interfering RNA (siRNA) Transfection—Elicited PMs were isolated and cultured as described above. The medium was replaced with antibiotic-free complete medium, and the incubation was continued overnight, after which the control or gene-specific siRNA (Abca1, CD14, or MyD88; 50 nm) was transfected into macrophages with DharmaFECT reagent (Dharmacon) according to the manufacturer's protocol. To evaluate silencing efficiency, RT-PCR of cellular RNA and Western blot analysis of cell lysates harvested 48–72 h after transfection were performed. siRNA-treated macrophages were then incubated for 6 or 8 h with or without 100 ng/ml LPS before analysis of cytokine expression by RT-PCR or ELISA as described above, apoA-I-induced FC efflux, and sterol content by gas-liquid chromatography.
Flow Cytometry—Exudates were harvested from peritoneal cavities of mice 4 days after intraperitoneal injection of 10% thioglycolate. After blocking the Fcγ receptor with purified anti-mouse CD16/CD32 antibody (Fcγ receptor III/II; BD Biosciences), peritoneal cells were incubated at 4 °C for 30 min with rat IgG2a isotype control (phycoerythrin or fluorescein isothiocyanate; eBioscience), anti-mouse F4/80 antibody (phycoerythrin; eBioscience), anti-mouse CD14 antibody (fluorescein isothiocyanate; eBioscience), or both anti-mouse CD14 and anti-mouse F4/80 antibodies. To visualize lipid rafts, peritoneal cells were first labeled with anti-mouse F4/80 antibody as described above and then rapidly stained with 10 ng of Alexa Fluor 488-labeled cholera toxin B (CT-B) from Vybrant lipid raft labeling kits (Invitrogen) for 10 min at 4 °C. Cell fluorescence was determined using a FACSCalibur flow cytometer (BD Biosciences) and analyzed with FlowJo software (Version 7.2.2).
Confocal Microscopy of Lipid Rafts in PMs—Lipid rafts in PMs were stained using the Vybrant lipid raft labeling kit. Briefly, elicited PMs were seeded onto circular coverslips (Fisher) at a cell density of 0.6 × 105 macrophages/coverslip in RPMI 1640 complete medium (plus 10% FBS and antibiotics). Two hours later, non-adherent cells were removed by washing, and macrophages were incubated for 24 h in RPMI 1640 complete medium. After cells were washed once with chilled RPMI 1640 complete medium, 150 μl of chilled Alexa Fluor 488-labeled CT-B conjugate (1 μg/ml) was added to each coverslip. Cells were incubated for 15 min at 4 °C. After washing with chilled PBS, 150 μl of chilled anti-CT-B antibody was added to each coverslip to cross-link the CT-B lipid rafts, and cells were then incubated for 15 min at 4 °C. After washing with chilled PBS, cells were fixed in 4% paraformaldehyde for 20 min at 4 °C. Fixed cells were mounted with a Prolong antifade kit (Molecular Probes) before examination under a 20× objective using a Model 510 laser scanning confocal microscope (Carl Zeiss AG).
Cholesterol Depletion and Repletion of Macrophages—Cholesterol depletion and repletion of macrophages with methyl-β-cyclodextrin (MβCD) were performed as described previously (36). Briefly, BMDMs were incubated with or without prewarmed 10 mm MβCD at 37 °C for 30 min to deplete cholesterol. Macrophages were then washed with PBS and incubated in the absence or presence of cholesterol (80 μg/ml) and 1.5 mm MβCD at 37 °C for 1 h to reload macrophages with cholesterol. Then, macrophages were extracted with isopropyl alcohol to measure cholesterol content by gas-liquid chromatography as described above or incubated with or without 100 ng/ml LPS for 8 h before quantification of cytokine gene expression by ELISA.
Expression of Sterol-responsive Genes—After overnight culture in RPMI 1640 medium containing 10% FBS, elicited PMs were serum-starved in serum-free RPMI 1640 medium for 2 h and then incubated with 10 μm T0901317 or vehicle (Me2SO) for 20 h. Cellular RNA was harvested, and RT-PCR was performed to measure expression of liver X receptor (LXR) and several LXR-responsive genes such as ABCG1 (ATP-binding cassette transporter G1), phospholipid transfer protein, and apoE. In separate experiments, elicited PMs were cultured overnight in RPMI 1640 medium containing 10% FBS and then incubated in serum-free RPMI 1640 medium for 8 h before RNA was harvested for RT-PCR quantification of SREBP2 (sterol regulatory element-binding protein-2), 3-hydroxy-3-methylglutaryl-CoA reductase, 3-hydroxy-3-methylglutaryl-CoA synthase, and low density lipoprotein receptor mRNAs. Primer sequences are listed in supplemental Table 1.
Analysis of the ER Stress Pathway—After isolated and overnight culture as described above, elicited PMs were serum-starved for 2 h and then treated with or without 10 μg/ml tunicamycin for 0–10 h. Expression of the ER stress proteins in the treated cells was determined by Western blot analysis as described above using antibodies to CHOP (C/EBP homologous protein; Santa Cruz Biotechnology) and BiP and IRE1α (Cell Signaling Technology, Inc.).
Statistics—Differences were compared with two-tailed Student's t test or one-way analysis of variance using GraphPad Prism software. p < 0.05 was considered statistically significant. Data are presented as the means ± S.D. unless indicated otherwise.
RESULTS
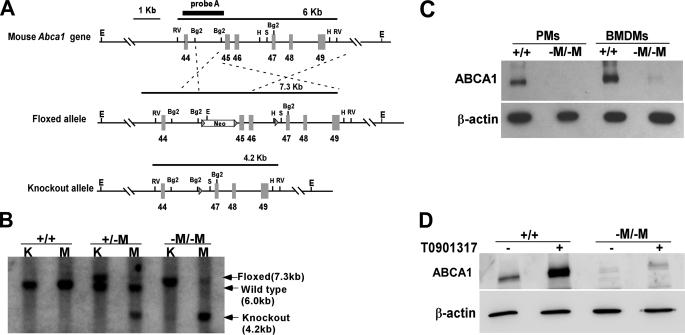
Creation of Macrophage-specific Abca1 KO Mice—Mice that specifically lack Abca1 in macrophages and neutrophils were generated by crossing Abca1 floxed mice with mice expressing Cre recombinase under the control of the macrophage/neutrophil-specific lysozyme M promoter (37). Conditional targeting of the mouse Abca1 gene was achieved by flanking exons 45–46, which encode the second nucleotide-binding cassette, with LoxP sites (designated by the arrowheads in the floxed allele in Fig. 1A). Cre recombinase-mediated recombination resulted in a KO allele missing exons 45 and 46 only in macrophages. Fig. 1B shows results from Southern blot analysis of EcoRV-digested genomic DNA from kidney (control tissue) and elicited PMs isolated from Abca1+/+ and Abca1–M/–M littermates. Cre recombinase-mediated deletion of Abca1 was evident in the macrophages from Abca1–M/–M mice, but not in kidney tissue from these mice (Fig. 1B). A near-complete deletion of ABCA1 protein expression in both freshly isolated elicited PMs and BMDMs from Abca1–M/–M mice was also confirmed by Western blot analysis (Fig. 1C). Because ABCA1 is highly responsive to transcriptional activation by LXR/retinoid X receptor transcription factors (38, 39), we treated PMs with an LXR agonist (T0901317) to maximize ABCA1 protein expression. Fig. 1D shows that 10 μm T0901317 treatment markedly increased ABCA1 expression in WT macrophages, but induced only a very low level of ABCA1 expression in Abca1–M/–M macrophages, supporting the conclusion that ABCA1 expression in Abca1–M/–M macrophages was nearly completely eliminated. In addition, resident macrophages in Abca1–M/–M mouse liver also lacked ABCA1 expression compared with WT liver, as shown by immunofluorescent staining of frozen liver sections (supplemental Fig. S1).
FIGURE 1.
