Abstract
The propensity of T cells to generate coordinated cytokine responses is critical for the host to develop resistance to pathogens while maintaining the state of immunotolerance to self-antigens. The exact mechanisms responsible for preventing the overproduction of proinflammatory cytokines including interferon (IFN)-γ are not fully understood, however. In this study, we examined the role of a recently described Ras GTPase effector and repressor of the Raf/MEK/ERK cascade called impedes mitogenic signal propagation (Imp) in limiting the induction of T-cell cytokines. We found that stimulation of the T cell receptor complex leads to the rapid development of a physical association between Ras and Imp. Consistent with the hypothesis that Imp inhibits signal transduction, we also found that disengagement of this molecule by the RasV12G37 effector loop mutant or RNA interference markedly enhances the activation of the NFAT transcription factor and IFN-γ secretion. A strong output of IFN-γ is responsible for the distinct lymphocyte traffic pattern observed in vivo because the transgenic or retroviral expression of RasV12G37 caused T cells to accumulate preferentially in the lymph nodes and delayed their escape from the lymphoid tissue, respectively. Together, our results describe a hitherto unrecognized negative regulatory role for Imp in the production of IFN-γ in T cells and point to Ras-Imp binding as an attractive target for therapeutic interventions in conditions involving the production of this inflammatory cytokine.
The small GTPase Ras is a potent signaling molecule that can bind with numerous downstream effector molecules including the protein kinase Raf (1, 2). Raf in turn activates the mitogen-activated protein kinase (MAPK)2 kinase (MEK)/extracellular signal-regulated kinase (Erk) cascade (3, 4). This signaling pathway controls many pivotal functions in T cells. The ERK kinases overcome SHP-1 phosphatase blockade of proximal T cell receptor (TCR) signaling (5), and more globally, influence the maturation of T cells in the thymus (6–9) and production of interleukin-2 (IL-2) in the post-thymic peripheral T cells (10–14). The observation that a relative lack of Ras/ERK signal is associated with inability of T cells to respond to an antigen further underscores the fundamental role of this signaling pathway in T cell stimulation (15–17). It is not surprising to find therefore that T cells and other cell types have developed inhibitors to control the magnitude and duration of the ERK signaling. Among the most prominent endogenous repressors of the Ras/ERK signaling are GTPase-activating proteins (18, 19), downstream of tyrosine kinase (Dok) adaptor proteins (20–22), members of the Sprouty protein family (23, 24), diacylglycerol kinases (DGK-ζ and DGK-α) (25, 26), ERK-specific dual specificity phosphatases (27), and a molecule with an E3 ubiquitin ligase activity described recently as impedes mitogenic signal propagation (Imp) (28).
Imp, also known as BRCA1-associated protein (BRAP2), was originally identified as a predominantly cytoplasmic protein that recognizes the nuclear localization signals of BRCA1, SV40 large T antigen, and myosin (29). It has been proposed that BRAP2 masks nuclear localization signal motifs causing the mislocalization of specific nuclear proteins and thus serving as a cytoplasmic retention protein (29, 30). Interestingly, White and collaborators (28) demonstrated that Imp can also disrupt the Ras/ERK pathway possibly by uncoupling Raf kinase from MEK through the inactivation of a scaffolding protein of the Ras pathway called the kinase suppressor of Ras (KSR). The exact mechanism of this inhibition of KSR function is unknown, however. Importantly, Imp is regulated by activated Ras. Specifically, binding of GTP-loaded Ras to a region encompassing the ubiquitin-protease-like zinc finger (UJBP-ZnF) domain of Imp/BRAP targets this molecule for ubiquitination and possible degradation by the proteasome. In summary, activated Ras relays a signal to the Erk cascade in two ways: (i) by recruiting Raf kinase to the plasma membrane and (ii) by relieving Imp-mediated inhibition of KSR-dependent formation of the Raf-MEK complex.
It is unknown whether Imp, by providing an additional layer of resistance to the Erk pathway prevents the unnecessary activation of T cells. However, the amplitude and kinetics of ERK activation have been associated with different outcomes for T cells including survival or apoptosis in “young” T cells in the thymus (31) and the generation of helper T cell (Th1 or Th2) cytokines in mature T cells (32), Thus, it is not unlikely that Imp influences these processes by limiting Erk pathway activity. Consequently, we designed this study to define overall sensitivity of T cells to repressive function of Imp.
EXPERIMENTAL PROCEDURES
Molecular Constructs—cDNAs encoding H-Ras and Ras effector loop mutants (RasV12G35, RasV12G37, and RasV12C40) were described previously (33). N17Ras in pcDNA3 was a gift from K. L. Guan (University of Michigan). K-Ras, N-Ras, and wild type Imp were generated by polymerase chain reaction amplification from the mouse cDNA library. ImpC264A was created using oligonucleotides carrying the desired point mutation and PCR amplification with high-fidelity polymerase. A construct encoding a membrane-targeted variant of Imp was generated by introducing the sequence encoding the 19 C-terminal residues of K-Ras4B (MSKDGKKKKKKSKTKCVIM) into the C terminus of full-length Imp (designated Imp-CAAX) (34). All new constructs were cloned into pMSCV, which contains a cassette consisting of the green fluorescent protein (GFP) and an internal ribosome entry site that permits the translation of two open reading frames from a single messenger RNA. In the pMIGR2 retroviral plasmid, tailless human CD2 replaces GFP.
Antibodies—Anti-CD3 mAb OKT3 (eBiosciences, San Diego, CA) and C363.29B followed by goat anti-rat polyclonal antibody (MP Biomedical, Solon, OH) were used to stimulate, respectively, Jurkat human T cells and CD4 T cells, that had been isolated from B10.BR mice. Polyclonal antibodies, anti-Imp (The Biodesign Institute, Arizona State University, Tempe, AZ) and anti-FLAG (Sigma), and mAbs, anti-ERK (clone D-2; Santa Cruz Biotechnology, Santa Cruz, CA), anti-phospho-ERK (clone E-4; Santa Cruz Biotechnology, Santa Cruz, CA), and anti-Ras (clone Ras10; Upstate Biotechnology, Lake Placid, NY) were used in a Western blot analysis. Anti-LFA-1 (M17/4; BD Biosciences) and anti-VLA-4 (R1–2) mAbs were used in adoptive transfer experiments to measure lymphocyte egress from the lymph nodes (35). Phycoerythrin (PE)-conjugated anti-IFN-γ, biotin-conjugated anti-human CD2, antigen-presenting cell (APC), anti-CD4, PE anti-Vβ5 TCR, and biotin anti-Vα2 TCR mAbs were used to stain CD4 T cells and analyze them using fluorescent-activated cell sorter (FACS); all of these antibodies were from BD Biosciences. mAbs, 11B11 (anti-IL4), and XMG1.2 (anti-IFN-γ) were used to make sure that Th1 and Th2 skewing took place during primary stimulation of CD4 T cells.
Cell Culture and Activation—Jurkat human T cells and mouse CD4 T cells were maintained in RPMI1640 and Bruff culture medium, respectively. Culture media were supplemented with 10% fetal calf serum, 10 mm HEPES, and antibiotics. Jurkat cells were activated in 96-well plates (EIA/RIA plates, Costar Corporation, Cambridge, MA) precoated overnight with OKT3 mAb at concentrations as indicated in the legend to Fig. 1d. Alternatively, Jurkat cells were incubated with 1.0 μg/ml OKT3 mAb on ice and then treated for 5 min at 37 °C with goat anti-rat secondary antibody. For stimulation with APCs, we isolated CD4 T cells from 6–8-week-old B10.BR and B10.5R mice that were transgenic for AND TCR. This TCR is specific for the moth cytochrome c peptide (VFAGLKKANERADLIAYLKQATK). CD4 T cells were grown in the presence of mitomycin C-treated T cell-depleted splenocytes. The APCs were pulsed with either 5 μg/ml pMCC or 15 μg/ml of a pMCC variant in which arginine had replaced lysine (K99R). For stimulation with phorbol esters, CD4 T cells were incubated with phorbol 12-myristate 13-acetate (100 ng/ml) at 37 °C.
