Abstract
Besides acting as molecular chaperones, the amphitropic small heat shock proteins (sHsps) are suggested to play an additional role in membrane quality control. We investigated sHsp membrane function in the model cyanobacterium Synechocystis sp. PPC 6803 using mutants of the single sHsp from this organism, Hsp17. We examined mutants in the N-terminal arm, L9P and Q16R, for altered interaction with thylakoid and lipid membranes and examined the effects of these mutations on thylakoid functions. These mutants are unusual in that they retain their oligomeric state and chaperone activity in vitro but fail to confer thermotolerance in vivo. We found that both mutant proteins had dramatically altered membrane/lipid interaction properties. Whereas L9P showed strongly reduced binding to thylakoid and model membranes, Q16R was almost exclusively membrane-associated, properties that may be the cause of reduced heat tolerance of cells carrying these mutations. Among the lipid classes tested, Q16R displayed the highest interaction with negatively charged SQDG. In Q16R cells a specific alteration of the thylakoid-embedded Photosystem II (PSII) complex was observed. Namely, the binding of plastoquinone and quinone analogue acceptors to the QB site was modified. In addition, the presence of Q16R dramatically reduced UV-B damage of PSII activity because of enhanced PSII repair. We suggest these effects occur at least partly because of increased interaction of Q16R with SQDG in the PSII complex. Our findings further support the model that membrane association is a functional property of sHsps and suggest sHsps as a possible biotechnological tool to enhance UV protection of photosynthetic organisms.
Small heat shock proteins (sHsps)2 are a ubiquitous family of ATP-independent chaperones defined by their conserved α-crystallin domain, which is flanked by an N-terminal arm of variable length and sequence and by a short C-terminal extension (1, 2). In their native state the majority of sHsps are found as large oligomers with 12 to greater than 32 subunits dependent on the sHsp (3, 4). sHsps can capture unfolding proteins to form stable complexes and prevent their irreversible aggregation. The coaggregation of sHsps with aggregation-prone proteins facilitates subsequent efficient disaggregation (5) and release of substrate proteins by the ATP-dependent (DnaK) chaperone systems (6). Besides acting as molecular chaperones, sHsps also exhibit widely documented membrane interactions in prokaryotes, plants, and mammalian systems (7), which can profoundly affect various membrane functions. Lipid-specific interactions of sHsps were shown not only to modulate the molecular order and polymorphism of the membrane lipid phase (8–10) but were also accompanied by a subtle conformational change of sHsp (11) that may influence its cellular functions.
In the cyanobacterium Synechocystis sp. strain PCC 6803, a well characterized photosynthetic model organism, the single sHsp Hsp17 (also known as Hsp16.6), is essential for tolerance to high temperature (12), and its amount greatly increases during UV-B stress (13). Consistent with the chaperone model of sHsp activity, Hsp17 interacts with a large number of heat-labile proteins in vivo and can prevent irreversible protein aggregation in vitro (5). However, Synechocystis Hsp17 possesses not only protein-protective activity but also the ability to stabilize the lipid phase of membranes (9), and a portion of Hsp17 is typically found in the thylakoid-containing fraction of cell extracts. Thus, this sHsp appears to play an additional role in membrane quality control, potentially contributing to the maintenance of thylakoid integrity, especially under stress conditions (12). A link to membrane function is also reflected in the observation that transcription of the hsp17 gene is strongly regulated by subtle changes in the physical order of the membranes (14).
Analysis of Synechocystis hsp17 deletion and wild type cells provided evidence that heat-induced membrane dynamical changes are causally related to the membrane lipid-Hsp17 interaction. The more heat-sensitive Synechocystis Δhsp17 strain displayed a significantly reduced stability of thylakoid electron transfer during heat stress in parallel with a strong increase in membrane fluidity as measured by 1,6-diphenyl-1,3,5-hexatriene (DPH) anisotropy (8). Differential scanning calorimetry studies revealed that in vitro Hsp17 strongly stabilizes the lamellar liquid-crystalline phase at the expense of the nonlamellar lipid phase (HII), which is known to disrupt membranes under severe heat or osmotic stress (9). Recently a novel “heat shock lipid,” the highly saturated MGlcDG, was identified as rapidly accumulating in Synechocystis cells exposed to combined heat and light stress (10). A preferential interaction of Hsp17 with this nonbilayer phase-forming MGlcDG together with MGDG is consistent with the hypothesis that optimization of the membrane physical state by thylakoid-associated sHsps is likely mediated via specific lipid-sHsp interactions.
Additional evidence for the physiological relevance of thylakoid-associated Hsp17 was presented by Lee et al. (12), who found that inactivation of hsp17 results in greatly reduced photosynthetic oxygen evolution in heat-stressed Synechocystis. Studies with Synechococcus PCC 7942 strain further demonstrated that constitutive expression of HspA, a sHsp from the thermophilic Synechococcus vulcanus, confers cellular thermotolerance and greatly increases the thermostability of both photosystem II (PSII) electron transport and light-harvesting phycocyanins (15).
It is shown that UV-B radiation led to the induction of the heat shock response in Synechocystis. Heat shock regulon members whose mRNAs were elevated include dnaJ, dnaK, groEL, groES, clpB, htpG, and hspA. The htpA gene encoding Hsp17 was up-regulated ∼20-fold compared with ∼3-fold for the other chaperones (13). Because a subset of Hsp17 was shown to be thylakoid-associated (14), we cannot exclude the possibility that it plays a more specific, unexplored function in UV-B stress. Besides DNA damage the main target site of UV-B radiation is the PSII complex of the photosynthetic apparatus, which is a thylakoid bound water-plastoquinone oxido-reductase. UV-B inactivates the catalytic site of the water-oxidizing complex in PSII and leads to the degradation of the D1 (and to a lesser extent of D2) subunit of the PSII reaction center (16). Restoration of PSII activity occurs via subsequent de novo synthesis of the UV-B damaged D1 (and D2) subunits (17).
To define more precisely those features of Synechocystis Hsp17 that are essential for its in vivo function, the effects of Hsp17 mutations have been tested in systematic studies both in vivo and in vitro (18–20). The majorities of mutations studied were in the conserved α-crystallin domain and were found to disrupt Hsp17 oligomerization and to reduce both in vitro chaperone activity and in vivo thermotolerance. However, interestingly, mutations in the N terminus (L9P and Q16R), a region known to play a role in sHsp oligomerization and proposed to be important for substrate binding, were found to exert only minor effects on the oligomeric state of Hsp17, with L9P oligomers being somewhat less stable and Q16R more stable than wild type. Most surprisingly, these mutants could be shown to act as chaperones in vitro in protection of firefly luciferase but did not support in vivo cellular thermotolerance equivalent to wild type (20). These results indicate that the general chaperone action, as defined by in vitro biochemical assays, does not adequately explain the complete role of sHsps in vivo.