Generation of macrophage-specific Abca1 knock-out (Abca1–M/–M) mice. A, targeting strategy of Abca1–M/–M mice. The schematic of the 3′-region (exons 44–49) of the Abca1 gene shows WT (upper), floxed (middle), and KO (lower) Abca1 alleles. Cre recombinase-mediated deletion of exons 45 and 46 eliminates the second ATP-binding cassette, resulting in a KO allele. E, EcoRI; Rv, EcoRV; Bg2, BglII; H, HindIII; S, SacI. B, Southern blot of genomic kidney (K) and peritoneal macrophage (M) DNAs from mice with WT (+/+) or floxed (–M/–M) alleles in the presence of lysozyme M Cre recombinase. DNA was digested with EcoRV and hybridized with a probe to the genomic region between exons 44 and 45 in the Abca1 gene to produce the 6-kb WT, 7.3-kb floxed, or 4.2-kb KO band. C, Western blot of cell lysates from cultured elicited PMs or BMDMs from WT (+/+) and Abca1–M/–M (–M/–M) mice with antibodies against mouse ABCA1 and β-actin (loading control). D, Western blot of cell lysates from cultured thioglycolate-elicited PMs after 18 h of incubation with or without 10 μm T0901317 (LXR agonist).
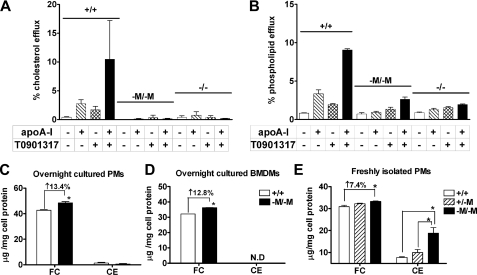
Macrophage-specific Deletion of Abca1 Eliminates Lipid Efflux to Lipid-free ApoA-I—ABCA1 mediates the essential initial step in HDL particle formation by promoting the efflux of cellular FC and PL to apoA-I (40, 41). To determine whether this essential function is eliminated in macrophages from Abca1–M/–M mice, we studied lipid efflux in the presence or absence of T0901317 using elicited PMs from WT, Abca1–M/–M, and Abca1 total KO mice. As expected, in the absence of apoA-I, FC (Fig. 2A) and PL (Fig. 2B) efflux from macrophages was low and similar for all three genotypes. However, in the presence of apoA-I or apoA-I and T0901317, macrophages from WT mice exhibited a substantial increase in FC and PL efflux, whereas macrophages from Abca1–M/–M or total KO mice demonstrated only background efflux, demonstrating that lipid efflux was completely eliminated in Abca1–M/–M macrophages.
FIGURE 2.
Decreased lipid efflux and increased cholesterol accumulation in cultured macrophages from Abca1–M/–M mice. A and B, thioglycolate-elicited PMs were isolated from WT (+/+), Abca1–M/–M (–M/–M), and Abca1 total KO (–/–) mice; radiolabeled with [3H]cholesterol or [14C]choline chloride for 24 h; and then incubated with 10 μm T0901317 or vehicle (Me2SO) for an additional 24 h. Lipid efflux was initiated by incubating macrophages with or without 20 μg/ml apoA-I ± T0901317. Radiolabel in the medium and the cellular isopropyl alcohol extracts were quantified, and percentage efflux was calculated as the ratio of radioactivity in the medium divided by the total radioactivity (cells + medium) × 100%. C–E, after the specified incubation conditions, cells were extracted with isopropyl alcohol containing 5α-cholestane as an internal standard, and free and total cholesterol were quantified by gas-liquid chromatography. CE was calculated as (total – free cholesterol) × 1.67. Cell protein was determined using the Lowry protein assay. C, after 2 h of incubation and removal of non-adherent cells, PMs were cultured in RPMI 1640 medium overnight before cholesterol mass measurement (n = four to five dishes of cells/genotype). D, after 7 days of culture in bone marrow medium, BMDMs were incubated in 1% Nutridoma-SP medium overnight before measurement of cholesterol (n = four to five dishes of cells/genotype). Data are the means ± S.D. (assayed in triplicate). ND, not detectable. E, elicited peritoneal cells were isolated from individual mice (n = 3 for WT and Abca1–M/–M and 2 for Abca1+/–M), incubated in 1% Nutridoma-SP medium for 2 h, and washed to remove non-adherent cells before cholesterol measurement (n ≥ four dishes of cells/mouse). *, p < 0.05 compared with +/+.
Increased FC Accumulation in Cultured Abca1–M/–M Macrophages—ABCA1 is an important determinant of macrophage sterol homeostasis. The severe impairment of cholesterol efflux to lipid-free apoA-I in Abca1-deficient macrophages (Fig. 2A) would be expected to result in cholesterol accumulation. To determine whether this was the case, we measured FC and CE content in 1) elicited PMs after overnight culture in medium supplemented with or without 10% FBS, 2) BMDMs after overnight incubation in RPMI 1640 medium with 1% Nutridoma-SP, and 3) freshly isolated elicited PMs (i.e. 2 h post-harvest). Fig. 2 (C and D) shows that Abca1–M/–M macrophages had a slightly but statistically significant higher amount of FC (∼13%) compared with WT macrophages after overnight culture (PMs, 48.4 ± 1.0 versus 42.7 ± 0.4 μg of FC/mg of cell protein, and BMDMs, 36.3 ± 0.6 versus 32.1 ± 0.2 μg of FC/mg of cell protein, respectively). CE content was either undetectable or very low in overnight cultured PMs or BMDMs regardless of genotype (Fig. 2, C and D). Freshly isolated Abca1–M/–M PMs also had significantly higher FC content (∼7.4%) compared with WT PMs (33.2 ± 0.5 versus 30.9 ± 1.2 μg of FC/mg of cell protein) (Fig. 2E), and there was a gene dosage-dependent increase in FC content with successive deletion of Abca1 alleles (i.e. Abca1–M/–M > Abca1+/–M > Abca1+/+). However, unlike macrophages cultured overnight, freshly isolated Abca1–M/–M PMs had a 2.4-fold higher CE content compared with WT macrophages and exhibited a gene dosage-related increase in CE content (7.7 ± 0.5, 10.0 ± 2.0, and 18.8 ± 4.3 μg of CE/mg of cell protein for WT, Abca1+/–M, and Abca1–M/–M, respectively) (Fig. 2E). Thus, overnight incubation of macrophages resulted in a shift of cellular cholesterol from CE to FC stores, presumably due to CE hydrolysis.
Plasma Lipid, Lipoprotein, and ApoA-I Concentrations in Chow-fed Abca1–M/–M Mice—Fasting total plasma lipid, lipoprotein, and apoA-I concentrations in Abca1+/+, Abca1+/–M, and Abca1–M/–M mice were not different among the three genotypes (Table 1). Fast protein liquid chromatography fractionation of whole plasma also showed a similar plasma lipoprotein profile among these three genotypes of mice (supplemental Fig. S2). These data support the conclusion of previous bone marrow transplant studies (18, 19) that macrophage ABCA1 expression contributes minimally to the plasma HDL pool.
TABLE 1.
Plasma lipid and apolipoprotein concentrations for chow-fed WT and macrophage-specific Abca1 KO mice
Values are the means ± S.D. Blood was obtained from chow-fed mice after a 4-h fast. The number of mice is indicated in parentheses. +/+, WT; +/–M, heterozygous macrophage-specific Abca1 KO; –M/–M, homozygous macrophage-specific Abca1 KO; –/–, Abca1 total KO; TPC, total plasma cholesterol; HDL-C, HDL cholesterol; TG, triglyceride; ND, not detectable.
