Abstract
The flagellar calcium-binding protein (FCaBP) of the protozoan Trypanosoma cruzi is targeted to the flagellar membrane where it regulates flagellar function and assembly. As a first step toward understanding the Ca2+-induced conformational changes important for membrane-targeting, we report here the x-ray crystal structure of FCaBP in the Ca2+-free state determined at 2.2Å resolution. The first 17 residues from the N terminus appear unstructured and solvent-exposed. Residues implicated in membrane targeting (Lys-19, Lys-22, and Lys-25) are flanked by an exposed N-terminal helix (residues 26-37), forming a patch of positive charge on the protein surface that may interact electrostatically with flagellar membrane targets. The four EF-hands in FCaBP each adopt a “closed conformation” similar to that seen in Ca2+-free calmodulin. The overall fold of FCaBP is closest to that of grancalcin and other members of the penta EF-hand superfamily. Unlike the dimeric penta EF-hand proteins, FCaBP lacks a fifth EF-hand and is monomeric. The unstructured N-terminal region of FCaBP suggests that its covalently attached myristoyl group at the N terminus may be solvent-exposed, in contrast to the highly sequestered myristoyl group seen in recoverin and GCAP1. NMR analysis demonstrates that the myristoyl group attached to FCaBP is indeed solvent-exposed in both the Ca2+-free and Ca2+-bound states, and myristoylation has no effect on protein structure and folding stability. We propose that exposed acyl groups at the N terminus may anchor FCaBP to the flagellar membrane and that Ca2+-induced conformational changes may control its binding to membrane-bound protein targets.
Flagellar calcium-binding protein (FCaBP)3 is a 24-kDa highly immunogenic protein found in the flagellum of the protozoan parasite Trypanosoma cruzi (1). FCaBP contains four EF-hand calcium binding motifs (2, 3) (see Fig. 1), the third and fourth (EF-3 and EF-4) of which bind calcium (4). The protein is modified at the N terminus by covalent attachment of myristate at Gly-2 and palmitate at Cys-4, both of which are required for association with the inner leaflet of the flagellar membrane (5). Calcium is required for stable flagellar localization as well, as FCaBP can be washed out of detergent-permeabilized trypanosomes if calcium chelators are included in the wash solutions. The N-terminal acylation and calcium-dependent membrane localization of FCaBP suggested that the protein may possess a functional calcium-acyl switch, similar to the Ca2+-myristoyl switch observed previously for recoverin (6, 7) and other members of the neuronal calcium sensor family (8). Acyl switch proteins undergo calcium-dependent membrane association by virtue of calcium-regulated extrusion or sequestration of a myristate moiety that mediates membrane binding (9). However, an FCaBP mutant unable to bind calcium still maintains its flagellar localization, suggesting that FCaBP may not cycle the membrane on and off like some calcium acyl switch proteins (4).
FIGURE 1.
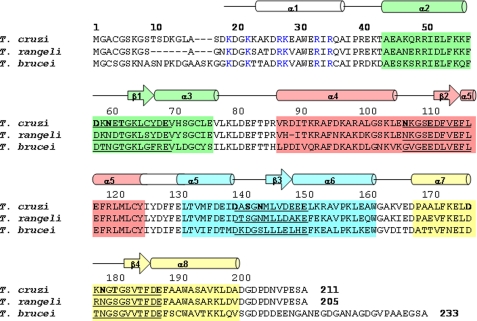
Primary and secondary structure features of FCaBP. Alignment of the primary sequence of T. cruzi FCaBP with that of T. rangeli, and T. brucei. Secondary structural elements indicated schematically were derived from the x-ray structure and analysis of NMR data (3JHNHα, chemical shift index (59), and sequential NOE patterns). The four EF-hands (EF1, EF2, EF3, and EF4) are highlighted in green, salmon, cyan, and yellow, respectively. Residues in the 12-residue Ca2+ binding loops are underlined, and chelating residues are highlighted in bold. Invariant basic residues on the protein surface and implicated in membrane binding are colored blue.
The best studied calcium myristoyl switch protein is recoverin, a calcium-binding protein in retinal rod cells that inhibits rhodopsin kinase only at high Ca2+ levels (10, 11) and regulates the recovery phase of phototransduction (12, 13). In the resting dark state, recoverin binds two calcium ions and associates with retinal rod outer segment membranes through its exposed myristoyl group (6, 7). Photoexcitation of the rod cell results in a lowering of cytosolic calcium (14), causing recoverin to lose its bound calcium and adopt a conformation in which the myristoyl group becomes sequestered within a hydrophobic cleft in the protein (9). Ca2+-free recoverin then dissociates from rhodopsin kinase at the membrane, allowing rhodopsin kinase to phosphorylate light-excited rhodopsin and promote receptor inactivation. The calcium-myristoyl switch mechanism allows calcium regulation of two proteins (rhodopsin kinase and rhodopsin) that do not themselves bind calcium. One major difference between FCaBP and recoverin is the presence of palmitate in FCaBP, which may or may not participate in a potential switch mechanism.
A variety of FCaBP-like proteins are found in other trypanosomes: Trypanosoma rangeli, Trypanosoma lewisi, and Trypanosoma brucei (Fig. 1). A flagellar calmodulin has also been characterized in T. brucei (15). These Ca2+-binding proteins all localize to the flagellum, a unique organelle that has many functions, including motility, chemotaxis, and cell signaling. In addition to the traditional “9 + 2” microtubule structure of the axoneme, there is a structure known as the paraflagellar rod that runs alongside the axoneme. The axoneme, paraflagellar rod, and flagelloplasm are encased by the flagellar membrane. It has been shown by freeze-fracture analysis that the flagellar membrane contains a higher concentration of sterols than does the pellicular (cell body) membrane (16), and the flagellum of T. brucei is highly enriched in lipid rafts. Flagellar membrane stability and function is also controlled by the recruitment of FCaBP to the membrane surface, where it is believed to interact with a variety of membrane-bound protein targets (4).