In the present study we examined whether the two N-terminal mutations of Hsp17 altered the extent and/or mode of thylakoid and lipid association as well as thylakoid function during stress. Interestingly, the mutant L9P protein showed reduced association with thylakoid and lipid membranes, whereas the Q16R mutant protein was almost exclusively associated with the thylakoid and liposome fraction. Cells carrying Q16R showed specific effects of Hsp17 with the PSII complex impacting PQ binding to the QB site and reducing the extent of UV-B damage. This effect is related to significant improvement of PSII repair and suggests a novel and specific function for thylakoid-associated sHsp. In addition, these results suggest a potential role for sHsps as biotechnological tools to enhance UV protection in photosynthetic organisms.
EXPERIMENTAL PROCEDURES
Plasmids, Strains, and Culture Conditions—Synechocystis 6803 hsp17 and its mutants were subcloned into the HpaI and ApaI sites of the plasmid pNaive containing a spectinomycin resistance marker for transformation into Synechocystis. For expression in Escherichia coli, the genes were inserted into the same sites in pJC20/Hpa, a modified version of pJC20 described previously (18). Pools of pNaive containing mutated hsp17 were isolated from E. coli and transformed into the Synechocystis strain HK-1/ΔClpB1, which is null for both hsp17 and clpB1 (18). The cells were maintained in an illuminated 30 °C incubator on BG-11 agar plates, buffered with 10 mm TES, pH 8.2, supplemented with 5 mm glucose, 50 μg/ml kanamycin sulfate, 100 μg/ml spectinomycin dihydrochloride, and 100 μg/ml erythromycin sulfate, as appropriate (18). Suspension cultures were grown photoautotrophically (35 μE/m2/s) in BG-11, buffered with 5 mm HEPES, pH 7.8, supplemented with 5 mm glucose. The cells in exponential growth phase (A800 = 1.5 ± 0.3) were used in the experiments. In all of the experiments, the cells were treated at 42 °C for 3 h to induce Hsp production.
Lipid Binding Assay—Large unilamellar vesicles were made of total polar lipids isolated from cells grown at 30 °C and then treated at 42 °C for 3 h by the extrusion method using polycarbonate filters of 200-nm pore size. Liposomes and purified proteins at a final concentration of 1.0 and 0.5 mg/ml, respectively, were incubated in 20 mm sodium phosphate (pH 7.0) containing 20 mm sodium chloride for 1 h at room temperature. The protein-lipid mixture was pelleted at 60,000 rpm (Beckman, TLA-100.3) and separated to lipid-bound and supernatant fractions. Soluble samples were concentrated by acetone precipitation, and then both fractions were solubilized in Laemmli sample buffer. After boiling, equal portions of soluble and lipid-bound proteins were analyzed by SDS-PAGE and revealed by Coomassie staining.
Radioactively labeled lipids were prepared by metabolic incorporation of [14C]carbonate into fatty acid backbones. Briefly, 100 ml of cells were grown on 3.4 mm sodium [14C]bicarbonate (0.9 mCi), yielding a total polar lipid mixture with a specific activity of 0.4 mCi/mg. Lipid binding experiments were repeated with liposomes made of 14C-labeled lipids as described above. The radioactive lipid content of protein samples separated by SDS-PAGE was detected by fluorography (21) and by liquid scintillation counting.
Monolayer Experiments—Interaction of sHsps with monomolecular lipid layers was measured using a KSV3000 Langmuir Blodget instrument (KSV Instruments, Helsinki). Briefly, isolated lipids dissolved in chloroform at 0.4 mg/ml concentration were directly spread onto the surface of 20 mm sodium phosphate, pH 7.0, and 20 mm sodium chloride buffer at room temperature. Initial surface pressure of the lipid layer was adjusted to 22.0 ± 0.1 mN/m at a constant surface area. The sHsp of interest was injected underneath the monolayer, and subsequent changes in surface pressure, which are proportional to the strength of the interaction, were monitored. The extent of interaction was determined by calculating the maximal changes in surface pressure caused by the insertion of the actual sHsps as indicated. Kinetics of protein-lipid interaction was followed by monitoring the equilibration of 4.2 μg/ml sHsp with the monomolecular lipid layer (22).
UV-B Treatment—Heat-treated Synechocystis cells were exposed to UV-B radiation in open, rectangular glass containers, in which 100 ml of culture formed a 1-cm-deep layer. The containers were immersed into a temperature-controlled incubator to keep the suspension at 30 °C during the treatment. The cells, at a chlorophyll concentration of 6.5 μg of Chla/ml, were maintained in suspension by magnetic stirring. UV-B light was provided by a Vilbert-Lourmat VL-215 M lamp in combination with a 0.1-mm cellulose acetate filter (Clarfoil; Courtaluds Chemicals). The UV intensity at the surface of the cell suspension was 4.5 W/m2, which corresponds to 12 μmol/m2 s photon flux density at 312 nm. During recovery, the cells were illuminated by 40–50 μmol/m2 s visible light by an array of halogen spot lamps.
Oxygen Evolution Measurements—Steady state rates of oxygen evolution were measured using a Hansatech DW2 O2 electrode at a light intensity of 1000 μmol·m–2·s–1 of photosynthetically active radiation (400–700 nm) in the presence of 0.5 mm DMBQ. For some measurements DCBQ or pBQ was also used as electron acceptors. Usually, 1 ml of heat-treated cells at 6.5 μg of Chla/ml were used in each measurement. The results are expressed as percentages of the oxygen evolution rate measured at time 0.
Flash Fluorescence Measurements—Flash-induced increase and the subsequent decay of chlorophyll fluorescence yield were measured by a double-modulation fluorometer (PSI Instruments, Brno, Czech Republic) in the 150 μs to 100 s time ranges. Heat-treated samples were dark adapted for 3 min, and measurements were performed as described earlier (23). Multicomponent deconvolution of the curves was done using a fitting function with three components: two exponentials and one hyperbolic as described earlier (23): F(t) – F0 = A1 exp(–t/T1) + A2 exp(–t/T2) + A3/(1 + t/T3), where F(t) is the variable fluorescence yield, F0 is the basic fluorescence level before the flash, A1–A3 are the amplitudes, and T1–T3 are the time constants. The nonlinear correlation between the fluorescence yield and the redox state of QA was corrected by using the Joliot model with a value of 0.5 for the energy transfer parameter between PSII subunits (24).