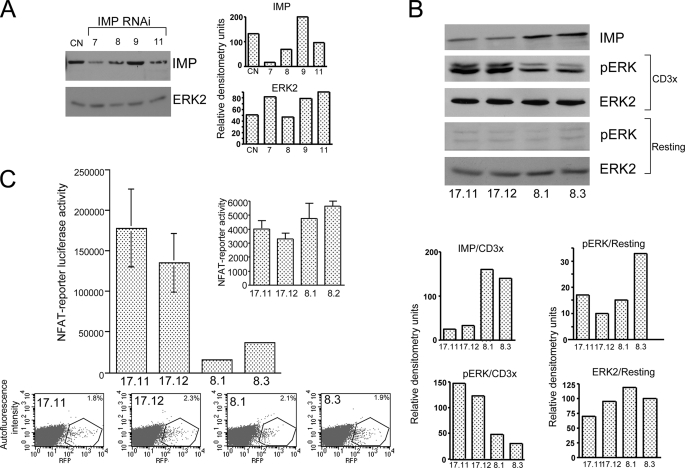
FIGURE 1.
Imp represses the transcriptional response in human T cells. A, Western blot analysis of Jurkat T cells expressing individual RNAi designed against human Imp. Jurkat T cells were transiently electroporated with individual RNAi subcloned into the pSilcencer™ 3.1-H1 hygro and pMSCV, which expresses GFP. After 48 h, the cells were sorted for GFP expression and then treated with the 1% Nonidet P-40 lysis buffer. The cell extracts were analyzed on Western blots using anti-Imp and anti-Erk2 antibodies (see “Experimental Procedures”). B, Western blot analysis of the Imp protein level and activated ERK in individual Jurkat T clones expressing Imp-specific RNAi (number 7) or control RNAi. After a resting period in RPMI1640 medium supplemented with 0.1% fetal calf serum, the cells were left unstimulated (resting) or stimulated with anti-CD3 mAb (CD3x) (1.0 μg/ml) followed by cross-linking with 100 μg/ml goat anti-rat secondary antibody (5 min at 37 °C). Next, equal numbers of cells were lysed and the materials collected were run on the 9% acrylamide gel and analyzed by Western blotting, as indicated. C, transactivation of the NFAT reporter in Jurkat T cells expressing reduced levels of Imp. Jurkat T cell clones stably expressing either Imp-specific RNAi number 7 (clones 17.11 and 17.12) or CD8-specific RNAi (8.1 and 8.3 controls) were transfected with NFAT-luciferase reporter (top) and red fluorescent protein (RFP) to measure transfection efficiency by FACS (x axis, expression level of red fluorescent protein; y axis, autofluorescence intensity, bottom). After 48 h the cells were stimulated for an additional 18 h with the plate-bound anti-CD3 mAb (1 μg/ml) or left unstimulated (inset), harvested, and analyzed using the dual-luciferase reporter assay system (36). One representative of three experiments is shown (the data are expressed as mean ± S.D. of triplicate cultures (S.D. values for 17.11, 17.12, 8.1, and 8.2 cells are 48,386, 36,608, 1,785, and 2,024, respectively).
Transfections—To obtain Jurkat cells exhibiting stable repression of Imp protein, we electroporated the cells with Imp RNAi expressed from pSilcencer™ 3.1-H1 hygro (Ambion, Foster City, CA) and GFP as a marker for early selection and then subcloned them by limited dilution in the presence of hygromycin as a selection drug. Individual clones were subsequently electroporated with an NFAT-luciferase reporter and a plasmid expressing red fluorescence protein to monitor transfection efficiency. Alternatively, Jurkat T cells were electroporated with the PathDetect trans-reporting system (Elk1-GAL4 transactivator and 5xGAL4-luciferase reporter at a ratio of 1:50) (Stratagene, La Jolla, CA) (36), Imp-CAAX construct, and pRL-CMV plasmid containing the Renilla luciferase under control of the cytomegalovirus immediate-early enhancer-promoter region (Promega Corporation, Madison, WI). Reporters were detected using the dual-luciferase reporter assays system (Promega). To normalize the data, Renilla luciferase activity was measured in unstimulated cells. All electroporations (2 × 107 cells per cuvette) were carried out at 250 V and 960 μF on a Gene Pulser™ (Bio-Rad). Lipofectamine 2000 reagent (Invitrogen) was used to transfect retroviral and lentiviral packaging cell lines, Phoenix Ecotropic cells and 293 T cells.
Viral Transductions—Two types of viral infection were performed. For experiments involving the use of a retrovirus, the supernatants were obtained from cultures of Phoenix Ecotropic packaging cells transfected with Ras effector loop mutants or Imp constructs cloned into pMSCV, and applied directly to CD4 T cells. For experiments with lentivirus, 293T cells were cotransfected with pLL3.7 and packaging vectors (38), and collected after 60 h. The lentivirus was recovered by ultracentrifugation for 2 h at 23,000 × g and resuspension in culture medium. The viral preparations were delivered to CD4 T cells in the presence of Polybrene (final concentration: 8 μg/ml) by centrifugation for 1.5 h at 2,000 × g (37).
RNA Interference—The oligonucleotide sequences used to generate Imp RNAi number 7 were as follows: 5′-GATCCGGGAAGTCAGCCGGGGAGATTCAAGAGATCTCTCCTGGTGACTTGCCTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAAGGCAAGTCACCAGGAGAGATCTCTTGAATCTCTCCTGGTGACTTGCCG. The sequences targeted by RNAi 8, 9, and 11 were GTCCAACCCAGATGAACTA, GTAAAGATCACAGTAAGGA, and GACAAATAAGATGACCTCC, respectively. To generate Imp that is RNAi-resistant, the target sequence was changed from GGGAAGTCAGCCGGGGAGA to GGTAAATCGGCAGGAGAAA. The sequences of primers used to generate CD8-specific RNAi were exactly the same as those described elsewhere (38).
Quantitative PCR—CD4 T cells were isolated from B10.BR mice and sorted for a naïve population expressing CD62Lhi CD44lo. RNA was prepared using a TRIzol reagent and the reverse transcription was carried out using oligo(dT) as a primer and SuperScript™ II Reverse Transcriptase (Invitrogen) as described in the manufacturer's protocol (Invitrogen). Quantitative PCR assays were performed on a Mx3000P® platform (Stratagene) using 25 μl of a reaction mixture that contained 8 to 10 ng of cDNA, SYBR Green, dNTP, primers, and platinum Taq polymerase (Invitrogen). Cycling conditions were as follows: incubation at 95 °C for 3 min, and 40 cycles at 94 °C for 10 s, 60 °C for 15 s, and 72 °C for 27 s. A melting curve was added between 55 and 95 °C. Amplification of hypoxanthine phosphoribosyltransferase gene mRNA was used as an internal control to normalize the data. For Imp the forward primer was 5′-GTGGAAAGGAAGTGTACCCAG-3′ and the reverse primer was 5′-CTCCGCAGGCAGGTGGCTGATCTG-3′. For S1P1 the forward primer was 5′-GACAACCCAGAGACCATTATG-3′ and the reverse primer was 5′-GATCAAGTCAGAATGCTTCCCTCA-3′. For S1P4 the forward primer was 5′-CGTGTTCAACTCAGCCATTAATCC-3′ and the reverse primer was 5′-CTGAAGCTGAGTGACCGAGAAGTC-3′. For hypoxanthine phosphoribosyltransferase the forward primer was 5′-CCAGCAAGCTTGCAACCTTAACCA-3′ and the reverse primer was 5′-ATGATCAGTCAACGGGGGAC-3′.
Mice—AND TCR B10.BR, AND TCR B10.A (5R), and OT2 TCR C57BL/6 mice were used as donors of TCR transgenic CD4 T cells. B10.BR, B10.A (5R), and C57BL/6 mice were used as donors of APCs. In adoptive transfer experiments, C57BL/6 and C57BL/6 IFNγR knock-out mice were also used as recipients of RasV12G37 or GFP control CD4 T cells. All animal experiments in this work were done in accordance with the institutional guidelines of the Yale Animal Resources Center (YARC).
