Abstract
Formation and degradation of SsrA-tagged proteins enable ribosome recycling and elimination of defective products of incomplete translation. We produced an antibody against the SsrA peptide and used it to measure the amounts of SsrA-tagged proteins in Escherichia coli cells without interfering with tagging or altering the context of the tag added at the ends of nascent polypeptides. SsrA-tagged proteins were present in very small amounts unless a component of the ClpXP protease was missing. From the levels of tagged proteins in cells in which degradation is essentially blocked, we calculate that ≥1 in 200 translation products receives an SsrA tag. ClpXP is responsible for ≥90% of the degradation of SsrA-tagged proteins. The degradation rate in wild type cells is ≥1.4 min–1 and decreases to ∼0.10 min–1 in a clpX mutant. The rate of degradation by ClpXP is decreased ∼3-fold in mutants lacking the adaptor SspB, whereas degradation by ClpAP is increased 3–5-fold. However, ClpAP degrades SsrA-tagged proteins slowly even in the absence of SspB, possibly because of interference from ClpA-specific substrates. Lon protease degrades SsrA-tagged proteins at a rate of ∼0.05 min–1 in the presence or absence of SspB. We conclude that ClpXP, together with SspB, is uniquely adapted for degradation of SsrA-tagged proteins and is responsible for the major part of their degradation in vivo.
Recycling of ribosomes at the completion of translation is a regulated process requiring specific interaction of release factors with stop codons (1). When protein translation does not proceed to completion or terminate properly, one alternative mechanism used to disassemble ribosomal complexes and to degrade defective or incomplete polypeptides employs the trans-translation system (2, 3), the centerpiece of which is the transfer mRNA (tmRNA) (also known as 10 S RNA or SsrA RNA), the product of the ssrA or sipB gene (4, 5). tmRNA is a small, stable RNA containing an alanyl-tRNA domain and an mRNA domain encoding an open reading frame of 10 amino acids followed by a stop codon (6). tmRNA, together with the RNA chaperone, SmpB, is recruited to stalled ribosomes (7), where the charged alanyl-tRNA domain enters the A-site. The nascent chain is transferred to the Ala-tmRNA, and the mRNA domain takes the place of the previous message allowing the short open reading frame to be translated (8, 9) followed by termination at the stop codon. In the process, the polypeptide is extended by 11 amino acids, AANDENYALAA (6, 10, 11). Proteins with this C-terminal extension are referred to as SsrA-tagged proteins. SsrA-tagged proteins are recognized by ClpXP and other proteases and rapidly degraded in vivo (12, 13). Thus, the SsrA-tagging serves to rescue ribosomes from stalled translation complexes and to remove incomplete polypeptides from the cell (3, 9).
Translating ribosomes can stall at the ends of mRNAs that are missing in-frame stop codons, which can occur with partially degraded or incompletely synthesized mRNAs (3, 12), or at the ends of normal mRNAs, when the ribosome reads through a stop codon, as occurs in the presence of suppressor tRNAs (14). Stalling can also occur when two or more in-frame rare codons appear in succession (15) or when rare codons closely precede a stop codon (16, 17). Other conditions, such as loss of fidelity of translation in the presence of antibiotics, also cause translational pausing, recruitment of tmRNA, and production of SsrA-tagged proteins (18–20). Some full-length or nearly full-length proteins acquire SsrA tags. Designed translational pausing near the end of the open reading frame of the lactose repressor, LacI, or the phage Mu repressor, MuC, results in a fraction of the repressor proteins acquiring an SsrA tag (21, 22). SsrA tagging of LacI affects the efficiency of repression and the timing of induction of the lac operon (21). Mu repressor variants that acquire an SsrA tag near their C terminus have weaker DNA binding properties and interact with wild type repressor targeting it for degradation by ClpXP (22–24).
The extensive production of SsrA-tagged proteins was shown by Moore and Sauer (25) using a tmRNA mutated to replace a portion of the SsrA tag with a polyhistidine, which results in tagged proteins that are not degraded by ClpXP. They estimated that 0.4% of translation initiations end with SsrA tagging. Two-dimensional gel analysis of the tagged proteins purified by metal chelate chromatography revealed a heterogeneous mixture of proteins, implying that some degree of SsrA tag incorporation occurs during translation of many proteins (26). Recovery of peptides very close to the C terminus of five proteins that were identified suggested that the SsrA tag had been added after translation was complete or nearly complete (26), consistent with a mechanism favoring recruitment of tmRNA at the end of translation, perhaps occasioned by delays in ribosome release (27).
ClpXP degrades model SsrA-tagged proteins rapidly both in vivo and in vitro (28–31). ClpXP can directly degrade SsrA-tagged proteins in vitro, but the adaptor protein SspB, which recognizes a different portion of the SsrA tag from that recognized by ClpX, contributes to degradation by delivering the tagged proteins to ClpX. SspB lowers the Michaelis constant for SsrA-tagged proteins (32, 33), and thus has its greatest activating effect at subsaturating concentrations of tagged proteins. SspB is especially effective in targeting proteins bearing mutant SsrA tags that are not well recognized by ClpX alone or tags in which the portions recognized by SspB and ClpX are separated, allowing both components to bind the tag efficiently (34). A possible implication of this result is that SspB is also involved in targeting Escherichia coli proteins that bear SsrA-like tags; the contribution of SspB to degradation of SsrA-tagged proteins might vary depending on the abundance of competing substrates and of the substrates themselves. ClpAP also degrades SsrA-tagged proteins in vitro but appears to have lower activity against these proteins in vivo (12, 35–37). One possibility for the lower activity of ClpA in vivo is competition from SspB (36). SspB and ClpA bind overlapping portions of the SsrA tag and compete with each other in vitro (30). In addition, Lon protease has also been shown to degrade SsrA-tagged proteins (37), suggesting that multiple protease systems can contribute to degradation of these proteins, perhaps dependent on their abundance and the presence of competing substrates.
To gain more insight into the number of proteins tagged with SsrA and to determine the relative contributions of the different ATP-dependent proteases to their degradation, we produced an antibody that specifically recognizes the wild type SsrA tag and used it to measure the accumulation and degradation of the bulk population of SsrA-tagged proteins in E. coli cells. We have found that SsrA tags are incorporated into ∼0.5% proteins during translation, and all tagged proteins are degraded by ClpXP, ClpAP, and Lon. ClpXP is responsible for ≥90% of the degradation of SsrA-tagged proteins, whereas ClpAP and Lon contribute about 5 and 2% to the degradation.
EXPERIMENTAL PROCEDURES
Antibody Production and Characterization—Details of the production of anti-SsrA antibody, quantitation of SsrA-tagged proteins, and details of strain and plasmid construction are provided in the supplemental material.
