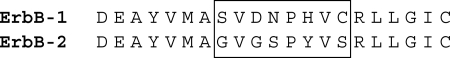In the two decades that have elapsed since the molecular chaperone Hsp902 was shown to regulate the function of steroid receptors (1), >200 signaling proteins have been found to be regulated by Hsp90 (2). These Hsp90 “client” proteins form complexes containing Hsp90 and Hsp70 that are assembled by a multichaperone machinery (3), with Hsp90 regulating both signaling protein function and turnover. Degradation of these Hsp90-regulated signaling proteins occurs via the ubiquitin-proteasome pathway (4), which in this case is initiated by Hsp70-dependent E3 ligases, such as CHIP and parkin (5). Formation of a complex with Hsp90 stabilizes the client signaling protein, and treatment with a specific inhibitor of Hsp90, such as geldanamycin, triggers its rapid degradation (6). Because many of the Hsp90-regulated signaling proteins are involved in cancer cell growth, Hsp90 inhibitors have emerged as a promising new class of anticancer drugs (7).
In this Minireview, we provide a mechanistic basis for understanding how the abundant and ubiquitous chaperones Hsp90 and Hsp70 function together as essential components of the Hsp90 chaperone machinery to regulate signaling protein function and turnover. Like other chaperones, Hsp90 alone has been shown in vitro to assist the refolding of partially unfolded proteins to a properly folded, active conformation. However, Hsp90 is not required for de novo protein folding (8), and it is likely that in cells Hsp90 acts only in concert with Hsp70 in the multichaperone machinery. In contrast to the in vitro experiments on unfolded substrates, this Hsp90 machinery acts on proteins that are in their native conformations to assist the opening of ligand binding clefts.
These clefts are hydrophobic clefts that must open to allow access of ligands, such as steroids, ATP, and heme, to their binding sites within the protein's interior. In the absence of the chaperone machinery, ligand binding clefts are dynamic, shifting to varying extents between closed and open states. When clefts open, hydrophobic residues of the protein's interior are exposed to solvent, and continued opening may progress to protein unfolding. Therefore, the extent to which the ligand binding cleft is open determines ligand access and thus protein function, but clefts are inherent sites of conformational instability. The chaperone machinery assists cleft opening, and Hsp90 binding stabilizes the open state of the cleft, preventing further unfolding and Hsp70-dependent ubiquitination.
The Hsp90 client proteins are assembled into complexes with the chaperone that are stable enough to be isolated and analyzed biochemically. Although we will refer to these as “stable” Hsp90 complexes, they are constantly undergoing cycles of assembly and disassembly in the cytoplasm and nucleoplasm (3). We will refer to this client protein cycling with Hsp90 as stable cycling. As we will show, a variety of manipulations, including mutations of the LBD or ligand binding itself, result in heterocomplexes that very rapidly disassemble such that no (or only trace amounts of) Hsp90 heterocomplexes can be observed in cell lysates. This rapid complex disassembly we define as “dynamic” Hsp90 cycling, and some signaling proteins naturally interact with Hsp90 in this dynamic cycling mode. Because the function and turnover of these proteins are not as affected by Hsp90 inhibitors as proteins undergoing stable Hsp90 complex assembly, they have not been considered as Hsp90-regulated client proteins, but they are nevertheless Hsp90 substrates. There are several examples where the LBDs of signaling proteins with this dynamic “kiss-and-run” interaction with Hsp90 have been converted by mutation to metastable clefts that undergo stable Hsp90 complex assembly. This conversion of signaling protein-Hsp90 interaction is associated with the acquisition of stringently Hsp90-regulated behavior, typical of client proteins.
As Neckers and colleagues have noted (9), many “nodes” in overlapping signaling pathways involved in cancer cell growth are subject to stringent Hsp90 regulation. These Hsp90 client proteins may have evolved from a wide variety of signaling proteins that undergo a more common dynamic cycling of Hsp90 with ligand binding clefts. However, there is no motif for Hsp90 binding, and the basis for its interaction with proteins to form stable or dynamic complexes has not been defined. Here we will present selected examples of Hsp90 effects on signaling protein function and turnover to develop a model in which ligand binding clefts are the common feature determining the interaction with the chaperone. Additional examples in support of the model are cited elsewhere (10).