| Abca1 genotype | TPC | HDL-C | FC | PL | TG | ApoA-I |
|---|---|---|---|---|---|---|
| mg/dl | mg/dl | mg/dl | mg/dl | mg/dl | mg/dl | |
| +/+ | 98 ± 16 (18) | 86 ± 17 (13) | 21 ± 6 (12) | 214 ± 37 (12) | 29 ± 14 (12) | 73 ± 21 (7) |
| +/-M | 98 ± 20 (30) | 83 ± 12 (11) | 19 ± 4 (23) | 212 ± 60 (25) | 22 ± 9 (24) | 77 ± 25 (9) |
| -M/-M | 97 ± 24 (17) | 82 ± 18 (11) | 20 ± 8 (12) | 221 ± 64 (11) | 32 ± 19 (24) | 68 ± 19 (8) |
| -/- | 13 ± 0.5 (3) | 8 ± 1 (3) | ND | ND | ND | ND |
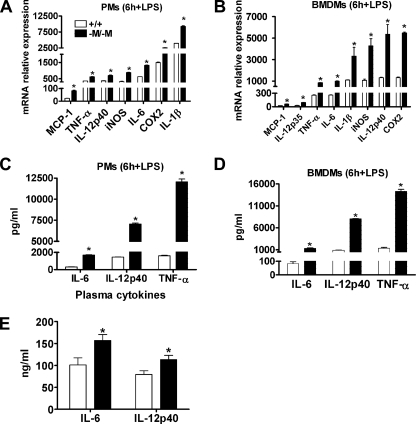
ABCA1-deficient Macrophages Are Hypersensitive to LPS—Macrophages from Abca1 total KO mice are pro-inflammatory and hypersensitive to LPS stimulation (20, 30). To determine the activation state of macrophages from Abca1–M/–M mice compared with those from WT mice, we performed in vitro studies using LPS to activate cultured macrophages. The results in Fig. 3 (A and B) show a significant (p < 0.05) increase in mRNA expression of MCP-1 and cytokines such as IL-1β, TNF-α, IL-6, IL-12, inducible nitric-oxide synthase, and COX2 (cyclooxygenase-2) in both PMs and BMDMs from Abca1–M/–M mice in response to 6 h of LPS treatment compared with WT mice. Similar results were obtained at earlier and later time points (1, 3, and 18 h) (data not shown). Consistent with the elevated mRNA levels, ELISA results demonstrated significantly higher concentrations of pro-inflammatory cytokines (IL-12p40, TNF-α, and IL-6) in culture supernatants from LPS-activated macrophages from Abca1–M/–M mice compared with WT mice (Fig. 3, C and D). The increased secretion of pro-inflammatory cytokines (IL-12p40, TNF-α, and IL-6) from Abca1–M/–M macrophages peaked at 6 h of LPS stimulation compared with 1- and 3-h treatments (data not shown). No significant differences in pro-inflammatory cytokine expression prior to LPS stimulation were observed between the two genotypes of macrophages (data not shown). Taken together with the data in Fig. 2 (C and D), the results demonstrate that loss of ABCA1 in macrophages results in hypersensitivity to LPS with a minor but significant elevation in macrophage FC content and very low or undetectable CE content.
FIGURE 3.
Abca1–M/–M macrophages are hypersensitive to LPS. After overnight culture in 10% FBS containing RPMI 1640 medium, PMs and BMDMs were incubated for 2 h in serum-free medium and then treated with 100 ng/ml LPS for 6 h. Cytokine and chemokine mRNA expression and protein secretion were analyzed by real-time PCR and ELISA, respectively. LPS stimulated higher mRNA (A and B) and protein (C and D) expression of cytokines and chemokines in Abca1–M/–M (–M/–M) PMs (A and C) and BMDMs (B and D) relative to WT (+/+) macrophages. Data are the means ± S.D. (assayed in triplicate). Experiments were repeated at least twice. iNOS, inducible nitric-oxide synthase. E, plasma levels of inflammatory cytokines in LPS-injected mice. Mice were given an intraperitoneal injection of pyrogen-free PBS or 3 mg of LPS/kg of body weight and killed 3 h after injection. Plasma cytokine levels were determined using commercially available ELISA kits as described under “Experimental Procedures.” Data are the means ± S.D. for LPS-injected mice (n = four to five mice/genotype). Values for PBS-injected mice were below the detection limits of the assay (16.5 pg/ml). *, p < 0.05 compared with +/+.
Elevated Plasma Levels of Pro-inflammatory Cytokines in LPS-treated Abca1–M/–M Mice—To evaluate the effect of an acute pro-inflammatory stimulus in vivo, mice were injected in the peritoneal cavity with 3 mg of LPS/kg of body weight, and plasma levels of several pro-inflammatory cytokines were measured 3 h post-injection. Plasma IL-6 and IL-12p40 concentrations were significantly higher in LPS-stimulated Abca1–M/–M mice compared with WT mice (IL-6, 156.8 ± 13.9 versus 101.1 ± 16.2 ng/ml, p < 0.05; and IL-12p40, 113.3 ± 9.6 versus 79.6 ± 8.2 ng/ml, p < 0.05) (Fig. 3E), supporting the in vitro data that macrophages lacking ABCA1 are hypersensitive to LPS.
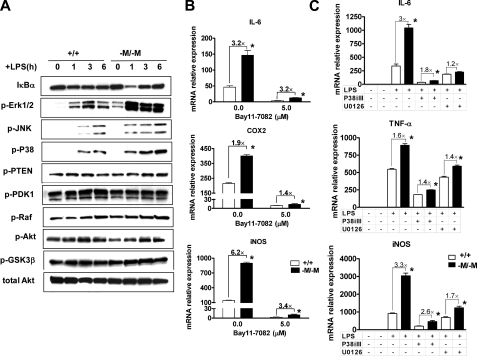
NF-κB and MAPK Signaling Pathways Are Involved in Hypersensitivity of Abca1–M/–M Macrophages to LPS Stimulation—The NF-κB (42), MAPK (43), and phosphatidylinositol 3-kinase/Akt (44) pathways are three major signaling pathways involved in TLR4 (Toll-like receptor 4)-mediated pro-inflammatory cytokine expression in macrophages. To determine the activation state of these pathways in Abca1–M/–M versus WT macrophages, BMDMs from each genotype of mice were stimulated with 100 ng/ml LPS for 1, 3, or 6 h before isolating cell lysates for Western blot analysis of key intermediates in these signaling pathways. As shown in Fig. 4A, the extent of degradation of IκBα (inhibitory κB protein α) was much greater at 1 h in Abca1–M/–M macrophages compared with WT macrophages, indicative of increased NF-κB pathway activation, and there was also a considerable increase in phospho-ERK1/2, phospho-JNK (c-Jun N-terminal kinase), and phospho-P38 during the time course of LPS stimulation, suggesting that both the NF-κB and MAPK pathways are involved in the pro-inflammatory activation of macrophages in the absence of ABCA1. In contrast, the active phosphatase or kinases in the phosphatidylinositol 3-kinase/Akt pathway (phospho-PTEN, phospho-PDK1, phospho-Raf, phospho-GSK3β, and phospho-Akt) did not show any apparent differences between the two genotypes. Additionally, pretreating both WT and Abca1–M/–M macrophages with inhibitors of either the NF-κB or MAPK pathway (IκBα phosphorylation inhibitor Bay 11-7082, ERK1/2 inhibitor U0126, and p38 inhibitor p38 MAPK inhibitor III) before cells were challenged with LPS dramatically attenuated LPS-induced cytokine expression in macrophages from both genotypes and dampened the LPS-induced hypersensitivity of macrophages resulting from loss of ABCA1 (Fig. 4, B and C). The fact that no one inhibitor completely eliminated the hypersensitivity of Abca1–M/–M macrophages to LPS suggests that multiple pathways of TLR4 signaling are involved.
FIGURE 4.
NF-κB and MAPK pathways are involved in the hypersensitivity of Abca1–M/–M macrophages to LPS. A, BMDMs from WT (+/+) or Abca1–M/–M (–M/–M) mice were treated for 0, 1, 3, or 6 h with 100 ng/ml LPS. Cell lysates were harvested, followed by immunoblotting with the indicated antibodies. B, elicited PMs from mice were pretreated for 1 h with the IκBα inhibitor Bay 11-7082 (5.0 μm) before challenge with 100 ng/ml LPS for 6 h. Cellular RNA was then harvested to quantify inflammatory gene expression by quantitative RT-PCR. iNOS, inducible nitric-oxide synthase. C, BMDMs were pretreated for 1 h with 25 μm U0126 (MEK/ERK inhibitor) or 2 μm p38 MAPK inhibitor III (P38iIII) and then treated with 100 ng/ml LPS for 3 h before quantification of inflammatory gene expression by RT-PCR. Data are presented as the means ± S.D. (assayed in triplicate). Experiments were repeated twice. *, p < 0.05 compared with +/+.