We report here the x-ray crystal structure of Ca2+-free FCaBP as a first step toward understanding the Ca2+-induced conformational changes that control membrane targeting. This is only the second atomic resolution structure of a Ca2+-acyl switch protein in the Ca2+-free state. The structure of Ca2+-free recoverin first showed its covalently attached myristoyl group to be highly sequestered and buried deep inside the protein (17). Also, the recent crystal structure of Ca2+-bound GCAP1 indicates a sequestered myristoyl group (18). By contrast, the structure of Ca2+-free FCaBP in this study suggests a solvent-exposed N terminus, making its covalently attached acyl groups accessible to interact with membrane targets. We propose that the exposed acyl groups and N-terminal positively charged residues (Lys-19, -22, and -25) may promote the binding of FCaBP to the flagellar membrane and that Ca2+-induced protein conformational changes may control its binding to membrane-bound protein targets.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification—To prepare recombinant unmyristoylated FCaBP (and Se-methionine-labeled FCaBP), FCaBP tagged with a C-terminal His6 tract was expressed in Escherichia coli strain BL21(DE3) carrying a derivative of the pET23d vector (Novagen) as previously described (4) grown in M9 minimal medium supplemented with or without Se-methionine, according to well established procedures (19-21).
Recombinant unmyristoylated FCaBP for the native crystal was expressed as above in LB media and was initially purified via nickel affinity chromatography. Peak fractions were pooled and diluted 3-fold with buffer containing 20 mm Tris, pH 7.5, 1 mm EDTA, 1 mm dithiothreitol, and applied to a Q-HP HiTrap column (Amersham Biosciences) at 5 ml/min. FCaBP was eluted with a 200-ml linear KCl gradient to a final concentration of 250 mm KCl. Peak fractions were pooled, and purified FCaBP was dialyzed and concentrated to 10 mg/ml in 20 mm Tris, pH 7.5, 1 mm dithiothreitol in a centrifugal filter device with a 10,000 molecular weight cutoff (Amicon).
Recombinant myristoylated FCaBP was generated by co-expressing FCaBP and N-myristoyl CoA transferase (pBB131-NMT) in BL21(DE3) cells grown on M9 medium supplemented with myristic acid (10 mg/liter). Myristoylated and non-myristoylated FCaBP was purified from the soluble fraction of bacterial cell lysates using Ni2+-chelate affinity chromatography on a nitrilotriacetate resin (Qiagen) according the manufacturer's instructions. Peak fractions were then applied to an anion-exchange column (Hi-Trap DEAE-FF, GE Healthcare) equilibrated in buffer A (1 mm EDTA, 1 mm dithiothreitol, 10 mm Tris-HCl, pH 7.4) and eluted with a linear salt gradient (0-0.2 m KCl) at flow rate of 5 ml min-1 over the course of 150 min. Peak fractions were concentrated to 5 ml and subjected to size exclusion chromatography (Sephacryl S-100, GE Healthcare) in buffer B (1 mm dithiothreitol, 2 mm CaCl2, 50 mm HEPES, pH 7.4). Final purity was greater than 98%, as judged by SDS-PAGE. Protein Crystallization—Two different native crystals were grown by the hanging drop method (22). Crystal form #1 was obtained by combining 900 μl of original well solution (3.2 m ammonium sulfate and 0.1 m MES, pH 6.0) with 100 μl of Opti-Salts (Qiagen) optimization solution (0.75 m magnesium chloride and 0.1 m sodium acetate, pH 4.6). Crystal form #2 was obtained using a well solution containing solely 3.2 m ammonium sulfate and 0.1 m MES, pH 6.0. The selenomethionine protein crystal was grown in a sitting drop with the well solution containing 3.2 m ammonium sulfate, 0.1 m MES, pH 5.7. The drop was made with 6 μl of protein (20 mg/ml) and 3 μl of well solution. All crystallizations were at room temperature.
X-ray Crystallography—Diffraction data for the native structure (from crystal #1) were collected using a Rigaku Micro Max 007 rotating anode generator (Rigaku/MSC, The Woodlands, TX). The crystal was cooled to 100 K with a Cryocool low temperature probe (Cryo Industries of America) and was cryoprotected by pulling the loop mounted crystal through immersion oil. The diffraction data were processed with CrystalClear/d*Trek (23). Statistics are shown in Table 1 (crystal #1). Single wavelength anomalous dispersion data for the selenomethionine protein were collected at Brookhaven National Laboratory on beamline X29 using the wavelength 0.9790 Å and processed with HKL2000 (24). When the anomalous data were processed in space group P212121, 15,413 measurements were rejected. With space group P1, only 442 reflections were rejected, and the 33 Å axis was identified as the screw axis. When processed in space group P21, 504 reflections were rejected. The anomalous signal was weak but usable to 2.6 Å. The software program, SHELXC/D/E (25, 26) was used to find eight selenium sites and examine the hand of the sites. A reasonable value for the Matthews coefficient (1.8) indicated that there were two molecules in the asymmetric unit for space group P21 or one for P212121. The selenium sites were used in SOLVE/RESOLVE (27, 28) to produce an electron density map, and the map was examined in COOT (29) and looked good. Using the iterative script, which includes cycles of density modification and automated model building by RESOLVE and molecular refinement by REFMAC5 (30), 271 of the 422 residues were built, and 104 side chains were placed. The model was completed by iteratively building in COOT and refining in REFMAC5. The model was transferred to the native data and refined in both space group P21 and P212121. The model and the statistics were essentially the same. There was no compelling reason to use P21 for the native data so the final statistics for the P212121 space group are shown in Table 2. The final model includes residues 17-208. The side chains of 11 residues were modeled with double conformations, and Cys-66 was modeled as s-oxy cysteine. The sequence has four methionines, but the selenium sites seen were 122M, 145m, and a double conformation for 135M. The loop region including residues 102-113 has minimal density, but the path is clear. In a Ramachandran plot, 97.8% of the residues fall into the preferred regions, and 2.2% fall into the allowed regions as analyzed by COOT.
TABLE 1.
Data statistics table
| Native | SeMet | |
|---|---|---|
| Space group | P212121 | P21 |
| Cell parameters (a, b, c) (Å) | 32.91, 37.67, 141.33 | 37.71, 32.91, 140.88 |
| Cell parameters (α, β, γ) (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Wavelength of data collection (Å) | 1.541 | 0.979 |
| No. of measured intensities | 53,305 | 52,133 |
| No. of unique reflections | 12,558 | 18,011 |
| Resolution of data (Å) | 29.8-2.0 | 30.0-2.1 |
| Highest resolution shell (Å) | 2.07-2.00 | 2.18-2.10 |
| Rsym (overall/high resolution shell) | 0.070/0.308 | 0.059/0.261 |
| Completeness (%) (overall/high resolution shell) | 99.8/97.9 | 87.3/45.6 |
| Redundancy (overall/high resolution shell) | 4.2/4.1 | 5.9/2.4 |
| Mean I/σ (overall/high resolution shell) | 10.9/4.1 | 11.1/2.5 |
TABLE 2.