Cell Growth and Sample Preparation—Samples of overnight cultures grown at 37 °C were diluted 1 to 1000 in LB in a 125-ml Erlenmeyer flask. The culture was incubated at 37 °C with shaking at 250 rpm. At times during the growth, with cell densities between A600 0.1 and 4.0, samples were withdrawn, and 100% (w/v) trichloroacetic acid was added to give a final concentration of 5% trichloroacetic acid. After incubation on ice for 30 min, precipitated protein was collected by centrifugation (14,000 rpm, 5 min, 4 °C), washed three times with cold acetone, and dried in air. SDS-PAGE sample buffer containing 5% (v/v) β-mercaptoethanol was added to the dried samples to give a protein concentration of 0.7–1.4 mg/ml. After heating at ∼99 °C for 5 min, the samples were centrifuged in a bench top centrifuge for 5 min to remove insoluble debris. To confirm uniform recovery of cell proteins, 10-μl aliquots of the solubilized protein were separated by SDS-PAGE and stained with Coomassie Blue.
Western Blot Analysis—Samples containing ∼10 μg of protein in SDS sample buffer were loaded into lanes of a 10% BisTris2 NuPAGE gel (Invitrogen) in MES running buffer. A control lane was loaded with 1–2 ng of GFP-SsrA. After electrophoresis, proteins were transferred to 0.2-μm nitrocellulose membranes in Tris-glycine buffer containing 10% (v/v) methanol. The membrane was blocked for 1 h in 25 mm Tris/HCl, pH 7.5, 0.1 m NaCl, and 0.025% (v/v) Tween 20 (TBS-T) containing 5% (w/v) nonfat dried milk. Membranes were transferred to fresh blocking buffer containing an appropriate dilution of anti-SsrA IgG (1/500 to 1/3000, depending on the preparation) and incubated at room temperature with gentle shaking for 1 h. The membrane was washed three times at room temperature with blocking buffer without milk, after which it was treated with blocking buffer containing a 3000-fold dilution of donkey anti-rabbit IgG conjugated to horseradish peroxidase (GE Healthcare). After shaking for 1 h at room temperature, the membrane was washed as above, and the ECL plus detection kit (GE Healthcare) was used to visualize the cross-reactive bands. Membranes were sometimes blocked overnight at 4 °C, and immunochemical detection was conducted the next day. On occasion blocked membranes were stored wet at –20 °C and blocked again at room temperature prior to immunochemical detection.
Protein Degradation Rates Measured by Antibiotic Chase—Overnight cultures were inoculated into fresh LB and grown as described above. At mid-exponential phase (A600 ∼ 0.5), spectinomycin was added to a final concentration of 200 μg/ml from a freshly prepared stock solution. Samples (0.2 ml) were removed between 0 and 60 min, treated with trichloroacetic acid, and analyzed by SDS-PAGE and Western blotting as described above. Control samples were removed just prior to addition of antibiotic and treated along with test samples. Optical density readings were used to monitor cell growth throughout the chase period. To measure protein degradation in stationary phase cells, chloramphenicol (100 μg/ml) was added to cells at A600 ∼ 1.0, and samples were taken at 1-h intervals between 0 and 4 h and treated as above.
RESULTS
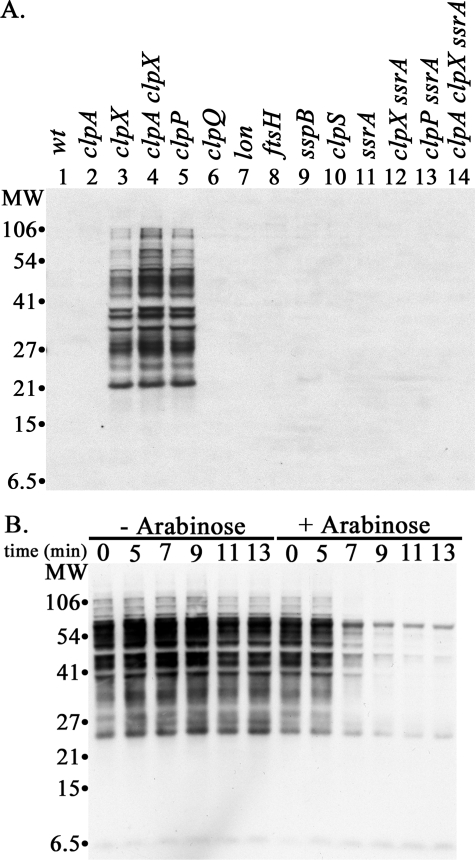
Detection of SsrA-tagged Proteins in E. coli Cells Using Anti-SsrA Antibody—SsrA-tagged proteins in E. coli cell extracts were monitored by SDS-PAGE and Western blotting using affinity-purified anti-SsrA antibody. No specific bands were detected in lanes containing 20 μg of protein from wild type cells (Fig. 1A, lane 1). In contrast, a large number of protein bands, ranging in size from 20 to 200 kDa, were readily detected by the antibody in clpX or clpP mutant strains (Fig. 1A, lanes 3–5). These protein bands were not seen in mutant strains that also carried null mutations in ssrA (Fig. 1A, lanes 11–14, and Fig. 5B, below) confirming that they require the presence of a functional ssrA gene. We refer to these bands as SsrA-tagged proteins throughout the paper. In some blots with high protein loadings, bands of ∼7 and ∼80 kDa were observed; the levels of these proteins did not change in any of the protease mutants and were present in ssrA mutants, indicating they are not SsrA-tagged proteins.
FIGURE 1.
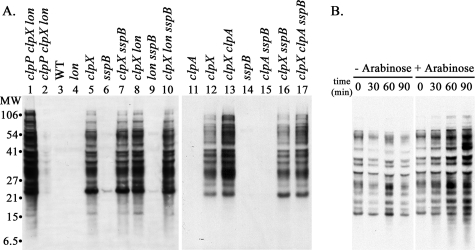
SsrA-tagged proteins accumulate in E. coli MG1655 cells with mutations in clpX or clpP. A, accumulation of SsrA-tagged proteins. Cells were grown to mid-exponential phase (A600 ∼ 0.5). Proteins were isolated by trichloroacetic acid precipitation, and 20 μg of protein from each culture was run on an SDS gel. SsrA-tagged proteins were detected by Western blotting with anti-SsrA antibody. A composite image containing normalized exposures of selected lanes from different blots is shown to allow comparison between the mutant strains. Duplicate gels run at the time of the blots were stained with Coomassie Blue to confirm equal sample loading. The genotypes of the strains are listed above the lanes. The strains used for the samples in each lane were as follows: lane 1, wild type (MG1655); lane 2, clpA (SG30069); lane 3, clpX (ML30035); lane 4, clpA clpX (ML30060); lane 5, clpP (YN-594); lane 6, clpQ (ML30009); lane 7, lon (ML30008); lane 8, ftsH (CAG39502); lane 9, sspB (ML30018); lane 10, clpS (SK1002); lane 11, ssrA (ML30020); lane 12, clpX ssrA (ML30037); lane 13, clpP ssrA (SG30088); and lane 14, clpA clpX ssrA (ML30039). B, induced expression of ClpX leads to rapid degradation of accumulated SsrA-tagged proteins. ML30035 (clpX) was transformed with plasmid pBAD33-ClpX, which has wild type clpX under control of the arabinose promoter. At mid-exponential phase, the culture was split, and arabinose was added to one of the cultures to induce ClpX synthesis (right). SsrA-tagged proteins remaining in the culture were assayed at timed intervals. For the control culture (left), sample loading was adjusted so that each lane has equal protein (∼10 μg); for the samples from the ClpX-induced cultures, the initial samples were adjusted to 10 μg, and subsequent samples contained equivalent volumes of culture.
