Abstract
Tumor necrosis factor (TNF) receptor 1 (TNFR1, p55) and 2 (TNFR2, p75) are characterized by several cysteine-rich modules in the extracellular domain, raising the possibility that redox-induced modifications of these cysteine residues might alter TNFR function. To test this possibility, we examined fluorescence resonance energy transfer (FRET) in 293T cells transfected with CFP- and YFP-tagged TNFRs exposed to the thiol oxidant diamide. Treatment with high concentrations of diamide (1 mm) resulted in an increase in the FRET signal that was sensitive to inhibition with the reducing agent dithiothreitol, suggesting that oxidative stress resulted in TNFR self-association. Treatment of cells with low concentrations of diamide (1 μm) that was not sufficient to provoke TNFR self-association resulted in increased TNF-induced FRET signals relative to the untreated cells, suggesting that oxidative stress enhanced ligand-dependent TNFR signaling. Similar findings were obtained when the TNFR1- and TNFR2-transfected cells were pretreated with a cell-impermeable oxidase, DsbA, that catalyzes disulfide bond formation between thiol groups on cysteine residues. The changes in TNFR self-association were functionally significant, because pretreating the HeLa cells and 293T cells resulted in increased TNF-induced NF-κB activation and TNF-induced expression of IκB and syndecan-4 mRNA levels. Although pretreatment with DsbA did not result in an increase in TNF binding to TNFRs, it resulted in increased TNF-induced activation of NF-κB, consistent with an allosteric modification of the TNFRs. Taken together, these results suggest that oxidative stress promotes TNFR receptor self-interaction and ligand-independent and enhanced ligand-dependent TNF signaling.
Tumor necrosis factor (TNF)2 is a potent cytokine that is involved in a wide range of biological activities, ranging from host defense to inflammation. TNF exerts its biological activities by binding to the extracellular domains of two distinct cell surface receptors termed tumor necrosis factor 1 (TNFR1 or p55) and tumor necrosis factor 2 (TNFR2 or p75, Ref. 1). Both TNFRs are type 1 membrane proteins that belong to the nerve growth factor receptor family, which includes CD27, CD40, FAS, and RANK. There are four cysteine-rich domains (CRDs) within the extracellular domains of TNFR1 and TNFR2, each of which contains six cysteine residues (2). The first three CRDs are characteristic of the TNFR superfamily, whereas the 4th CRD, which is membrane proximal, is less well conserved (1). The first CRD of the TNFRs is essential for the formation of homotypic, ligand-independent receptor complexes through the pre-ligand assembly domain (PLAD) (3). The ligand-binding pocket for TNF is formed mainly by CRD2 and CRD3 of the TNFRs (1). The current model of TNF signaling suggests that TNF binding leads to signal transduction by allowing the cytoplasmic tails of the intracellular domains to assemble together in the correct steric orientation to initiate cell signaling (1).
Oxidative stress is increasingly recognized as an underlying cause of a broad variety of inflammatory diseases (4, 5). Although reactive oxygen species (ROS) produce deleterious effects on cell activity because of their ability to damage cell structures, such as membrane lipids and proteins, it is also well recognized that ROS-induced post-translational modification of proteins may also play an important role in cell signaling (6-8). Germane to the present discussion, cysteine residues in receptor proteins have been shown to be the target of such redox modulation, resulting in changes of their ligand binding affinity and activation status (9-11). The observation that TNFR1 and TNFR2 have numerous cysteine residues in the extracellular domain, together with the prior observation that prooxidant conditions are sufficient to modify the number of reduced thiol groups of the extracellular cysteines of TNFR1 (12), raised the intriguing possibility that oxidative stress might influence TNFR signaling. Accordingly, in the present study we sought to determine whether prooxidant conditions were sufficient to modulate oligomerization between TNF receptors, as well as influence TNF signaling. Here, we report that oxidative stress is sufficient to promote self-interaction of the TNF receptors, as well as enhance TNF signaling.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
To determine whether oxidative stress promoted self-interaction between type 1 TNF receptors (p55, TNFR1) and/or type 2 TNF receptors (p75, TNFR2) in living cells, we used flow cytometry to analyzed fluorescence resonance energy transfer (FRET) between TNFR subunits that had been fused at their C termini to a cyan fluorescent protein (CFP) or a yellow fluorescent protein (YFP). The TNFR chimeric fusion proteins used herein were a generous gift from Dr. Francis K. Chan. The 293T cells used for these studies were maintained at 37 °C with 5% CO2 using Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Transient transfection with chimeric TNFR1 and chimeric TNFR2 CFP and YFP fusion proteins was performed with polyethylenimine (PEI) as described (13), by adding 2 μg of DNA encoding the CFP and YFP fusion proteins to each well. The DNA-PEI mixture was incubated at room temperature for 20 min, and the cells were analyzed for FRET or Western blotting 36-48 h after transfection, as described below.
Effect of Oxidative Stress on TNF Receptor Self-interaction
Measurement of FRET—Transfected 293T and HeLa cells were treated as described below and analyzed using a FACSAriaJ sell sorter (Becton Dickinson, San Jose CA) equipped with 3 lasers (488-nm Argon Blue laser, 633-nm red, and 407-nm violet lasers). Briefly, cells expressing only CFP- or only YFP-tagged proteins were used to set compensation values for spectral overlap on the flow cytometer so that cells expressing only CFP- or YFP-tagged receptors produced no signal in the FRET channel. The excitation and emission peaks of CFP (donor) are 434 and 476 nm, respectively, whereas the excitation and emission peaks for YFP (acceptor) are 514 and 527 nm, respectively. Data were analyzed using Becton Dickinson FACSDiva software. Fluorescence data were collected using the following filter sets: CFP emission was detected with a 510-nm (DF21) filter using 405-nm excitation, and non-FRET YFP emission with a 530/30 bandpass filter at 488-nm excitation. The FRET signal was detected using a 550-nm (DF30) filter.
Effect of Diamide on TNF Receptor Self-interaction—Transfected 293T cells were treated with diluent (PBS) or diamide (1 mm) for 15 min at 37 °C, in the presence and absence of 100 μm DTT. As a positive control, transfected 293T cell cultures were treated with TNF (200 units/ml) for 60 min at 4 °C. In separate experiments we examined the effects of TNF-induced TNFR oligomerization following treatment with a concentration of diamide (1 μm) that had no discernable effect of TNFR self-interaction. Briefly, 293T cells were pretreated for 15 min either with 1 μm diamide or 1 μm diamide and 100 μm DTT; the cultures were then washed with PBS and treated with TNF (100 units/ml) for an additional 1 h at 4 °C. Group data were analyzed as the mean fluoresence intensity in the FRET channel.