The Hsp90 Chaperone Machinery
The concept of an Hsp90 chaperone machinery evolved from studies of Hsp90 regulation of steroid receptors. The GR must be in a complex with Hsp90 to have high affinity steroid binding activity, and it is the most studied example of Hsp90 regulation (1, 3). Incubation of Hsp90-free GR with concentrated eukaryotic cell lysates results in ATP-dependent formation of GR·Hsp90 complexes and restoration of high affinity steroid binding activity. During the 1990s, the active components for generating ligand binding activity of steroid receptors were isolated and reconstituted to yield a minimal and efficient Hsp90 complex assembly system of five purified proteins: Hsp90, Hsp70, Hop, Hsp40, and p23 (3).
The two essential proteins for cleft opening, Hsp90 and Hsp70, possess nucleotide-binding sites that regulate their conformations. For both chaperones, the ATP-bound conformation has a low affinity for binding hydrophobic peptide, and the ATPase activity of the chaperone yields an ADP-bound conformation with high affinity for binding hydrophobic peptide (11). Hop is not essential for cleft opening, but it binds independently via an N-terminal TPR domain to Hsp70 and via a central TPR domain to Hsp90 (12). Immunoadsorption of Hop from reticulocyte lysates yields an Hsp90·Hop·Hsp70 complex with a stoichiometry of 2:1:1 that converts the GR to the high affinity steroid binding state (13). Small amounts of the Hsp70 cochaperone Hsp40 are also recovered with this chaperone machinery, and Hsp40 increases the efficiency of cleft opening by the purified proteins (3). This Hsp90 chaperone machinery does not contain p23, and addition of p23 is essential for stable receptor·Hsp90 heterocomplex assembly (14). Plant orthologs exist for all of these proteins, and there is conservation of function throughout eukaryotes (3).
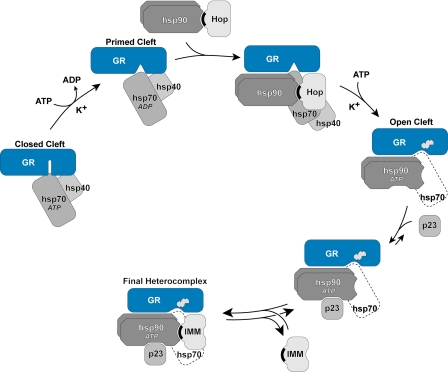
A precise study of progesterone receptor·Hsp90 complex assembly by reticulocyte lysates showed that Hsp70 was bound before there was stable assembly with Hsp90 (15). Stepwise assembly of receptor·Hsp90 complexes by the purified proteins of the machinery led to the assembly scheme shown in Fig. 1 (16). In the first, rapid, ATP-dependent step, the LBD of the receptor is bound by 1 molecule of Hsp70 (17) and 1 molecule of Hsp40 (18), which “primes” the receptor for a second ATP-dependent interaction with Hsp90 that is rate-limiting and yields an open cleft with high affinity ligand binding activity. During this second step, the receptor-bound Hsp90 must pass through at least one complete ATPase cycle, and continued ATPase activity of Hsp70 is required (3), suggesting a cooperative action of the substrate-bound chaperones. To have an open cleft, the receptor-bound Hsp90 has to be in its ATP-dependent conformation (19). This is the conformation of Hsp90 that binds p23 (20), which now acts dynamically to stabilize the complex.
FIGURE 1.