FIGURE 5.
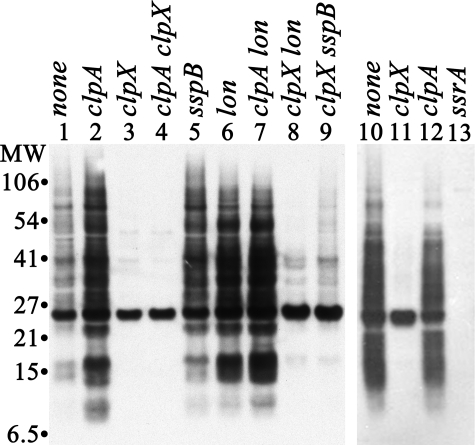
ClpX promotes efficient trapping of SsrA-tagged proteins by an inactive mutant of ClpP in vivo. Plasmid pBAD24-His-ClpPin, which expresses the proteolytically inactive mutant, His-ClpP-S97A, under control of the arabinose promoter, was transformed into a clpP mutant host strain and into its isogenic derivatives carrying mutations in clpA, clpX, sspB, or lon. Cells were grown to low density (A600 ∼ 0.2) in the presence of 50 μg/ml ampicillin, and synthesis of His-ClpPin was induced by addition of arabinose. After 40 min, cells were collected. His-ClpPin was purified from cell extracts by metal chelate chromatography on a Talon resin. Protein (5 μg) from the peak fraction containing purified His-ClpPin was loaded in each lane and separated by SDS-PAGE. SsrA-tagged proteins were detected by Western blotting. Left panel, lanes are labeled with the relevant genotypes of the strains used: lane 1, clpP (YN594); lane 2, clpP clpA (SG30071); lane 3, clpP clpX (SG30070); lane 4, clpP clpA clpX (SG30073); lane 5, clpP sspB (ML30122); lane 6, clpP lon (ML30010); lane 7, clpP clpA lon (ML30220); lane 8, clpP clpX lon (ML30014); lane 9, clpP clpX sspB (ML30316). Right panel, partial repeat of the previous experiment including an additional control strain carrying a mutation in ssrA. Lane 10, clpP (YN594); lane 11, clpP clpX (SG30070); lane 12, clpP clpA (SG30071); lane 13, clpP ssrA (SG30088).
Null mutations in clpQ, lon, or ftsH, the genes for other ATP-dependent proteases, did not give rise to detectable amounts of SsrA-tagged proteins (Fig. 1A, lanes 6–8) nor did a mutation in clpA (Fig. 1A, lane 2), indicating that ClpXP is the major protease involved in the degradation of endogenous SsrA-tagged proteins. SsrA-tagged proteins also did not accumulate in an sspB mutant (Fig. 1A, lane 9). SspB facilitates interaction of ClpX with SsrA-tagged proteins and promotes their degradation in vitro and in vivo (30, 32). We show below that SspB does affect the rate of degradation of SsrA-tagged proteins, but degradation in the absence of SspB is rapid, and the steady-state levels of tagged proteins are below the detection limit of our antibody. To confirm that the SsrA-tagged proteins that accumulated in the clpX mutant were substrates for ClpX, we introduced a plasmid expressing ClpX under control of the arabinose promoter. The SsrA-tagged proteins were abundant before induction of ClpX (Fig. 1B, –arabinose) but declined rapidly 5 min after induction (Fig. 1B, +arabinose).
To estimate the amounts of SsrA-tagged proteins, we used purified GFP-SsrA as a standard. The limit of detection of GFP-SsrA was in the range of 1 ng (<40 fmol) in most blots; no cross-reaction was seen with untagged GFP (data not shown). We then compared the intensities of various dilutions of GFP-SsrA to the total intensities in each lane after densitometric scanning. Lanes loaded with 20 μg of protein from a clpX mutant contained the equivalent of ∼22 ng of GFP-SsrA. As GFP-SsrA (Mr 27,264) is only slightly smaller than the average E. coli protein (Mr ∼ 33,000), we calculate that ∼0.11% of the proteins carry an SsrA tag in clpX mutant cells. The rate of incorporation of SsrA tags into newly synthesized nascent chains must be considerably higher than 0.11%, because the tagged proteins are not completely stable in clpX mutants, and <30% accumulates in the steady state (see below).
An estimate of the minimum degradation rate of SsrA-tagged proteins in wild type cells can be obtained by comparison of the accumulation of the tagged proteins in wild type and mutant cells.3 When clpX cell extracts were serially diluted, SsrA-tagged proteins were visible when as little as 2–4 μgof protein was loaded in the lane (see supplemental Fig. S2). In contrast, no tagged proteins were visible even with 100 μgof wild type cell extract, indicating that ≤3% of the tagged proteins accumulate in the steady state in wild type cells. If tagging occurs at the same rate in the wild type and mutant cells, this low level accumulation must reflect a degradation rate in wild type cells of ≥1.0 min–1 (t½ ≤ 1 min).4 Below, we show that direct measurements are in agreement with this estimate.
ClpA Makes a Minor Contribution to the Degradation of SsrA-tagged Proteins—Purified SsrA-tagged proteins are efficiently degraded by both ClpXP and ClpAP in vitro (28, 29, 38), and both proteases target overexpressed or engineered SsrA-tagged proteins in vivo, although ClpX is more efficient in promoting their degradation (12, 35, 36). No SsrA-tagged proteins were detectable in wild type or ΔclpA mutant cells during exponential or stationary phase (see supplemental Fig. S5), indicating that ClpA cannot be responsible for eliminating a large fraction of the SsrA-tagged proteins. ClpA expression levels were comparable with those of ClpX under the conditions of our experiments (Ref. 39 and data not shown). Thus, ClpX activity is much higher than ClpA activity against SsrA-tagged proteins in wild type cells.