Effect of DsbA on TNF Receptor Self-interaction—Because diamide can penetrate cell membranes, and might therefore lead to oxidative modifications of the cytoplasmic tails of the TNF receptor CYP and YFP fusion constructs, we employed DsbA, a cell-impermeant oxidoreductase, that catalyzes thiol: disulfide exchange reactions between thiol groups on cysteine residues and oxidized gluthathione (GSSG) (14). Transfected 293T cells were treated overnight in serum-free medium with DsbA (0.01 μg/ml) in the presence and absence of 500 μm glutathione disulfide (GSSG). In separate experiments, we examined the effects of TNF (100 units/ml for 1 h at 4 °C) on TNFR2 oligomerization in 293T cells that had been treated overnight with 0.01 μg/ml DsbA and 500 μm GSSG. For each set of experiments, the cells were studied using flow cytometry, and the data were analyzed as the mean fluoresence intensity measured in the FRET channel.
Western Blotting—Cell cultures were treated with diamide (1 mm) for 15 min or DsbA (1 μg/ml) plus GSSG (500 μm) for 15 min, as described above, and Western blotting for TNFR1 and TNFR2 was performed (3). The TNFR1 or TNFR2 complexes were detected using an anti-HA antibody (clone HA-7, Sigma Aldridge) diluted 1:10000, followed by goat anti-mouse secondary antibody diluted 1:10,000 (Sigma Aldridge), and the protein complexes were visualized by enhanced chemiluminescence (ECL, Amersham Biosciences).
Assessment of Free Sulhydryl Groups on TNFR1—To determine whether the concentrations of DsbA that led to TNFR self-association were sufficient to modify free thiol groups on the cysteine residues of TNFR1, we exposed transfected 293T cells to 4-acetamido-4′-maleimidylstilbene-2,2′ disulfonate (AMS). AMS is a membrane-impermeable maleimide that forms covalent bond with proteins containing free thiol groups. Given that AMS has a molecular mass of ∼500 Da, proteins that react with AMS have retarded mobility on electrophoretic gels. Briefly, 293T cells expressing the TNFR1 fusion protein were treated overnight in serum-free medium with DsbA (1 μg/ml) in the presence and absence of GSSG (500 μm). Following treatment with DsbA, the cells were washed with PBS, harvested, and the proteins precipitated with 5% trichloroacetic acid and centrifuged at 4 °C at 10,000 rpm for 5 min. The supernatants were removed, and the pellets were washed with acetone and dissolved in a buffer containing 100 mm Tris and 1% SDS, pH 7.4. The samples were then incubated with AMS (20 mm) for 30 min at room temperature, and the reaction halted by adding reducing SDS buffer. N-Ethylmaleimide (NEM, 20 mm), which blocks free thiol groups, was used as a mock control. Samples were separated on a 10% SDS-Tricine gel, followed by Western blotting with anti-HA primary antibody diluted 1:10,000 and a goat anti-mouse secondary antibody diluted 1:10,000, followed by ECL.
Effect of Oxidative Stress on TNF-induced NF-κB Activation
HeLa cell cultures were maintained at 37 °C with 5% CO2 using Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. HeLa cells, grown to 70-80% confluence, were treated for 15 min at 37 °C with diamide (1 μm) in the presence and absence of DTT (100 μm). The cells were then washed with PBS and challenged with TNF (100 units/ml) for an additional 15 min. For the experiments involving DsbA, HeLa cells were treated with DsbA (0.001-10 μm) to determine a concentration of DsbA that resulted in submaximal activation of NF-κB. Next, the cells were treated overnight in serum-free medium with 0.1 μg/ml DsbA, in the presence and absence of GSSG (500 μm). After treatment with DsbA, the cells were stimulated with TNF (100 units/ml) or diluent for 15 min at 37 °C, and then washed with ice-cold PBS. The cells were collected, and nuclear extracts were prepared using reagents (Pierce) according to the manufacturer's instructions (15). The oligonucleotide containing the NF-κB consensus sequence (5′-AGTTGAGGGGACTTTCCCAGGC-3′; Santa Cruz Biotechnology) was labeled using T4 polynucleotide kinase and [γ-32P]ATP. Nuclear extracts (15 μg) were incubated with the labeled NF-κB oligonucleotide as described previously (15). The DNA-protein complexes were separated electrophoretically on 4% polyacrylamide gels, and the radiographic images of the dried gels captured using a STORM 860 imager (Molecular Dynamics).
Effect of Oxidative Stress on TNF-induced Gene Expression
HeLa cells were grown to 70-80% confluence in 100-mm plates and were treated for 15 min at 37 °C with diamide (1 μm) or diluent, washed with PBS, and then challenged with TNF (100 units/ml) for 1 h in serum-free medium. For the experiments involving DsbA, HeLa cells were treated overnight in serum-free medium with DsbA (0.1 μg/ml) in the presence and absence of GSSG (500 μm). Following treatment with DsbA, the cells were stimulated for 1 h (37 °C) with TNF (100 units/ml) in serum-free medium, and the cells washed three times with ice-cold PBS. Total cellular RNA was extracted using the RNA STAT-60 reagent (Tel-Test, Inc., Friendswood, TX) according to the manufacturer's instructions. We measured gene expression of two TNF-sensitive genes, IκB and sydecan-4, using an RNase protection assay, as previously described (16). Autoradiography was performed, and the gel images were quantified using laser densitometry and ImageQuanT software (Storm 860, Molecular Dynamics), and the resulting data normalized to levels of L32 mRNA, which was used as an internal control to normalize the degree of loading in each lane.
Effect of Oxidative Stress on TNF Ligand-TNF Receptor Interactions
To determine whether oxidative stress resulted in modifications of TNF binding to its cognate receptors, we performed TNF saturation/competition studies using a modification of the method of Ding et al. (17). Briefly, TNF saturation/competition studies assays were performed in 293T cells that had been transfected with the chimeric TNFR1 or TNFR2 constructs. The cultures of 293T cells were seeded in 24-well plates, and the cells treated overnight with DsbA (0.1 μg/ml) or diluent, as described above. The cells were then washed twice with PBS containing 1% BSA (PBSA), and incubated for 2 h at 4 °C with 0.5 ml of 0.01, 0.03, 0.1, 0.3, 1, or 3 nm 125I-TNF in PBSA, in the presence and absence of 100 nm of unlabeled TNF. TNF binding was terminated by aspirating the labeling medium and rapidly washing the cells with ice-cold PBSA. The cells were then solubilized with 0.3 ml of 1 n NaOH for 30 min at room temperature, transferred to scintillation vials, and the γ-emissions determined by scintillation counting. To determine whether oxidative stress influenced TNF signaling, we measured NF-κB activation in TNF-stimulated cells that had been pretreated with DsbA. Briefly, HeLa cells were grown as described above and were treated overnight in serum-free medium with diluent or DsbA (0.01 μg/ml) in the presence and absence of GSSG (500 μm). After the DsbA treatment, cell cultures were washed with PBS and treated for 15 min at 37 °C with different concentrations of TNF (0.01, 0.1, 1, 10, and 100 pm). Following this, the nuclear extracts were prepared, and electrophoretic mobility assays performed as described above. Autoradiography was performed and the gel images were quantified using laser densitometry and ImageQuanT software (Storm 860, Molecular Dynamics).