Generation of RasV12G37 Transgenic Mice—FLAG-tagged RasV12G37 was subcloned into pBI-EGFP plasmid (Clontech) containing a bidirectional tetracycline-responsive promoter. The transgenic expression cassette was subsequently cut out using AseI and PshAI restriction enzymes, then purified, and injected into the embryos of F2(C57BL/6JxSJL/J) mice at Yale Animal Genomic Services. The transgene-positive weanling mice were identified by PCR analysis of the tail biopsies. The following primers were used to amplify a 225-bp fragment containing the 5′ sequence of FLAG-Ras: 5′-CCAGCCTCCGCGGCCCCGAATTCG-3′ and 5′-GATGACCACCTGCTTCCGGTAGG-3′, and a 296-bp fragment containing the 5′ sequence of the EGFP gene: 5′-GCTCGTTTAGTGAACCGTCAGATC-3′ and 5′-GCTTGCCGGTGGTGCAGATGAAC-3′. To identify the founders, the animals that belonged to the F1 generation of a breeding cross between 296-bp positive/225-bp positive and M2 mice (these mice express reverse tetracycline-controlled transactivator), were fed a grain-based doxocycline diet (2.3 g/kg) (BioServ, Laurel, MD) for 2 weeks and then screened by FACS for the presence of EGFP-positive CD4 T cells in the peripheral blood and lymph nodes.
Adoptive Cell Transfer Experiments—C57BL/6 or C57BL/6 IFNγR knock-out mice were injected with ∼5 × 106 CD4 T cells transduced with either RasV12G37 or an empty vector control. After 48 h, the mice were sacrificed and lymphocytes were isolated from their inguinal lymph nodes for staining with a mixture of anti-CD4 and anti-TCR mAbs (see below) and for FACS analysis. In some experiments, we blocked further entry of transferred cells by injecting the mice intraperitoneally with 100 μg of anti-LFA1 and VLA-4 mAbs (33), and performing analysis after 15 h, as described above.
Migration Assay—Transwell chemotaxis assays were performed using 24-well plates and 3-μm pore width filters (BD Biosciences). The lower chamber of the transwell chamber contained 0.6 ml of SP diluted in chemotaxis medium (RPMI1640, 0.5% bovine serum albumin, 10 mm Hepes). The cells were diluted in chemotaxis medium, and 0.1 ml (3 × 105 cells) was added to the top chamber. After 8 h at 37 °C, the cells that had migrated into the lower chamber were collected and counted in four high-power fields using a hemocytometer. The assays were performed in duplicate for each SP concentration.
Intracellular Cytokine Staining and FACS Analysis—GFP-sorted CD4 T cells expressing both siRNA from the pLL3.7 lentiviral vector and Ras effector loop mutants from a pMIGR2 retroviral plasmid were stimulated with the antigen for 12 h and cultured for an additional 4 h in the presence of GolgiStop reagent (BD Biosciences) to trap cytokines in the endoplasmic reticulum. Cells were then processed with biotin-conjugated anti-human CD2 mAb followed by streptavidin-Alexa 647 (Invitrogen) for extracellular staining, and PE-labeled anti-IFN-γ mAb for intracellular staining. In adoptive transfer experiments, GFP-positive CD4 T cells were stained with APC anti-CD4, PE anti-Vβ5 TCR, and biotin anti-Vα2 TCR followed by Per-CP streptavidin.
Western Blots and Immunoprecipitation—T cells were lysed in a buffer containing 1% Nonidet P-40, 150 mm NaCl, 50 mm Tris (pH 7.4), 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride and protease inhibitor (Roche Diagnostics). Protein samples from precleared cell lysates were fractioned under reducing conditions on a sodium dodecyl sulfate-9% polyacrylamide gel. To verify expression of Imp constructs, proteins samples were fractioned on a SDS 6–18% gradient polyacrylamide gel. After electrophoresis, proteins were electroblotted onto nitrocellulose membranes (Bio-Rad), blocked with either 5% nonfat dry milk or 3% bovine serum albumin in phosphate-buffered saline (staining with p-ERK mAb), probed with the first antibody, and incubated with the electrochemiluminescent (ECL) anti-mouse IgG-horseradish peroxidase-linked antibody (Amersham Biosciences). The immunoblots were developed using an ECL detection system (Amersham Biosciences). For immunoprecipitation, immune complexes were recovered with protein A/G-conjugated-Sepharose beads (Amersham Biosciences) and further analyzed by the immunoblot procedure exactly as described above.
Yeast Two-hybrid Assay—We evaluated the relationship between Imp and Ras using the yeast two-hybrid assay, H-RasV12 or H-RasV12 effector loop mutants were cloned inframe with the GAL4 DNA-binding domain within pGBKT7, and a region of Imp encompassing residues 273 to 377 was cloned in-frame with a 768–881-amino acid fragment of the GAL4 activation domain within the pGAD vector. Next, AH109- and Y187-competent yeast cells transformed with pGAD and pGBKT7 plasmids, respectively, were mated and transferred onto plates with SD minimal media containing a –leucine (Leu)/-tryptophan (Trp) dropout supplement. Positive clones were transferred onto new plates with a quadruple dropout supplement (-adenine (Ade)/-histidine(His)/-Leu/-Trp) and incubated at 30 °C for 5 days until colonies appeared.
Cytokine Assays—IFN-γ and IL-4 levels from cell supernatants were determined by enzyme-linked immunosorbent assay (ELISA, Endogen, Cambridge, MA). The lower limit of sensitivity for the ELISA for IFN-γ and IL-4 was 0.6 ng/ml and 10 pg/ml, respectively.
Statistical Analysis—Mann-Whitney analysis was performed to assess influence of RasV12G37 on T cell retention in the lymph nodes. The level of significance was accepted at p < 0.05.
RESULTS
Imp Is a Potent Down-regulator of T Cells Responses—In our first attempt to examine the potential role of Imp in T cell activation, we used human Jurkat T cells that had been transfected with anti-Imp-specific RNAi. We used RNAi 7 because it had the strongest inhibitory effect on Imp (Fig. 1A). Two independent Jurkat clones (17.11 and 17.12) that had been stably transfected with this RNAi demonstrated up-regulation of the phosphorylated form of ERK following stimulation with anti-CD3 mAb, compared with control clones (8.1 and 8.3) carrying CD8a-specific RNAi (Fig. 1B). In addition, in 17.11 and 17.12 Jurkat clones the NFAT reporter demonstrated a markedly increased transcriptional response, compared with control clones (Fig. 1C). Of note, unstimulated Jurkat T cells expressing distinct levels of Imp demonstrated similar levels of p-ERK (Fig. 1B) and NFAT reporter activity (Fig. 1C, inset).
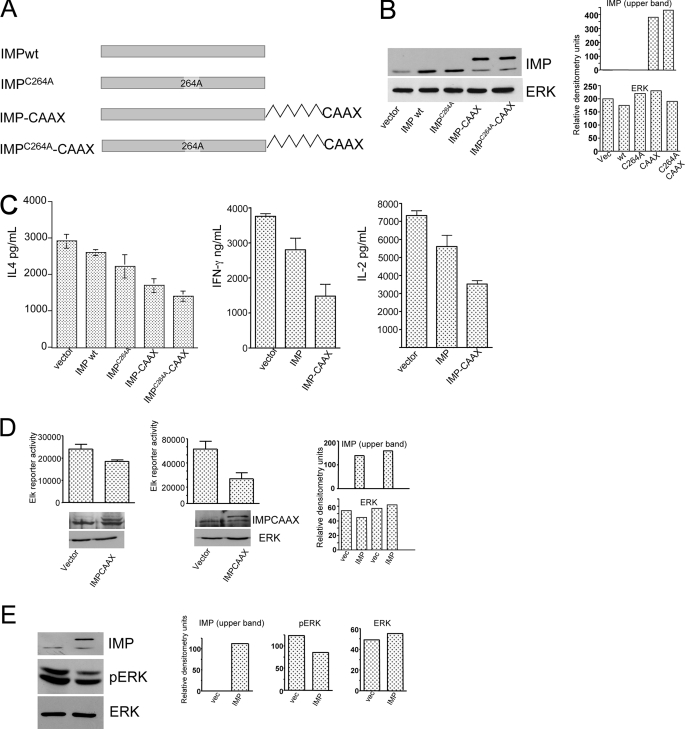
We then explored the possibility that overexpression of Imp inhibits T cells from inducing a cytokine response. We found that eptopically expressed wild type Imp has a relatively weak suppressive effect in wild type Jurkat T cells (data not shown). Rapid degradation or lack of Imp in close proximity to Ksr in the overexpression system may be responsible for this insufficiency. Therefore, we generated a membrane-targeted form of full-length Imp in which a consensus C-terminal CAAX (C-cysteine, A = aliphatic amino acid, and X = any amino acid) and the sequence encoding the 19 C-terminal residue of K-Ras4B were introduced (Imp-CAAX) (34) (Fig. 2A). In addition, we generated another Imp mutant in which the first cysteine in RING-H2 was changed to alanine (C264A) (Fig. 2A). This Imp variant is thought to be resistant to degradation (28). In contrast to wild type Imp, the Imp-CAAX variant appeared to be more effective in repressing the IL-2, IL-4, and IFN-γ response in normal CD4 T cells (Fig. 2C). Similar effects were seen with overexpression of Imp-CAAX, which partially blocked NFAT reporter activity in Jurkat T cells. Interestingly, Jurkat cells (e.g. 17.11), which expressed lowered levels of Imp, were more sensitive to the repressing activity of Imp-CAAX (Fig. 2D, right panel) compared with Jurkat cells (e.g. 8.1) expressing normal levels of this molecule (Fig. 2D, left panel). Additionally, a biochemical analysis of the phosphorylated form of ERK revealed Imp-dependent down-regulation of this kinase in antigen-stimulated CD4 T cells (Fig. 2E). Thus, our observations suggest that Imp is a negative regulator of T cell activation.