Refinement statistics table
| Resolution limits (Å) | 20.0-2.0 |
| Number of reflections used/for Rfree | 11,896/600 |
| R-factor (overall/high resolution shell) | 0.211/0.251 |
| Rfree (overall/high resolution shell) | 0.289/0.339 |
| Number of water molecules | 65 |
| r.m.s.d. bond length (Å) | 0.019 |
| r.m.s.d. angle (°) | 1.73 |
| Average B main chain/side chain/water (Å2) | 30.3/31.7/32.4 |
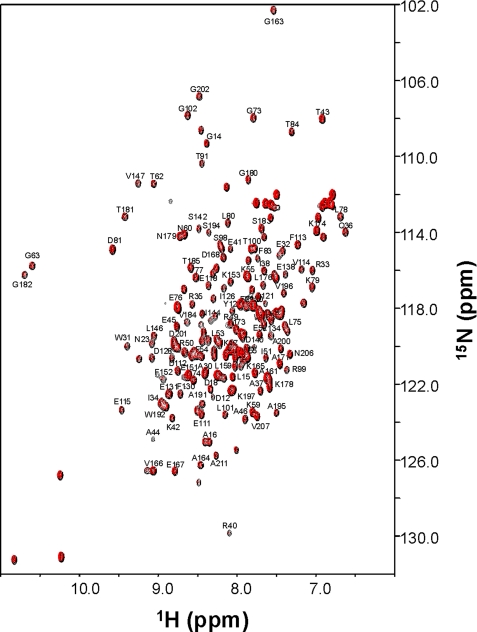
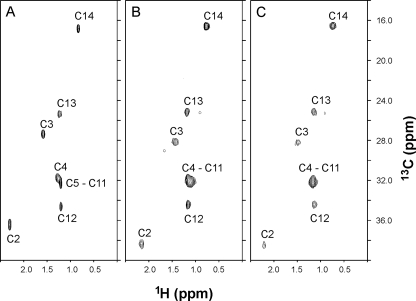
NMR Spectroscopy—Samples for NMR analysis consisted of 15N-labeled FCaBP with and without a covalently attached, 13C-labeled myristoyl group. FCaBP protein for NMR studies (1.0 mm) was dissolved in 0.3 ml of a 95% H2O, 5% [2H]H2O solution containing 10 mm Tris-HCl, pH 7.4, and 2 mm EDTA (Ca2+-free) or 2 mm CaCl2 (Ca2+-bound). All NMR experiments were performed at 25 °C on a Bruker DRX-600 spectrometer equipped with a four-channel interface and triple-resonance probe with triple-axis pulsed field gradients and a DRX-600 spectrometer equipped with an Ultrashield Bruker magnet, a three-channel interface, and a cryo-probe with z axis pulsed field gradients. The 1H,15N HSQC spectra (see Fig. 4) were recorded on a sample of 15N-labeled FCaBP (in 95% H2O, 5% 2H2O). The number of complex points and acquisition times were: 256, 180 ms (15N(F1)); 512, 64 ms (1H(F2)). The 13C,1H HMQC spectra (see Fig. 5) and 13C (F1)-edited, 13C (F3)-filtered NOESY-HMQC spectra were recorded on a sample of unlabeled FCaBP protein attached to 13C-labeled myristate (31) as well as 13C-labeled FCaBP bound to unlabeled myristate (data not shown). Intermolecular NOESY experiments were performed as described previously (32).
FIGURE 4.
NMR spectral analysis of myristoylated and unmyristoylated FCaBP. Two-dimensional (1H,15N HSQC) NMR spectra of 15N-labeled unmyristoylated (black) and myristoylated FCaBP (red) in the Ca2+-free state. Sequence-specific assignments are indicated.
FIGURE 5.
The N-terminal myristoyl group of FCaBP appears to be solvent-exposed. Two-dimensional (1H,13C HMQC) NMR spectra of free myristic acid dissolved in chloroform (A) and the myristoyl group of Ca2+-free (B) and Ca2+-bound (C) FCaBP. The myristoyl group was labeled with carbon-13, and the protein was unlabeled. The peaks in the spectrum represent protons attached to 13C-labeled fatty acyl chain.
15N{1H} NOE data were measured using two-dimensional 15N,1H HSQC-based experiments as described previously (33). Saturation was carried out with a series of 120° 1H pulses separated by 5 ms delays applied during the interscan delay (3 s).
RESULTS AND DISCUSSION
Protein Crystallization—Recombinant FCaBP crystallized in the orthorhombic space group, P212121, with one molecule of FCaBP in the asymmetric unit. It is seen to be monomeric in the crystal, as measured in solution by NMR and dynamic light scattering analysis. The polypeptide structure derived from diffraction data on crystal #1 (in the presence of saturating Mg2+) and crystal #2 (apo-form) appear virtually identical except for very minor differences in the EF2 loop structure (residues, 102-113), suggesting that Mg2+ has almost no effect on the overall structure. In each crystal form, the first 17 residues from the N terminus appear disordered. Amino acid sequencing of the protein from dissolved crystals showed that all 17 N-terminal residues remained intact in both crystal forms and, therefore, were not cleaved by proteolysis during crystallization.
Three-dimensional Structure—The x-ray crystal structure of Ca2+-free FCaBP was solved at 2.2 Å resolution, with a final R-factor of 16.95% and R-free of 21.97% (Fig. 2). The entire polypeptide chain has been traced except for the first 17 residues from the N terminus. High temperature factors for the N-terminal residues suggest they may be dynamically disordered. NMR relaxation studies and heteronuclear 15N nuclear Overhauser effect (NOE) analysis on FCaBP also confirms that the first 17 N-terminal residues are indeed unstructured (data not shown) and the N-terminal myristoyl group is solvent-exposed (see below). The overall main chain structure for residues 17-208 of FCaBP determined by x-ray crystallography was consistent with and nearly identical to a much lower resolution NMR solution structure (not shown).