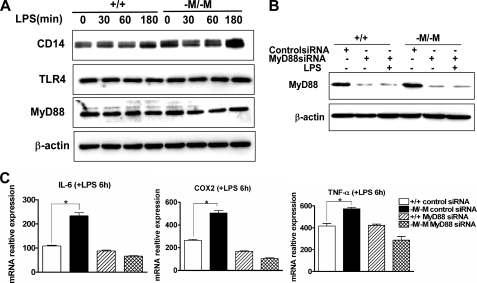
Hyperactivation of ABCA1-deficient Macrophages Is Mediated through a MyD88-dependent Pathway—The binding of LPS to the TLR4/CD14 receptor cluster initiates the recruitment of downstream adaptor proteins such as MyD88 (myeloid differentiation primary-response protein 88), followed by activation of MyD88-dependent and MyD88-independent pathways that ultimately lead to the release of inflammatory mediators (45). Our data showed that enhanced inflammatory activation of Abca1–M/–M macrophages by LPS was independent of cellular CD14 expression (supplemental Fig. S3). To determine whether the hypersensitivity of Abca1–M/–M macrophages to LPS is mediated through a MyD88-dependent pathway, we examined basal and LPS-stimulated MyD88 and cytokine expression before and after siRNA transfection to silence MyD88. MyD88 expression before and after LPS stimulation was not different between WT and Abca1–M/–M macrophages, demonstrating the ABCA1 deletion did not affect MyD88 expression (Fig. 5A). Silencing of MyD88 resulted in >85% reduction of expression in both genotypes of mice (Fig. 5B) and eliminated the hypersensitivity of Abca1–M/–M macrophages to LPS stimulation (Fig. 5C), demonstrating that the hyper-responsiveness of Abca1–M/–M macrophages to LPS was mediated through a MyD88-dependent pathway.
FIGURE 5.
Enhanced LPS-stimulated activation of Abca1–M/–M macrophages is MyD88-dependent. A, shown are the results from Western blot analysis of CD14, TLR4, and MyD88 protein expression in BMDMs from WT (+/+) and Abca1–M/–M (–M/–M) mice after in vitro treatment with 100 ng/ml LPS for 0, 30, 60, and 180 min. B, MyD88 expression was silenced by transfecting elicited PMs from WT or Abca1–M/–M mice with the indicated siRNAs for 72 h before challenging the cells with 100 ng/ml LPS for 6 h. Western blotting was performed to determine protein silencing efficiency. C, quantitative RT-PCR was performed to quantify cytokine mRNA expression after MyD88 silencing. Data are presented as the means ± S.D. (assayed in triplicate). *, p < 0.05 compared with +/+.
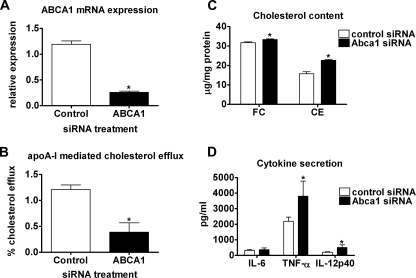
Acute Silencing of Macrophage ABCA1 Results in Reduced FC Efflux, Increased FC Content, and Increased Sensitivity to LPS—To determine whether acute silencing of ABCA1 in macrophages results in increased sterol content and LPS hypersensitivity, we transfected WT macrophages with control or ABCA1 siRNA. ABCA1 mRNA expression was decreased by ∼80% in cells transfected with ABCA1 siRNA relative to the control (Fig. 6A). Acute silencing of ABCA1 expression in WT macrophages resulted in a 70% reduction of apoA-I-mediated FC efflux (Fig. 6B), a statistically significant 5% increase in FC content (31.7 ± 1.0 versus 33.3 ± 0.9 μg/mg of protein, p < 0.05), and a 45% increase in CE content (15.7 ± 2.6 versus 22.7 ± 1.3 μg/mg of protein, p < 0.05) (Fig. 6C). Furthermore, silencing of ABCA1 in macrophages led to a significantly higher level of secretion of TNF-α and IL-12p40, but not IL-6, after LPS stimulation compared with control siRNA-transfected macrophages (Fig. 6D). These data demonstrate that acute silencing of ABCA1 in macrophages results in a pro-inflammatory phenotype that is similar to that of macrophages from Abca1–M/–M mice and support an important role of macrophage ABCA1 expression in regulating macrophage cholesterol homeostasis and innate immune response.
FIGURE 6.
Acute silencing of Abca1 in WT macrophages results in increased free cholesterol accumulation and pro-inflammatory response to LPS. PMs from WT mice were transfected with 50 nm control or ABCA1 siRNA for 48 h. A, quantitative RT-PCR was performed to quantify ABCA1 mRNA expression in control and ABCA1 siRNA-transfected cells. B, transfected macrophages were radiolabeled with [3H]cholesterol for 24 h and then equilibrated with bovine serum albumin for an additional 2 h. FC efflux was initiated by incubating macrophages with or without 20 μg/ml apoA-I for 8 h. Radiolabel in the medium and the cellular isopropyl alcohol extracts were quantified, and percentage efflux was calculated as the ratio of radioactivity in the medium divided by the total radioactivity (cells + medium) × 100%. C, cellular lipid was extracted with isopropyl alcohol to measure FC and CE content by gas-liquid chromatography (means ± S.D., n = four to five dishes of cells/group). D, macrophages were incubated with or without 100 ng/ml LPS for 8 h, and pro-inflammatory gene expression was measured by ELISA (means ± S.D., n = 4). *, p < 0.05 compared with control siRNA.
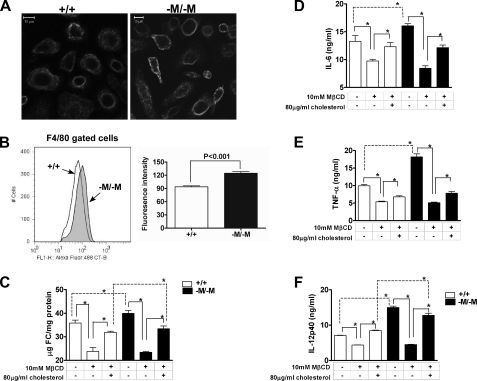
Abca1–M/–M Macrophages Have an Increase in Membrane Lipid Rafts—Specialized regions of the membrane known as lipid rafts are enriched in cholesterol and sphingomyelin and form platforms for extracellular signal transduction. To determine whether the increase in cellular FC in Abca1–M/–M macrophages results in an increase in membrane lipid raft content, we stained live PMs from WT and Abca1–M/–M mice with Alexa Fluor 488-labeled CT-B, which binds to the pentasaccharide chain of ganglioside GM1, a raft-associated lipid. Fig. 7A shows the confocal laser scanning images of PMs visualized with CT-B. There was a trend toward brighter fluorescence and more punctuate staining in macrophages with deletion of ABCA1 relative to WT macrophages, suggesting an increase in lipid rafts (Fig. 7A). To investigate this trend quantitatively, we stained thioglycolate-elicited total peritoneal cells with phycoerythrin-labeled anti-mouse F4/80 antibody and Alexa Fluor 488-labeled CT-B and then analyzed the cells by flow cytometry. As shown in Fig. 7B, F4/80-gated cells (macrophage population) from Abca1–M/–M mice had a significantly higher CT-B staining intensity compared with cells from WT mice, supporting the conclusion of increased lipid rafts.
FIGURE 7.
Macrophages from Abca1–M/–M mice have increased membrane lipid rafts. A, lipid raft staining of elicited PMs was performed with fluorescently labeled CT-B as described under “Experimental Procedures.” Images were captured under a 20× objective using a Zeiss Model 510 laser scanning confocal microscope. Images are representative at least 50 fields of three coverslips/genotype. Scale bars = 10 μm. B, shown are the results from flow cytometric analysis of macrophage lipid rafts. Cells were isolated from peritoneal cavity fluid of mice 4 days after intraperitoneal injection of thioglycolate and were incubated with phycoerythrin-labeled anti-mouse F4/80 antibody and Alexa Fluor 488-labeled CT-B. A representative intensity plot for F4/80-gated cells from both genotypes of mice is shown in the left panel, and the average data (means ± S.D.) from WT (+/+; n = 4) and Abca1–M/–M (–M/–M; n = 3) mice are shown in the right panel. C–F, cholesterol depletion and repletion using MβCD abolished and partially restored the hypersensitivity of Abca1–M/–M macrophages to LPS, respectively. BMDMs from WT and Abca1–M/–M mice were incubated with or without 10 mm MβCD at 37 °C for 30 min to deplete cells of cholesterol. Some dishes of cells were then repleted with cholesterol by incubation with cholesterol-loaded MβCD (80 μg/ml FC solubilized with 1.5 mm MβCD; 1 h at 37 °C). Cells were then harvested for cholesterol analysis by gas-liquid chromatography (C) or incubated with or without LPS (100 ng/ml; 8 h) before measuring cytokine concentration in the medium by ELISA (D–F). *, p < 0.05 for the indicated comparisons.