Mechanism of cleft opening and GR·Hsp90·immunophilin heterocomplex assembly. The ATP-dependent conformation of Hsp70 binds initially to the GR, and in an ATP-, K+-, and Hsp40-dependent step, a GR·Hsp70 complex is formed that is primed to interact with Hsp90. After Hsp90 binding, there is a second ATP- and K+-dependent step that is rate-limiting and leads to opening of the steroid binding cleft, enabling access of the steroid (indicated by the steroid structure). During GR·Hsp90 heterocomplex assembly in cells and cell lysates, Hop and some of Hsp70 dissociate during or at the end of the cleft opening step. The GR-bound Hsp90 is now in its ATP-dependent conformation and can be bound by p23, which stabilizes the chaperone in that conformation, preventing disassembly of the GR·Hsp90 heterocomplex. When Hop dissociates, TPR domain proteins, such as immunophilins (IMM), can bind reversibly to the TPR acceptor site on GR-bound Hsp90. TPR domains are indicated by black crescents.
In addition to its effects on ligand binding and turnover that are the focus of this review, Hsp90 has other effects on signaling protein function. These other effects result from the interaction of the signal protein-bound Hsp90 with a variety of Hsp90-binding proteins (2, 3). In the case of the GR, for example, TPR domain immunophilins bind dynamically to Hsp90 when heterocomplex assembly is complete (Fig. 1) to link the receptor to the dynein motor system involved in its retrograde trafficking to the nucleus (21).
Hsp90 Cycling with the GR
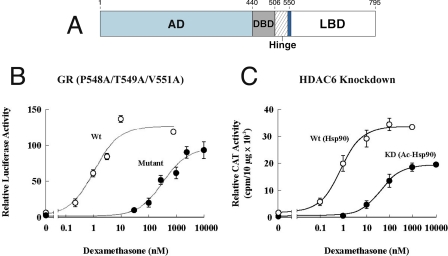
Although the Hsp90 contact sites on the surface of the GR LBD have not been determined, a seven-amino acid segment lying at the N terminus of the LBD (Fig. 2A) was found to be required for stable Hsp90 heterocomplex assembly and high affinity steroid binding activity (22). This seven-amino acid segment (positions 547–553, rat) lies in the middle of helix 1 of the GR LBD. Some mutations within this segment have produced GRs that cycle dynamically with Hsp90 and have the phenotype shown in Fig. 2B, where the dose-response curve for transactivation is shifted ∼100-fold to the right (23). Mutations of leucines in an LXXLL motif that lies within this segment have yielded GRs that assemble biochemically stable Hsp90 heterocomplexes that have low ligand binding affinity due to rapid dissociation of steroid (24, 25). Thus, although this seven-amino acid segment on the GR lies outside the predicted steroid binding cleft, it clearly affects the steroid binding properties of the cleft.
FIGURE 2.
Dexamethasone-dependent transactivation under conditions of stable and dynamic GR·Hsp90 heterocomplex cycling. A, the seven-amino acid segment (dark blue band) in the LBD of the GR (numbers for rat). AD, activation domain; DBD, DNA binding domain. B, dexamethasone stimulation of transcription from a luciferase reporter in cells with wild-type (wt) and P458A/T549A/V551A (Mutant) GRs (23). C, dexamethasone stimulation of transcription from a chloramphenicol acetyltransferase (CAT) reporter in wild-type (non-acetylated Hsp90) and HDAC6 knockdown (KD; acetylated Hsp90) cells (27).
Assembly of the GR complexes with Hsp90 can be converted from the stable to dynamic mode by altering the acetylation state of Hsp90. Hsp90 is normally deacetylated by HDAC6, a cytoplasmic HDAC. In HDAC6 knockdown cells, Hsp90 is hyperacetylated (26). The acetylated Hsp90 does not interact properly with p23, is capable only of dynamic heterocomplex assembly with the GR, and has the phenotype shown in Fig. 2C (27).