FIGURE 2.
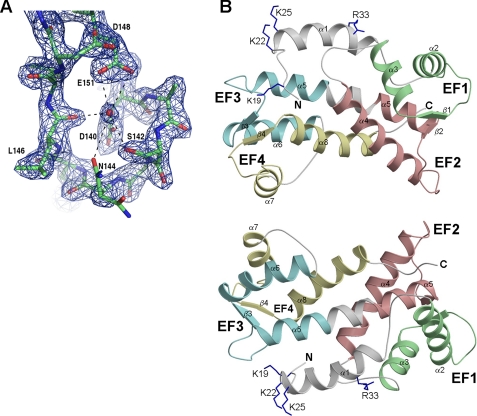
X-ray crystal structure of FCaBP (PDB code 3CS1). A, representative electron density map of FCaBP, showing the Ca2+ binding loop of EF3. A bound water molecule (red sphere) is present in the EF3 Ca2+ binding site of apoFCaBP. B, ribbon diagrams depicting the main chain structure of FCaBP viewed from the membrane binding interface (top) and rotated 180° (bottom). The EF-hands are colored as in Fig. 1.
The protein structure visible in the crystal has a globular fold with overall dimensions of 32 × 23 × 21 Å (Fig. 2). The first structured residue in the crystal begins at Ala-18 that marks the start of an exposed, 5-residue conformation (residues 18-23), resembling a hook that forms a right angle with an adjacent α-helix (residues 24-37). After this N-terminal helix is a compact arrangement of four EF-hands: EF1 (residues 49-77, green); EF2 (residues 98-126, red); EF3 (residues 131-159, cyan); EF4 (residues 168-196, yellow). Overall, FCaBP contains a total of eight α-helices and four β-strands: α1 (residues 24-37), α2 (residues 44-57), α3 (residues 67-76), α4 (residues 87-105), α5 (residues 116-139), α6 (149-162), α7 (residues 169-176), α8 (residues 186-201), β1 (residues 64-66), β2 (residues 113-115), β3 (residues 146-148), and β4 (residues 183-185) (Fig. 1). The four EF-hands of FCaBP associate into two pairs through a characteristic, short, two-stranded β-sheet arrangement, EF1 (green) pairs with EF2 (red), whereas EF3 (cyan) pairs with EF4 (yellow). The two pairs of EF-hands are connected by a long, central helix (α5) formed by merging the exiting helix of EF2 into the entering helix of EF3. This topology causes three long, central helices (α4, α5, and α8) to interact closely as a twisted, vertical bundle. As a consequence, EF2 and EF4 make close contact with one another. This globular arrangement of EF-hands is most similar to that seen for the first four EF-hands in the penta-EF-hand proteins, grancalcin (37) and domain IV of calpain (38). However, unlike penta-EF-hand proteins, FCaBP lacks a fifth EF-hand and is not dimeric.
EF-hands and Ca2+ Binding—The individual metal-free EF-hands in FCaBP each consist of a helix-turn-helix structure similar to the closed conformation of EF-hands seen in previous Ca2+-free structures of calmodulin (39, 40), troponin C (41), and grancalcin (37). The interhelical angles for the “closed” EF-hands in FCaBP are 119° (EF1), 128° (EF2), 122° (EF3), and 120° (EF4). The binding of Ca2+ to each EF-hand in calmodulin causes a marked decrease in interhelical angle that promotes formation of an open conformation, leading to the exposure of many hydrophobic residues that interact with protein targets (42, 43). Functional Ca2+ binding to EF3 and EF4 in FCaBP (4) may cause a similar Ca2+-induced exposure of hydrophobic residues and might explain the observed Ca2+-induced binding of FCaBP to protein phosphatase 2A4 and various other protein targets (4). Indeed, Ca2+-dependent membrane binding by FCaBP might arise in part by its Ca2+-induced binding to protein targets localized on the flagellar membrane surface.
The 12-residue Ca2+ binding loop of EF2 in FCaBP appears loosely structured in crystal form #1 and adopts an unusual, distorted conformation in crystal form #2, which may explain why EF2 is not able to bind Ca2+ (4). The instability and/or structural distortion of the EF2 loop may be due in part to the presence of G109 at position-3 in the binding loop, which lacks an acidic side chain required for Ca2+ chelation at this key position. The lack of Ca2+ binding at EF2 may also explain why Ca2+ is not able to bind to EF1, as the helices of EF1 and EF2 interlock to form a four-helix bundle, causing both EF-hands to be cooperatively locked in the Ca2+-free closed conformation. In addition, Cys-66 at position 9 in the EF1 loop is not able to form a hydrogen bond with Glu-69, which may destabilize the binding-loop structure and prevent Ca2+ binding at EF1.
By contrast, EF3 and EF4 both adopt a more favorable conformation for binding Ca2+, consistent with functional Ca2+ binding measured at these sites (4). In the Ca2+ binding loop of EF3, side-chain oxygen atoms from residues Asp-140, Ser-142, Asn-144, and Glu-151 are held in a cage-like arrangement surrounding a bound H2O molecule (Fig. 2A) that resembles the binding of Ca2+ with the familiar pentagonal bipyramid geometry (3). For EF4, side-chain oxygen atoms from residues Asp-177, Asn-179, Thr-181, and Glu-188 are also pre-arranged with a similar geometry that surrounds a bound H2O. In addition, the main-chain nitrogen of the sixth residue in the Ca2+ binding loops of EF3 (M145) and EF4 (G182) both form hydrogen bonds with aspartate at the one position, forming a loop conformation characteristic of Ca2+-bound calmodulin and troponin C. Therefore, the Ca2+ binding loops in EF3 and EF4 both adopt a favorable, preformed local conformation that should promote rapid and functional Ca2+ binding.