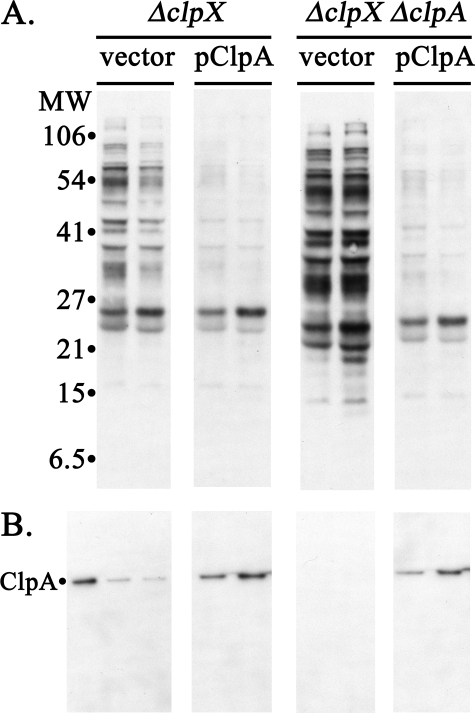
The double clpA clpX mutant has as much as two times the level of SsrA-tagged proteins as the clpX mutant (compare vector lanes in Fig. 2A and see supplemental Fig. S5), suggesting that ClpA is capable of degrading these proteins. To confirm that ClpA can promote degradation of SsrA-tagged proteins that accumulate in the absence of ClpX, the clpX mutant and the clpX clpA mutant were transformed with a plasmid expressing ClpA under control of the arabinose promoter. SsrA-tagged proteins were abundant in the mutant cells containing the empty vector (Fig. 2A, top left), whereas they were absent in cells expressing ClpA from the plasmid (Fig. 2A, top right). ClpA is expressed at a low level without induction with this clone, producing about three times the endogenous ClpA level (Fig. 2, A and B, lower panels). This small increase was sufficient to allow degradation of the SsrA-tagged proteins, which were reduced to essentially undetectable levels. We conclude that ClpA can recognize SsrA-tagged proteins in vivo, but endogenous ClpA either does not have access to the SsrA-tagged proteins or is occupied by other protein substrates limiting its availability.
FIGURE 2.
Increased expression of ClpA from a plasmid suppresses accumulation of SsrA-tagged proteins. A clpX mutant and an isogenic clpX clpA double mutant were transformed with a pBAD33 plasmid expressing clpA under the arabinose promoter or with the pBAD33 vector. Cells were grown to mid-exponential phase without arabinose, and equal aliquots were removed at two times 60 min apart and processed for Western blotting. A, blots with anti-SsrA antibody. The genotypes of the host strains, ML30035 (ΔclpX) and ML30060 (ΔclpX ΔclpA), are indicated at the top, and the lanes are labeled with the plasmid present in that host. B, blots with anti-SsrA antibody. Samples from the above culture were run on a separate gel and blotted with anti-ClpA antibody. The 1st lane contains purified ClpA as a reference. Other lanes were loaded with protein from equal volumes of the cultures indicated above. ClpA is expressed from the plasmid without induction at 2–3 times the endogenous level seen in clpA+ cells. The vector control samples for the clpX clpA mutant were taken from a parallel culture that was induced with 0.1% arabinose.
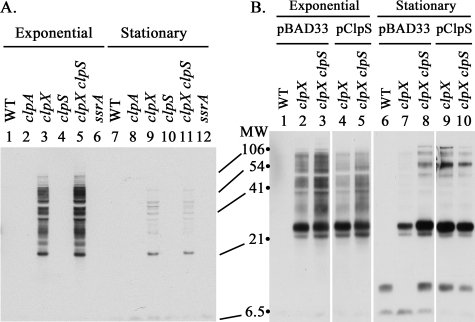
Stability of SsrA-tagged Proteins Is Not Because of Inhibition of ClpA by ClpS—ClpS is an adaptor protein that targets substrates bearing N-degrons to ClpA (40) and inhibits ClpA activity against other substrates in vitro (41). To determine whether ClpS prevented ClpA from degrading SsrA-tagged proteins in vivo, we compared accumulation of SsrA-tagged proteins in a clpX and a clpX clpS double mutant during exponential growth. Increased activity of ClpA in the absence of ClpS should have reduced the accumulation of SsrA-tagged proteins; however, the tagged protein levels were not lower but were slightly higher instead (Fig. 3A, lane 3 versus lane 5). We have found that the loss of ClpS leads to a 20–30% reduction in ClpA levels (data not shown), which could explain the small increase in tagged proteins. Because ClpA has been shown to make a larger contribution to degradation of GFP-SsrA during stationary phase (36), we also checked SsrA-tagged protein levels during stationary phase in the clpS mutant. SsrA-tagged proteins accumulated during growth and then declined during stationary phase (Fig. 3A). The decrease during stationary phase was similar in clpX and clpX clpS mutant strains (Fig. 3A, lane 9 versus lane 11), in contrast to the sharper decline in GFP-SsrA observed during stationary phase with the clpS mutant (36). The clpS mutation also had no effect on the loss of tagged proteins in stationary phase in a clpX lon strain (data not shown). These results indicate that inhibition of ClpA by endogenous ClpS is not responsible for the accumulation of SsrA-tagged proteins in cells with active ClpAP.
FIGURE 3.
Effects of ClpS on the degradation of SsrA-tagged proteins. A, wild type levels of ClpS have little effect on accumulation of SsrA-tagged proteins. A null mutation in clpS was introduced into a clpX mutant, and the SsrA-tagged proteins present in mid-exponential and stationary phase cells were probed as described above. The lanes contained samples from the following strains: lanes 1 and 7, wild type (MG1655); lanes 2 and 8, clpA (SG30069); lanes 3 and 9, clpX (ML30035); lanes 4 and 10, clpS (SK1002); lanes 5 and 11, clpX clpS (ML30037); and lanes 6 and 12, ssrA (ML30020). B, overexpression of ClpS has a minimal effect on accumulation of SsrA-tagged proteins. Transformants carrying pBAD33 with or without clpS under control of the arabinose promoter were grown in LB with 0.1% arabinose. Samples were removed during exponential growth and stationary phase and probed for SsrA-tagged proteins. The expression of ClpS was confirmed by direct visualization with Coomassie Blue staining after SDS-PAGE. The genotypes of the host strains are listed above the lanes: lanes 1 and 6, wild type (MG1655); lanes 2, 4, 7, and 9, clpX (ML30035); lanes 3, 5, 8, and 10, clpX clpS (ML30037).
We next asked whether overexpression of ClpS would affect accumulation of SsrA-tagged proteins, because excess ClpS was shown to inhibit degradation of GFP-SsrA (36). When ClpS was expressed from a plasmid in clpX cells, there was a modest decrease in the amount of tagged proteins during exponential phase (Fig. 3B, lanes 1–5) and a small increase during stationary phase (Fig. 3B, lanes 6–10). The presence of the pBAD plasmid during stationary phase also gave rise to a few specific SsrA-tagged proteins whose identities are not known at present, but induction of ClpS produced only a slight increase in the bulk of SsrA-tagged proteins. In contrast, overexpression of ClpS led to a 10-fold increase in GFP-SsrA in stationary phase cells (36). These results suggest that ClpS does not effectively block ClpAP-dependent degradation of SsrA-tagged proteins that accumulate in vivo under these conditions and that other conditions or factors affect the ability of ClpAP to degrade these proteins.