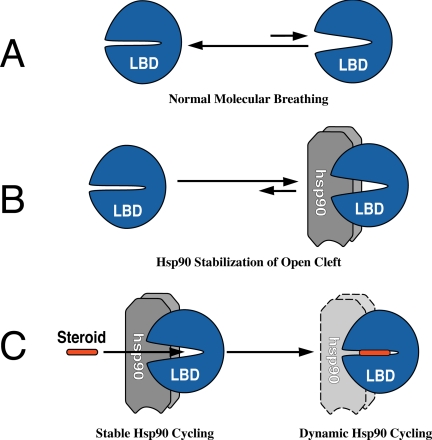
GRs that are not associated with Hsp90 bind steroids in vitro with a very low affinity (28), and a similar right shift in the steroid dose-response curve was observed in yeast engineered to express a very low level of Hsp90 (29). As illustrated in the schematics in Fig. 3, in the absence of Hsp90, the cleft in the GR LBD is predominantly closed, opening only very transiently during the course of normal molecular breathing (Fig. 3A); thus, high concentrations of steroid are required to initiate the hormone effect. When stable complexes are assembled with Hsp90 (Fig. 3B), nearly all the ligand binding clefts are open at any time, and low concentrations of steroid are now sufficient for binding. Under conditions of dynamic cycling, Hsp90 dissociates very rapidly, and the open and closed states of the cleft are more like the non-Hsp90-bound receptor, so higher concentrations of steroid are required for the hormone effect.
FIGURE 3.
States of the steroid binding cleft and Hsp90 cycling. The open versus closed states of the cleft are shown in the absence of Hsp90 (A) or when there is stable complex assembly with Hsp90 (B). Steroid binding promotes closing of the cleft, converting the receptor from stable to dynamic cycling with Hsp90 (C).
As illustrated in the schematic in Fig. 3C, steroid binding within the cleft promotes a temperature-dependent collapse of the cleft around the ligand to the closed state (30). Receptors that have bound steroid under physiological conditions in the cell are not recovered in association with Hsp90, and originally, it was thought that the liganded receptor dissociated from Hsp90 and no longer cycled into complexes with the chaperone (1). This model in which steroid binding triggers Hsp90 dissociation is widely accepted and is presented in basic science textbooks as the initial step in steroid hormone action. However, the model needs to be modified because the ligand-bound transformed receptor undergoes dynamic cycling with Hsp90 that is important for receptor trafficking to and within the nucleus (21).
Hsp90 Cycling with ErbB-1 and ErbB-2
ErbB-1 and ErbB-2 (HER2) are receptor tyrosine kinases that are often overexpressed in cancers of epidermal and neuronal origin (31). ErbB-1 is the epidermal growth factor receptor, and ErbB-2 functions as a ligandless coreceptor that heterodimerizes with other members of the ErbB family to amplify signaling. Hsp90 regulates ErbB-2 function by limiting heterodimer formation (32). Like the Src family kinases, the kinase domain of ErbB-2 is assembled into a stable complex with Hsp90, whereas little or no Hsp90 is recovered with ErbB-1 (33). Upon Hsp90 inhibition by geldanamycin, ErbB-2 is polyubiquitinated and rapidly degraded, whereas ErbB-1 is modestly ubiquitinated and slowly degraded (33, 34). Thus, ErbB-2 undergoes stable cycling with Hsp90, whereas ErbB-1 undergoes dynamic cycling. The difference in geldanamycin sensitivity is accounted for by a short segment within the highly homologous kinase domains (32, 35). Fig. 4 shows this motif, which lies in close association with the ATP binding cleft and the αC helix, a region regulating kinase activity (36). Swapping the eight-amino acid segments shown in the box in Fig. 4 between ErbB-1 and ErbB-2 yields the appropriate exchange of dynamic versus stable cycling with Hsp90 and the corresponding change in geldanamycin sensitivity (32). Simply changing a glycine in the ErbB-2 motif to the aspartate of ErbB-1 results in decreased Hsp90 association and a substantial decrease in geldanamycin-induced degradation (37).
FIGURE 4.
Segment of the ErbB kinase domains determining geldanamycin sensitivity.