Statistical Analysis
Values are presented as mean ± S.E. One-way analysis of variance (ANOVA) was used to test for mean differences in the FRET mean fluorescence intensity or the optical density of the bands from RNase protection assays in the different treatment groups. Where appropriate, posthoc ANOVA testing (Tukey's test) was used to assess mean differences between groups. An unpaired Student's t test was used to test for mean differences between vehicle and DsbA-treated groups for the TNF dose response curves of NF-κB activation. Significant differences were said to exist at a value of p < 0.05.
RESULTS
Effect of Oxidative Stress on TNF Receptor Self-interaction
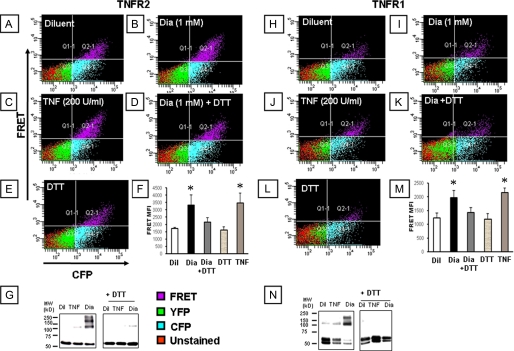
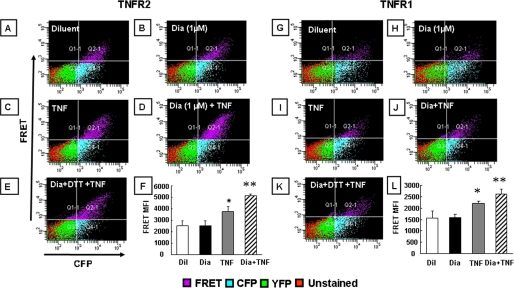
Effect of Diamide on TNF Receptor Self-association—Fig. 1, panels A-E and H-L show, respectively, representative FACS analyses of 293T cells transfected with the chimeric TNFR2 and TNFR1 fusion constructs (n = 3-6 experiments/group). Fig. 1, panels F and M depict the summary of group data for mean fluorescence intensity in the FRET channel (quadrant 2 (upper right hand panel) for TNFR2 and TNFR1, respectively). With respect to TNFR2, treatment with diamide (1 mm) provoked an increase in the mean fluoresence intensity in the FRET channel (Fig. 1, panels B and F) when compared with diluent-treated control cells (Fig. 1, panels A and F), consistent with a closer physical association of the cytoplasmic CFP and YFP moieties of the TNFR2 fusion protein. The observation that a small percentage of diluent-treated cells emit a FRET signal is consistent with the view that TNFRs pre-associate as oligomers on the cell surface (3). The diamide-induced increase in the FRET signal was abolished by simultaneous treatment with DTT (Fig. 1, panel F), a thiol-reducing agent, whereas DTT alone had no effect on the baseline mean fluoresence intensity in the FRET channel (Fig. 1, panels E and F), suggesting that under the conditions used herein, the majority of the TNFR2 receptors exist in a reduced state. Treatment with TNF (200 units/ml), which was used as a positive control, resulted in a similar mean fluorescence in the FRET channel (Fig. 1, panels C and F) when compared with diamide-treated cells. The results with FRET were confirmed using Western blot analysis of 293T cells transfected with the TNFR2 fusion proteins. The left panel of Fig. 1G shows that diamide induced the formation of TNFR2 complexes with molecular masses that were 2-3× that of monomeric TNFR2, consistent with the self-association of TNFR2 into dimers and trimers. In contrast, the diamide-induced increase in TNFR2 oligomerization was not evident when the gels were run under reducing conditions using DTT. Fig. 1, panels H-M depicts the results of similar experiments in which TNFR1-transfected 293T cells were treated with diamide and then analyzed by FACS. As shown, the results were qualitatively similar to those described above for TNFR2. The mean fluoresence intensity of the FRET channel for the diamide and TNF-treated TNFR1-transfected cells was less than we observed with TNFR2-transfected cells, which may relate to transfection efficiency of the TNFR1 chimeric fusion proteins. Taken together, these results suggest that oxidative stress, in the absence of ligand, is sufficient to increase the self-interaction of both TNF receptors. To determine whether lower levels of oxidative stress were sufficient to augment TNF-induced TNFR association, we examined the FRET signal in TNFR2-transfected cells in the presence and absence of TNF, after pretreating the 293T cells with a concentration of diamide (1 μm) that had no discernable effect of the FRET signal (compare Fig. 2, panels A and B). Fig. 2, panel F, which summarizes the results of group data (n = 3-6 experiments/group), illustrates two important findings. First, low (1 μm) concentrations of diamide had no effect of the FRET signal when compared with diluent. Second, pretreatment with 1 μm diamide resulted in a statistically significant (p < 0.05) increase in the FRET signal in TNF-treated cells, when compared with TNF-stimulated cells that had been pretreated with diluent. Importantly, the diamide-induced increase in the FRET signal in TNF-treated cells was abolished by treatment with DTT (Fig. 2, panel E). Fig. 2, panels G-L summarizes the results of parallel experiments conducted in TNFR1-transfected 293T cells. As shown by the group data in Fig. 2, panel L (n = 6 experiments/group), the results were similar to those observed for TNFR2. That is, pretreatment with 1 μm diamide, which had no discernable effect on the FRET signal, resulted in a significant (p < 0.05) enhancement of the FRET signal in TNF-stimulated cells when compared with TNF treatment alone. Taken together, these results suggest that oxidative stress, at levels that do not promote self-association of either TNFR1 or TNFR2, is sufficient to augment TNF-induced oligomerization of TNF receptors.
FIGURE 1.