Effects of SspB on Degradation by ClpXP and ClpAP—SspB facilitates degradation of model SsrA-tagged proteins by ClpXP and blocks their degradation by ClpAP in vivo and in vitro (30, 36). SspB and ClpA recognize overlapping regions of the SsrA peptide (30), allowing SspB to compete with ClpA for binding to SsrA-tagged proteins. We first asked whether SspB is needed for ClpXP to prevent accumulation of SsrA-tagged proteins. No tagged proteins were detected in cells carrying a null mutation in sspB alone (Fig. 4A, lanes 6 and 14) or the sspB mutation in combination with mutations in lon (lane 9) or clpA (lane 15). Thus, ClpXP can degrade the bulk of the SsrA-tagged proteins on its own. Next, we asked if removing SspB would allow ClpAP to degrade a larger fraction of the SsrA-tagged proteins in a clpX mutant. However, the clpX sspB mutant (Fig. 4A, lanes 7 and 16) had essentially the same amount of SsrA-tagged proteins as the clpX mutant (lanes 5 and 12), and the same was true comparing the clpX lon sspB (lane 10) and clpX lon (lane 8) mutants, indicating that ClpAP activity is not significantly blocked by endogenous levels of SspB. The presence of SspB exerts a slightly inhibitory effect on degradation by Lon, as indicated by the decrease in SsrA-tagged proteins in the clpX clpA sspB mutant (Fig. 4A, lane 17) compared with the clpX clpA mutant (lane 13). To confirm that SspB is capable of competing with ClpA or other proteases for endogenous SsrA-tagged proteins, we expressed high levels of SspB from a plasmid. Fig. 4B shows that the pools of SsrA-tagged proteins in a clpX lon mutant increase after induction of SspB, indicating that when higher levels of SspB are present, more SsrA-tagged proteins are bound and protected from degradation by ClpAP. We conclude that the endogenous levels of SspB are not sufficient to sequester a large fraction of the SsrA-tagged proteins and that another mechanism is responsible for the low activity of ClpA on these proteins.
FIGURE 4.
Effects of SspB on the degradation of SsrA-tagged proteins. A, ClpXP, but not Lon or ClpAP, is sufficient to eliminate SsrA-tagged proteins in the absence of SspB. A null mutation in sspB was introduced into strain MG1655 and isogenic derivatives lacking ClpX or Lon (left panel) or ClpA (right panel). Accumulation of SsrA-tagged proteins was measured in mid-exponential phase by Western blotting. Lanes 3–17 were loaded with 20 μg of total cell protein. To compare accumulation in the various mutants to the maximum amount observed when degradation was completely blocked, lanes 1 and 2 were loaded with 20 and 2 μg protein, respectively, from the triple mutant clpP clpX lon (ML30014). The other lanes contained samples from the following strains: lane 3, wild type (MG1655); lane 4, lon (ML30015); lanes 5 and 12, clpX (ML30035); lanes 6 and 14, sspB (ML30038); lanes 7 and 16, clpX sspB (ML30054); lane 8, clpX lon (ML30064); lane 9, lon sspB (ML30264); lane 10, clpX lon sspB (ML30260); lane 11, clpA (SK1004); lane 13, clpX clpA, (ML30060); lane 15, clpA sspB (ML30055); lane 17, clpX clpA sspB (ML30061). B, overexpressed SspB blocks degradation of SsrA-tagged proteins by ClpAP in a clpX lon mutant. A clpX lon mutant (ML30064) was transformed with pBAD24-SspB and grown to mid-exponential phase, and the culture was split. Arabinose (0.1% (w/v)) was added to one-half; the other half was untreated, and the cultures were continued. Samples from the control culture (–arabinose) and from the culture in which SspB was induced (+arabinose) were collected at the times indicated above the lanes. SsrA-tagged proteins were detected by Western blotting; each lane contained 20 μg of protein. Increased SspB expression after arabinose addition was confirmed by Western blotting (data not shown).
Trapping of SsrA-tagged Proteins in ClpPin Is Dependent on ClpX but Not ClpA—As an alternative way of estimating the relative contributions of ClpX and ClpA to targeting of SsrA-tagged proteins, we trapped endogenous SsrA-tagged proteins in the chamber of an inactive mutant of ClpP (His-ClpPin) and isolated the trapped complexes by metal chelate affinity chromatography. His-ClpPin was expressed in cells deleted for endogenous clpP and carrying mutant alleles of clpX or clpA in combination with other mutations. After isolation, the trapped SsrA-tagged proteins were detected by Western blotting with anti-SsrA antibody. An abundance of tagged proteins was trapped in cells wild type for ClpX and ClpA (Fig. 5, lanes 1 and 10). No cross-reactive proteins were trapped in the ssrA mutant strain confirming that these are SsrA-tagged proteins (Fig. 5, lane13), and no proteins were trapped in the clpX clpA double mutant confirming that trapping was dependent on ClpX or ClpA (lane 4). Only trace amounts of SsrA-tagged proteins were pulled down with ClpPin from the clpX mutant (Fig. 5, lanes 3 and 11), indicating that ClpA alone does not target SsrA-tagged proteins efficiently. In the clpA mutant, the amount of trapped proteins was not reduced but rather was higher than in wild type strains (e.g. Fig. 5, lane 2 versus lane 1). Because the levels of ClpPin in the cell limit the total amount of tagged proteins that can be trapped and ClpPin is shared between ClpX and ClpA, we conclude that the absence of ClpA makes more ClpPin available to ClpX and allows targeting of additional SsrA-tagged proteins.
The amount of trapped proteins in the clpX sspB mutant was ∼2 times that seen in the clpX mutant (Fig. 5, lanes 3 and 9), showing that competition from SspB does reduce ClpA activity against these proteins, although this effect cannot fully account for the low levels of ClpA-dependent trapping, as the amount of trapped tagged proteins was still quite low. As we show below, Lon protease also degrades SsrA-tagged proteins, and we measured trapping of SsrA-tagged proteins in lon mutants. More tagged proteins were trapped in the lon mutant (Fig. 5, lane 1 versus lane 6) and in the lon clpA double mutant (Fig. 5, lane 3 versus 7). Based on this result, we conclude that Lon degrades the same SsrA-tagged proteins targeted by ClpX and that the increased pools of tagged proteins in the lon mutant permit ClpX to deliver more proteins to ClpP. On the other hand, trapping by ClpA in the clpX lon mutant was slightly increased over the clpX mutant (Fig. 5, lane 3 versus lane 8), indicating that ClpA has low activity even when SsrA-tagged proteins are present at elevated levels in the absence of ClpX and Lon.