This segment, which lies within the αC-β4 loop region of the catalytic domain of many protein kinases, is proposed to define a common surface with which Hsp90 interacts (38). However, direct binding of Hsp90 to this surface has not been demonstrated, and there is no clear similarity between this loop and the region of the GR that defines stable Hsp90 complex assembly. An alternative explanation is that the segments in both the kinases and steroid receptors are features that allow the ligand binding clefts to open more readily, conferring the metastability required for the chaperone machinery to produce a stable complex with Hsp90. That Hsp90 complex assembly is related to the state of the ATP binding cleft is suggested by the fact that an irreversible inhibitor that covalently modifies a cysteine residue in the cleft causes a decrease in ErbB-2·Hsp90 complex assembly and an increase in ErbB-2 ubiquitination and proteasomal degradation (34).
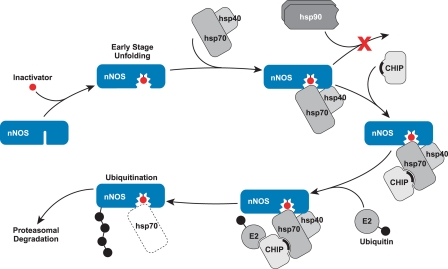
Cleft Modification and Turnover of nNOS
nNOS is the best described example of how distortion of a ligand binding cleft through specific chemical attack leads to chaperone-dependent ubiquitination and proteasomal degradation (39). The nitric-oxide synthases are cytochrome P450-like hemoprotein enzymes that catalyze the conversion of l-arginine to citrulline and nitric oxide by a process that requires NADPH and molecular oxygen (40). nNOS undergoes dynamic cycling with Hsp90, and Hsp90 inhibition leads to nNOS degradation via the ubiquitin-proteasome pathway (41). The apo-nNOS monomer is the form of the enzyme that is ubiquitinated (42). Hsp90 inhibitors also prevent heme binding by heme-deficient apo-nNOS in Sf9 insect cells, which have a low level of endogenous heme (43, 44).
Certain mechanism-based inactivators, such as NG-methyl-l-arginine and the antihypertensive drug guanabenz, cause accelerated nNOS degradation (45). These inactivators cross-link heme to the enzyme (46), a modification that has been shown in a myoglobin model to cause opening of the heme binding cleft (47) to yield a more unfolded state of the protein (48). As diagramed in Fig. 5, the mechanism-based inactivation triggers nNOS ubiquitination and proteasomal degradation (42, 45). As reported for several Hsp90 client proteins, including the GR and ErbB-2 (10), CHIP appears to be an important E3 ligase for nNOS ubiquitination (49), although it is clear that parkin also directs ubiquitination, suggesting redundancy of Hsp70-dependent E3 ligase action.
FIGURE 5.
Mechanism-based inactivators trigger nNOS degradation via Hsp70-dependent ubiquitination. Mechanism-based inactivation of nNOS by certain substrates leads to unfolding of the heme/substrate binding cleft to a degree that Hsp90 cannot cycle with the enzyme to inhibit ubiquitination by Hsp70-dependent E3 ligases, such as CHIP. The solid crescent represents the CHIP TPR domain.
Concluding Remarks
Using techniques that detect stable cycling, it is estimated that ∼10% of the yeast proteome is regulated by Hsp90 (50). Although most research has involved signaling proteins, a wide variety of proteins involved in housekeeping functions are also regulated by Hsp90 (2, 3, 50). When dynamic cycling with Hsp90 is considered, it may be that the Hsp90 chaperone machinery modulates ligand binding clefts in a majority of proteins. Proteins with clefts that are more in a closed state are inherently more stable and undergo dynamic cycling with Hsp90, whereas proteins with metastable clefts are inherently less stable and require stable cycling with Hsp90 to inhibit Hsp70-dependent ubiquitination. The interaction of the chaperone machinery with ligand binding clefts of properly folded proteins determines Hsp90 effects on protein function and protein stability, as well as Hsp90 requirements for protein trafficking and protein complex (e.g. the proteasome) assembly (10).