Effect of diamide (1 mm) on TNF receptor self-association. A flow cytometric analysis (FACS) of 293T cells co-expressing CFP- and YFP-tagged TNFR2 (panels A-F) or TNFR1 (panels H-M) was performed in the presence and absence of diamide. FACS analysis of cells with CFP and FRET fluorescence channels are shown on the x and y axes, respectively. The mean fluoresence intensity (MFI) of cells emitting a FRET signal is depicted in the top right quadrant of each FACS analysis (quadrant 2); the respective treatments are indicated for each panel: (panels A, H) control; (panels B, I) diamide (1 mm); (panels C, J) TNF (200 units/ml); (panels D, K) 1 mm diamide and 100 μm DTT; (panels E, L) DTT (100 μm). Panels F and L, group data (mean ± S.E.) for MFI for each treatment are shown in panels F and M, respectively for TNFR2 and TNFR1. Panels G and N depict the Western blot analyses of the TNFR2 (panel G) or TNFR1 (panel N) under non-reducing (left panel) and reducing conditions (right panel). Key: Dil, diluent; Dia, diamide; *, p < 0.05 compared with diluent). The respective position of TNFR monomers (M), dimers (D), and trimers (T) are shown. The above results are representative of a minimum of three different experiments (n = 3-6 experiments/group).
FIGURE 2.
Effect of diamide (1 μm) on TNF-induced TNF receptor oligomerization. A flow cytometric analysis (FACS) analysis of 293T cells co-expressing CFP- and YFP-tagged TNFR2 (panels A-F) or TNFR1 (panels G-L) was performed. FACS analysis of cells with CFP and FRET fluorescence channels are shown on the x and y axes, respectively. The mean fluoresence intensity (MFI) of cells emitting a FRET signal is depicted in the top right quadrant of each FACS analysis (quadrant 2); the respective treatments are presented on the top left of each analysis. Panels A and G, control; panels B and H, diamide (1 μm); panels C and I, TNF (100 units/ml); panels D and J, 100 units/ml TNF in diamide (1 μm)-pretreated cells; panels E and K, 100 units/ml TNF in cells pretreated with diamide (1 μm) and DTT (100 μm). The group data (mean ± S.E.) for MFI for each treatment is shown in panels F and L, respectively, for TNFR2 and TNFR1. These results are representative of a minimum of least three different experiments (n = 3-6 experiments/group). Key: Dil, diluent; Dia, diamide; *, p < 0.05 versus diluent; **, p < 0.05 versus TNF.
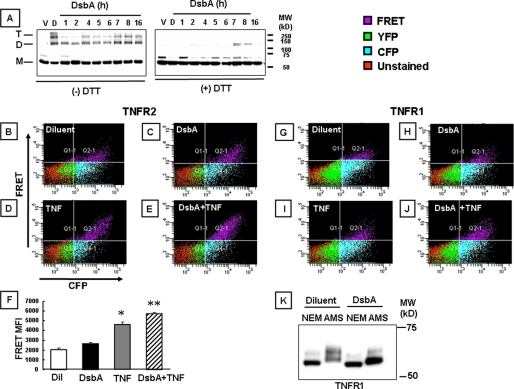
Effect of DsbA on TNF Receptor Self-association—Fig. 3 summarizes the results of the experiments wherein we used DsbA in 293T cells transfected with the chimeric TNFR1 and TNFR2 fusion constructs. The left panel of Fig. 3A shows that under nonreducing conditions, DsbA induced the formation of the TNFR2 complexes with molecular weights that were 2-3× the molecular weight of the TNFR2 monomers. As shown, the effect of DsbA was detectable as early as 4 h and was a maximum at 7-16 h. In contrast, TNFR2 receptor dimers and trimers were not detected in vehicle-treated samples (up to 16 h), whereas they were readily detected in diamide (positive control)-treated samples within 15 min. The right panel of Fig. 3A shows that DsbA-induced multimeric TNFR2 receptor complexes were not detected when the samples were electrophoretically separated under reducing conditions. FRET experiments were performed in 293T cells transfected with the chimeric TNFR1 and TNFR2 fusion constructs to determine whether oxidative stress-induced modifications of the extracellular domains of the TNFRs was sufficient to facilitate self-association. With respect to TNFR2, Fig. 3C shows that treatment with DsbA (0.1 μg/ml) for 16 h led to small increases in the FRET signal when compared with diluent-treated controls (Fig. 3B). However, the important finding was that pretreatment with DsbA (0.1 μg/ml) for 16 h enhanced the FRET signal in TNF (100 units/ml)-treated cells (Fig. 3, panel E), when compared with TNF (100 units/ml)-treated cells that had not been pretreated with DsbA (Fig. 3, panel D). Fig. 3, panel F, which summarizes the results of group data, shows that DsbA significant increased (p < 0.05) the FRET signal of the TNFR2-transfected cells in comparison to diluent, and that pretreatment with DsbA significantly increased (p < 0.05) the FRET signal in TNF-treated cells when compared with TNF stimulation in the absence of DsbA. Treatment with DsbA had similar qualitative effects on the FRET signal in 293T cells that had been transfected with TNFR1 fusion proteins, as depicted in Fig. 3, panels G-J. Thus, the findings in the DsbA-treated 203T cells are internally consistent with the results obtained with diamide, and suggest that the oxidative stress-induced self-association of TNFR1 and TNFR2 is secondary to modification of the extracellular domains of the TNF receptors. However, the results of these studies do not allow us to determine which cysteine residues were modified by diamide or DsbA.
FIGURE 3.
Effect of DsbA on TNF receptor oligomerization. Panel A, Western blot analysis of TNFR2 under non-reducing (left panel) and reducing conditions (right panel). 293T cells expressing TNFR2 fusion proteins were treated with DsbA (1 μg/ml) for 1-16 h; the position of TNFR2 monomers (M), dimers (D), and trimers (T) are depicted. Panels B-E depict the FACS analysis of 293T cells co-expressing CFP- and YFP-tagged TNFR2 constructs in the presence of diluent (500 μm GSSG (panel B)), 1 μg/ml DsbA (panel C) for 16 h, 100 units/ml TNF (panel D), and 100 units/ml TNF and 1 μg/ml DsbA for 16 h (panel E). Panels G-J depict FACS analysis of 293T cells co-expressing CFP- and YFP-tagged TNFR1 in the presence of diluent (500 μm GSSG (panel G)), 1 μg/ml DsbA (panel H) for 16 h, 100 units/ml TNF (panel I), and 100 units/ml TNF and 1 μg/ml DsbA for 16 h (panel J). The group data (mean ± S.E.) for mean fluoresence intensity for each treatment is shown in panel F for TNFR2. Panel K illustrates the binding of AMS and NEM to TNFR1 in transfected 293T cells in the presence and absence DsbA (1 μg/ml). Key: Dil, diluent; *, p < 0.05 versus diluent; **, p < 0.05 versus TNF; *, p < 0.05 versus control; **, p < 0.05 versus TNF.