When trapping was measured in cells carrying only an sspB mutation, the amount of SsrA-trapped protein was nearly the same as in wild type cells (Fig. 5, lane 5) despite the expectation that there would be smaller amounts, as SspB promotes targeting of SsrA-tagged proteins to ClpXP. We think it is unlikely that increased activity of ClpA is enough to offset the decreased ClpX-dependent activity, because the sspB mutation did not lead to increased ClpA-dependent trapping in the clpX background (above). Although we do not have a definitive explanation for why mutating sspB had so little effect on trapping, we should emphasize that the trapping assay reflects a cumulative translocation activity rather than a rate of translocation and that even slower translocation by ClpX in the absence of SspB might be sufficient to saturate the ClpP.
Steady-state Levels of an Endogenous SsrA-tagged Protein in the Presence of ClpXP—We observed in many experiments that a significant fraction of proteins expressed from multicopy plasmids get tagged with SsrA (e.g. ClpP in Fig. 5). The tagged proteins are concentrated into one or more large nascent chains or nearly full-length proteins and are detectable in cells with active ClpXP. Fig. 6A shows blots of wild type and clpX cells carrying a plasmid expressing E. coli glutamate dehydrogenase (GDH). Significant amounts of SsrA-tagged GDH were present in wild type cells and very large amounts accumulated in the clpX mutant (Fig. 6A). No bands were seen when GDH was expressed in a clpX ssrA mutant (data not shown), confirming that the proteins are SsrA-tagged, and we refer to them cumulatively as GDH-SsrA. Note that the amount of cell extract loaded was less, and the exposure for chemiluminescent detection was shorter, which explains why the endogenous SsrA-tagged proteins are less visible in these blots.
FIGURE 6.
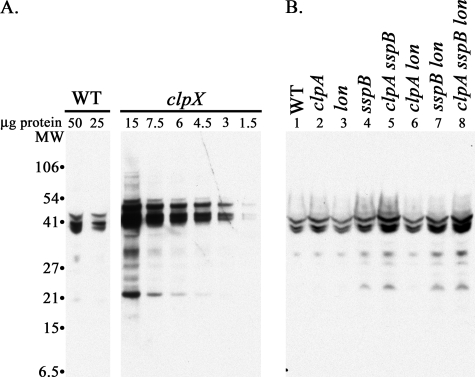
Decreased accumulation of SsrA-tagged GDH in the presence of SspB. GDH was expressed from the plasmid pBAD24-GDH. Cells were grown in LB with 50 μg/ml ampicillin. A and B, 0.2% (w/v/) arabinose was present throughout growth to induce expression of GDH. Samples were removed at mid-exponential phase, and 50 μg of total protein from each sample was loaded in separate wells of an SDS-polyacrylamide gel, transferred to nitrocellulose, and probed with anti-SsrA antibody. A, accumulation of SsrA-tagged GDH in a clpX mutant (ML30013). The 1st two lanes contain 50 and 25μg of protein from wild type cells. The next 6 lanes contain a serial dilution of cellular protein from the clpX mutant as indicated. B, accumulation of SsrA-tagged GDH in the absence of ClpA, Lon, and SspB during exponential phase. The lanes are labeled with the genotypes of the strains used as follows: lane 1, wild type (WT) (MG1655); lane 2, clpA (SK1004); lane 3, lon (ML30015); lane 4, sspB (ML30018); lane 5, clpA sspB (ML30055); lane 6, clpA lon (ML30065); lane 7, sspB lon (ML30264); lane 8, clpA sspB lon (ML30268).
Tagging of overexpressed GDH allowed us to examine the effects of other components and proteases on accumulation in the presence of an active ClpXP. Fig. 6B shows that mutating clpA (lane 2) or lon (lane 3) alone or in combination (lane 6) had almost no effect on the levels of GDH-SsrA. A mutation in sspB led to a 2-fold increase in GDH-SsrA (Fig. 6B, lane 4), as expected because SspB facilitates degradation by ClpXP (32). The increase in GDH-SsrA in an sspB mutant should be attenuated somewhat if degradation by ClpAP or Lon increases in the absence of SspB, and indeed, we observed significant increases in GDH-SsrA when mutations in clpA (Fig. 6B, lane 5), lon (lane 7), or both clpA and lon (lane 8) were introduced into the sspB mutant background, in contrast to the minimal effects seen in SspB+ cells (lanes 2, 3, and 6). The level of GDH-SsrA in sspB clpA or sspB lon mutants was still considerably lower (<25%) than those present in the clpX mutant (Fig. 6A), emphasizing that ClpXP has the predominant role in degradation. Thus, SspB preferentially binds to SsrA-tagged proteins, presenting them to ClpXP and excluding ClpA and Lon from access to the tagged proteins.
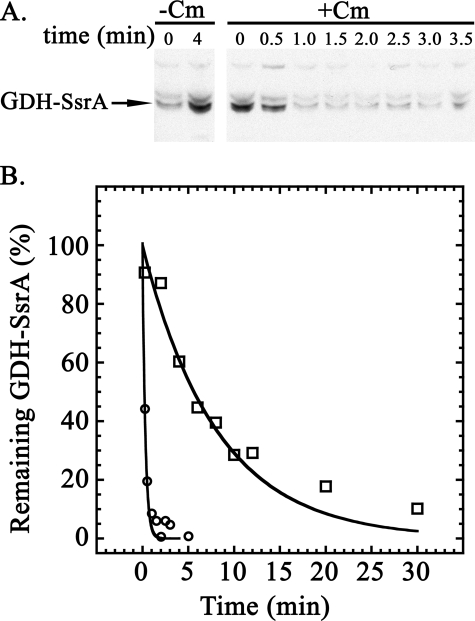
Half-life of Endogenously SsrA-tagged GDH in Vivo—Because endogenous SsrA-tagged proteins were not visible in cells with active ClpXP, we could not measure the half-lives by blocking new protein synthesis and following the decrease in cross-reactive bands by Western blotting. Overexpression of GDH allowed us to overcome this problem of detection. Fig. 7A shows the result of an experiment in which we used a chloramphenicol chase to measure the half-life of the GDH-SsrA in exponentially growing cells. GDH-SsrA degradation was very fast in wild type cells, and we estimated a half-life of ≤0.5 min (kdeg ∼ 1.4 min–1) (Table 1). The half-life of GDH-SsrA in the clpX mutant was ∼5 min (kdeg = 0.14 min–1) (Table 1). Variable lags in blocking protein synthesis with chloramphenicol introduce potential errors in measuring half-lives when degradation rates are very high. To confirm the rapid degradation, we used anti-SsrA antibody to immunoprecipitate GDH-SsrA and measured degradation by a pulse-chase procedure with [35S]methionine. Fig. 7B shows the decay of labeled GDH-SsrA in wild type and clpX mutant cells. GDH-SsrA was degraded with a half-life of ≤0.3 min (kdeg ≥ 2 min–1) in wild type cells, and the half-life increased to ∼6 min (kdeg ∼ 0.12) in the clpX mutant. There was good agreement between the two procedures in the rates determined for both rapid degradation seen in wild type cells and the slower degradation observed in the clpX mutant.