FIGURE 2.
Imp inhibits cytokine output and ERK activation in primary CD4 T cells. A, schematic display of Imp variants. In the ImpC264A mutant the first cysteine in the RING-H2 motif has been replaced by alanine. The Imp-CAAX variant contains a stretch of 19 C-terminal residues of K-Ras4B, as described (30). B, Western blot verification of Imp expression in T cells transduced with single variants of Imp. Aliquots of GFP-positive CD4 T cells were sorted as in C and analyzed on Western blots with anti-Imp and anti-ERK2 antibodies. C, ELISA of IL-4 (left), IFN-γ (middle), and IL-2 (right) cytokine production in CD4 T cells expressing single variants of Imp. CD4 T cells were isolated from AND TCR transgenic B10.5R mice and stimulated with 5R APCs and 15 μg/ml of pMCC K99R peptide for 48 h. The cells were spin infected with individual Imp constructs or an empty vector (control) in that order, as indicated. Three days after the infection, GFP-positive CD4 T cells were sorted and restimulated with the same combination of APCs and the peptide that was used during the priming phase. After 24 h, the cell culture supernatants were harvested and analyzed for the presence of cytokines. The data are expressed as mean ± S.D. of triplicate cultures. One representative of three experiments is shown. D, transactivation of the Elk luciferase reporter in Jurkat T cells transfected with Imp-CAAX. 8.1 (left panel) and 17.11 (right panel) Jurkat T cells were electroporated using the Elk-1 PathDetect transreporting system (see “Experimental Procedures”), pRL-CMV as an internal control, and either an RNAi-resistant form of Imp-CAAX (right bar in each panel) or an empty vector (left bar in each panel). After 48 h, the cells were stimulated with plate-bound anti-CD3 mAb (0.75 μg/ml), harvested, and analyzed for luciferase activity, as described in A. To normalize the data, Renilla luciferase activity was measured in unstimulated cells. The data are expressed as mean ± S.D. One representative of three experiments is shown. E, Western blot analysis of activated ERK in CD4 T cells transduced with ImpC264A-CAAX. Sorted GFP-positive CD4 T cells carrying either ImpC264A-CAAX or an empty vector (control) were processed to obtain cell extracts and analyzed by staining Western blots with anti-p-ERK and anti-Imp antibodies.
p21 Ras-mediated Repression of the Inhibitory Effect of Imp Is Responsible for Enhanced Cytokine Output by CD4 T Cells—According to a recent study (28) Imp is a novel effector of Ras. Therefore, we decided to explore the possibility that these two molecules associate with each other in T cells. As shown in Fig. 3A, stimulation of primary CD4 T cells through the TCR complex resulted in a rapid physical interaction between wild type Ras and endogenous Imp. Importantly, this binding activity was not observed in resting T cells.
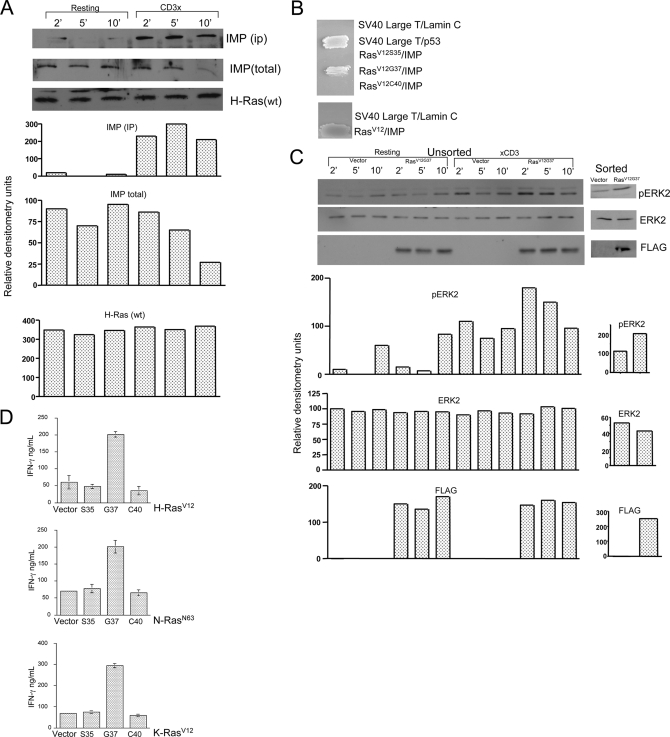
FIGURE 3.
Ras-mediated repression of Imp contributes to ERK activation and increased IFN-γ output in CD4 T cells. A, Imp Western blot analysis of anti-H-Ras immunoprecipitates. CD4 T cells isolated from AND TCR B10.BR mice and transduced with wild type FLAG-Ras in pMSCV were sorted for GFP expression. Following an 8-h resting period in 0.5% fetal calf serum, the cells were incubated on ice for 30 min with 1.0 μg/ml of anti-CD3 mAb (clone C363.29B) or left alone and then treated for 2, 5, and 10 min at 37 °C with 100 μg/ml of goat anti-rat secondary antibody. Next, the cells were lysed and Ras-associated Imp was isolated by incubation of the lysates with protein A-bound anti-FLAG antibody. The materials collected were run on the 9% acrylamide gel and analyzed by Western blotting for the levels of FLAG-Ras, associated Imp, and total Imp. B, yeast two-hybrid analysis of the association between H-RasV12 effector loop mutants and a region of Imp encompassing amino acid residues 273 to 377 (see “Experimental Procedures”). Cotransformants were patched onto plates lacking Trp, Leu, His, and Ade. Streaks of the colonies carrying SV40 large T and p53, or SV40 large T and laminin were included in the assay as positive and negative controls, respectively. C, Western blot analysis of activated ERK in CD4 T cells transduced with RasV12G37. Sorted (right) or unsorted (∼15% GFP-positive cells, left) AND TCR CD4 T cells that expressed either GFP alone or RasV12G37 were rested for 8 h in 0.5% fetal calf serum. The cells were then stimulated with anti-CD3 mAb, as described in A, and then analyzed by Western blotting for the presence of phosphorylated ERK, total ERK, and FLAG-RasV12G37. D, RasV12G37 selectively augments the production of IFN-γ in CD4 T cells. AND TCR-transgenic CD4 T cells were stimulated with pMCC (5μg/ml)-loaded APCs and then transduced with effector loop mutants (Ser35, Gly37, or Cys40) of H-(top), N-(middle), and K-Ras (low), as indicated. At 72 h posttransduction, the cells were sorted for GFP fluorescence, then restimulated with pMCC/APCs and finally analyzed for IFN-γ secretion by ELISA (plotted as mean ± S.D.).