Cell Fractionation and Immunoblotting—A thylakoid membrane fraction was prepared by breakage of heat-treated cells with glass beads (150–200 μm in diameter) at 4 °C followed by differential centrifugation according to Ref. 25. Soluble fractions were concentrated by acetone precipitation and used as cytosolic samples. Protein composition was assessed by electrophoresis in a denaturing 12% polyacrylamide gel. For D1 immunoblotting the thylakoid membrane fraction was solubilized in Tris/HCl buffer as in Ref. 26, adjusted at 0.2 μg of Chla/lane and separated overnight at 18 °C. The proteins were transferred onto a nitrocellulose membrane (0.45 μm; Schleicher & Schuell), and the membrane was probed with a global PsbA antibody (chicken anti PsbA). The secondary antibody was alkaline phosphatase-conjugated anti-chicken IgY. The antigen-antibody complexes were visualized by colorimetric reaction using a BCIP-NBT system. The linearity of the immunoresponse was verified by loading a dilution series of samples. Protein levels were quantified using the National Institutes of Health program ImageJ. sHsp content of thylakoid and soluble fractions were determined by immunoblotting. Protein loads of 20 μg were separated by gel electrophoresis, and the proteins were transferred onto an Immobilon™-P membrane (polyvinylidene difluoride, 0.45 μm; Millipore). Antigens were revealed by anti-Hsp16.6 rabbit antiserum, created against purified recombinant Hsp16.6 (i.e. Hsp17) and horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Sigma), and then visualized by ECL detection (Amersham Biosciences).
Other Methods—Lipids were isolated from heat-treated Synechocystis and analyzed as described earlier (27). Large unilamellar vesicles were prepared as detailed previously (9). The concentration of WT and mutant proteins was assayed by the microBCA method.
RESULTS
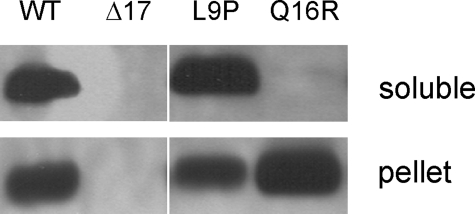
Mutations Affecting Membrane Association of Hsp17—It has been shown that point mutations in Synechocystis Hsp17 can affect the physical properties as well as in vitro and in vivo chaperone activity of these sHsps (5, 20). Interestingly, two mutations, both of which are located in the N-terminal arm of Hsp17 (L9P and Q16R), influenced neither the oligomeric state nor the in vitro chaperone activity of Hsp17, although they impaired heat tolerance in vivo. Because in vivo malfunctioning of L9P and Q16R mutants could not be interpreted on the basis of the current chaperone activity models, we investigated whether the mutated proteins have altered cellular localization that could lead to loss of thermotolerance. Exponentially growing Synechocystis cells were heat-treated for 3 h at 42 °C in the light to allow Hsp17 to accumulate and then thylakoid and cytosolic fractions were isolated. Immunoblot analysis revealed (Fig. 1) that WT Hsp17 is equally distributed between thylakoid and cytosolic fractions. In contrast, the L9P protein appears primarily in the soluble, whereas the Q16R mutant is found exclusively in thylakoid membrane fraction. These results clearly demonstrate that the N-terminal point mutations of Hsp17 protein, although they do not modify the overall oligomerization state, result in altered cellular localization compared with WT. We have attempted to learn whether the Q16R mutant protein is permanently associated with any of the photosynthetic complexes. To this end thylakoids isolated from the cells were solubilized with 0.5% β-dodecyl maltoside, and protein complexes were separated by blue native PAGE. Lanes were cut from the blue native gel and migrated into a second dimension SDS gel. After blotting, specific antibodies were used to identify D1 and Hsp17 proteins. We found that Hsp17 did not migrate together with the proteins of PSII or any other complexes (data not shown), but it behaved rather as a “free” protein. Thus, although the Q16R-Hsp17 protein is found on thylakoid membrane, its association to any photosystem complex, as predictable from results presented in this paper, is not “strong enough” to withstand separation on the gel system.
FIGURE 1.
Subcellular distribution of WT and mutant Hsp17 proteins in heat acclimated cells. Cells grown photoautrophically at 30 °C were exposed to 42 °C for 3 h, and then cytosolic (soluble) and thylakoid (pellet) fractions were isolated and subjected to Western analysis with anti-Hsp17 antibody.
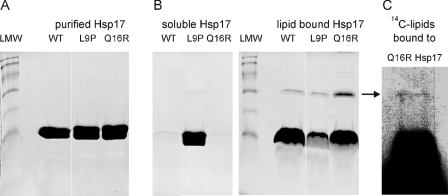
Interaction of Mutant Hsp17 Proteins with Lipid Membranes—According to our previous studies, thylakoid association of Hsp17 may be established via its interaction with membrane lipids (8–10). Thus, next we addressed how the above mutations altered the lipid binding activity of Hsp17. Liposomes made of total polar lipids isolated from Synechocystis cells grown at 30 °C and heat shocked at 42 °C for 3 h were incubated with purified recombinant proteins and then fractionated to soluble and liposome-associated, pellet fractions. Equal portions of soluble and sedimented samples were solubilized under reducing conditions and analyzed by SDS-PAGE with Coomassie blue staining (Fig. 2). Tested at a protein to lipid mass ratio of 1–2, both the WT and Q16R mutant Hsp17 were associated completely with the liposomes, whereas the L9P protein was almost exclusively localized to the soluble fraction (Fig. 2B).
FIGURE 2.
Binding of Hsp17 proteins to liposomes made of total polar lipids isolated from heat-treated cells. A, purity of recombinant proteins was assayed by SDS-PAGE and revealed by Coomassie blue staining. Low molecular weight markers (LMW) display protein bands referring to molecular masses of 94, 67, 43, 30, 20, and 14 kDa, respectively. B, purified proteins were incubated with liposomes for 1 h and then fractionated into lipid bound (pellet) and soluble (supernatant) fractions. The samples were solubilized under reducing conditions and analyzed by SDS-PAGE and Coomassie blue staining. C, the binding assay was repeated with vesicles made of 14C-labeled lipids. The arrow indicates the band corresponding to the Hsp17 associated with 14C-labeled lipids. Lipid content of the pelleted Q16R Hsp17 fraction solubilized under reducing conditions was revealed by fluorography.
Remarkably, despite solubilization of the lipid-bound sHsps under harsh reducing conditions, some portion of Hsp17 displayed a lower mobility on the gel, migrating at a position corresponding to the molecular mass of an Hsp17 dimer (∼30 kDa). Purified recombinant Hsp17 proteins showed no protein band in the corresponding region as visible by Coomassie staining (Fig. 2, A and B). The appearance of this ∼30-kDa form could result from the presence and/or consequence of lipids bound to Hsp17 proteins. Although this band was observed for all three proteins incubated with liposomes, it was especially prominent in samples with the Q16R mutant. To test whether the dimerization, especially in the case of Q16R mutant, was initiated by lipids associated with the dimer form or whether the liposome treatment resulted in change of protein structure without lipid binding, we repeated the binding experiment using a radioactively 14C-labeled Synechocystis lipid mixture. The gel was exposed to fluorography to visualize any radioactive lipids bound to Q16R protein. As Fig. 2C shows, a radioactive band is detected at the ∼30-kDa regions that can originate only from the dimer bound lipids. Radioactivity of the excised ∼30-kDa band was confirmed by liquid scintillation counting compared with an inactive gel region of similar size, yielding activities of 189.0 ± 25.7 and 63.6 ± 5.1 dpm, respectively.