FIGURE 7.
Degradation of GDH-SsrA in wild type cells. A, antibiotic chase to measure degradation of GDH-SsrA. MG1655 carrying the plasmid pBAD24-GDH was grown in LB supplemented with 50 μg/ml ampicillin and 0.2% (w/v) arabinose to induce expression of GDH. At mid-exponential phase, the culture was divided and incubation was continued. Chloramphenicol (final concentration 0.1 mg/ml) was added to one culture, and the other was untreated. Aliquots of equal volume were withdrawn at the times indicated and probed for SsrA-tagged proteins. The lanes with the initial samples contain 25 μgof protein, and the other lanes were loaded with protein from an equal volume of culture. –Cm, samples from untreated cells; +Cm, samples from cells treated with chloramphenicol. B, pulse-chase to measure degradation of GDH-SsrA. Separate cultures of wild type and clpX mutant cells carrying the plasmid pBAD24-GDH were grown in defined medium (see “Experimental Procedures”). When cells reached A600 of 0.3, 50 μCi/ml [35S]methionine was added, and after 1 min, 1 mm nonradioactive methionine was added. Samples were removed at intervals and quenched with cold 5% trichloroacetic acid. After solubilization of the protein, GDH-SsrA was immunoprecipitated as described under “Experimental Procedures” and run on an SDS gel. Radioactive bands were detected and quantitated by scanning the dried gel using the STORM PhosphorImaging system from GE Healthcare. The graph shows the fraction of radioactive GDH-SsrA remaining in wild type cells (MG1655) (open circles) and in the clpX mutant (ML30013) (open squares).
TABLE 1.
Degradation rates for GDH-SsrA
The degradation rates for GDH-SsrA in exponential phase cells were obtained by fitting the data in Fig. 7B to a first-order exponential, ΔA = ΔAmax·e–k·t. Degradation rates shown in parentheses were obtained with exponential phase cells using the chloramphenicol chase procedure. Data are from single experiments. The rates in stationary phase cells were obtained by the chloramphenicol chase procedure. The error reported is the standard deviation of two independent experiments (σ = √(1/N)Σ(xi — x)2, where xi is an individual value, and x is the mean of N determinations). The results for the three lon mutants are from single experiments.
| Strain | Degradation rate | Half-life |
|---|---|---|
| min-1 | min | |
| Exponential phase | ||
| Wild type | ∼1.7 (∼2.3) | 0.4 (0.3) |
| clpX | 0.14 (0.12) | 5.0 (6.0) |
| Stationary phase | ||
| Wild type | 0.063 ± 0.006 | 11 |
| clpX | 0.056 ± 0.003 | 12 |
| clpA | 0.058 ± 0.001 | 12 |
| clpX clpA | 0.043 ± 0.02 | 16 |
| lon | 0.10 | 6.7 |
| clpA lon | 0.14 | 5.0 |
| clpX lon | 0.043 | 16 |
Half-lives of the Endogenous SsrA-tagged Proteins in Cells Lacking Multiple Protease Components—Although ClpXP makes the largest contribution to the degradation of SsrA-tagged proteins, degradation by other proteases in the absence of ClpX (kdeg ∼ 0.07 min–1) results in steady-state accumulation of ≤30% of the proteins tagged with SsrA. Elimination of the other proteolytic activities (mostly ClpAP and Lon) results in higher accumulation and darker staining lanes on the Western blots (see supplemental Fig. S2 for a Discussion). To directly measure the contribution of these other activities to overall degradation, we used the antibiotic chase procedure to monitor turnover of SsrA-tagged proteins in strains missing ClpX or ClpP and one or more other protease components. Supplemental Fig. S3 shows the results when cells carrying clpX and clpP mutations were chased with spectinomycin. The cross-reactive bands in all size ranges decreased in parallel, which allowed us to calculate a half-life for the pool of tagged proteins. The average of 3–4 decay curves (supplemental Fig. S4) gave a half-life of ∼9.9 min in clpX cells, which corresponds to a degradation rate by proteases other than ClpXP of ∼0.070 min–1. When both ClpX and ClpA were absent, the degradation rate dropped to ∼0.049 min–1 (Table 2). The contribution from ClpAP obtained from the difference between these two rates is ∼0.021 min–1, assuming that the relative activities of the other proteases were unchanged in the double mutant. When we measured degradation in a clpX lon mutant, in which ClpAP should be the major protease acting on SsrA-tagged proteins, the rate of degradation was ∼0.037 min–1 (Table 2), which was in good agreement with the calculated rate. This ClpA-dependent rate is significantly lower than the ClpX-dependent rate, which is ∼2 min–1 (Table 1).
TABLE 2.
Degradation rates for endogenous SsrA-tagged proteins
Degradation of the endogenous pool of SsrA-tagged proteins was followed by blocking protein synthesis and measuring the remaining density following Western blotting as described under “Experimental Procedures.” The rates shown are the averages of the rates obtained in N independent experiments. Rates during exponential phase were obtained after fitting the graphs shown in supplemental Fig. S4 to a first-order exponential. Rates for stationary phase were derived from graphs of density data obtained from two independent experiments; representative blots are shown in Fig. 9, A—D. Standard deviations were determined as in Table 1.
| Strain | Rate | Half-life | N |
|---|---|---|---|
| min-1 | min | ||
| Exponential phase | |||
| clpX | 0.070 ± 0.01 | 9.9 | 4 |
| clpX clpA | 0.049 ± 0.01 | 14 | 4 |
| clpX lon | 0.037 ± 0.0004 | 19 | 2 |
| clpX clpA lon | 0.022 ± 0.0007 | 32 | 2 |
| clpX clpS lon | 0.043 ± 0.006 | 16 | 2 |
| clpX sspBa | 0.19 ± 0.004b | 3.6 | 1 |
| clpX clpA sspB | 0.067 ± 0.02 | 10 | 2 |
| clpX sspB lon | 0.11 ± 0.02b | 6.1 | 1 |
| clpP | 0.081 ± 0.020 | 8.6 | 4 |
| clpP lon | 0.0059 ± 0.0007 | 120 | 4 |
| clpP clpQ | 0.093 ± 0.01 | 7.5 | 4 |
| clpP ftsHc | 0.068 ± 0.005 | 10 | 2 |
| Stationary phase | |||
| clpX | 0.0068 | 100 | 2 |
| clpX clpA | 0.0142 | 50 | 2 |
| clpX lon | 0.0068 | 100 | 2 |
| clpX clpA lon | 0.0051 | 140 | 2 |
This rate was obtained in one experiment. In other experiments (not shown) the rates were ∼0.07 min-1 (see supplemental Fig. S4).