Cholesterol Depletion and Repletion in Macrophages Correlate with Macrophage Hypersensitivity to LPS—MβCD has been shown to disrupt lipid rafts by extracting membrane cholesterol, whereas cholesterol-loaded MβCD can replete the plasma membrane with cholesterol (36, 46, 47). To determine whether the pro-inflammatory state of the Abca1–M/–M macrophages could be reduced by depleting FC, we pretreated WT and Abca1–M/–M BMDMs with 10 mm MβCD for 30 min at 37 °C before stimulating cells with LPS (100 ng/ml) for 8 h. Fig. 7C shows that incubation with 10 mm MβCD significantly lowered cellular FC content in both WT macrophages (35.8 ± 1.9 to 23.8 ± 1.6 μg/mg of protein) and Abca1–M/–M macrophages (40.0 ± 1.2 to 23.4 ± 0.4 μg/mg of protein), resulting in similar FC content between the two genotypes. Cholesterol depletion with MβCD significantly reduced LPS-induced BMDM secretion of inflammatory cytokines (TNF-α, IL-6, and IL-12p40) to equivalent levels for WT and Abca1–M/–M BMDMs (Fig. 7, D–F). Furthermore, incubation of cholesterol-depleted BMDMs with cholesterol-loaded MβCD partially restored the FC difference between WT macrophages (23.8 ± 1.6 to 31.9 ± 0.5 μg/mg of protein) and Abca1–M/–M macrophages (23.4 ± 0.4 to 33.4 ± 1.2 μg/mg of protein), resulting in a statistically significant higher level of FC in Abca1–M/–M BMDMs compared with WT BMDMs (Fig. 7C). Cholesterol repletion also resulted in increased cytokine secretion for macrophages from both genotypes compared with cholesterol-depleted BMDMs, and the difference in cytokine secretion between WT and Abca1–M/–M BMDMs was recovered for IL-12p40 (Fig. 7F), even though the FC enrichment was only 5% higher in Abca1–M/–M BMDMs compared with WT BMDMs after repletion. These data establish a direct relationship between macrophage FC content and hypersensitivity to LPS and suggest that the hypersensitivity of Abca1–M/–M BMDMs to LPS is due to the increase in membrane FC and lipid rafts.
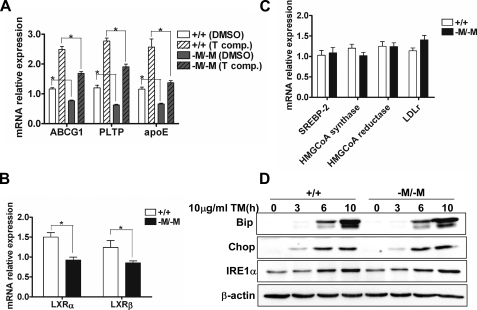
FC Accumulation in Abca1–M/–M Macrophages Fails to Activate LXR-responsive Genes and Cholesterol Biosynthetic Genes or Exacerbates the Unfolded Protein Response (UPR)—The increased lipid raft content of Abca1–M/–M macrophages suggested that the increase in FC was localized primarily to the plasma membrane. To obtain additional support for this concept, we measured the expression of LXR-responsive genes, which are activated by oxysterols when FC accumulation occurs in cells. Expression of LXR-responsive genes was less in Abca1–M/–M macrophages compared with WT macrophages, and the difference in response between the two genotypes was maintained with synthetic LXR agonist stimulation (Fig. 8A). The decrease in LXR-responsive genes may be related to the lower expression of LXRα and LXRβ in Abca1–M/–M macrophages (Fig. 8B). FC accumulation in the ER results in down-regulation of the cholesterol biosynthetic pathway through retention of SREBP2 in the ER (48). However, we did not see a decrease in mRNA for SREBP2, its target cholesterol biosynthetic genes such as 3-hydroxy-3-methylglutaryl-CoA reductase and 3-hydroxy-3-methylglutaryl-CoA synthase, or its cholesterol uptake genes such as the low density lipoprotein receptor (Fig. 8C). Finally, FC accumulation in the ER can also cause ER stress, resulting in activation of the UPR (49). To determine whether activation of the UPR is enhanced in Abca1–M/–M macrophages, we analyzed the expression of BiP, IRE1α, and CHOP by Western blot analysis after tunicamycin treatment of WT and Abca1–M/–M macrophages to induce ER stress. As expected, tunicamycin treatment of macrophages resulted in induction of the UPR with increased expression of BiP, CHOP, and IRE1α (Fig. 8D). However, there was no evidence that the UPR was enhanced in Abca1–M/–M compared with the WT, as expression of the three proteins was similar between the two genotypes. Taken together, these results suggest that the FC enrichment in Abca1–M/–M macrophages was not being sensed by the ER, but more likely was confined to the plasma membrane.
FIGURE 8.
Free cholesterol accumulation in macrophages from Abca1–M/–M mice is not sensed by the ER. A and B, shown is the lower expression of LXR target genes and LXRs in Abca1–M/–M (–M/–M) macrophages. Elicited PMs were treated with Me2SO (DMSO; vehicle) or 10 μm T0901317 (T comp.) for 20 h. Cellular RNA was harvested, and RT-PCR was performed to measure expression of LXR-responsive genes (A) and expression of LXRα/β (B). Assays were performed in triplicate, and data were normalized to the level of gene expression in WT (+/+) macrophages treated with vehicle. PLTP, phospholipid transfer protein. C, expression of sterol-sensing genes was unaltered in Abca1–M/–M macrophages. Elicited PMs were isolated and cultured overnight and then incubated in serum-free RPMI 1640 medium for 8 h. Gene expression was measured by RT-PCR after RNA isolation. Assays were performed in triplicate, and the data were normalized to the gene expression level in WT macrophages. HMGCoA, 3-hydroxy-3-methylglutaryl-CoA; LDLr, low density lipoprotein receptor. D, ER stress was not enhanced in Abca1–M/–M macrophages. Elicited PMs (n = three dishes of cells/genotype) were treated with 10 μg/ml tunicamycin (TM) for 0–10 h. The ER stress proteins were analyzed by Western blotting. *, p < 0.05 for the indicated comparisons.
DISCUSSION
Macrophages are key players in the pathogenesis of atherosclerosis (50). They are an integral cell type in the innate immune response and are involved in processing of excess lipid from phagocytosis of apoptotic/necrotic cells or from uptake of modified lipoproteins at sites of inflammation. A recent study has suggested that whole body deletion of ABCA1 in Ldlr–/– mice results in massive macrophage cholesterol accumulation and a pro-inflammatory phenotype (20). However, the association between macrophage cholesterol accumulation and the pro-inflammatory state is poorly understood. Using a unique animal model with targeted deletion of Abca1 in macrophages, we have documented that macrophages lacking ABCA1 are hypersensitive to LPS stimulation in vivo and in vitro via a MyD88-dependent signaling pathway in the absence of changes in plasma lipid and lipoprotein concentrations. The hypersensitivity to LPS is dependent on subtle increases in cell membrane cholesterol and lipid raft content, suggesting an important role of ABCA1 in the regulation of innate immunity through modulation of plasma membrane cholesterol and lipid rafts. Thus, macrophage ABCA1 expression may protect against atherosclerosis by at least two mechanisms, facilitating the net removal of excess lipid from macrophages and dampening pro-inflammatory MyD88-dependent signaling pathways by reduction of cellular lipid raft content.
Since its discovery, ABCA1 has been shown to be absolutely essential for plasma HDL formation and critical in preventing the accumulation of excess lipid in macrophages (5, 6, 8). Recent studies involving cell-specific Abca1 KO mouse models have shown that liver and intestine are the two most important organs contributing to the plasma HDL pool (16, 17). However, ABCA1 is variably expressed in most cells of the body (1, 2), including macrophages. Bone marrow transplantation studies have shown that bone marrow ABCA1 expression does not affect plasma HDL cholesterol concentrations (18, 19, 51). In agreement with these studies, we have shown that the plasma lipid and lipoprotein phenotype is indistinguishable between WT and Abca1–M/–M mice on a chow diet (Table 1 and supplemental Fig. S2). Our data were generated from a novel mouse model with the deletion of Abca1 restricted to macrophages and neutrophils, in which ABCA1 protein expression was <5% of the WT, even after maximal up-regulation of gene transcription with a synthetic LXR agonist (Fig. 1). Taken together, these studies prove that macrophage ABCA1 has a minimal role in determining plasma HDL concentrations.