It is known that lipids unusually resistant to harsh solubilization of the membrane-protein sample could be buried in the protein structure and inaccessible for detergent solvation (28). Considering the small fraction of sHsp and lipids involved in the formation of SDS-resistant dimers implies an altered interaction of the Q16R Hsp17 with certain specific lipids.
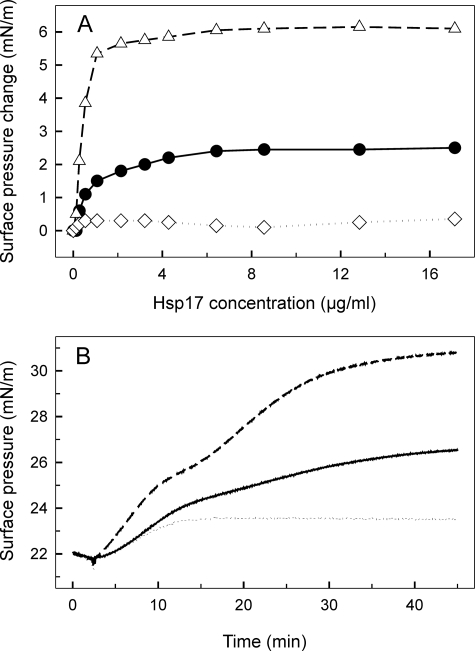
Lipid interaction of the Hsp17 proteins was further studied with the monolayer technique. WT and mutant proteins at different concentrations were injected underneath monomolecular layers of total polar lipids originating from the same source as used in the binding experiments described above. Following changes in surface pressure upon sHsp penetration into the lipid layer, a large difference between the avidity of the three sHsps to interact with the monomolecular layer was observed. As compared with the WT protein, which reached equilibrium surface pressure at ∼6 μg/ml, the Q16R Hsp17 protein displayed a 3-fold higher surface pressure increase and showed saturation at ∼2.5 μg/ml. In contrast L9P was unable to interact (Fig. 3A). Because it is very unlikely that all lipid classes are equally involved in the Hsp17 monolayer insertion (and in the formation of Hsp17 dimers), individual lipid classes (MGDG, digalactosyl diacylglycerol, SQDG, phosphatidyl glycerol, and MGlcDG) were isolated, and each of them was probed for their sHsp interacting ability. We obtained a notable difference in the surface pressure profiles resulting from the insertion of sHsps into different lipid monolayers (data not shown). The Hsp17 proteins displayed a prominent level of binding to monolayers formed from the heat shock lipid MGlcDG, but the increase of surface pressure revealed no selectivity for the individual proteins tested. Q16R interacted more than the WT but still moderately with glycolipids (MGDG and digalactosyl diacylglycerol) and phosphatidyl glycerol, whereas we obtained hardly measurable insertion of L9P with these lipids. There was a striking, biphasic nature to protein interaction with the negatively charged sulfoquinovosyl diacylglycerol SQDG (Fig. 3B), with which Q16R showed by far the highest interaction among lipid classes.
FIGURE 3.
Interaction of WT and mutant Hsp17 proteins with monomolecular lipid layers of total polar lipids (A) and SQDG (B) isolated from heat-treated cells. A, increasing concentrations of Hsp17 proteins (WT, Q16R, and L9P as circles, triangles, and diamonds, respectively) were injected underneath lipid layers, and equilibrium surface pressures were recorded. B, 4.2 μg/ml of each sHsp (WT, Q16R, and L9P as solid, dashed, and dotted lines, respectively) was injected into the buffer beneath the lipid layer, and development of protein-lipid binding was monitored by following the surface pressure increase.
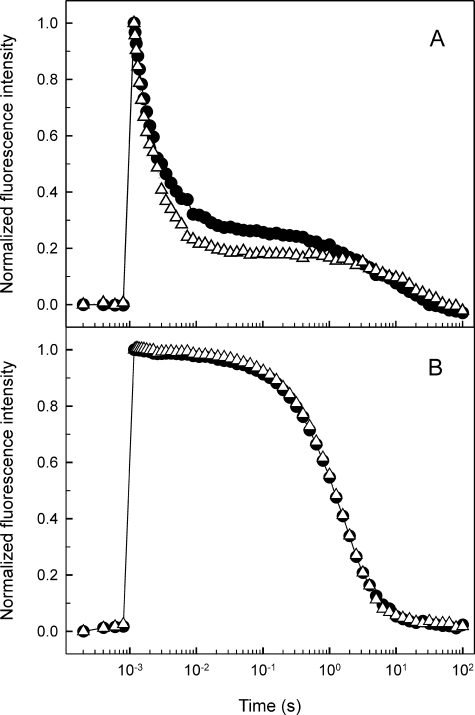
Effect of the Q16R Mutation on PSII Electron Transport—To explore the possible consequence of the exclusive thylakoid association and enhanced lipid binding properties of Hsp17 in the Q16R strain on PSII electron transport characteristics, we measured the kinetics of flash-induced chlorophyll fluorescence, which monitors forward and backward electron transfer processes at the reducing side of PS II. Illumination of Synechocystis cells with a short light pulse reduces the first quinone electron acceptor of the PSII complex, QA, which leads to increased fluorescence yield. Subsequent reoxidation of Q–A in the dark results in the relaxation of fluorescence yield exhibiting three main decay phases (23). The fast (a few hundred microseconds) phase reflects electron transfer from Q–A to QB, which is a PQ molecule occupying the QB plastoquinone-binding site. The middle (a few milliseconds) phase also arises from electron transfer from Q–A to QB, but in such PSII centers that bind PQ molecules after the light pulse, i.e. the time constant of this phase shows the rate of PQ binding to the QB site. Finally the slow (a few seconds) phase arises from backward electron transport from Q–B to the oxidized S2 state of the water-oxidizing complex (23). In the Q16R mutant the fast and middle phases were faster than in the WT (Fig. 4 and Table 1), indicating faster oxidation of Q–A by bound QB and faster binding of PQ to the QB site in the Q16R mutant.
FIGURE 4.
Relaxation of flash induced chlorophyll fluorescence in heat preconditioned (42 °C, 3 h) WT (circles) and Q16R (triangles) cells. Fluorescence excitation was achieved with a single turnover saturating flash at t = 1 ms. Subsequent fluorescence relaxation was measured in the absence of electron transport inhibitors (A) and in the presence of 10 μm (3-(3,4-dichlorophenyl)-1, 1-dimethylurea (DCMU) (B). The curves shown were normalized to the same initial amplitude and corrected according to Joliot and Joliot (24) to consider nonlinear relationships of fluorescence yield and Q–A concentration.