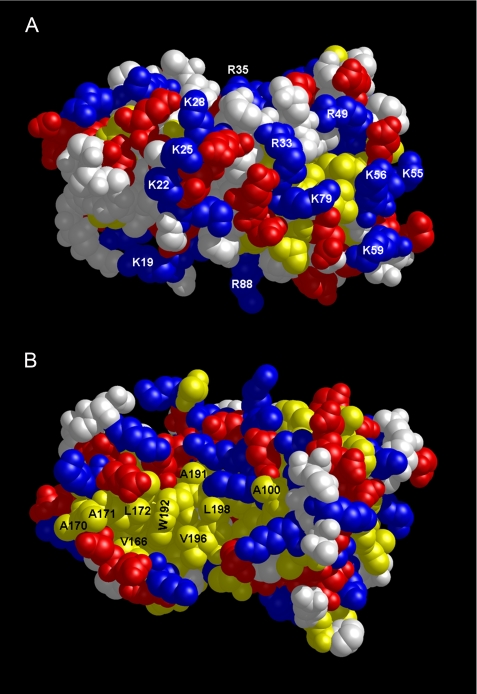
Surface Properties of FCaBP—A space-filling representation of FCaBP reveals multiple charged residues in FCaBP unevenly distributed on one side of the protein surface (Fig. 3A) with an exposed hydrophobic patch on the opposite side (Fig. 3B). Most striking is the arrangement of N-terminal Arg and Lys residues (Lys-19, -22, -25, -28 and Arg-33 and -35) whose side chains form a cluster of positive charge on the protein surface (highlighted in blue in Fig. 3A). Some of these N-terminal basic residues (Lys-19, Lys-22, and Lys-25) have been implicated previously in membrane binding.4 We propose that the exposed patch of N-terminal Arg and Lys residues may promote electrostatic interactions with negatively charged head groups on the surface of the flagellar membrane. A similar electrostatic membrane interaction has also been proposed for the myristoyl switch proteins, recoverin (44) and the Src protein (45). The additional electrostatic attraction helps augment membrane anchoring by the N-terminal myristoyl group and can increase the membrane binding affinity by more than 10-fold (45, 46).
FIGURE 3.
Surface representation of FCaBP protein structure. Space-filling models of FCaBP depicting the positively charged, membrane-binding interface (A) and exposed hydrophobic surface (B). The orientations are similar to those in Fig. 2B. Exposed hydrophobic, basic, and acidic residues are highlighted in yellow, blue, and red, respectively.
The protein surface on the opposite face (Fig. 3B) contains an exposed patch of hydrophobic residues near the C terminus (Ala-170, Ala-171, Leu-172, Ala-176, Ala-191, Trp-192, Ala-195, Val-196, Ala-198, and Ala-200, highlighted in yellow in Fig. 3B). The exposed hydrophobic residues are primarily located on the helices of EF4 (α7 and α8). Ca2+-induced conformational changes expected in EF4 therefore might alter the arrangement and environment of these hydrophobic residues. We propose that the exposed hydrophobic patch in Fig. 3B may represent a binding site for the interaction with membrane-bound protein targets.
NMR Analysis of Myristoylation—The absence of detectable electron density from the first 17 residues of FCaBP suggested that the N-terminal region and myristoyl group may be structurally disordered. To more directly examine the structural role of the myristoyl group, we recorded two-dimensional (1H,15N HSQC) NMR spectra of both myristoylated and unmyristoylated FCaBP, demonstrating that both spectra are nearly identical (Fig. 4). Hence, the presence of the myristoyl group does not appear to affect the overall structure of Ca2+-free FCaBP, in contrast to the large structural changes in Ca2+-free recoverin that result from N-terminal myristoylation (17, 31). The similarity in the spectra of FCaBP and myr-FCaBP suggests that the fatty acyl chain may be exposed to solvent and, hence, not interacting significantly with the protein.
NMR experiments were also performed on FCaBP containing a 13C-labeled myristoyl group to directly probe the structural environment and disposition of the N-terminal myristoyl group. Previously, two-dimensional (13C,1H HMQC) and three-dimensional (13C-filtered NOESY-HMQC) NMR experiments on samples of recoverin that contained a 13C-labeled myristoyl group were used to selectively probe the chemical environment around the N-terminal myristoyl group (31, 34). These studies revealed that the covalently attached fatty acyl chain in recoverin is sequestered in a hydrophobic pocket in the Ca2+-free protein and that binding of Ca2+ leads to conformational changes that extrude the N-myristoyl group into solvent.
Similar NMR experiments were performed on samples of FCaBP that contained a 13C-labeled myristoyl group (Fig. 5). Because the HMQC experiment selectively probes protons that are covalently attached to 13C, only the methylene and methyl proton resonances of the fatty acyl chain appear in these spectra. Weak extraneous peaks near 0.9, 1.4, and 1.7 ppm (1H dimension) are due to natural abundance 13C signals from the protein. The spectrum of the N-myristoyl group in Ca2+-free FCaBP (Fig. 5B) looks quite similar to the spectrum of free myristic acid in solution (Fig. 5A), and the resonance frequencies of corresponding peaks are nearly identical. Assignments of the myristoyl group resonances for Ca2+-free FCaBP (Fig. 5B) were, therefore, derived from assignments of those for free myristic acid, which were determined previously (31). The similarity in the spectrum of the myristoyl group in Ca2+-free FCaBP to that of free myristic acid in solution indicates that in Ca2+-free FCaBP the fatty acyl chain is solvent-exposed, in contrast to Ca2+-free recoverin where the myristoyl chain is deeply buried inside the core of the protein (9).
To further test whether the myristoyl group of FCaBP is solvent-exposed, three-dimensional (13C/F1)-edited and (13C/F3)-filtered NOESY experiments (31) were performed on unlabeled FCaBP protein containing a 13C-labeled myristate. These spectra selectively probed atoms of residues in the protein that lie within 5 Å of the labeled CH3 group of the myristoyl chain. NOE dipolar interactions between the myristate methyl group and the protein could not be detected (data not shown). The lack of observable NOEs in this experiment suggests that the methyl group of the fatty acyl chain is more than 5 Å away from atoms in the protein, providing additional support for the conclusion that the N-myristoyl group of FCaBP is solvent-exposed and does not interact intimately with the protein.
The addition of Ca2+ to myr-FCaBP caused no discernable change in the NMR spectrum of the myristoyl group (Fig. 5C). In addition, NOE interactions between the myristate and protein could not be detected (data not shown) in three-dimensional (13C/F1)-edited and (13C/F3)-filtered NOESY spectra of Ca2+-bound [3C]Myr-FCaBP, suggesting that the methyl group of the fatty acyl chain does not interact closely with the Ca2+-bound protein. Hence, the myristoyl group of Ca2+-bound FCaBP appears solvent-exposed and does not undergo a calcium-induced change in environment, in contrast to what has been observed previously in recoverin (31).