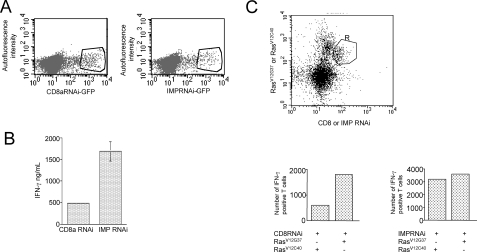
We also examined the physiological consequences of T cell transduction using the H-Ras effector loop mutant, RasV12G37. In contrast to the RasV12S35 and RasV12C40 (these mutants selectively bind Raf and phosphoinositide 3-kinase, respectively, but fail to interact physically with other effector molecules that RasV12G37 is able to recruit) RasV12G37 selectively interacted with a region of Imp encompassing residues 273–377 (Fig. 3B), increased the amplitude and duration of Erk activation (Fig. 3C), and strongly potentiated IFN-γ production in T cells (Fig. 3D). To provide formal proof that Imp is positioned downstream of Ras in CD4 T cells, we transduced these cells with either Imp- or CD8-specific RNAi and then with RasV12G37 or RasV12C40 as a control. As shown in Fig. 4B, T cells expressing Imp RNAi demonstrated marked improvement in their ability to produce IFN-γ, compared with the control RNAi. Similarly, retroviral transduction of control RNAi-positive T cells with RasV12G37 resulted in increased IFN-γ output compared with transduction with RasV12C40 (Fig. 4C, lower panel, left). Importantly, the level of enhancement by RasV12G37 was markedly diminished in T cells treated with Imp-specific RNAi (Fig. 4C, lower panel, right). These findings support the hypothesis that negative regulation of signal transduction by Imp is controlled by activated Ras in T cells.
FIGURE 4.
Imp is positioned downstream of Ras in the regulation of IFN-γ response. A, FACS analysis of AND TCR CD4 T cells lentivirally transduced with CD8a-(left) or Imp-(right) specific RNAi. The cells were stimulated with APCs and pMCC (5 μg/ml) for 48 h and then spin-infected with lentiviral preps. After an additional incubation period (96 h), GFP-positive T cells were sorted and used for further analysis as depicted in B and C. B, analysis of IFN-γ secretion in CD4 T cells transduced with Imp RNAi. CD4 T cells expressing CD8- or Imp-specific RNAi were sorted for GFP expression and restimulated for 24 h with APCs plus pMCC (5 μg/ml). After restimulation, the culture supernatants were harvested from triplicate cultures and analyzed by ELISA. One representative of three experiments is shown. C, FACS analysis of intracellular IFN-γ in CD4 T cells transduced with RasV12G37 and Imp-specific RNAi. Top, AND TCR CD4 T cells lentivirally transduced with CD8 or Imp RNAi were restimulated with pMCC and APCs for 48 h and then transduced a second time with RasV12G37 or RasV12C40 (control) in a pMIGR2 retroviral plasmid in which human tailless CD2 has replaced GFP. After an additional 48-h rest, the cells were rested and briefly stimulated again as described under “Experimental Procedures.” Finally, the cells were stained for the extracellular portion of human CD2 and intracellular accumulation of IFN-γ. The cells that were positive for both GFP and Ras construct (e.g. CD8RNAi/RasV12G37, CD8RNAi/RasV12C40, ImpRNAi/RasV12G37 and ImpRNAi/RasV12C40) were gated (R) and analyzed for IFN-γ-production (bottom). The data are expressed as total number of cells that stained positive for intracellular expression of IFN-γ. One representative of three experiments is shown.
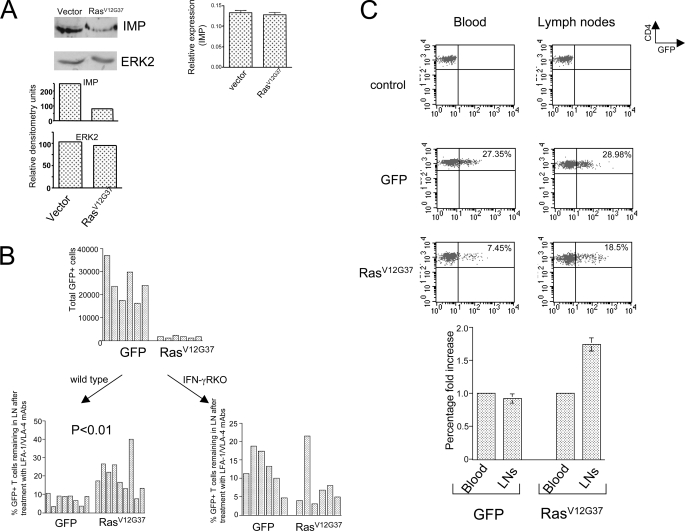
Ras-dependent Knockdown of Imp Is Responsible for Delayed Egress of T Cells from Lymph Nodes—It has been proposed that Th1 immunity involves a potential feedback loop in which IFN-γ induces the production of chemokines by APCs, which then recruit chemokine receptor-positive Th1 cells that produce IFN-γ (39). We decided to explore the possibility that the increase in IFN-γ output resulting from Ras-mediated inhibition of Imp (Fig. 5A) leads to the accumulation of T cells in the peripheral lymphoid organs. We addressed this issue by examining RasV12G37-positive T cells because Imp protein levels are markedly diminished in these cells (Fig. 5A, left panel)(de novo synthesis is not affected, however (Fig. 5A, right panel)). In our first attempt to determine the influence of Imp deficiency on T-cell accumulation in the lymphoid tissue, we generated RasV12G37-positive T cells and GFP-positive T cells (controls) and transferred these cell pools into normal mice. As shown in Fig. 5B (top), the number of RasV12G37-positive T cells in the inguinal lymph nodes was much lower than the number of control T cells. These findings suggest that most RasV12G37-positive T cells migrate preferentially to the peripheral tissues other than lymph nodes. This behavior may be explained by the findings of a previous study indicating that RasV12G37-positive cells express relatively low levels of CD62L, a homing receptor for lymphocyte to enter lymph nodes (33).
FIGURE 5.
Ras/Imp signaling pathway regulates in vivo traffic of CD4 T cells. A, Western blot analysis of Imp in T cells with RasV12G37. CD4 T cells isolated from the spleens of mice expressing either inducible RasV12G37-IRES-GFP or GFP (vector) were processed to obtain cell extracts and analyzed by RT-PCR (right panel) or staining Western blots with anti-Imp and anti-Erk (control) antibodies (left panel). One representative of three experiments is shown. B, top, analysis of the distribution of GFP-positive, RasV12G37-positive CD4 T cells, and GFP-positive CD4 T cells (controls) in recipient mice after adoptive transfer. B6 mice were injected with aliquots containing 5 × 106 CD4 T cells of which ∼25% cells were transduced with either RasV12G37 or GFP alone. Then 48 h after adoptive transfer, the animals were sacrificed and their inguinal lymph nodes were analyzed for the total cell number and percentage of GFP-positive CD4 T cells. Bottom, analysis of egress of RasV12G37 + CD4 T cells from the lymph nodes. Wild-type (left) or IFN-γR knock-out (right) B6 recipient mice were treated as described in the top part, or in addition received an intraperitoneal injection of phosphate-buffered saline or 100 μg of LFA-1 and VLA-4 mAbs 48 h after the adoptive transfer, as described previously (35). After an additional 15 h, the mice were sacrificed and their inguinal lymph nodes were processed as described above. The data are expressed as percentages of an average GFP- or RasV12G37-IRES-GFP-positive CD4 T cell count in mice that received phosphate-buffered saline. Each bar represents a single animal. C, top, FACS analysis of CD4 T cells isolated from transgenic mice expressing inducible RasV12G37. Wild-type mice (upper) and transgenic mice expressing either GFP alone (middle) or RasV12G37 GFP (lower) were subjected to a doxocycline diet for 2 weeks and sacrificed for an analysis of GFP-positive CD4 T cells isolated from their inguinal lymph nodes (right) and peripheral blood (left). Bottom, the average fold change of the percentage of GFP-positive CD4 T cells in the lymph nodes (LNs) compared with the peripheral blood. The mean ± S.D. are shown (n = 3).