TABLE 1.
Characteristics of chlorophyll fluorescence yield relaxation in WT and Q16R mutant cells
Heat-treated samples were dark-adapted for 3 min and excited with a single turnover flash, and fluorescence relaxation was measured. Relaxation curves were analyzed as detailed under “Experimental Procedures.” Standard errors show the reliability of the obtained kinetic parameters.
| Strain | Fast phase (T1/amplitude) | Middle phase (T2/amplitude) | Slow phase (T2/amplitude) |
|---|---|---|---|
| ms/% | ms/% | ms/% | |
| No addition | |||
| WT | 0.67 ± 0.04/52 ± 2 | 5.67 ± 0.4/28 ± 2 | 8.8 ± 1/19 ± 2 |
| Q16R | 0.43 ± 0.04/53 ± 3 | 3.46 ± 0.2/30 ± 3 | 17 ± 2/16 ± 0.6 |
| With DCMU | |||
| WT | -/0 | -/0 | 1.15 ± 0.05/100 ± 0.4 |
| Q16R | -/0 | -/0 | 1.14 ± 0.03/100 ± 0.4 |
Together these findings point to an alteration of the QB site in the mutant, which could be a consequence of a structural change induced by the association (or the different mode of association) of mutated Hsp17 with the PSII complex.
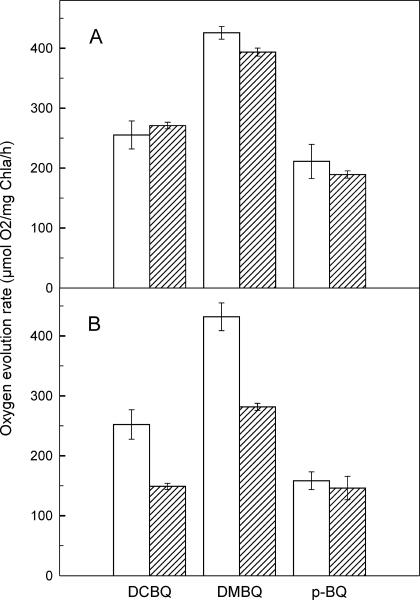
The modification of the QB site was further investigated by measuring PSII activity detecting oxygen evolution in the presence of various artificial acceptors. The rate of oxygen evolution was almost the same in WT and Q16R mutant cells when measured in the presence of DMBQ (∼400 μm O2/μg Chla/h), DCBQ (∼ 250 μm O2/μg Chla/h), and pBQ (∼ 200 μm O2/μg Chla/h) prior to heat treatment (Fig. 5A). Following 3 h of exposure to 42 °C, which is required for the induction of Hsp17, the oxygen evolving activity remained unchanged in the WT but decreased significantly in the Q16R mutant cells when assayed in the presence of DMBQ and DCBQ (Fig. 5B). In contrast, when tested using pBQ as acceptor, the PSII activity was unchanged after heat treatment both in Q16R and WT cells. To rule out the possibility that heat treatment resulted in a decreased amount of functional PSII centers in Q16R cells, the initial amplitude of the flash-induced chlorophyll fluorescence signal, which reflects the amount of active PSII centers, was measured. Heat treatment caused no significant change in the initial amplitude of flash-induced fluorescence signal either in WT or in Q16R cells. Accordingly, the amplitude ratios (non-treated versus heat-treated) were 0.91 ± 0.14 for WT and 1.04 ± 0.03 for Q16R cells.
FIGURE 5.
PSII activity measured as the rate of oxygen evolution in the WT (white bars) and Q16R mutant (striped bars) cells before (A) and after heat treatment (42 °C, 3 h) (B). The activity was tested in the presence of different electron acceptors such as 2,5-DMBQ, 2,5-DCBQ, and pBQ (0.5 mm each). The error bars represent standard errors obtained from three repetitions.
The benzoquinone electron acceptors can take electrons via direct binding to the QB site and/or via interacting with the PQ molecules dissolved in the lipid phase of the thylakoid membrane. The three acceptors used in our measurements have different affinities for the QB site, in the following order: DCBQ > DMBQ ≫ pBQ (29). The decreased oxygen evolution rates in the heat-treated Q16R-Hsp17 cells relative to the WT (and non-heat-treated Q16R), when measured in the presence of DCBQ and DMBQ, indicate altered accessibility of these acceptors to the QB site, possibly because of the association of Q16R Hsp17 protein to the thylakoid. The lack of this effect in the presence of pBQ can be explained by the low affinity of pBQ to the QB site and preferential uptake of electrons via interaction with the PQ molecules in the lipid phase of the membrane (29). Taken together, the fluorescence and oxygen evolution data demonstrate that the accessibility of the QB site to plastoquinone and benzoquinone molecules is significantly altered because of the mutation and the consequent thylakoid association of Q16R.
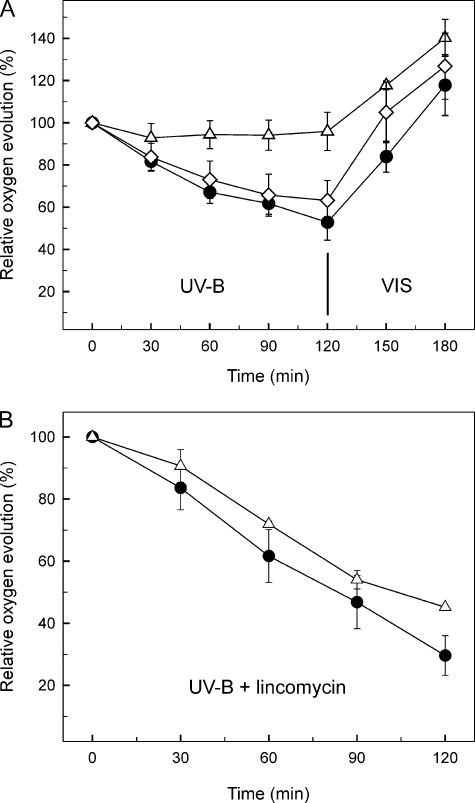
Effect of the Q16R Mutation on the UV-B Sensitivity of Oxygen Evolving Activity—UV-B radiation induces a well characterized inhibition of the PSII complex, which can be repaired via de novo synthesis of the D1 (and D2) reaction center subunit(s) (17), a process that is strongly influenced by chaperones (30) and the physical state of the thylakoid membrane (31, 32). Therefore, we addressed whether the Hsp17 mutants with modified thylakoid association possess an altered UV-B sensitivity of PSII. Heat-hardened cells (42 °C, 3 h) of the WT and mutant strains were exposed to UV-B radiation for 2 h at 30 °C and then shifted back to visible light for 1 h to recover. As a consequence of UV-B induced damage, the oxygen evolving activity in the WT and L9P strains decreased to around 60% of the initial values within 2 h. Astonishingly, the rate of oxygen evolution in the Q16R mutant showed no significant decrease during the 2 h of UV-B treatment (Fig. 6A). When cells were shifted back to normal growth conditions in the light, the oxygen evolving ability recovered completely within 1 h in WT. Simultaneous assessment of PSII activity by fluorescence amplitude measurement confirmed the results of oxygen evolution (data not shown).