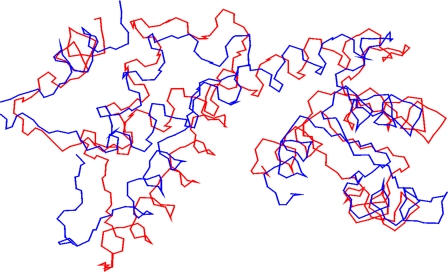
Structural Relationship with Penta EF-hand (PEF) Proteins—The N-terminal myristoylation and four EF-hands in FCaBP would suggest that it might be structurally similar to recoverin (9, 47) and related Ca2+-myristoyl switch proteins (48, 49). However, the overall three-dimensional structures of FCaBP and recoverin are surprisingly quite different and unrelated. Instead, the overall main chain fold of Ca2+-free FCaBP is most similar to that of apograncalcin (37) and related proteins that belong to the PEF family (50). Interestingly, FCaBP bears very little sequence homology to the PEF proteins (<20% identity). The root mean square deviation (r.m.s.d.) was 3.5 Å when comparing the main chain atoms of the four EF-hands of FCaBP with those of the first four EF-hands of grancalcin (Fig. 6). The structural similarity is even more striking when comparing the main chain atoms from EF1, EF3, and EF4 (r.m.s.d. = 2.9 Å), whereas the structure of EF2 was much more divergent between the two (r.m.s.d. = 4.5 Å). The second helix of EF2 and first helix of EF3 in FCaBP are merged into a long, 23-residue helix (α5) that is flanked in an antiparallel fashion by helices α4 and α8, forming a twisted bundle. A similar helical bundle and overall topology is also seen in grancalcin and seems to be a structural hallmark of all PEF proteins. The similarity of the main chain conformations between FCaBP and PEF proteins suggests that their functions might be related. Indeed, grancalcin and PEF proteins exhibit Ca2+-induced membrane binding (51-53) and interact with target proteins such as L-plastin (54), annexins (55), and integrins (56) implicated in cytoskeletal dynamics. FCaBP also binds to membranes and has been shown recently to interact with several candidate target proteins (4). Future studies are needed to determine whether any of the FCaBP target proteins might be related to L-plastin or other PEF protein targets that regulate cytoskeletal adhesion and migration.
FIGURE 6.
Structural comparison of FCaBP with penta EF-hand proteins. The main chain atoms of the EF-hand regions of FCaBP (red, PDB code 3CS1) and first four EF-hands of apograncalcin (blue, PDB code 1F4Q) are superimposed (3.5 Å r.m.s.d.).
An important structural difference between FCaBP and PEF proteins is that FCaBP is monomeric in solution and does not have a fifth EF-hand like the dimeric PEF proteins. The fifth EF-hand of FCaBP may have been deleted during evolution perhaps to allow for the exposure of hydrophobic residues in EF4 (Fig. 3B) that otherwise would be inaccessible if the fifth EF-hand were present. Another difference is the presence of the lipophilic N-terminal extension region in PEFs that have variable Gly-Pro-Ala sequences implicated in membrane targeting (57). By contrast, FCaBP has conserved basic residues (Lys-19, Lys-22, Lys-25) in the N-terminal region that we suggest might be important for membrane binding in addition to N-terminal myristoylation and palmitoylation sites (Figs. 1 and 7). We propose that the four EF-hands of FCaBPs are evolutionarily related to the first four EF-hands of PEF proteins and that FCaBPs may represent a sub-branch of the PEF family that lacks a fifth EF-hand and have N-terminal myristoylation and palmitoylation membrane-targeting motifs instead. It will be interesting to look for other examples of truncated PEF proteins that are structurally related to FCaBP and perhaps serve as myristoylated, membrane-targeting calcium sensors.
FIGURE 7.
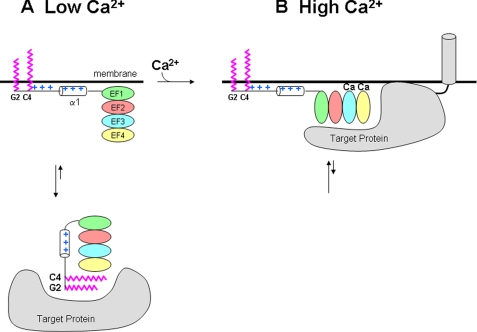
Model of membrane-target recognition by FCaBP. Schematic diagram of FCaBP binding to membrane and protein targets at low (A) and high (B) Ca2+ levels. Myristoyl and palmitoyl groups highlighted in magenta; EF-hands are colored as in Fig. 1; N-terminal Arg and Lys residues are colored blue, and cytosolic and membrane-bound target proteins shown in gray.
Membrane-targeting Mechanism—The crystal structure of FCaBP (Fig. 2) has implications for its Ca2+-induced membrane-targeting mechanism (Fig. 7). The solvent-exposed myristoylation (and presumably palmitoylation (5)) sites at the N terminus suggest that the covalently attached fatty acyl groups serve as membrane anchors that insert inside the lipid bilayer. In addition, exposed basic residues in the N-terminal region (Fig. 3A) are suggested to provide additional binding energy by interacting electrostatically with negatively charged head groups on the membrane surface. A similar combination of electrostatic effects and myristoylation was seen previously for membrane binding by the myristoyl-electrostatic switch of Src and MARCKS (myristoylated alanine-rich C kinase substrate) protein (45).