Cholesterol accumulation in macrophages is associated with a pro-inflammatory phenotype (22). Macrophages have multiple ways to protect against accumulation of toxic levels of FC, including increased esterification of FC to form relatively inert CE, increased PL synthesis to solubilize excess FC, and increased efflux of FC (52). An increase in FC and oxygenated derivatives (i.e. oxysterols) results in activation of LXR, which increases transcription of multiple genes involved in cholesterol efflux, including Abca1, Abcg1, phospholipid transfer protein, and apoE (53). These multiple systems protect against the accumulation of FC in the ER, which can activate the UPR and ultimately lead to apoptosis (49). A previous study with Abca1–/– mice in the atherosclerosis-prone Ldlr–/– background documented the existence of pro-inflammatory macrophages compared with Ldlr–/– mice, suggesting a role for ABCA1 expression in the innate immune response to the TLR4 agonist LPS (20). Although the molecular explanation for the hyper-responsiveness to LPS in that study is not known, it likely was related to an 80-fold increase in CE, the lack of plasma HDL, or both. Our study shows that macrophages from Abca1–M/–M mice are hyper-responsive to LPS in vivo and in vitro compared with macrophages from WT mice, with modest accumulation of CE (2.4-fold in freshly isolated PMs) (Fig. 2E) and no change in plasma lipoprotein concentration. Acute silencing of ABCA1 in vitro also results in significant increases in FC and CE accumulation and hypersensitivity to LPS (Fig. 6). The extent to which the higher cellular CE content contributes to the hyper-responsiveness of Abca1–M/–M macrophages to LPS in vivo and in vitro is unknown. However, hyper-responsiveness to LPS stimulation was observed for freshly isolated PMs as well as PM and BMDMs cultured for extended periods of time, even though CE content was non-detectable or very low after extended culture, suggesting that elevated FC, not CE, was responsible for the hypersensitivity (Fig. 2). The increase in FC and the MyD88 dependence of the hypersensitivity to LPS suggested an alteration at the proximal part of the TLR4 signaling pathway, which was verified by direct experimentation. 1) Freshly isolated PMs had a significant increase in lipid raft content (Fig. 7, A and B). 2) Depletion of membrane lipid raft cholesterol with MβCD dampened the heightened inflammatory response of macrophages to LPS and eliminated the difference in responsiveness to LPS between Abca1–M/–M and WT macrophages (Fig. 7, C–F). 3) Cholesterol repletion increased FC accumulation in macrophages, enhanced the pro-inflammatory response of macrophages to LPS (Fig. 7, C–F), and also partially restored their hypersensitivity to LPS in Abca1–M/–M macrophages (shown by significantly increased IL-12p40 expression in Fig. 7F) relative to WT macrophages. Overall, our data strongly support the concept that increased macrophage cellular FC and lipid raft content, not CE accumulation, is the primary explanation for Abca1–M/–M macrophage hypersensitivity to LPS.
Several recent studies have shown that ABCA1 expression affects cellular lipid raft content and distribution. One study demonstrated that overexpression of ABCA1 in baby hamster kidney cells resulted in the redistribution of FC from lipid rafts to non-raft regions of the cell membrane (29). Another study showed that cultured macrophages from Abca1 total KO mice had increased staining for lipid rafts, increased raft FC, and increased secretion of TNF-α in response to LPS (30). Our results in Abca1–M/–M mice document increased lipid raft staining in macrophages from freshly isolated elicited peritoneal cells, suggesting that the increase in FC and lipid raft content occurs in vivo and is not an artifact of tissue culture. The increase in FC sensitized the proximal steps of TLR4 signaling as MyD88 silencing eliminated the hypersensitivity to LPS (Fig. 5, B and C). Our results also suggest that the FC enrichment in Abca1–M/–M macrophages is largely restricted to the plasma membrane based on the following observations: 1) the lack of stimulation of LXR-responsive genes (Fig. 8A), 2) the lack of stimulation of genes involved in cholesterol biosynthesis (Fig. 8C), and 3) the lack of stimulation of the UPR (Fig. 8D). Previous studies have shown that an alteration in membrane FC content affects cellular signaling (54, 55), suggesting that control of membrane FC and lipid raft distribution is a critical component of this regulation. Taken together, these data suggest that ABCA1 may play a novel role in regulating signaling events through control of lipid raft composition and distribution.
What is the physiological significance of ABCA1 regulation of lipid rafts? Excessive accumulation of lipid by macrophages through phagocytosis of apoptotic/necrotic cells or uptake of oxidized lipoproteins results in activation of LXR by oxysterols. The activation of LXR results in increased lipid efflux and transrepression of pro-inflammatory gene expression (56, 57). This concerted response protects against overstimulation of the macrophage inflammatory activation upon lipid loading, allowing an orderly processing of excess cellular lipid by ABCA1- and ABCG1-mediated efflux to apoA-I and HDL, respectively (58). Abca1 is a highly responsive gene to LXR activation (53), and our data suggest a second mechanism by which ABCA1 may dampen the inflammatory response by decreasing TLR4-induced transactivation of the NF-κB and MAPK signaling pathways. LPS activation of macrophages occurs when LPS binds to CD14, which is a glycosylphosphatidylinositol-linked receptor and is exclusively located in lipid rafts (26). However, TLR4 and other adaptor proteins must be recruited to lipid rafts upon LPS binding to CD14 to activate the canonical signaling through NF-κB and MAPK. Any process that disrupts or disperses lipid rafts would lessen signaling potential by dispersing receptors/co-receptors involved in transducing the extracellular signal across the plasma membrane. Thus, cholesterol loading of macrophages activates LXR, leading to transrepression of pro-inflammatory genes and increased expression of ABCA1, resulting in decreased cellular lipid raft content and diminished transactivation of pro-inflammatory genes. A recent study has also shown that the pro-inflammatory cytokine TNF-α induces ABCA1 expression through an NF-κB-mediated pathway, suggesting an additional pathway that could limit pro-inflammatory pathway activation by further dispersion of lipid rafts and diminished TLR4 signaling by increased ABCA1 expression (59). Our conclusion of a novel role for ABCA1 in regulation of membrane signaling is also supported by our recent demonstration of defective insulin release from pancreatic islet cells of beta cell-specific Abca1 KO mice (60).
There is mounting evidence that human ABCA1 gene polymorphisms are associated with altered glucose metabolism, onset of obesity, and insulin resistance in Mexican (61, 62), French (63), and Japanese (64) populations, which were independent of HDL formation in the latter study. In addition, increasing evidence highlights that insulin resistance, a key component of type 2 diabetes, is caused by chronic low-grade inflammation (65, 66). For instance, macrophage-specific disruption of the pro-inflammatory NF-κB activation pathway eliminates insulin resistance in liver and skeletal muscle induced by feeding mice a high fat/high sucrose diet (67). Although polymorphisms in the human population likely affect ABCA1 function in all tissues, our data suggest that loss of ABCA1 function in macrophages may lead to increased low-grade chronic inflammation in vivo, which may predispose one to insulin resistance. Because saturated fatty acids can stimulate inflammation in macrophages by activating TLR4 receptors (68), we hypothesize that consumption of a high saturated fat diet by Abca1–M/–M mice compared with WT mice may result in increased obesity and insulin resistance due to chronic low-grade inflammation. Studies are ongoing to test this hypothesis. If true, individuals with polymorphisms in ABCA1 that compromise function may be at increased risk for developing obesity and insulin resistance when dietary consumption of saturated fatty acids is high.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. Linda Curtis (Scripps Institute) for providing the protocol for isolation and culture of BMDMs, Ken Grant for technical assistance with laser scanning confocal microscopy, Dr. Ira Tabas (Columbia University) for providing antibodies to CHOP, and Karen Klein for editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants HL49373, HL54176, and AT27820 (to J. S. P.) and HL07115 (cardiovascular pathology training grant; to J. M. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1–S3, and Table S1.
Footnotes
The abbreviations used are: FC, free cholesterol; PL, phospholipid; HDL, high density lipoprotein; CE, cholesteryl ester; KO, knock-out; WT, wild-type; LPS, lipopolysaccharide; ER, endoplasmic reticulum; TNF-α, tumor necrosis factor-α; PM, peritoneal macrophage; BMDM, bone marrow-derived macrophage; FBS, fetal bovine serum; PBS, phosphate-buffered saline; ELISA, enzyme-linked immunosorbent assay; IL, interleukin; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase; ERK, extracellular signal-regulated kinase; MAPK, mitogen-activated protein kinase; RT, reverse transcription; siRNA, small interfering RNA; CT-B, cholera toxin B; MβCD, methyl-β-cyclodextrin; LXR, liver X receptor; NF-κB, nuclear factor-κB; UPR, unfolded protein response; GM1, monosialotetrahexosylganglioside.