FIGURE 6.
Effect of Hsp17 mutations on oxygen evolving activity. Heat-preconditioned (42 °C, 3 h) WT (circles), Q16R (triangles), and L9P (diamonds) strains were exposed to UV-B radiation for 2 h followed by a recovery period in visible light (VIS) for 1 h. Oxygen evolving activities were measured at the indicated time points. The results are expressed as percentages of the oxygen evolution rate measured at time 0. The error bars represent standard errors obtained from three repetitions. The experiments were performed in the absence (A) or presence (B) of the protein synthesis inhibitor lincomycin.
To determine the origin of the resistance of Q16R mutants to UV-B-induced damage, the above experiment was repeated in the presence of lincomycin, a protein synthesis inhibitor, which prevents resynthesis of the UV-B-damaged reaction center subunits of the PSII complex. Under these conditions both the WT and the Q16R mutant showed an accelerated loss of oxygen evolution to 30–40% of their original activity as a consequence of UV-B treatment (Fig. 6B). The similar extent of activity loss in WT and Q16R in the presence of lincomycin demonstrates that the resistance of the Q16R strain against UV-B damage must arise from an enhanced rate of PSII repair.
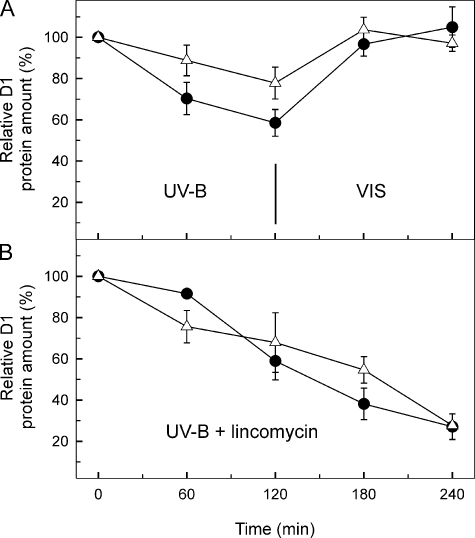
The repair cycle of PSII involves the degradation of the damaged D1 protein subunit of the reaction center and its replacement by a new copy (33). Immunoblot analysis revealed a higher steady state level of D1 protein in the Q16R mutant than in the WT throughout the period of UV-B photodamaging illumination (Fig. 7A). This finding, in parallel with the oxygen evolution results, points to an involvement of the Q16R Hsp17 mutant protein in PSII repair. The higher steady state level of D1 protein and the protected PSII function in the Q16R mutant could originate either from an inhibition of the damage or from an enhanced repair of D1 protein. Experiments in the presence of protein synthesis inhibitor lincomycin showed identical kinetics for D1 degradation in the WT and Q16R mutant (Fig. 7B), indicating that the Q16R mutation in Hsp17 does not affect the rate of D1 protein damage; instead it must enhance the rate of D1 repair.
FIGURE 7.
Change in the total D1 protein content in heat preconditioned WT (circles) and Q16R (triangles) cells challenged by UV-B stress. D1 content in samples was followed during UV-B stress and subsequent recovery (A) or tested upon prolonged UV-B treatment in the presence of lincomycin (B). The data are obtained by densitometry of the Western blots and are the means of three parallel experiments. The results are expressed as percentages of the initial D1 protein level. VIS, visible light.
DISCUSSION
We have exploited our established genetic system in the cyanobacterium Synechocystis sp. PCC 6803 to study the impact of point mutations of Hsp17 on lipid and membrane interaction properties of the sHsp along with alterations of membrane function as assessed by measurements of PSII activity. This system provides the ability to correlate in vivo functional studies to in vitro biochemical experiments. We focused on mutants in the N-terminal domain that exhibited reduced thermotolerance (L9P and Q16R) but that retained essentially normal oligomeric structure and were active in chaperone assays in vitro (20). Considering the membrane binding properties of Hsp17, we hypothesized that the in vivo thermosensitivity of the mutants is due to an altered cytosol/membrane redistribution of mutant proteins. We documented that Hsp17 in heat-pretreated WT cells is equally partitioned in the soluble and a thylakoid-containing insoluble fraction. Introducing the Q16R mutation shifted this equilibrium completely toward the thylakoid fraction, whereas the L9P mutation caused an almost exclusive cytosolic localization. Because the two mutants did not differ essentially in oligomeric structure, these data support the conclusion that properties of the N-terminal arm impact membrane interactions and that these alterations in membrane interaction properties contribute to the phenotypic defects observed in vivo.
Our earlier studies revealed that the nature of lipid-sHsp interactions is highly specific and depends on the sHsp, the membrane lipid head group, and the extent of fatty acid unsaturation (8–10). In the present work we found that both WT Hsp17 and Q16R sediment with liposome membranes made of Synechocystis membrane lipids, indicating that these two proteins bind to lipids. In this experiment, L9P was mainly localized in the supernatant fraction, strengthening our idea that thylakoid membrane-sHsp interaction is governed by lipids.
Interestingly, despite the harsh reducing solubilization of the liposome bound sHsps, some portion of Hsp17 showed a lower mobility on SDS gel corresponding to the molecular weight of dimers. The appearance of the dimer band was especially pronounced in the Q16R mutant, which may reflect its preferable and/or more specific lipid interaction compared with WT. We assume that certain lipids that were unusually resistant to solubilization of the membrane-protein sample are buried in the protein structure and thereby inaccessible for detergent solvation (28). These integrative lipids are distinct from bulk lipids in terms of exerting preferential and/or specific interaction with certain protein-binding sites. Proteins in a biological membrane are known to be surrounded by a shell or annulus of “solvent” lipid molecules. As discussed by Lee (34), whereas these lipid molecules in general interact rather nonspecifically with the protein molecules, a few “hot spots” can be present on the protein where mostly anionic lipids bind with high affinity and are essential for activity of the protein.