Our NMR analyses (Figs. 4 and 5) indicate that the fatty acyl group is exposed regardless of Ca2+ concentration, suggesting that both the Ca2+-free and Ca2+-bound forms of FCaBP may be anchored to the flagellar membrane (Fig. 7). But how then is membrane binding by FCaBP regulated by calcium? We propose that the Ca2+ sensitive localization of FCaBP at the flagellar membrane could be controlled by Ca2+-dependent binding of FCaBP to various target proteins. Previous studies have identified candidate target proteins that differentially bind to either the Ca2+-free or Ca2+-bound forms of FCaBP (4). The binding of Ca2+ to calmodulin and other EF-hand proteins generally leads to the exposure of hydrophobic residues that promotes their binding to protein targets (42, 43). A similar Ca2+-dependent exposure of hydrophobic residues in EF4 (Fig. 3B) might promote association of Ca2+-bound FCaBP with a membrane-bound target protein and preferentially localize FCaBP at the flagellar membrane at high Ca2+ levels (Fig. 7B). Alternatively, the Ca2+-free state of FCaBP might preferentially bind to a cytosolic target protein that would localize FCaBP in the cytosol at low Ca2+ levels (Fig. 7A). The binding of a cytosolic target protein to Ca2+-free FCaBP might help to sequester the N-terminal fatty acyl groups inside a protein environment (perhaps involving exposed hydrophobic residues in EF4 (Fig. 3B)) like that seen for Ca2+-free recoverin (17) or GCAP1 (18). Sequestration of the myristoyl group would stabilize cytosolic FCaBP and, therefore, promote its membrane dissociation only at low Ca2+ levels. Last, reversible cleavage of the labile palmitoyl group attached to FCaBP (by a thioester linkage) may further modulate its membrane anchoring capacity. The enzyme-catalyzed cleavage of palmitate may also be regulated by Ca2+.
In short, both the Ca2+-free and Ca2+-bound FCaBP have exposed myristoyl groups (Fig. 5) that we propose anchor the protein to the flagellar membrane. Thus, FCaBP does NOT exhibit Ca2+-induced extrusion of the myristoyl group to control membrane localization like what is seen for recoverin (6) and related Ca2+-myristoyl switch proteins (58). Instead, we propose that Ca2+-induced protein conformational changes in FCaBP (4) may modulate its interaction with target proteins that control membrane localization as depicted in Fig. 7. Future structural studies on Ca2+-bound FCaBP bound to various target proteins are needed to further test and refine the proposed membrane-targeting mechanism.
Acknowledgments
We thank Jeff de Ropp for help with NMR experiments, Dr. Frits Abildgaard for providing NMR pulse-sequence programs, John Cieslak for producing the E. coli strains coexpressing FCaBP and yeast N-myristoyltransferase, and Frank Delaglio and Dan Garrett for writing computer software for NMR data processing and analysis.
The atomic coordinates and structure factors (code 3CS1) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grants EY012347 and NS045909 (to J. B. A.), RR11973 (to the University of California Davis NMR Facility), and AI46781 (to D. M. E.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: FCaBP, flagellar calcium-binding protein; HSQC, heteronuclear single quantum coherence; HMQC, heteronuclear multiple quantum coherence; NOE, nuclear Overhauser effect; NOESY, NOE spectroscopy; r.m.s.d., root-mean-squared deviation; MES, 4-morpholineethanesulfonic acid; PEF, penta EF-hand.
D. Engman, unpublished results.
References
- 1.Engman, D. M., Krause, K. H., Blumin, J. H., Kim, K. S., Kirchhoff, L. V., and Donelson, J. E. (1989) J. Biol. Chem. 264 18627-18631 [PubMed] [Google Scholar]
- 2.Moncrief, N. D., Kretsinger, R. H., and Goodman, M. (1990) J. Mol. Evol. 30 522-562 [DOI] [PubMed] [Google Scholar]
- 3.Ikura, M. (1996) Trends Biochem. Sci. 21 14-17 [PubMed] [Google Scholar]
- 4.Buchanan, K. T., Ames, J. B., Asfaw, S. H., Wingard, J. N., Olson, C. L., Campana, P. T., Araujo, A. P., and Engman, D. M. (2005) J. Biol. Chem. 280 40104-40111 [DOI] [PubMed] [Google Scholar]
- 5.Godsel, L. M., and Engman, D. M. (1999) EMBO J. 18 2057-2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zozulya, S., and Stryer, L. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 11569-11573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dizhoor, A. M., Chen, C. K., Olshevskaya, E., Sinelnikova, V. V., Phillipov, P., and Hurley, J. B. (1993) Science 259 829-832 [DOI] [PubMed] [Google Scholar]
- 8.Burgoyne, R. D. (2004) Biochim. Biophys. Acta 1742 59-68 [DOI] [PubMed] [Google Scholar]
- 9.Ames, J. B., Ishima, R., Tanaka, T., Gordon, J. I., Stryer, L., and Ikura, M. (1997) Nature 389 198-202 [DOI] [PubMed] [Google Scholar]
- 10.Kawamura, S. (1993) Nature 362 855-857 [DOI] [PubMed] [Google Scholar]
- 11.Klenchin, V. A., Calvert, P. D., and Bownds, M. D. (1995) J. Biol. Chem. 270 16147-16152 [DOI] [PubMed] [Google Scholar]
- 12.Erickson, M. A., Lagnado, L., Zozulya, S., Neubert, T. A., Stryer, L., and Baylor, D. A. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 6474-6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray-Keller, M. P., Polans, A. S., Palczewski, K., and Detwiler, P. B. (1993) Neuron 10 523-531 [DOI] [PubMed] [Google Scholar]