References
- 1.Langmann, T., Klucken, J., Reil, M., Liebisch, G., Luciani, M. F., Chimini, G., Kaminski, W. E., and Schmitz, G. (1999) Biochem. Biophys. Res. Commun. 257 29–33 [DOI] [PubMed] [Google Scholar]
- 2.Wellington, C. L., Walker, E. K., Suarez, A., Kwok, A., Bissada, N., Singaraja, R., Yang, Y. Z., Zhang, L.-H., James, E., Wilson, J. E., Francone, O., McManus, B. M., and Hayden, M. R. (2002) Lab. Investig. 82 273–283 [DOI] [PubMed] [Google Scholar]
- 3.Oram, J. F., and Lawn, R. M. (2001) J. Lipid Res. 42 1173–1179 [PubMed] [Google Scholar]
- 4.Attie, A. D., Kastelein, J. P. P., and Hayden, M. R. (2001) J. Lipid Res. 42 1717–1726 [PubMed] [Google Scholar]
- 5.Bodzioch, M., Orsó, E., Klucken, J., Langmann, T., Böttcher, A., Diederich, W., Drobnik, W., Barlage, S., Buchler, C., Porsch-Ozcurumez, M., Kaminski, W. E., Hahmann, H. W., Oette, K., Rothe, G., Aslanidis, C., Lackner, K. J., and Schmitz, G. (1999) Nat. Genet. 22 347–351 [DOI] [PubMed] [Google Scholar]
- 6.Brooks-Wilson, A., Marcil, M., Clee, S. M., Zhang, L.-H., Roomp, K., van Dam, M., Yu, L., Brewer, C., Collins, J. A., Molhuizen, H. O. F., Loubser, O., Ouelette, B. F. F., Fichter, K., Ashbourne-Excoffon, K. J. D., Sensen, C. W., Scherer, S., Mott, S., Denis, M., Martindale, D., Frohlich, J., Morgan, K., Koop, B., Pimstone, S., Kastelein, J. J. P., Genest, J., and Hayden, M. R. (1999) Nat. Genet. 22 336–345 [DOI] [PubMed] [Google Scholar]
- 7.Clee, S. M., Kastelein, J. J. P., van Dam, M., Marcil, M., Roomp, K., Zwarts, K. Y., Collins, J. A., Roelants, R., Tamasawa, N., Stulc, T., Suda, T., Ceska, R., Boucher, B., Rondeau, C., DeSouich, C., Brooks-Wilson, A., Molhuizen, H. O. F., Frohlich, J., Genest, J., and Hayden, M. R. (2000) J. Clin. Investig. 106 1263–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rust, S., Rosier, M., Funke, H., Real, J., Amoura, Z., Piette, J. C., Deleuze, J. F., Brewer, H. B., Duverger, N., Denefle, P., and Assmann, G. (1999) Nat. Genet. 22 352–355 [DOI] [PubMed] [Google Scholar]
- 9.Schaefer, E. J., Zech, L. A., Schwartz, D. E., and Brewer, H. B., Jr. (1980) Ann. Intern. Med. 93 261–266 [DOI] [PubMed] [Google Scholar]
- 10.van Dam, M. J., de Groot, E., Clee, S. M., Hovingh, G. K., Roelants, R., Brooks-Wilson, A., Zwinderman, A. H., Smit, A. J., Smelt, A. H. M., Groen, A. K., Hayden, M. R., and Kastelein, J. J. P. (2002) Lancet 359 37–42 [DOI] [PubMed] [Google Scholar]
- 11.McNeish, J., Aiello, R. J., Guyot, D., Turi, T., Gabel, C., Aldinger, C., Hoppe, K. L., Roach, M. L., Royer, L. J., de Wet, J., Broccardo, C., Chimini, G., and Francone, O. L. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 4245–4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulligan, J. D., Flowers, M. T., Tebon, A., Bitgood, J. J., Wellington, C., Hayden, M. R., and Attie, A. D. (2003) J. Biol. Chem. 278 13356–13366 [DOI] [PubMed] [Google Scholar]
- 13.Attie, A. D., Hamon, Y., Brooks-Wilson, A. R., Gray-Keller, M. P., MacDonald, M. L. E., Rigot, V., Tebon, A., Zhang, L.-H., Mulligan, J. D., Singaraja, R. R., Bitgood, J. J., Cook, M. E., Kastelein, J. J. P., Chimini, G., and Hayden, M. R. (2002) J. Lipid Res. 43 1610–1617 [DOI] [PubMed] [Google Scholar]
- 14.Christiansen-Weber, T. A., Voland, J. R., Wu, Y., Ngo, K., Roland, B. L., Nguyen, S., Peterson, P. A., and Fung-Leung, W.-P. (2000) Am. J. Pathol. 157 1017–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, J.-Y., Timmins, J. M., Mulya, A., Smith, T. L., Zhu, Y., Rubin, E. M., Chisholm, J. W., Colvin, P. L., and Parks, J. S. (2005) J. Lipid Res. 46 2233–2245 [DOI] [PubMed] [Google Scholar]
- 16.Brunham, L. R., Kruit, J. K., Iqbal, J., Fievet, C., Timmins, J. M., Pape, T. D., Coburn, B. A., Bissada, N., Staels, B., Groen, A. K., Hussain, M. M., Parks, J. S., Kuipers, F., and Hayden, M. R. (2006) J. Clin. Investig. 116 1052–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Timmins, J. M., Lee, J.-Y., Boudyguina, E., Kluckman, K. D., Brunham, L. R., Mulya, A., Gebre, A. K., Coutinho, J. M., Colvin, P. L., Smith, T. L., Hayden, M. R., Maeda, N., and Parks, J. S. (2005) J. Clin. Investig. 115 1333–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haghpassand, M., Bourassa, P. A., Francone, O. L., and Aiello, R. J. (2001) J. Clin. Investig. 108 1315–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Eck, M., Bos, I. S. T., Kaminski, W. E., Orsó, E., Rothe, G., Twisk, J., Böttcher, A., Van Amersfoort, E. S., Christiansen-Weber, T. A., Fung-Leung, W.-P., Van Berkel, T. J. C., and Schmitz, G. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 6298–6303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francone, O. L., Royer, L., Boucher, G., Haghpassand, M., Freeman, A., Brees, D., and Aiello, R. J. (2005) Arterioscler. Thromb. Vasc. Biol. 25 1198–1205 [DOI] [PubMed] [Google Scholar]
- 21.Feng, B., Yao, P. M., Li, Y., Devlin, C. M., Zhang, D., Harding, H. P., Sweeney, M., Rong, J. X., Kuriakose, G., Fisher, E. A., Marks, A. R., Ron, D., and Tabas, I. (2003) Nat. Cell Biol. 5 781–792 [DOI] [PubMed] [Google Scholar]
- 22.Li, Y., Schwabe, R. F., Vries-Seimon, T., Yao, P. M., Gerbod-Giannone, M. C., Tall, A. R., Davis, R. J., Flavell, R., Brenner, D. A., and Tabas, I. (2005) J. Biol. Chem. 280 21763–21772 [DOI] [PubMed] [Google Scholar]
- 23.Vries-Seimon, T., Li, Y., Yao, P. M., Stone, E., Wang, Y., Davis, R. J., Flavell, R., and Tabas, I. (2005) J. Cell Biol. 171 61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kay, J. G., Murray, R. Z., Pagan, J. K., and Stow, J. L. (2006) J. Biol. Chem. 281 11949–11954 [DOI] [PubMed] [Google Scholar]
- 25.Olsson, S., and Sundler, R. (2006) Mol. Immunol. 43 607–612 [DOI] [PubMed] [Google Scholar]
- 26.Triantafilou, M., Miyake, K., Golenbock, D. T., and Triantafilou, K. (2002) J. Cell Sci. 115 2603–2611 [DOI] [PubMed] [Google Scholar]
- 27.Pike, L. J. (2003) J. Lipid Res. 44 655–667 [DOI] [PubMed] [Google Scholar]
- 28.Simons, K., and Toomre, D. (2000) Nat. Rev. Mol. Cell Biol. 1 31–39 [DOI] [PubMed] [Google Scholar]
- 29.Landry, Y. D., Denis, M., Nandi, S., Bell, S., Vaughan, A. M., and Zha, X. (2006) J. Biol. Chem. 281 36091–36101 [DOI] [PubMed] [Google Scholar]