To determine whether DsbA modified the free sulfhydryl groups on cysteine residues on the extracellular domains of TNFR1 under the experimental conditions used herein, we treated TNFR1-transfected 293T cells with DsbA, as described above, and then exposed the cells to AMS. As shown in Fig. 3K the Western blot for TNFR1 in the diluent-treated cells exposed to NEM showed a single band with a lower molecular mass, when compared with the multiple bands of higher molecular weight in the diluent-treated cells exposed to AMS, consistent with the presence of free sulfhydryl groups in the extracellular domain of the TNFR1. However, the important finding shown by Fig. 3, panel K is that treatment with DsbA resulted in a shift in the pattern of AMS binding to lower molecular proteins, suggesting that there were less free sulfhydryl groups available for AMS binding following DsbA-induced oxidation of the free sulfhydryl groups, consistent with a prior study that showed that TNFR1 contains ∼15-17 redox-sensitive thiol groups (12). As expected, DsbA had no effect on the molecular weight of the cells exposed to NEM. This gel is representative of four experiments.
Effect of Oxidative Stress on TNF-induced NF-κB Activation
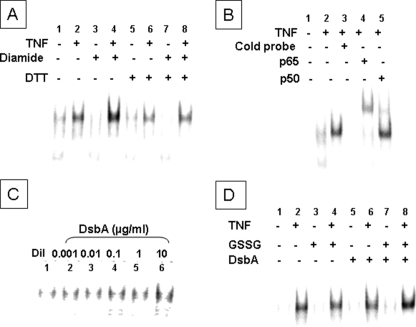
To determine whether oxidative stress influenced TNF-mediated cell signaling, we measured NF-κB activation in the presence and absence of diamide and DsbA. Fig. 4A shows that treating HeLa cells with TNF (100 units/ml) led to NF-κB activation (lane 2) and that the degree of NF-κB activation detected by EMSA was enhanced by pretreatment with diamide (lane 4). Importantly, diamide did not enhance TNF-induced NF-κB activation (lane 8) in the presence of DTT, whereas DTT itself had no effect on TNF-induced NF-κB activation (lane 6). The specificity of the NF-κB signal was determined in several ways, including a 20× molar excess of unlabeled NF-κB consensus sequence, which completely abrogated binding of the labeled oligonucleotide (Fig. 4B, lane 3), as well as by supershift assays, which showed that the NF-κB complexes were supershifted by polyclonal antibodies directed against the p65 and p50 components of NF-κB (Fig. 4B, lanes 4 and 5). The above experiments were repeated using the cell-impermeant oxidoreductase DsbA. As shown in Fig. 4C, increasing concentrations of DsbA resulted in a progressive increase in NF-κB activation. To delineate the effect of DsbA on TNF receptor signaling, we measured TNF-induced NF-κB activation with and without pretreatment with DsbA (0.1 μg/ml). Fig. 4D shows that the degree of TNF-induced NF-κB activation (lane 8) was more robust in the presence of DsbA pretreatment than without (lane 7). Neither basal NF-κB activation (lane 3) nor TNF-induced NF-κB activation (lane 4) was affected by GSSG. Further, DsbA had no affect on basal (lane 5) or TNF-induced (lane 6) NF-κB activation in the absence of GSSG.
FIGURE 4.
Effect of diamide and DsbA on TNF-induced NF-κB activation. A, HeLa cells were treated with diamide (1 μm) or TNF (100 units/ml), and NF-κB activation was determined by EMSA in the absence and presence of DTT (100 μm). B, to determine the specificity of DNA-protein binding, nuclear extracts from TNF-treated HeLa cells were treated with a 20× excess of unlabeled oligonucleotides. Supershift assays were performed by adding p50 and p65 antibodies to the nuclear extracts. C, HeLa cells were treated with increasing concentrations of DsbA (0.001-10 μg/ml) + 500 μm GSSG, and NF-κB activation was determined by EMSA. D, HeLa cells were treated with DsbA (1 μg/ml) and GSSG (500 μm) overnight followed by treatment with TNF (100 units/ml) for 15 min, and NF-κB activation was assessed by EMSA. The results in each panel are representative of at least three different experiments.
Effect of Oxidative Stress on TNF-induced Gene Expression
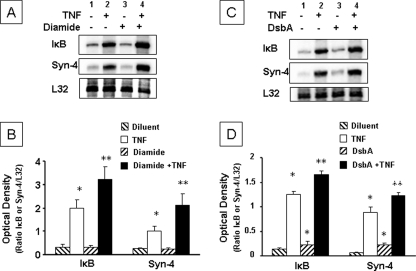
To determine whether oxidative stress influenced TNF-mediated gene expression, we measured the mRNA levels of two TNF-sensitive NF-κB-dependent genes, IκB, and syndecan-4 (18) in the presence and absence of diamide (1 μm) and DsbA (0.1 μg/ml). Fig. 5A shows representative RNase protection assays for IκB and syndecan-4, whereas Fig. 5B summarizes the results of group data. The salient finding shown by Fig. 5 is that TNF-induced gene expression for IκB and syndecan-4 was greater in diamide-pretreated cells (lane 4, p < 0.05) than in diluent-pretreated cells (lane 2). Importantly, diamide alone had no effect on the basal levels (lane 3) of IκB and syndecan-4 gene expression. Similarly, TNF-induced expression of IκB and syndecan-4 mRNA levels were increased significantly (p < 0.05) by DsbA pretreatment when compared with vehicle-pretreated cells (Fig. 5, C and D). As shown, pretreatment with DsbA resulted in a small but significant increase in IκB and syndecan-4 mRNA levels when compared with vehicle, consistent with DsbA-induced TNF receptor self-association in the absence of TNF (Fig. 3).
FIGURE 5.
Effect of diamide and DsbA on TNF-induced gene expression. A, representative RNase protection assay showing IκB and syndecan-4 mRNA levels in HeLa cells treated with diluent or TNF (100 units/ml) in the presence and absence of diamide (1 μm). B, group data (mean ± S.E.) for IκB and syndecan-4 mRNA levels in HeLa cells treated with diluent or TNF (100 units/ml) (lane 2) in the presence and absence of diamide (1 μm) pretreatment. C, representative RNase protection assay showing IκB and syndecan-4 mRNA levels in HeLa cells treated with diluent (GSSG (500 μm)), TNF (100 units/ml), DsbA (0.1 μg/ml) + GSSG (500 μm), and TNF + DsbA (0.1 μg/ml) + GSSG (500 μm). D, group data (mean ± S.E.) for IκB and syndecan-4 mRNA levels in HeLa cells treated with diluent, TNF (100 units/ml), DsbA (0.1 μg/ml) + GSSG (500 μm), and TNF + DsbA (0.1 μg/ml) + GSSG (500 μm). Group data are expressed as the ratio of the optical density of the ratio IκB or syndecan-4 mRNA level to the L32 mRNA level. *, p < 0.05 versus diluent; **, p < 0.05 versus TNF.