To investigate the trafficking capabilities of RasV12G37-positive T cells further, we transferred these cells into mice that were then treated with a combination of antibodies against integrins VLA-4 and LFA-1, which are essential for lymphocyte entry into lymophoid tissue. In the presence of anti-VLA-4 and anti-LFA-1, lymphocyte entry into lymph nodes is almost completely blocked, but relatively little change occurs in the rate of lymphocyte egress from lymph nodes (35). We found that when mice are treated with a mixture of anti-VLA4 and anti-LFA-1, the RasV12G37-positive T cells that initially migrated toward lymph nodes are significantly delayed in exiting this tissue (p < 0.01) (Fig. 5B, bottom, left panel). Importantly, this delay was not observed in antibody-treated mice that lacked IFN-γ receptors (Fig. 5B, bottom, right panel). These data suggest that the ability of Ras-mediated inhibition of Imp to slow T cell transit through lymphoid tissue varies with IFN-γ levels secreted by the cell.
To address the role of Ras-induced Imp inhibition in T cell migration further, we used transgenic mice in which CD2-positive hematopoietic cells express an inducible form of RasV12G37. We found that the lymph nodes in these animals were rich in RasV12G37-positive CD4 T cells, compared with peripheral blood (Fig. 5C). This difference was not seen in animals expressing GFP alone. These findings suggest that Ras-mediated repression of Imp, at least partially prevents T cells from leaving the lymph nodes.
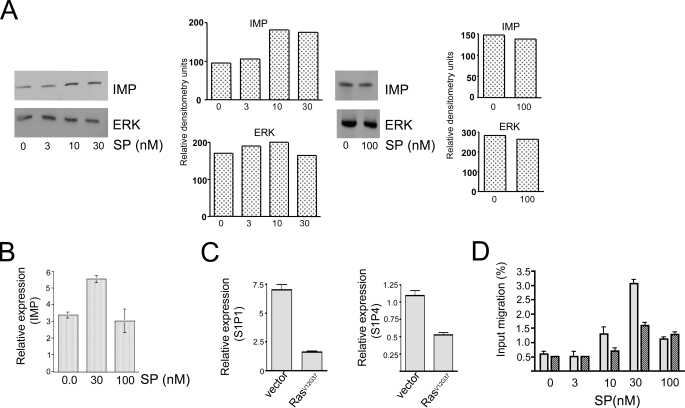
Finally, because Imp appears to be involved in the regulation of T cell traffic, we reasoned that spingosine phosphate (SP), which promotes lymphocyte egress from the lymphoid nodes (40), may somehow regulate Imp. To explore this possibility, we grew CD4 T cells in vitro with increasing doses of SP. As shown in Fig. 6, A and B, low to intermediate concentrations of SP that previously were reported to induce the egress of lymphocytes exit from the lymph nodes (40) resulted in markedly increased levels of Imp. By contrast, higher concentrations of SP (e.g. 100 nm) failed to enhance Imp levels. To determine whether the functional relationship between SP and Imp is reciprocal, we analyzed the activity of SP receptors in RasV12G37-positive T cells, in which the levels of Imp are reduced (Fig. 5A). We found that overexpression of RasV12G37 in T cells resulted in the diminished expression of SP receptors S1P1 and S1P4 (Fig. 6C). Moreover, RasV12G37-positive T cells migrated at a slower rate in response to SP compared with control T cells expressing normal levels of Imp (Fig. 6D). Together, our findings lend support to the hypothesis that Imp is involved in migration and maintaining the balance between T cell retention and loss in lymph nodes.
FIGURE 6.
Imp signaling opposes T cell migration induced by sphingosine phosphate. A, Western blot, and B, reverse transcriptase-PCR analyses of Imp induction by sphingosine phosphate. CD62hi-CD44lo CD4 T cells were isolated from 6-week-old B10BR mice and cultured for 24 h in the presence of increasing concentrations of sphingosine phosphate, as indicated. PCR data are expressed as mean ± S.D. The analyses were performed exactly as described under “Experimental Procedures.” One representative of three experiments is shown. C, reverse transcriptase-PCR analysis of S1P1 (left) and S1P4 (right) in CD4 T cells transduced with empty vector or RasV12G37. The cells were sorted for GFP expression at 96 h after spin infection and immediately used for the analysis. The data are expressed as mean ± S.D. D, analysis of chemotactic response of CD4 T cells to SP. Following treatment of transgenic mice with a doxocycline diet for 2 weeks, RasV12G37-positive (dark) or wild type (light) CD4 T cells were isolated from mice and used in a Transwell migration assay exactly as described under “Experimental Procedures.” The data are expressed as mean ± S.D. One representative of 3 experiments is shown.
DISCUSSION
It had been assumed that the Raf/MAP kinase cascade is the primary effector mechanism for Ras activity. However, it is now well appreciated that this small GTPase mediates its biological activity through a multitude of downstream effectors (41, 42). Nevertheless, the role of Ras effectors other than Raf in lymphocytes has been examined in only a handful of studies. In this study, we focused on a molecule called Imp, which unlike many other Ras effectors is a negative regulator of signal transduction (28, 43). Consistent with the model of Imp as a Ras effector, we found that stimulation of T cells through the TCR complex induces the rapid formation of an association between these molecules. Moreover our observation that the enhancing effect of RasV12G37 on cytokine output is impaired in T cells expressing decreased levels of Imp supports the hypothesis that this signaling molecule is positioned downstream to Ras rather than just in parallel. Of note, we found that overexpression of RasV12G37 alone in resting CD4 T cells failed to induce IFN-γ secretion (data not shown), which suggests that signaling events that are controlled by this variant of Ras require additional input through the TCR complex.
Our work does not exclude the possibility that there may be other non-Raf signaling molecules (in addition to Imp) that are controlled by Ras in T cells. The findings of one study utilizing the RasV12G37 mutant suggested that Ras via members of the Ral GEF family signals to Ral GTPase and influences the production of reactive oxygen species in Jurkat T cells (44). Also, previous work in our laboratory showed that a novel member of the phospholipase C protein family can imitate the effects of Ras by enhancing cytokine- and NFAT-induced transcriptional responses in CD4 T cells (33). In yet another study, Ras appeared to interact with phosphatidylinositol 3-kinase p110δ in bone marrow-derived mast cells and activate VLA-5 integrin; however, it is not known whether this mechanism of integrin activation exists in T cells (45). Thus, it is likely that Imp is just one of several molecules that are controlled by Ras in antigen-stimulated CD4 T cells.
In addition to experiments with RasV12G37, we studied T cells in which the Imp level had been modified. Quite surprisingly, we found that overexpression of wild type Imp (a predominantly cytosolic protein) failed to suppress IFN-γ and IL-4 production, which suggests that physiological levels of this signaling molecule are sufficient to control the cytokine responses mentioned above. Alternatively, it is possible that retrovirally delivered Imp is unstable or expressed in an intracellular compartment that is devoid of Ksr. Indeed, targeting of Imp to the plasma membrane, a compartment for which Ksr exhibits specificity, may have placed these molecules in close proximity thereby improving overall ability of the Imp-CAAX construct to inhibit T-cell activation. Moreover, another Imp variant that lacks E3 ligase activity and is thought to be resistant to proteasome degradation (ImpC264A) (28) also inhibited T-cell cytokine responses with somewhat better efficiency than wild type Imp.
In contrast to experiments based on protein overexpression, studies using RNAi to block Imp expression have provided more striking results. Imp inhibition markedly enhanced both the NFAT transcriptional response and IFN-γ secretion in CD4 T cells when they were stimulated through the TCR. Interestingly, however, Imp-specific RNAi induced a relatively moderate increase in the production of IL-2 (data not shown). In a similar fashion, it was demonstrated in one of our previous studies that RasV12G37-induced potentiation of IFN-γ is much stronger than its potentiation of Il-2 (33). Preferential sensitivity of the IFN-γ response to Imp-specific siRNA and RasV12G37, compared with the Il-2 response may be attributed to the fact that both constructs were delivered into T cells 48 h after antigen stimulation and therefore could not influence the initial phase of antigenic stimulation, when the IL-2 gene is induced. An alternative explanation may be the relative resistance of IL-2 response to inhibitory Imp activity because of the compartmentalization of Ras signaling (46–48) and possibly the participation of several distinct Ras pools in the induction of the IL-2 cytokine response. Accordingly, it could be hypothesized that Imp, which has a negative effect on the Ksr scaffold and on Ras effector signaling in the plasma membrane, fails to down-regulate other scaffolds in the Ras/ERK pathway (49) that may be important for the IL-2 response. This notion is supported by the following observations: 1) the Golgi apparatus is critical for Ras activation in T cells (46); 2) Sef (similar expression to fgf genes) protein, not Ksr, is the MEK/ERK scaffold that resides on the Golgi apparatus (50, 51); and 3) T cells isolated from Ksr knock-out mice display only partially defective proliferation following antigenic stimulation (52).