- 30.Koseki, M., Hirano, K., Masuda, D., Ikegami, C., Tanaka, M., Ota, A., Sandoval, J. C., Nakagawa-Toyama, Y., Sato, S. B., Kobayashi, T., Shimada, Y., Ohno-Iwashita, Y., Matsuura, F., Shimomura, I., and Yamashita, S. (2007) J. Lipid Res. 48 299–306 [DOI] [PubMed] [Google Scholar]
- 31.Schiller, N. K., Black, A. S., Bradshaw, G. P., Bonnet, D. J., and Curtiss, L. K. (2004) J. Lipid Res. 45 1398–1409 [DOI] [PubMed] [Google Scholar]
- 32.Furbee, J. W., Jr., Francone, O., and Parks, J. S. (2002) J. Lipid Res. 43 428–437 [PubMed] [Google Scholar]
- 33.Koritnik, D. L., and Rudel, L. L. (1983) J. Lipid Res. 24 1639–1645 [PubMed] [Google Scholar]
- 34.Furbee, J. W., Jr., Sawyer, J. K., and Parks, J. S. (2002) J. Biol. Chem. 277 3511–3519 [DOI] [PubMed] [Google Scholar]
- 35.Livak, K. J., and Schmittgen, T. D. (2001) Methods (San Diego) 25 402–408 [DOI] [PubMed] [Google Scholar]
- 36.Powers, K. A., Szaszi, K., Khadaroo, R. G., Tawadros, P. S., Marshall, J. C., Kapus, A., and Rotstein, O. D. (2006) J. Exp. Med. 203 1951–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clausen, B. E., Burkhardt, C., Reith, W., Renkawitz, R., and Forster, I. (1999) Transgenic Res. 8 265–277 [DOI] [PubMed] [Google Scholar]
- 38.Repa, J. J., Turley, S. D., Lobaccaro, J. A., Medina, J., Li, L., Lustig, K., Shan, B., Heyman, R. A., Dietschy, J. M., and Mangelsdorf, D. J. (2000) Science 289 1524–1529 [DOI] [PubMed] [Google Scholar]
- 39.Venkateswaran, A., Laffitte, B. A., Joseph, S. B., Mak, P. A., Wilpitz, D. C., Edwards, P. A., and Tontonoz, P. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 12097–12102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Francis, G. A., Knopp, R. H., and Oram, J. F. (1995) J. Clin. Investig. 96 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmitz, G., and Langmann, T. (2001) Curr. Opin. Lipidol. 12 129–140 [DOI] [PubMed] [Google Scholar]
- 42.de Winther, M. P., Kanters, E., Kraal, G., and Hofker, M. H. (2005) Arterioscler. Thromb. Vasc. Biol. 25 904–914 [DOI] [PubMed] [Google Scholar]
- 43.Pearson, G., Robinson, F., Beers, G. T., Xu, B. E., Karandikar, M., Berman, K., and Cobb, M. H. (2001) Endocr. Rev. 22 153–183 [DOI] [PubMed] [Google Scholar]
- 44.Ojaniemi, M., Glumoff, V., Harju, K., Liljeroos, M., Vuori, K., and Hallman, M. (2003) Eur. J. Immunol. 33 597–605 [DOI] [PubMed] [Google Scholar]
- 45.Akira, S., and Takeda, K. (2004) Nat. Rev. Immunol. 4 499–511 [DOI] [PubMed] [Google Scholar]
- 46.Furuchi, T., and Anderson, R. G. (1998) J. Biol. Chem. 273 21099–21104 [DOI] [PubMed] [Google Scholar]
- 47.Scheiffele, P., Roth, M. G., and Simons, K. (1997) EMBO J. 16 5501–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horton, J. D., Goldstein, J. L., and Brown, M. S. (2002) J. Clin. Investig. 109 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rutkowski, D. T., and Kaufman, R. J. (2004) Trends Cell Biol. 14 20–28 [DOI] [PubMed] [Google Scholar]
- 50.Lusis, A. J. (2000) Nature 407 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aiello, R. J., Brees, D., Bourassa, P. A., Royer, L., Lindsey, S., Coskran, T., Haghpassand, M., and Francone, O. L. (2002) Arterioscler. Thromb. Vasc. Biol. 22 630–637 [DOI] [PubMed] [Google Scholar]
- 52.Tabas, I. (2002) J. Clin. Investig. 110 905–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chawla, A., Repa, J. J., Evans, R. M., and Mangelsdorf, D. J. (2001) Science 294 1866–1870 [DOI] [PubMed] [Google Scholar]
- 54.Foster, L. J., de Hoog, C. L., and Mann, M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 5813–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhuang, L., Lin, J., Lu, M. L., Solomon, K. R., and Freeman, M. R. (2002) Cancer Res. 62 2227–2231 [PubMed] [Google Scholar]
- 56.Joseph, S. B., Castrillo, A., Laffitte, B. A., Mangelsdorf, D. J., and Tontonoz, P. (2003) Nat. Med. 9 213–219 [DOI] [PubMed] [Google Scholar]
- 57.Ghisletti, S., Huang, W., Ogawa, S., Pascual, G., Lin, M. E., Willson, T. M., Rosenfeld, M. G., and Glass, C. K. (2007) Mol. Cell 25 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yvan-Charvet, L., Ranalletta, M., Wang, N., Han, S., Terasaka, N., Li, R., Welch, C., and Tall, A. R. (2007) J. Clin. Investig. 117 3900–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gerbod-Giannone, M. C., Li, Y., Holleboom, A., Han, S., Hsu, L. C., Tabas, I., and Tall, A. R. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 3112–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brunham, L. R., Kruit, J. K., Pape, T. D., Timmins, J. M., Reuwer, A. Q., Vasanji, Z., Marsh, B. J., Rodrigues, B., Johnson, J. D., Parks, J. S., Verchere, C. B., and Hayden, M. R. (2007) Nat. Med. 13 340–347 [DOI] [PubMed] [Google Scholar]
- 61.Villarreal-Molina, M. T., Guilar-Salinas, C. A., Rodriguez-Cruz, M., Riano, D., Villalobos-Comparan, M., Coral-Vazquez, R., Menjivar, M., Yescas-Gomez, P., Konigsoerg-Fainstein, M., Romero-Hidalgo, S., Tusie-Luna, M. T., and Canizales-Quinteros, S. (2007) Diabetes 56 1881–1887 [DOI] [PubMed] [Google Scholar]
- 62.Villarreal-Molina, M. T., Flores-Dorantes, M. T., Rellano-Campos, O., Villalobos-Comparan, M., Rodriguez-Cruz, M., Miliar-Garcia, A., Huertas-Vazquez, A., Menjivar, M., Romero-Hidalgo, S., Wacher, N. H., Tusie-Luna, M. T., Cruz, M., Guilar-Salinas, C. A., and Canizales-Quinteros, S. (2008) Diabetes 57 509–513 [DOI] [PubMed] [Google Scholar]
- 63.Porchay, I., Pean, F., Bellili, N., Royer, B., Cogneau, J., Chesnier, M. C., Caradec, A., Tichet, J., Balkau, B., Marre, M., and Fumeron, F. (2006) Obesity (Silver Spring) 14 1874–1879 [DOI] [PubMed] [Google Scholar]
- 64.Daimon, M., Kido, T., Baba, M., Oizumi, T., Jimbu, Y., Kameda, W., Yamaguchi, H., Ohnuma, H., Tominaga, M., Muramatsu, M., and Kato, T. (2005) Biochem. Biophys. Res. Commun. 329 205–210 [DOI] [PubMed] [Google Scholar]
- 65.Eckel, R. H., Grundy, S. M., and Zimmet, P. Z. (2005) Lancet 365 1415–1428 [DOI] [PubMed] [Google Scholar]
- 66.Hotamisligil, G. S. (2006) Nature 444 860–867 [DOI] [PubMed] [Google Scholar]
- 67.Arkan, M. C., Hevener, A. L., Greten, F. R., Maeda, S., Li, Z. W., Long, J. M., Wynshaw-Boris, A., Poli, G., Olefsky, J., and Karin, M. (2005) Nat. Med. 11 191–198 [DOI] [PubMed] [Google Scholar]
- 68.Shi, H., Kokoeva, M. V., Inouye, K., Tzameli, I., Yin, H., and Flier, J. S. (2006) J. Clin. Investig. 116 3015–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.