Effect of Oxidative Stress on TNF Ligand-TNF Receptor Interactions
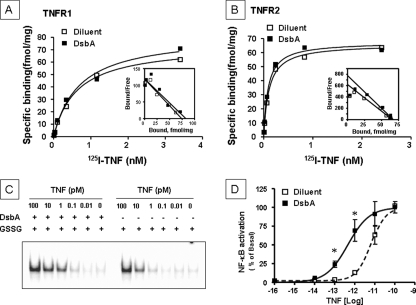
We considered two possible explanations for the effect of oxidative stress on enhanced TNF signaling (Fig. 4) and enhanced TNF induced gene expression (Fig. 5), namely increased TNF binding affinity for the TNF receptors and/or allosteric modifications of the TNF receptors. To determine whether oxidative modifications of the cysteine residues of the extracellular domains of the TNF receptors affected TNF binding we measured the binding affinities of TNF in 293T cells that had been transfected with chimeric TNFR1 and TNFR2 constructs. As shown in Fig. 6, A and B, respectively, treatment with DsbA did not lead to changes in the binding affinity of TNF for either TNFR1 or TNFR2, when compared with vehicle. Scatchard analysis of the data for TNFR1 (Fig. 6A, inset) indicated a Kd of 0.63 nm and 0.69 nm and a Bmaxof 74.16 fmol/mg and 82.53 mol/mg, respectively. Scatchard analysis of the data for TNFR2 (Fig. 6B, inset) indicated a Kd of 0.11 nm and 0.09 nm and a Bmax of 66.0 fmol/mg and 67.4 fmol/mg, respectively. Next, we examined the effect of increasing concentrations of TNF on NF-κB activation in HeLa cells, with and without DsbA pretreatment (Fig. 6, C and D). As shown in Fig. 6D, the dose response curve for TNF-induced activation of NF-κB was shifted leftward following pretreatment with DsbA, consistent with an increase in the relative potency of TNF. The log(EC50) of TNF-induced NF-κB in DsbA-treated cells (log10-12.4±0.14) was significantly (p < 0.05) less than the log(EC50) in vehicle-treated cells (log 10-11.2±0.1, n = 4 experiments). As shown in Fig. 6D, TNF-induced NF-κB activation at doses of 0.1 and 1 pm was significantly greater in DsbA-treated cells than that in vehicle-treated cells (p < 0.05, n = 4 experiments).
FIGURE 6.
TNF receptor binding and TNF-induced NF-κB activation. 125I-TNF saturation/competition studies were performed in 293T cells transfected with the chimeric TNFR1 (A) or TNFR2 (B) constructs and treated overnight with diluent (500 μm GSSG, open squares) or DsbA (0.1 μg/ml, closed squares) + 500 μm GSSG (representative of three separate experiments). C, representative EMSA showing the effect of increasing concentrations of TNF (0-100 pm) on NF-κB activation in the presence of DsbA (0.1 μg/ml) + GSSG (500 μm) or diluent (500 μm GSSG). This figure is representative of three separate experiments. D, dose response curve of TNF-induced NF-κB activation in the presence of DsbA (0.1 μg/ml) + GSSG (500 μm) or diluent (500 μm GSSG). *, p < 0.05 versus diluent.
DISCUSSION
The principal new finding of this study is that oxidative stress promotes self-interaction of the TNF receptors that leads to enhanced TNF signaling. The following lines of evidence support this statement. First, the thiol oxidant diamide induced TNF receptor self-oligomerization in the absence of exogenous TNF, as demonstrated by an increase in the FRET signal in 293T cells transfected with CFP- or YFP-tagged TNF receptors, as well as by the formation of multimeric TNFR complexes in the transfected 293T cells (Fig. 1, panels A-L). Of note, pretreating the 293T cells with concentrations of diamide that were not sufficient to provoke self-association of either TNFR1 or TNFR2, resulted in an increased FRET signal in TNF-stimulated 293T cells (Fig. 2, panels A-L), suggesting that oxidative stress augmented TNF-induced TNFR signaling. In addition, pretreatment with diamide resulted in increased TNF-induced NF-κB activation (Fig. 4A), as well as increased TNF-induced IκBα and syndecan-4 gene expression (Fig. 5, A and B) when compared with cells pretreated with diluent alone. Importantly, the concentrations of diamide that resulted in enhanced TNF signaling had no discernable effect on NF-κB activation or NF-κB-dependent gene expression. Further, the effects of diamide were completely abrogated in the presence of the reducing agent DTT. Given that diamide can readily diffuse through cell membranes, we considered the possibility that diamide might promote TNFR self-association through oxidative modifications of the cytoplasmic tails of either TNFR1 or TNFR2. Therefore, we repeated the above experiments with DsbA, a cell-impermeant oxidoreductase. Treatment with DsbA led to an increase in the FRET signal (Fig. 3, panels B-J) and increased formation of multimeric TNF receptor complexes in 293T cells transfected with CFP- or YFP-tagged TNF receptors (Fig. 3A). The DsbA-induced TNFR complexes were functionally important, insofar as DsbA pretreatment in the absence of TNF stimulation resulted in concentration-dependent activation of NF-κB (Fig. 4C) and increased expression of two NF-κB-sensitive genes, namely IκBα and syndecan-4 (Fig. 5, C and D). Furthermore, pretreatment with DsbA resulted in increased TNF-induced NF-κB activation and increased TNF-induced gene expression (Figs. 4D and 5, C and D). The concentration of DsbA that resulted in increased TNFR self-association was sufficient to oxidize the free thiol groups on the cysteine residues of TNFR1, as shown by the studies with AMS (Fig. 3, panel K), suggesting that modifications of the cysteine residues of TNFRs contributed to the enhanced TNF-induced TNFR signaling and gene expression. Unfortunately, these studies do not allow us to determine the specific nature (inter- or intramolecular) of the disulfide bonds that were formed following oxidation of the cysteine residues. To further explore the mechanism for the effects of oxidative stress on enhanced ligand-dependent and ligand-independent TNF signaling, we examined the effect of oxidative stress with respect to TNF binding affinity for TNFR1 and TNFR2. As shown in Fig. 6, A and B, respectively, treatment with DsbA had no effect on TNF binding affinity for either TNFR1 or TNFR2 when compared with vehicle alone. However, when we examined the effects of DsbA on TNF-induced NF-κB activation, we observed a leftward shift in the TNF dose response curve for NF-KB activation (Fig. 6, C and D), consistent with allosteric modification of the TNFRs. Taken together, these observations suggest that oxidative modifications of the cysteine residues of the TNFRs lead to conformational changes within the receptors that favors the formation of TNFR oligomers on the surface of the cell membrane, which in turn allows the cytoplasmic tails to come into close proximity, thereby fostering ligand-independent as well as enhanced TNF-induced signaling and gene expression.