The Hsp90 chaperone machinery may be the major mechanism for quality control of damaged proteins via the ubiquitin-proteasome pathway. We envision that, as proteins undergo toxic or oxidative damage, ligand binding clefts open to expose hydrophobic residues as the initial step in unfolding. When Hsp90 can no longer cycle with the protein to inhibit ubiquitination, E3 ligases interacting with substrate-bound Hsp70 target ubiquitin-charged E2 enzyme to the nascently unfolding substrate. In this manner, the Hsp90 chaperone machinery may function as a comprehensive protein management system for protein quality control.
This work was supported, in whole or in part, by National Institutes of Health Grants DA022354 and GM077430. This minireview will be reprinted in the 2008 Minireview Compendium, which will be available in January, 2009.
Footnotes
The abbreviations used are: Hsp, heat shock protein; E3, ubiquitin-protein isopeptide ligase; CHIP, C terminus of Hsc70-interacting protein; LBD, ligand binding domain; GR, glucocorticoid receptor; Hop, Hsp-organizing protein; TPR, tetratricopeptide repeat; HDAC, histone deacetylase; nNOS, neuronal nitric-oxide synthase.
References
- 1.Pratt, W. B., and Toft, D. O. (1997) Endocr. Rev. 18 306–360 [DOI] [PubMed] [Google Scholar]
- 2.Picard, D. (2002) CMLS Cell. Mol. Life Sci. 59 1640–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pratt, W. B., and Toft, D. O. (2003) Exp. Biol. Med. 222 111–133 [DOI] [PubMed] [Google Scholar]
- 4.Glickman, M. H., and Ciechanover, A. (2002) Physiol. Rev. 82 373–428 [DOI] [PubMed] [Google Scholar]
- 5.Cyr, D. M., Hohfeld, J., and Patterson, C. (2002) Trends Biochem. Sci. 27 368–375 [DOI] [PubMed] [Google Scholar]
- 6.Neckers, L., Schulte, T. W., and Mimnaugh, E. (1999) Investig. New Drugs 17 361–373 [DOI] [PubMed] [Google Scholar]
- 7.Sharp, S., and Workman, P. (2006) Adv. Cancer Res. 95 323–347 [DOI] [PubMed] [Google Scholar]
- 8.Nathan, D. F., Vos, M. H., and Lindquist, S. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 12949–12956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Isaacs, J. S., Xu, W., and Neckers, L. (2003) Cancer Cell 3 213–217 [DOI] [PubMed] [Google Scholar]
- 10.Pratt, W. B., Morishima, Y., and Osawa, Y. (2007) in Heat Shock Proteins in Cancer (Calderwood, S. K., Sherman, M. Y., and Ciocca, D. R., eds) pp. 1–30, Springer-Verlag, Dordrecht, The Netherlands
- 11.Hartl, F. U., and Hayer-Hartl, M. (2002) Science 295 1852–1858 [DOI] [PubMed] [Google Scholar]
- 12.Chen, S., Prapapanich, V., Rimerman, R. A., Honore, B., and Smith, D. F. (1996) Mol. Endocrinol. 10 682–693 [DOI] [PubMed] [Google Scholar]
- 13.Murphy, P. J. M., Kanelakis, K. C., Galigniana, M. D., Morishima, Y., and Pratt, W. B. (2001) J. Biol. Chem. 276 30092–30098 [DOI] [PubMed] [Google Scholar]