It is not unlikely that the inhibitory effect of Imp on the ERK cascade is relevant to the pathogenesis of autoimmune diseases and inflammatory conditions in several distinct ways. It is possible that Imp may help protect against autoimmune conditions by limiting the output of proinflammatory cytokines such as IFN-γ. However, it is equally possible that Imp-mediated repression of the ERK cascade increases the risk for autoimmune-like conditions. In a recent study, for example, Ksr was shown to promote antiapoptotic signals including Raf-1/MEK/ERK signal, and protects intestinal epithelial cells from TNF-induced apoptosis (53). By mediating disengagement of Ksr, Imp may interfere with Ksr-induced protection of the bowel mucosa, thereby permitting exacerbation of the inflammatory injury. Also, studies of gene profiles in peripheral blood mononuclear cells have shown that the expression of BRCA1, which is thought to associate with Imp, is down-regulated in families at risk for autoimmune diseases (54). Although the significance of this alteration is not clear, one may speculate that decreased expression of BRCA1 supports the interaction between Imp and Ksr and ultimately leads to the down-regulation of signal transduction in T cells. Consistent with this hypothetical model are published data suggesting that autoreactive T cells generate relatively weak cytokine and proliferative responses when they encounter with self-antigens (55).
Our study findings also suggest that in addition to TCR-mediated stimulation, Imp influences T cell migration. This conclusion is based on two observations: 1) RasV12G37, which inhibits the inhibitory function of Imp and induces the increased output of IFN-γ, is associated with a delay in T-cell egress from the lymphoid tissues. Second, Imp knockdown is associated in vitro with diminished expression levels of SP receptors, and consequently, results in delayed migration of T cells in response to the SP gradient. Interestingly, however, we also found that SP, which is known to reduce IFN-γ secretion (56) and guide the escape of T cells from the lymphoid tissues (40, 57), stimulated the de novo synthesis of Imp. Taken together, Ras-mediated inhibition of Imp activity and SP-mediated induction of this signaling molecule may work together to fine tune the balance between stimuli that increase the T cell IFN-γ response and retention in the lymph node and those that limit these changes in favor of T cell migration and egress from the lymph node. The notion that T cell motility versus TCR stop signal is clinically relevant was suggested in the report of a recent study on a CTLA4 costimulatory molecule. According to that report, protection against autoimmunity is linked with the ability of CTLA4 to exert a positive influence on the motility of T cells and reduce duration of contact with APCs (58). Analogous to the CTLA4 model, Imp signaling may also influence the motility of T cells. It will be interesting to determine whether the augmentation of Imp activity causes lymphocytes to escape from lymph nodes prematurely and, thus, leads to an increased risk of peripheral tissues being seeded with these cells.
Acknowledgments
We thank P. Ranney for assistance with mouse breeding, M. Piecychna and T. Taylor for experimental assistance, D. Amsen in the laboratory of Dr. Flavell for M2 mice, K. Murphy and A. Rudensky for the retroviral vectors, and L. van Parijs for PLL3.7 and packaging vectors.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 5R37A1026791-18 (to K. B.). This work was also supported by an American Cancer Society Research Scholar Grant (to J. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: MAPK, mitogen-activated protein kinase; NFAT, nuclear factor of activated T cells; TCR, T cell receptor; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase; ERK, extracellular signal-regulated kinase kinase; IL, interleukin; Imp, impedes mitogenic signal propagation; KSR, kinase suppressor of Ras; mAb, monoclonal antibody; PE, phycoerythrin; IFN, interferon; FACS, fluorescent-activated cell sorter; RNAi, RNA interference; EGFP, epidermal growth factor protein; APC, antigen-presenting cell; ELISA, enzyme-linked immunosorbent assay; SP, spingosine phosphate.
References
- 1.Stokoe, D., MacDonald, S. G., Cadwallader, K., Symons, M., and Hancock, J. F. (1994) Science 264 1463–1467 [DOI] [PubMed] [Google Scholar]
- 2.Vojtek, A. B., Hollenberg, S. M., and Cooper, J. A. (1993) Cell 74 205–214 [DOI] [PubMed] [Google Scholar]
- 3.Downward, J., Graves, J. D., Warne, P. H., Rayter, S., and Cantrell, D. A. (1990) Nature 346 719–723 [DOI] [PubMed] [Google Scholar]
- 4.Franklin, R. A., Tordai, A., Patel, H., Gardner, A. M., Johnson, G. L., and Gelfand, E. W. (1994) J. Clin. Investig. 93 2134–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stefanova, I., Hemmer, B., Vergelli, M., Martin, R., Biddison, W. E., and Germain, R. N. (2003) Nat. Immunol. 4 248–254 [DOI] [PubMed] [Google Scholar]
- 6.Alberola-Ila, J., and Hernandez-Hoyos, G. (2003) Immunol. Rev. 191 79–96 [DOI] [PubMed] [Google Scholar]
- 7.Alberola-Ila, J., Forbush, K. A., Seger, R., Krebs, E. G., and Perlmutter, R. M. (1995) Nature 373 620–623 [DOI] [PubMed] [Google Scholar]
- 8.Swan, K. A., Alberola-Ila, J., Gross, J. A., Appleby, M. W., Forbush, K. A., Thomas, J. F., and Perlmutter, R. M. (1995) EMBO J. 14 276–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swat, W., Shinkai, Y., Cheng, H. L., Davidson, L., and Alt, F. W. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 4683–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldari, C. T., Heguy, A., and Telford, J. L. (1993) J. Biol. Chem. 268 2693–2698 [PubMed] [Google Scholar]
- 11.Baldari, C. T., Macchia, G., and Telford, J. L. (1992) J. Biol. Chem. 267 4289–4291 [PubMed] [Google Scholar]
- 12.Izquierdo, M., Bowden, S., and Cantrell, D. (1994) J. Exp. Med. 180 401–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Owaki, H., Varma, R., Gillis, B., Bruder, J. T., Rapp, U. R., Davis, L. S., and Geppert, T. D. (1993) EMBO J. 12 4367–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rayter, S. I., Woodrow, M., Lucas, S. C., Cantrell, D. A., and Downward, J. (1992) EMBO J. 11 4549–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fields, P. E., Gajewski, T. F., and Fitch, F. W. (1996) Science 271 1276–1278 [DOI] [PubMed] [Google Scholar]
- 16.Li, W., Whaley, C. D., Mondino, A., and Mueller, D. L. (1996) Science 271 1272–1276 [DOI] [PubMed] [Google Scholar]
- 17.Zha, Y., Marks, R., Ho, A. W., Peterson, A. C., Janardhan, S., Brown, I., Praveen, K., Stang, S., Stone, J. C., and Gajewski, T. G. (2006) Nat. Immunol. 7 1166–1173 [DOI] [PubMed] [Google Scholar]