Our results applying monomolecular layers of Synechocystis lipids to further study the sHsp-lipid interaction also indicate that the strong association of Q16R Hsp17 with membranes can be linked to a specific lipid constituent of Synechocystis membranes. Compared with WT and L9P, Q16R displayed the highest interaction with monolayer formed both from TPL (Fig. 3A) and from each individual lipid class (Fig. 3B and data not shown). On the other hand, by far the highest degree of insertion of Q16R protein was recorded with a negatively charged lipid, SQDG. Intriguingly, the interaction of Q16R and WT proteins with SQDG monolayer showed a biphasic character. Our results, using stable dimer sHsp mutant proteins (L66A, F102S, and V143A) and a stable oligomer (S2Y) (19) (data not shown) indicate that dissociation of sHsp oligomers is a prerequisite for lipid binding, similar to the model for sHsp chaperone activity (18). We suppose that in the course of lipid membrane association, the pre-existing sHsp oligomers dissociate, enabling the otherwise nonaccessible N terminus of this chaperone to interact with membrane lipids (1). Accordingly, biphasic interaction of WT and Q16R sHsps allows us to reveal the kinetic properties of binding dimers and to unravel the beneficial effect of Q16R mutation on the insertion of Hsp17 dimers into lipid layers. Understanding whether the dimeric sHsps are the predominant membrane-associated species is key to defining their exact role in thylakoid function.
Photosynthetic electron transport between the thylakoid embedded PSII and cytb6f complexes is mediated by PQ molecules, which take up the electrons at the PQ (QB)-binding site of PSII and then move via diffusion in the membrane lipid phase. The chlorophyll fluorescence relaxation and oxygen evolution measurements in this study show that the accessibility of PQ and the artificial electron acceptor benzoquinone molecules to the QB sites is altered in the Q16R mutant. This finding indicates that the mutated sHsp interacts not only with the thylakoid membrane in general but specifically with the PSII complex. The QB site is partly constituted by a large cytosol exposed loop of the D1 protein (35). Thus, Q16R Hsp17, which is expected to bind at the cytosolic side of the thylakoid membrane, may affect this region. This idea is supported by recent structural data, which show the presence of protein-associated lipids, including negatively charged SQDG, at the cytosolic side of the PSII complex around the QB-binding site (35, 36). Because we have shown a preferential interaction of the Q16R Hsp17 protein with SQDG in vitro, similar interaction in the PSII complex is a possible explanation for the specific effect of the mutated protein on the QB electron acceptor observed in the mutant cells.
Photosynthesis is the most sensitive cellular function in photosynthetic organisms, and among the various components of the photosynthetic apparatus, the stability of the PSII complex is particularly sensitive to heat and other environmental stresses (37). Induction of Hsps in cyanobacterial cells occurs not only after heat stress but also after exposure to UV-B radiation or strong visible light (13, 14). This phenomenon indicates that Hsps may have an important role in protection against photo-oxidative stress. Despite the above reasoning, the heat-preadapted Δhsp17 Synechocystis strain (HK-1/ΔClpB1, which is null for both hsp17 and clpB) exhibited no reduced tolerance to UV-B stress when compared with WT (which is ΔClpB) (data not shown). We propose that removal of a potential protective component may be compensated during UV-B exposure by the involvement of the up-regulation of other defensive mechanisms. In line of this interpretation, a major chaperone, GroEL, was found to be expressed to a higher extent in the ΔHsp17 strain (data not shown).
The main consequences of UV radiation in PSII are the destruction of the manganese cluster of the water-oxidizing complex, as well as damage of the D1 (and to a lesser extent the D2) subunit(s), which form the protein core of the reaction center and bind or contain the redox cofactors of light-induced electron transport (16). An important defense mechanism of photosynthetic organisms against the adverse effects of UV-B radiation is the repair of damaged PSII centers (17), which is very similar to what occurs in response to photodamage by visible light (33). Crucial steps in the repair process are the degradation of the UV-damaged D1 (and D2), with involvement of the FtsH protease (38), followed by de novo synthesis and reinsertion of D1 (and D2) into the PSII complex. Our data demonstrate that association of Q16R Hsp17 with the thylakoid membrane does not prevent the UV induced damage of the already existing PSII complex; therefore, the protective effect of the Q16R protein must be exerted at the level of PSII repair. The functional PSII unit is a dimeric complex of two monomeric reaction centers (39). At the interface between the two monomeric PSII, there are four SQDG and two MGDG molecules, which are expected to have an important role in the monomer-monomer interaction (35). The dynamic process of D1 replacement was shown to involve the monomerization of the PSII dimer structure, and likewise, the dimerization of repaired PSII monomers to form fully functional PSII (40). The preferential in vitro interaction of the Q16R protein with SQDG lipids, besides altering the binding of PQ to the QB site, indicates that the Q16R Hsp17 may facilitate the dimer to monomer and/or the monomer to dimer reorganization of PSII during the repair cycle. However, we cannot exclude the possibility that the beneficial effect of the Q16R Hsp17 is related to other factors, such as chaperoning the insertion of newly synthesized D1 copies into the PSII complex or otherwise modulating overall protein synthesis.
In conclusion, here we used two mutants of Hsp17 of Synechocystis, the well established stress model (41) that showed loss of function in vivo in thermotolerance tests, although they were active as chaperones in vitro (20). We found that compared with WT Q16R had an enhanced lipid-mediated thylakoid membrane interaction, which affects directly the PSII complex and leads to a greatly enhanced resistance to UV-induced PSII inactivation via facilitating PSII repair.
The importance of our findings lies in the fact that this is the first documentation of the significant improvement of UV-B stress tolerance attained by targeting a mutant Hsp to the thylakoid membrane. In line with this concept, recently the carrot sHsp Hsp17.7 was introduced into potato, which exhibited significantly improved cellular membrane stability at high temperatures as evidenced by electrolyte leakage (42). Our findings imply the potential to genetically engineer enhanced stress tolerance in important agronomic crop plants by introducing an Hsp gene encoding Hsps with well defined membrane-interacting features.
Acknowledgments
We thankÉva Dobó for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant 1 R03 TW006154-01A1 (to E. V. and L. V.). This work was also supported by European Union Grant SOLAR-H2, 212508 (to O. C. and I. V.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Hsp, heat shock protein; sHsp, small Hsp; MGDG, monogalactosyl diacylglycerol; SQDG, sulfoquinovosyl diacylglycerol; MGlcDG, monoglucosyl diacylglycerol; PQ, plastoquinone; PSII, photosystem II; WT, wild type; TES, 2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}ethanesulfonic acid; DMBQ, 2,5-dimethyl-p-benzoquinone; DCBQ, 2,5-dichloro-p-benzoquinone; pBQ, p-benzoquinone.