- 14.Morishima, Y., Kanelakis, K. C., Murphy, P. J. M., Lowe, E. R., Jenkins, G. J., Osawa, Y., Sunahara, R. K., and Pratt, W. B. (2003) J. Biol. Chem. 278 48754–48763 [DOI] [PubMed] [Google Scholar]
- 15.Smith, D. F. (1993) Mol. Endocrinol. 7 1418–1429 [DOI] [PubMed] [Google Scholar]
- 16.Morishima, Y., Murphy, P. J. M., Li, D. P., Sanchez, E. R., and Pratt, W. B. (2000) J. Biol. Chem. 275 18054–18060 [DOI] [PubMed] [Google Scholar]
- 17.Murphy, P. J. M., Morishima, Y., Chen, H., Galigniana, M. D., Mansfield, J. E., Simons, S. S., and Pratt, W. B. (2003) J. Biol. Chem. 278 34764–34773 [DOI] [PubMed] [Google Scholar]
- 18.Hernandez, M. P., Chadli, A., and Toft, D. O. (2002) J. Biol. Chem. 277 11873–11881 [DOI] [PubMed] [Google Scholar]
- 19.Grenert, J. P., Johnson, B. D., and Toft, D. O. (1999) J. Biol. Chem. 274 17525–17533 [DOI] [PubMed] [Google Scholar]
- 20.Sullivan, W., Stensgard, B., Caucutt, G., Bartha, B., McMahon, N., Alnemri, E. S., Litwack, G., and Toft, D. (1997) J. Biol. Chem. 272 8007–8012 [DOI] [PubMed] [Google Scholar]
- 21.Pratt, W. B., Galigniana, M. D., Harrell, J. M., and DeFranco, D. B. (2004) Cell. Signal. 16 857–872 [DOI] [PubMed] [Google Scholar]
- 22.Xu, M., Dittmar, K. D., Giannoukos, G., Pratt, W. B., and Simons, S. S. (1998) J. Biol. Chem. 273 13918–13924 [DOI] [PubMed] [Google Scholar]
- 23.Kaul, S., Murphy, P. J. M., Chen, J., Brown, L., Pratt, W. B., and Simons, S. S. (2002) J. Biol. Chem. 277 36223–36232 [DOI] [PubMed] [Google Scholar]
- 24.Giannoukos, G., Silverstein, A. M., Pratt, W. B., and Simons, S. S. (1999) J. Biol. Chem. 274 36527–36536 [DOI] [PubMed] [Google Scholar]
- 25.Dong, D. D., Jewell, C. M., Bienstock, R. J., and Cidlowski, J. A. (2006) J. Steroid Biochem. Mol. Biol. 101 106–117 [DOI] [PubMed] [Google Scholar]
- 26.Kovacs, J. J., Murphy, P. J. M., Galliard, S., Zhao, X., Wu, J. T., Nicchitta, C. V., Yoshida, M., Toft, D. O., Pratt, W. B., and Yao, T. P. (2005) Mol. Cell 18 601–607 [DOI] [PubMed] [Google Scholar]
- 27.Murphy, P. J. M., Morishima, Y., Kovacs, J. J., Yao, T. P., and Pratt, W. B. (2005) J. Biol. Chem. 280 33792–33799 [DOI] [PubMed] [Google Scholar]
- 28.Nemoto, T., Ohara-Nemoto, Y., Denis, M., and Gustafsson, J. A. (1990) Biochemistry 29 1880–1886 [DOI] [PubMed] [Google Scholar]
- 29.Picard, D., Khursheed, B., Garabedian, M. J., Fortin, M. G., Lindquist, S., and Yamamoto, K. R. (1990) Nature 348 166–168 [DOI] [PubMed] [Google Scholar]
- 30.Gee, A. C., and Katzenellenbogen, J. A. (2001) Mol. Endocrinol. 15 421–428 [DOI] [PubMed] [Google Scholar]
- 31.Yarden, Y., and Sliwkowski, M. X. (2001) Nat. Rev. Mol. Cell Biol. 2 127–137 [DOI] [PubMed] [Google Scholar]
- 32.Citri, A., Gan, J., Mosesson, Y., Vereb, G., Szollosi, J., and Yarden, Y. (2004) EMBO Rep. 5 1165–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu, W., Minaugh, E., Rosser, M. F. N., Nicchitta, C., Marcu, M. Yarden, Y., and Neckers, L. (2001) J. Biol. Chem. 276 3702–3708 [DOI] [PubMed] [Google Scholar]