- 18.Boguski, M. S., and McCormick, F. (1993) Nature 366 643–654 [DOI] [PubMed] [Google Scholar]
- 19.Bos, J. L., Rehmann, H., and Wittinghofer, A. (1997) Cell 129, 865–877 [DOI] [PubMed] [Google Scholar]
- 20.Cristofano, A. D., Niki, M. M., Zhao, M., Karnell, F. G., Clarkson, B., Pear, W. S., Van Aelst, L., and Pandofili, P. P. (2001) J. Exp. Med. 194 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamanashi, Y, Tamura, T., Kanamori, T., Yamane, H., Nariuchi, H., Yamamoto, T., and Baltimore, D. (2000) Genes Dev. 14 11–16 [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao, M., Schmitz, A. A. P., Qin, Y., Di Cristofano, A., Pandolfi, P. P., and Van Aelst, L. (2001) J. Exp. Med. 194 265–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasaki, A., Taketomi, T., Kato, R., Saeki, K., Nonami, A., Sasaki, M., Kuriyama, M., Saito, N., Shibuya, M., and Yoshimura, A. (2003) Nat. Cell Biol. 5 427–432 [DOI] [PubMed] [Google Scholar]
- 24.Hanafusa, H., Torii, S., Yasunaga, T., and Nishida, E. (2002) Nat. Cell Biol. 4 850–858 [DOI] [PubMed] [Google Scholar]
- 25.Olenchock, B. A., Guo, R., Carpenter, J. H., Jordan, M., Topham, M. K., Koretzky, G. A., and Zhong, X. P. (2006) Nat. Immunol. 7 1174–1181 [DOI] [PubMed] [Google Scholar]
- 26.Zhong, X.-P., Hainey, E. A., Olenchock, B. A., Jordan, M. S., Maltzman, J. S., Nichols, K. E., Shen, H., and Koretzky, G. A. (2003) Nat. Immunol. 4 882–890 [DOI] [PubMed] [Google Scholar]
- 27.Li, Q. J., Chau, J., Ebert, P. J., Sylvester, G., Min, H., Liu, G., Braich, R., Manoharan, M., Soutschek, J., Skare, P., Klein, L. O., Davis, M. M., and Chen, C. Z. (2007) Cell 129 147–161 [DOI] [PubMed] [Google Scholar]
- 28.Matheny, S. A., Chen, C., Kortum, R. L., Razidlo, G. L., Lewis, R. E., and White, M. (2004) Nature 427 256–260 [DOI] [PubMed] [Google Scholar]
- 29.Li, S., Ku, C.-Y., Farmer, A. A., Cong, Y.-S., Chen, C.-F., and Lee, W.-H. (1998) J. Biol. Chem. 273 6183–6189 [DOI] [PubMed] [Google Scholar]
- 30.Asada, M., Ohmi, K., Delia, D., Enosawa, S., Suzuki, S., Yuo, A., Suzuki, H., and Mizutani, S. (2004) Mol. Cell. Biol. 24 8236–8243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daniels, M. A., Teixeiro, E., Gill, J., Hausmann, B., Roubaty, D., Holmberg, K., Werlen, G., Holländer, G. A., Gascoigne, N. R. J., and Palmer, E. (2006) Nature 444 724–729 [DOI] [PubMed] [Google Scholar]
- 32.Jorritsma, P. J., Brogdon, J. L., and Bottomly, K. (2003) J. Immunol. 170 2427–2434 [DOI] [PubMed] [Google Scholar]
- 33.Czyzyk, J., Brogdon, J. L., Badou, A., Henegariu, O., Preston Hurlburt, P., Flavell, R. A., and Bottomly, K. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 6003–6008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark, J. C., Quilliam, L. A., Hisaka, M. M., and Der, C. J. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 4887–4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lo, C. G., Xu, Y., Proia, R. L., and Cyster, J. G. (2005) J. Exp. Med. 201 291–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czyzyk, J., Leitenberg, D., Taylor, T., and Bottomly, K. (2000) Mol. Cell. Biol. 20 8740–8747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCarty, N., Paust, S., Ikizawa, K., Dan, I., Li, X., and Cantor, H. (2005) Nat. Immunol. 6 65–72 [DOI] [PubMed] [Google Scholar]
- 38.Rubinson, D. A., Dillon, C. P., Kwiatkowski, A. V., Sievers, C., Yang, L., Kopinja, J., Rooney, D. L., Zhang, M., Ihrig, M., and McManus, M. T. (2003) Nat. Genet. 33 401–406 [DOI] [PubMed] [Google Scholar]
- 39.Murphy, P. M. (2003) in Fundamental Immunology (Paul, W. E., ed) pp. 801–840, Lippincott Williams & Wilkins, Baltimore, MD
- 40.Matloubian, M., Lo, C. G., Cinamon, G., Lesneski, M. J., Xu, Y., Brinkmann, V., Allende, M. L., Proia, R. L., and Cyster, J. G. (2004) Nature 427 355–360 [DOI] [PubMed] [Google Scholar]
- 41.Campbell, S. L., Khosravi-Far, R., Rossman, K. L., Clark, G. L., and Der, C. J. (1998) Oncogene 17 1395–1413 [DOI] [PubMed] [Google Scholar]
- 42.White, M. A., Nicolette, C., Minden, A., Polverino, A., Van Aelst, L., Karin, M., and Wigler, M. H. (1995) Cell 80 533–541 [DOI] [PubMed] [Google Scholar]
- 43.Ory, S., and Morrison, D. K. (2004) Curr. Biol. 14 R277-R278 [DOI] [PubMed] [Google Scholar]
- 44.Remans, P. H. J., Gringhuis, S. I., Van Laar, J. M., Sanders, M. E., Papendrecht-van der Voort, E. A. M., Zwartkruis, F. J. T., Levarht, W. N., Rosas, M., Coffer, P. J., Breedveld, F. C., Bos, J. L., Tak, P. P., Verweij, C. L., and Reedquist, K. A. (2004) J. Immunol. 173 920–931 [DOI] [PubMed] [Google Scholar]
- 45.Kinashi, T., Katagiri, K., Watanabe, S., Vanhaesebroeck, B., Downward, J., and Takatsu, K. (2000) J. Biol. Chem. 275 22590–22596 [DOI] [PubMed] [Google Scholar]
- 46.Bivona, T. G., de Castro, I. P., Ahearn, I. M., Grana, T. M., Chiu, V. K., Lockyer, P. J., Cullen, P. J., Pellicer A., Cox, A. D., and Philips, M. R. (2003) Nature 424 694–698 [DOI] [PubMed] [Google Scholar]
- 47.Bivona, T. G., Quatela, S. E., Bodemann, B. O., Ahearn, I. M., Soskis, M. J., Mor, A., Miura, J., Wiener, H. H., Wright, L., Saba, S. G., and Philips, M. R. (2006) Mol. Cell 21 481–493 [DOI] [PubMed] [Google Scholar]
- 48.Chiu, V. K., Bivona, T., Hach, A., Sajous, J. B., Silletti, J., Wiener, H., Johnson, R. L., Cox, A. D., and Philips, M. R. (2002) Nat. Cell Biol. 4 343–350 [DOI] [PubMed] [Google Scholar]
- 49.Mor, A., and Philips, M. R. (2006) Annu. Rev. Immunol. 24 771–800 [DOI] [PubMed] [Google Scholar]
- 50.Torii, S., Kasakabe, M., Yamamoto, T., Maekawa, M., and Nishida, E. (2004) Dev. Cell 7 33–44 [DOI] [PubMed] [Google Scholar]
- 51.Morrison, D., and Davis, R. J. (2003) Annu. Rev. Cell Dev. Biol. 19 91–118 [DOI] [PubMed] [Google Scholar]
- 52.Nguyen, A., Burack, W. R., Stock, J. L., Kortum, R., Chaika, O. V., Afkarian, M., Muller, W. J., Murphy, K. M., Morrison, D. K., and Lewis, R. E. (2002) Mol. Cell. Biol. 22 3035–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan, F., John, S. K., Wilson, G., Jones, D. S., Washington, M. K., and Polk, D. B. (2004) J. Clin. Investig. 114 1272–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olsen, N. J., Moore, J. H., and Aune, T. M. (2004) Arthritis Res. Therapy 6 120–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kent, S. C., Chen, Y., Bregoli, L., Clemmings, S. M., Kenyon, N. S., Ricordi, C., Hering, B. J., and Hafler, D. A. (2005) Nature 435 224–228 [DOI] [PubMed] [Google Scholar]
- 56.Rosen, H., and Goetzl, E. J. (2005) Nat. Rev. Immunol. 5 560–570 [DOI] [PubMed] [Google Scholar]
- 57.Chi, H., and Flavell, R. A. (2005) J. Immunol. 174 2485–2488 [DOI] [PubMed] [Google Scholar]
- 58.Schneider, H., Downey, J., Smith, A., Zinselmeyer, B. H., Rush, C., Brewer, J. M., Wei, B., Hogg, N., Garside, P., and Rudd, E. C. (2006) Science 313 1972–1975 [DOI] [PubMed] [Google Scholar]