References
- 1.van Montfort, R. L. M., Basha, E., Friedrich, K. L., Slingsby, C., and Vierling, E. (2001) Nat. Struct. Biol. 8 1025–1030 [DOI] [PubMed] [Google Scholar]
- 2.De Jong, W. W., Caspers, G.-J., and Leunissen, J. A. M. (1998) Int. J. Biol. Macromol. 22 151–162 [DOI] [PubMed] [Google Scholar]
- 3.Waters, E. R., Lee, G. J., and Vierling, E. (1996) J. Exp. Bot. 47 325–338 [Google Scholar]
- 4.Stamler, R., Kappe, G., Boelens, W., and Slingsby, C. (2005) J. Mol. Biol. 353 68–79 [DOI] [PubMed] [Google Scholar]
- 5.Basha, E., Lee, G. J., Breci, L. A., Hausrath, A. C., Buan, N. R., Giese, K. C., and Vierling, E. (2004) J. Biol. Chem. 279 7566–7575 [DOI] [PubMed] [Google Scholar]
- 6.Lee, G. J., and Vierling, E. (2000) Plant Physiol. 122 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamoto, H., and Vigh, L. (2007) Cell Mol. Life Sci. 64 294–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Török, Z., Goloubinoff, P., Horváth, I., Tsvetkova, N. M., Glatz, A., Balogh, G., Varvasovszki, V., Los, D. A., Vierling, E., Crowe, J. H., and Vigh, L. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 3098–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsvetkova, N. M., Horváth, I., Török, Z., Wolkers, W. F., Balogi, Z., Shigapova, N., Crowe, L. M., Tablin, F., Vierling, E., Crowe, J. H., and Vigh, L. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13504–13509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balogi, Z., Török, Z., Balogh, G., Josvay, K., Shigapova, N., Vierling, E., Vigh, L., and Horváth, I. (2005) Arch. Biochem. Biophys. 436 346–354 [DOI] [PubMed] [Google Scholar]
- 11.Chowdary, T. K., Raman, B., Ramakrishna, T., and Rao, C. M. (2007) Biochem. J. 401 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, S., Owen, H. A., Prochaska, D. J., and Barnum, S. R. (2000) Curr. Microbiol. 40 283–287 [DOI] [PubMed] [Google Scholar]
- 13.Huang, L., McCluskey, M. P., Ni, H., and LaRossa, R. A. (2002) J. Bacteriol. 184 6845–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horváth, I., Glatz, A., Varvasovszki, V., Török, Z., Páli, T., Balogh, G., Kovács, E., Nádasdi, L., Benkö, S., Joó, F., and Vigh, L. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 3513–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamoto, H., Suzuki, N., and Roy, S. K. (2000) FEBS Lett. 483 169–174 [DOI] [PubMed] [Google Scholar]
- 16.Vass, I. (1996) Handbook of Photosynthesis, pp. 931–950, Marcel Dekker, Inc., New York
- 17.Sass, L., Spetea, C., Máté, Z., Nagy, F., and Vass, I. (1997) Photosynth. Res. 54 55–62 [Google Scholar]
- 18.Giese, K. C., and Vierling, E. (2002) J. Biol. Chem. 277 46310–46318 [DOI] [PubMed] [Google Scholar]
- 19.Giese, K. C., and Vierling, E. (2004) J. Biol. Chem. 279 32674–32683 [DOI] [PubMed] [Google Scholar]
- 20.Giese, K. C., Basha, E., Catague, B. Y., and Vierling, E. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 18896–18901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skinner, M. K., and Griswold, M. (1983) Biochem. J. 209 281–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torok, Z., Horvath, I., Goloubinoff, P., Kovacs, E., Glatz, A., Balogh, G., and Vigh, L. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 2192–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vass, I., Kirilovsky, D., and Etienne, A.-L. (1999) Biochemistry 38 12786–12794 [DOI] [PubMed] [Google Scholar]
- 24.Joliot, A., and Joliot, P. (1964) C. R. Acad. Sci. 258 4622–4625 [PubMed] [Google Scholar]
- 25.Komenda, J., and Barber, J. (1995) Biochemistry 34 9625–9631 [DOI] [PubMed] [Google Scholar]
- 26.Allahverdiyeva, Y., Deak, Zs., Szilárd, A., Diner, B. A., Nixon, P. J., and Vass, I. (2004) Eur. J. Biochem. 271 3523–3532 [DOI] [PubMed] [Google Scholar]
- 27.Vigh, L., Los, D. A., Horvath, I., and Murata, N. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 9090–9094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Y., Heitz, F., Le Grimellec, C., and Cole, R. B. (2004) Rapid Com. Mass Spectrom. 18 135–137 [DOI] [PubMed] [Google Scholar]
- 29.Satoh, K., Oh-hashi, M., Kashino, Y., and Koike, H. (1995) Plant Cell Physiol. 36 597–605 [Google Scholar]
- 30.Silva, P., Thompson, E., Bailey, S., Kruse, O., Mullineaux, C. W., Robinson, C., Mann, N. H., and Nixon, P. J. (2003) Plant Cell 15 2152–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanervo, E., Aro, E. M., and Murata, N. (1995) FEBS Lett. 364 239–242 [DOI] [PubMed] [Google Scholar]
- 32.Kanervo, E., Tasaka, Y., Murata, N., and Aro, E. M. (1997) Plant Physiol. 114 841–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vass, I., and Aro, E. M. (2007) Comprehensive Series in Photochemical and Photobiological Sciences, pp. 393–411, Royal Society of Chemistry, Cambridge, UK
- 34.Lee, A. G. (2005) Mol. Biosyst. 1 203–212 [DOI] [PubMed] [Google Scholar]
- 35.Loll, B., Kern, J., Saenger, W., Zouni, A., and Biesiadka, J. (2005) Nature 438 1040–1044 [DOI] [PubMed] [Google Scholar]
- 36.Loll, B., Kern, J., Saenger, W., Zouni, A., and Biesiadka, J. (2007) Biochim. Biophys. Acta 1767 509–519 [DOI] [PubMed] [Google Scholar]
- 37.Berry, J., and Björkman, O. (1980) Annu. Rev. Plant Physiol. 31 491–543 [Google Scholar]
- 38.Cheregi, O., Sicora, C., Kós, P. B., Barker, M., Nixon, P. J., and Vass, I. (2007) Biochim. Biophys. Acta 1767 820–828 [DOI] [PubMed] [Google Scholar]
- 39.Rhee, K. H., Morris, E. P., Zheleva, D., Hankamer, B., Kuhlbrandt, W., and Barber, J. (1997) Nature 389 522–526 [Google Scholar]
- 40.Barbato, R., Friso, G., Rigoni, F., Vecchia, F. D., and Giacometti, G. M. (1992) J. Cell Biol. 119 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glatz, A., Vass, I., Los, D. A., and Vigh, L. (1999) Plant Physiol. Biochem. 37 1–12 [Google Scholar]
- 42.Ahn, Y. J., and Zimmerman, J. L. (2006) Plant Cell Environ. 29 95–104 [DOI] [PubMed] [Google Scholar]