- 34.Citri, A., Alroy, I., Lavi, S., Rubin, C., Xu, W., Grammatikakis, N., Patterson, C., Neckers, L., Fry, D. W., and Yarden, Y. (2002) EMBO J. 21 2407–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tikhomirov, O., and Carpenter, G. (2003) Cancer Res. 63 39–43 [PubMed] [Google Scholar]
- 36.Huse, M., and Kuriyan, J. (2002) Cell 109 275–282 [DOI] [PubMed] [Google Scholar]
- 37.Xu, W., Yuan, X., Xiang, Z., Mimnaugh, E., Marcu, M., and Neckers, L. (2005) Nat. Struct. Mol. Biol. 12 120–126 [DOI] [PubMed] [Google Scholar]
- 38.Citri, A., Harari, D., Shohat, G., Ramakrishnan, P., Gan, J., Lavi, S., Eisenstein, M., Kimchi, A., Wallach, D., Pietrokovski, S., and Yarden, Y. (2006) J. Biol. Chem. 281 14361–14369 [DOI] [PubMed] [Google Scholar]
- 39.Osawa, Y., Lowe, E. R., Everett, A. C., Dunbar, A. Y., and Billecke, S. S. (2003) J. Pharmacol. Exp. Ther. 304 1–5 [DOI] [PubMed] [Google Scholar]
- 40.Marletta, M. A. (1993) J. Biol. Chem. 268 12231–12234 [PubMed] [Google Scholar]
- 41.Bender, A. T., Silverstein, A. M., Demady, D. R., Kanelakis, K. C., Noguchi, S., Pratt, W. B., and Osawa, Y. (1999) J. Biol. Chem. 274 1472–1478 [DOI] [PubMed] [Google Scholar]
- 42.Bender, A. T., Demady, D. R., and Osawa, Y. (2000) J. Biol. Chem. 275 17407–17411 [DOI] [PubMed] [Google Scholar]
- 43.Billecke, S. S., Bender, A. T., Kanelakis, K. C., Murphy, P. J. M., Lowe, E. R., Kamada, Y., Pratt, W. B., and Osawa, Y. (2002) J. Biol. Chem. 277 20504–20509 [DOI] [PubMed] [Google Scholar]
- 44.Billecke, S. S., Dragonov, D. I., Morishima, Y., Murphy, P. J. M., Dunbar, A. Y., Pratt, W. B., and Osawa, Y. (2004) J. Biol. Chem. 279 30252–30258 [DOI] [PubMed] [Google Scholar]
- 45.Noguchi, S., Jianmongkol, S., Bender, A. T., Kamada, Y., Demady, D. R., and Osawa, Y. (2000) J. Biol. Chem. 275 2376–2380 [DOI] [PubMed] [Google Scholar]
- 46.Vuletich, J. L., Lowe, E. R., Jianmongkol, S., Kamada, Y., Kent, U. M., Bender, A. T., Demady, D. R., Hollenberg, P. F., and Osawa, Y. (2002) Mol. Pharmacol. 62 110–118 [DOI] [PubMed] [Google Scholar]
- 47.Osawa, Y., Darbyshire, J. F., Steinbach, P. J., and Brooks, B. R. (1993) J. Biol. Chem. 268 2953–2959 [PubMed] [Google Scholar]
- 48.Osawa, Y., and Pohl, L. R. (1989) Chem. Res. Toxicol. 2 131–141 [DOI] [PubMed] [Google Scholar]
- 49.Peng, H. M., Morishima, Y., Jenkins, G. J., Dunbar, A. Y., Lau, M., Patterson, C., Pratt, W. B., and Osawa, Y. (2004) J. Biol. Chem. 279 52970–52977 [DOI] [PubMed] [Google Scholar]
- 50.Zhao, R., Davey, M., Hsu, Y.C., Kaplanek, P., Tong, A., Parsons, A. B., Krogan, N., Cagney, G., Mai, D., Greenblatt, J., Boone, C., Emili, A., and Houry, W. A. (2005) Cell 120 715–727 [DOI] [PubMed] [Google Scholar]