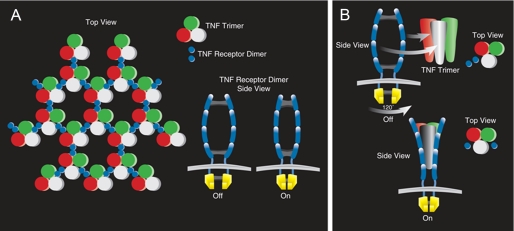
TNF Receptor Signaling—The classic model of TNF receptor signaling suggests that homotrimeric TNF recruits three separate chains of the TNF receptor through ligand-induced trimerization of monomeric receptor chains, which in turn leads to recruitment of signaling complexes to the cytoplasmic domains of these receptors (2, 19, 20). Support for the classic trimerization model is provided by the crystal structure of lymphtoxin-α with the extracellular domain of TNFR1, in which three receptor fragments crystallize with each homotrimer of LT-α. However, more recent studies suggest that many members within the TNFR family exist as pre-assembled oligomers prior to ligand stimulation (21). Indeed, relatively recent studies suggest that the first cysteine-rich domain of the TNF receptor is critical for forming a pre-ligand assembly domain (PLAD) that mediates TNF receptor self-association in the absence of ligand binding (22). Cross-linking experiments suggested that TNFR1 and TNFR2 trimers are the preferred configuration. However, crystallographic studies of unliganded TNFR1 performed at alkaline pH (8.0) suggest that the receptor exists as a dimeric subunit, in which each subunit is arranged head to head (23). A dimeric conformation of TNFR conformation would allow for a hexagonal array of dimer receptors and trimeric ligands (aggregation model), which could make contacts between their cytoplasmic domains (Fig. 7A). While the aggregation model is consistent with the observation that trimerization of the receptors is necessary for TNF signaling, there are no clear experimental data that demonstrate the formation of networks of ligands and receptors on the cell surface. An alternative explanation that has been proposed is that each receptor dimer acts as an independent molecular switch, in which ligand binding enforces displacement and rotation of the receptor units relative to one another, forcing a conformational change on the cytoplasmic side (Fig. 7B). However, although elegant, this latter model does not account for the observation that the cytoplasmic receptor tails of both TNFR1 and TNFR2 and their interacting signaling adaptors appear to require trimeric axis of symmetry to initiate downstream signaling (24). Although our studies do not allow us to formally exclude or embrace the aggregation and/or the molecular switch models of TNFR signaling presented above, the most parsimonious interpretation of the data set is that oxidative modifications of the cysteine residues allows for dimerization of the receptors, which then act as independent molecular units to initiate cell signaling, and that further ligand-induced oligomerization of the receptor dimers by TNF leads to enhanced signaling through the assembled dimers. One limitation of the present study is that we used exogenously supplied prooxidants to model redox changes of the TNFRs. Accordingly, it remains to be determined whether physiological oxidants will modulate the FRET signal and TNF signaling in a similar manner. Nonetheless, it bears emphasis that our findings are entirely consistent with a prior study, which demonstrated that the cysteine residues of TNFR1 undergo oxidative modification after up-regulation of membrane-bound γ-glutamyltransferase activity (an endogenous physiological source of prooxidants) (12). Further, redox priming has been observed with the insulin receptor, wherein oxidizing conditions resulted in enhanced insulin responsiveness in the absence of tyrosine autophosphorylation (9). Moreover, our data are consistent with studies which have shown that point mutations that create free cysteine residues in the extracellular domain of the fibroblast growth factor receptor and/or the erythropoietin receptor result in receptor dimerization that leads to constitutive receptor signaling (25, 26). In summary, this study shows that oxidative stress promotes TNFR receptor self-interaction, thus suggesting that TNFRs may exist in a constitutively active state depending on the redox milieu of the pericellular space. These studies further suggest that oxidative stress may lead to enhanced TNF signaling through allosteric modifications of the TNF receptors (i.e. TNF receptor priming). Because TNF is generally present at low concentrations physiologically, oxidative stress and or higher extracellular pH may ensure more efficient TNF signaling by permitting TNFRs to undergo conformational changes that facilitate TNF signaling at subnanomolar concentrations. Moreover, the observation that oxidative stress was sufficient to provoke ligand-independent signaling raises the interesting possiblity that redox-induced alterations in TNFR signaling may allow for extremely rapid TNF-like cellular responses during periods of cellular stress. Conversely, during periods of reductive stress (27) and/or lower extracellular pH, the TNFRs would be expected to undergo conformational changes that would dampen TNFR signaling at physiologic concentrations of TNF. Although speculative, these studies also raise the intriguing possibility that in addition to the classic ligand-receptor model of TNF signaling, an additional level of control of TNF signaling may reside at the level of the cell membrane wherein ectodomain-based prooxidant enzymes (e.g. γ-glutamyltransferase, Ref. 28) and oxidoreductases (e.g. protein-disulfide isomerase, Ref. 29) are capable of modulating the redox environment of the pericellular space.
FIGURE 7.
Theoretical models depicting TNF receptor activation. A, aggregation model. If aggregation of TNF receptors into trimers is required to elicit a biological response, it may involve cross-linking of receptor dimers (shown in the inset) by homotrimeric TNF ligand. B, molecular switch model. The molecular switch model suggests that single receptor dimers are engaged by a homotrimeric TNF ligand that promotes displacement of the receptor units relative to one another, forcing a conformational change on the cytoplasmic side. The second receptor subunit engaged would be displaced through 120° of rotation relative to the long-axis of symmetry of the ligand, and would then rotate 180° on it own axis to engage the receptor. (Modified from Bazzoni and Beutler, 30.)
Acknowledgments
We thank Dr. Richard Bond for critical review of the manuscript and Karen Ramirez for technical assistance with flow cytometry.