- 14.Koch, K. W., and Stryer, L. (1988) Nature 334 64-66 [DOI] [PubMed] [Google Scholar]
- 15.Ridgley, E., Webster, P., Patton, C., and Ruben, L. (2000) Mol. Biochem. Parasitol. 109 195-201 [DOI] [PubMed] [Google Scholar]
- 16.Tetley, L. (1986) Acta Trop. 43 307-317 [PubMed] [Google Scholar]
- 17.Tanaka, T., Ames, J. B., Harvey, T. S., Stryer, L., and Ikura, M. (1995) Nature 376 444-447 [DOI] [PubMed] [Google Scholar]
- 18.Stephen, R., Bereta, G., Golczak, M., Palczewski, K., and Sousa, M. C. (2007) Structure 15 1392-1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muchmore, D. C., McIntosh, L. P., Russell, C. B., Anderson, D. E., and Dahlquist, F. W. (1989) Methods Enzymol. 177 44-86 [DOI] [PubMed] [Google Scholar]
- 20.McIntosh, L. P., and Dahlquist, F. W. (1990) Q. Rev. Biophys. 23 1-38 [DOI] [PubMed] [Google Scholar]
- 21.Ames, J. B., Tanaka, T., Stryer, L., and Ikura, M. (1994) Biochemistry 33 10743-10753 [DOI] [PubMed] [Google Scholar]
- 22.Jancarik, J., and Kim, S. H. (1991) J. Appl. Crystallogr. 24 409-411 [Google Scholar]
- 23.Pflugrath, J. W. (1999) Acta Crystallogr. Sect. D Biol. Crystallogr. 55 1718-1725 [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 25.Schneider, T. R., and Sheldrick, G. M. (2002) Acta Crystallogr. Sect. D Biol. Crystallogr. 58 1772-1779 [DOI] [PubMed] [Google Scholar]
- 26.Sheldrick, G. M. (2002) Kristallogr. 217 644-650 [Google Scholar]
- 27.Terwilliger, T. C. (2003) Acta Crystallogr. Sect. D Biol. Crystallogr. 59 38-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terwilliger, T. C. (2003) Acta Crystallogr. Sect. D Biol. Crystallogr. 59 45-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 30.Murshudov, D. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 140-255 [DOI] [PubMed] [Google Scholar]
- 31.Ames, J. B., Tanaka, T., Ikura, M., and Stryer, L. (1995) J. Biol. Chem. 270 30909-30913 [DOI] [PubMed] [Google Scholar]
- 32.Lee, W., Revington, M. J., Arrowsmith, C., and Kay, L. E. (1994) FEBS Lett. 350 87-90 [DOI] [PubMed] [Google Scholar]
- 33.Farrow, N. A., Muhandiram, R., Singer, A. U., Pascal, S. M., Kay, C. M., Gish, G., Shoelson, S. E., Pawson, T., and Kay, L. E. (1994) Biochemistry 33 5984-6003 [DOI] [PubMed] [Google Scholar]
- 34.Tanaka, T., Ames, J. B., Kainosho, M., Stryer, L., and Ikura, M. (1998) J. Biomol. NMR 11 135-152 [DOI] [PubMed] [Google Scholar]
- 35.Deleted in proof
- 36.Deleted in proof
- 37.Jia, J., Han, Q., Borregaard, N., Lollike, K., and Cygler, M. (2000) J. Mol. Biol. 300 1271-1281 [DOI] [PubMed] [Google Scholar]
- 38.Blanchard, H., Grochulski, P., Li, Y., Arthur, J. S., Davies, P. L., Elce, J. S., and Cygler, M. (1997) Nat. Struct. Mol. Biol. 4 532-538 [DOI] [PubMed] [Google Scholar]
- 39.Schumacher, M. A., Crum, M., and Miller, M. C. (2004) Structure 12 849-860 [DOI] [PubMed] [Google Scholar]
- 40.Zhang, M., Tanaka, T., and Ikura, M. (1995) Nat. Struct. Biol. 2 758-767 [DOI] [PubMed] [Google Scholar]
- 41.Gagne, S. M., Tsuda, S., Li, M. X., Smillie, L. B., and Sykes, B. D. (1995) Nat. Struct. Biol. 2 784-789 [DOI] [PubMed] [Google Scholar]
- 42.Hoeflich, K. P., and Ikura, M. (2002) Cell 108 739-742 [DOI] [PubMed] [Google Scholar]
- 43.Vetter, S. W., and Leclerc, E. (2003) Eur. J. Biochem. 270 404-414 [DOI] [PubMed] [Google Scholar]
- 44.Valentine, K., Mesleh, M., Ikura, M., Ames, J. B., and Opella, S. (2003) Biochemistry 42 6333-6340 [DOI] [PubMed] [Google Scholar]
- 45.McLaughlin, S., and Aderem, A. (1995) Trends Biochem. Sci. 20 272-276 [DOI] [PubMed] [Google Scholar]
- 46.Resh, M. D. (1994) Cell 76 411-413 [DOI] [PubMed] [Google Scholar]
- 47.Flaherty, K. M., Zozulya, S., Stryer, L., and McKay, D. B. (1993) Cell 75 709-716 [DOI] [PubMed] [Google Scholar]
- 48.Bourne, Y., Dannenberg, J., Pollmann, V. V., Marchot, P., and Pongs, O. (2001) J. Biol. Chem. 276 11949-11955 [DOI] [PubMed] [Google Scholar]
- 49.Vijay-Kumar, S., and Kumar, V. D. (1999) Nat. Struct. Biol. 6 80-88 [DOI] [PubMed] [Google Scholar]
- 50.Maki, M., Kitaura, Y., Satoh, H., Ohkouchi, S., and Shibata, H. (2002) Biochim. Biophys. Acta 1600 51-60 [DOI] [PubMed] [Google Scholar]
- 51.Teahan, C. G., Totty, N. F., and Segal, A. W. (1992) Biochem. J. 286 549-554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meyers, M. B., Zamparelli, C., Verzili, D., Dicker, A. P., Blanck, T. J., and Chiancone, E. (1995) FEBS Lett. 357 230-234 [DOI] [PubMed] [Google Scholar]
- 53.Mellgren, R. L. (1987) FASEB J. 1 110-115 [DOI] [PubMed] [Google Scholar]
- 54.Lollike, K., Johnsen, A. H., Durussel, I., Borregaard, N., and Cox, J. A. (2001) J. Biol. Chem. 276 17762-17769 [DOI] [PubMed] [Google Scholar]
- 55.Brownawell, A. M., and Creutz, C. E. (1997) J. Biol. Chem. 272 22182-22190 [DOI] [PubMed] [Google Scholar]
- 56.Huttenlocher, A., Palecek, S. P., Lu, Q., Zhang, W., Mellgren, R. L., Lauffenburger, D. A., Ginsberg, M. H., and Horwitz, A. F. (1997) J. Biol. Chem. 272 32719-32722 [DOI] [PubMed] [Google Scholar]
- 57.Maki, M., Narayana, S. V., and Hitomi, K. (1997) Biochem. J. 328 718-720 [PMC free article] [PubMed] [Google Scholar]
- 58.Burgoyne, R. D., and Weiss, J. L. (2001) Biochem. J. 353 1-12 [PMC free article] [PubMed] [Google Scholar]
- 59.Wishart, D. S., Sykes, B. D., and Richards, F. M. (1992) Biochemistry 31 1647-1651 [DOI] [PubMed] [Google Scholar]