This work was supported, in whole or in part, by National Institutes of Health Grants P50 HL-O6H, R01 HL58081, R01 HL61543, and HL42250. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TNF, tumor necrosis factor; FRET, fluorescence resonance energy transfer; CRD, cysteine-rich domain; ROS, reactive oxygen species; CFP, cyan fluorescent protein; YFP, yellow fluorescent protein; PBS, phosphate-buffered saline; DTT, dithiothreitol; HA, hemagglutinin; FACS, fluorescence activated cell sorting; NEM, N-ethylmaleimide; EMSA, electrophoretic mobility shift assay; AMS, 4-acetamido-4′-maleimidylstilbene-2,2′ disulfonate.
References
- 1.Locksley, R. M., Killeen, N., and Lenardo, M. J. (2001) Cell 104 487-501 [DOI] [PubMed] [Google Scholar]
- 2.Banner, D. W., D'Arcy, A., Janes, W., Gentz, R., Schoenfeld, H. J., Broger, C., Loetscher, H., and Lesslauer, W. (1993) Cell 73 431-445 [DOI] [PubMed] [Google Scholar]
- 3.Chan, F. K., Chun, H. J., Zheng, L., Siegel, R. M., Bui, K. L., and Lenardo, M. J. (2000) Science 288 2351-2354 [DOI] [PubMed] [Google Scholar]
- 4.Conner, E. M., and Grisham, M. B. (1996) Nutrition 12 274-277 [DOI] [PubMed] [Google Scholar]
- 5.Droge, W. (2002) Physiol. Rev. 82 47-95 [DOI] [PubMed] [Google Scholar]
- 6.Dominici, S., Valentini, M., Maellaro, E., Del Bello, B., Paolicchi, A., Lorenzini, E., Tongiani, R., Comporti, M., and Pompella, A. (1999) Free Radic. Biol. Med. 27 623-635 [DOI] [PubMed] [Google Scholar]
- 7.Maellaro, E., Dominici, S., Del Bello, B., Valentini, M. A., Pieri, L., Perego, P., Supino, R., Zunino, F., Lorenzini, E., Paolicchi, A., Comporti, M., and Pompella, A. (2000) J. Cell Sci. 113 2671-2678 [DOI] [PubMed] [Google Scholar]
- 8.Moriarty-Craige, S. E., and Jones, D. P. (2004) Annu. Rev. Nutr. 24 481-509 [DOI] [PubMed] [Google Scholar]
- 9.Schmid, E., El Benna, J., Galter, D., Klein, G., and Droge, W. (1998) FASEB J. 12 863-870 [DOI] [PubMed] [Google Scholar]
- 10.Peus, D., Vasa, R. A., Beyerle, A., Meves, A., Krautmacher, C., and Pittelkow, M. R. (1999) J. Investig. Dermatol. 112 751-756 [DOI] [PubMed] [Google Scholar]
- 11.de Wit, R., Capello, A., Boonstra, J., Verkleij, A. J., and Post, J. A. (2000) Free Radic. Biol. Med. 28 28-38 [DOI] [PubMed] [Google Scholar]
- 12.Dominici, S., Pieri, L., Paolicchi, A., De, T. V., Zunino, F., and Pompella, A. (2004) Ann. N. Y. Acad. Sci. 1030 62-68 [DOI] [PubMed] [Google Scholar]
- 13.Gautam, A., Densmore, C. L., Golunski, E., Xu, B., and Waldrep, J. C. (2001) Mol. Ther. 3 551-556 [DOI] [PubMed] [Google Scholar]
- 14.Bardwell, J. C., McGovern, K., and Beckwith, J. (1991) Cell 67 581-589 [DOI] [PubMed] [Google Scholar]
- 15.Knuefermann, P., Chen, P., Misra, A., Shi, S. P., Abdellatif, M., and Sivasubramanian, N. (2002) J. Biol. Chem. 277 23888-23897 [DOI] [PubMed] [Google Scholar]
- 16.Baumgarten, G., Knuefermann, P., Nozaki, N., Sivasubramanian, N., Mann, D. L., and Vallejo, J. G. (2001) J. Infect. Dis. 183 1617-1624 [DOI] [PubMed] [Google Scholar]
- 17.Ding, A. H., Sanchez, E., Srimal, S., and Nathan, C. F. (1989) J. Biol. Chem. 264 3924-3929 [PubMed] [Google Scholar]
- 18.Zhou, A., Scoggin, S., Gaynor, R. B., and Williams, N. S. (2003) Oncogene 22 2054-2064 [DOI] [PubMed] [Google Scholar]
- 19.Smith, C. A., Farrah, T., and Goodwin, R. G. (1994) Cell 76 959-962 [DOI] [PubMed] [Google Scholar]
- 20.Hsu, H., Xiong, J., and Goeddel, D. V. (1995) Cell 81 495-504 [DOI] [PubMed] [Google Scholar]
- 21.Chan, K. F., Siegel, M. R., and Lenardo, J. M. (2000) Immunity 13 419-422 [DOI] [PubMed] [Google Scholar]
- 22.Siegel, R. M., Frederiksen, J. K., Zacharias, D. A., Chan, F. K., Johnson, M., Lynch, D., Tsien, R. Y., and Lenardo, M. J. (2000) Science 288 2354-2357 [DOI] [PubMed] [Google Scholar]
- 23.Naismith, J. H., Devine, T. Q., Brandhuber, B. J., and Sprang, S. R. (1995) J. Biol. Chem. 270 13303-13307 [DOI] [PubMed] [Google Scholar]
- 24.Ye, H., Park, Y. C., Kreishman, M., Kieff, E., and Wu, H. (1999) Mol. Cell 4 321-330 [DOI] [PubMed] [Google Scholar]
- 25.Watowich, S. S., Yoshimura, A., Longmore, G. D., Hilton, D. J., Yoshimura, Y., and Lodish, H. F. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 2140-2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson, S. C., Meyer, A. N., Hart, K. C., Galvin, B. D., Webster, M. K., and Donoghue, D. J. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 4567-4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayakawa, M., Miyashita, H., Sakamoto, I., Kitagawa, M., Tanaka, H., Yasuda, H., Karin, M., and Kikugawa, K. (2003) EMBO J. 22 3356-3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Behura, S. K. (2007) Insect Biochem. Mol. Biol. 37 3-9 [DOI] [PubMed] [Google Scholar]
- 29.Jordan, P. A., and Gibbins, J. M. (2006) Antioxid. Redox Signal. 8 312-324 [DOI] [PubMed] [Google Scholar]
- 30.Bazzoni, F., and Beutler, B. (1995) J. Inflamm. 45 221-238 [PubMed] [Google Scholar]