This value represents the error in the slope calculated using a general curve fitting algorithm provided by the Kaleidagraph program (Pearson's r value >0.95).
This strain grew poorly with a doubling time more than twice that of the other strains.
We compared degradation rates in the clpX lon strain and its isogenic derivative carrying a null mutation in clpS. The degradation rate increased very slightly from 0.037 to 0.043 min–1 in the absence of ClpS (Table 2), confirming the minor effect of ClpS on degradation of SsrA-tagged proteins. Our finding that ClpS is not responsible for the low activity of ClpA against these substrates in vivo is consistent with those of Farrell et al. (36), who also concluded that ClpS levels in growing cells are insufficient to completely inhibit ClpA activity.
Eliminating ClpX, SspB, and Lon should allow measurement of ClpA-dependent degradation in the absence of all competition. The rate of degradation in the clpX lon sspB mutant was ∼0.11 min–1 (Table 2), about three times that seen in the isogenic clpX lon strain. The rate is consistent with what we estimated from the accumulation data. Although it is not certain that all the degradation in this strain is because of ClpAP, we can conclude that the maximum ClpA-dependent degradation rate is <10% of the rate when ClpX is present in the cell.
SsrA-tagged proteins were degraded in clpP mutants about 50% faster (kdeg = 0.081 min–1) than in the clpX clpA mutant (kdeg = 0.049 min–1) (Table 2). One possibility for the faster degradation in clpP cells is that ClpX or ClpA can directly or indirectly increase degradation of SsrA-tagged proteins by other proteases. Similar effects of ClpX in the absence of ClpP were seen with GFP-SsrA (36). We do not know the mechanism of this effect, although it could be that, by preventing aggregation, ClpA and ClpX maintain the tagged proteins in a soluble state accessible to other proteases.
Lon Contributes to SsrA-tagged Protein Degradation—To identify the other proteases besides ClpXP and ClpAP involved in degradation of SsrA-tagged proteins, we measured accumulation and decay of SsrA-tagged proteins in clpP strains that carried additional mutations in lon, ftsH, or clpQ (hslV). Pools of tagged proteins were increased in clpP lon double mutant during exponential growth compared with the strain with only the clpP mutation, and the tagged proteins were still detectable in overnight cultures (Fig. 8A). There were no increases in accumulation of tagged proteins in the clpP strains carrying ftsH or clpQ mutations and no persistence of the tagged proteins into stationary phase (Fig. 8A). These results suggested that Lon degrades SsrA-tagged proteins, and that FtsH and ClpYQ (HslUV) have little or no role.
FIGURE 8.
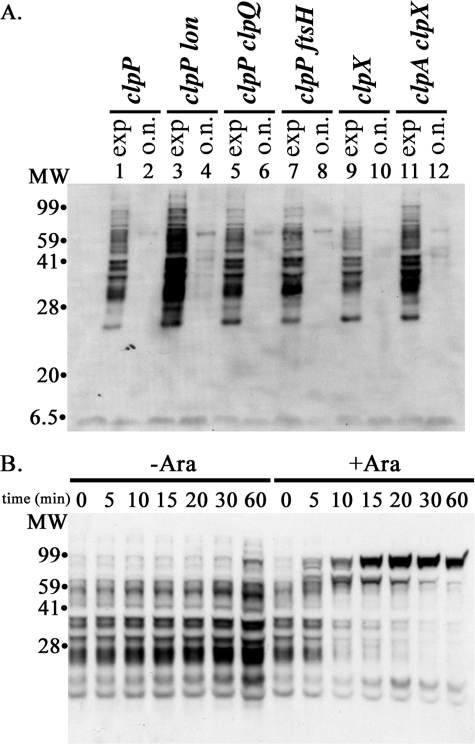
Lon protease contributes to degradation of SsrA-tagged proteins. A, increased accumulation of SsrA-tagged proteins in a clpP lon double mutant. Levels of SsrA-tagged proteins were measured in clpP mutant strains carrying a mutant allele of one of the other ATP-dependent proteases. Cells were grown in LB, and samples were taken at mid-exponential phase (A600 ∼ 0.5) (exp) and after overnight (>16 h) incubation (o.n.). The levels of SsrA-tagged proteins in each strain were compared by Western blotting with 10 μg of total cell protein from each culture. For comparison, clpX and clpX clpA mutants were analyzed in parallel. Lanes are labeled with the relevant genotype of the strains used: clpP (YN594); clpP lon (ML30010); clpP clpQ (ML30110); clpP ftsH (ML30102); clpX (ML30035); and clpX clpA (ML30060). B, induced Lon degrades accumulated SsrA-tagged proteins. Strain ML30014, clpP clpX lon, was transformed with plasmid pBAD-Lon, which has lon under control of the arabinose promoter (right) or with the pBAD33 vector (left). Lon was induced in mid-exponential phase, and SsrA-tagged proteins remaining in the culture were measured. The protein from samples taken at 0 min was dissolved in SDS sample buffer, and ∼10 μg of protein was loaded on the gel. Other samples were adjusted so that each lane received protein from an equivalent volume of culture.
To confirm that Lon can degrade SsrA-tagged proteins, we expressed Lon under control of an arabinose-inducible promoter and measured the degradation of accumulated SsrA-tagged proteins after induction. Fig. 8B shows that the SsrA-tagged proteins were rapidly degraded in cells after Lon was induced. The additional SsrA-tagged proteins accumulating at later times are most likely SsrA-tagged partially or completely synthesized Lon. As shown above for GDH and ClpP, SsrA-tagged forms of overexpressed proteins accumulate in cells lacking ClpXP. In the clpP lon strain, the half-life of SsrA-tagged proteins was ∼120 min (supplemental Fig. 4S), corresponding to a degradation rate of ∼0.006 min–1 (Table 2). The Lon-dependent degradation rate, calculated from the difference in rates between the clpP and clpP lon strains, is 0.075 min–1, indicating that Lon is responsible for >90% of the degradation in the absence of Clp proteases. The Lon-dependent rate calculated from the difference between the clpX clpA and the clpX clpA lon mutants is 0.027 min–1 (Table 2), significantly lower than this rate. The higher Lon-dependent degradation rates when ClpA and ClpX are present suggest that ClpX or ClpA might enhance Lon action on these substrates.
The degradation rate was essentially the same in the clpP and clpP clpQ strains (Table 2), indicating that ClpYQ does not degrade endogenous SsrA-tagged proteins under these conditions. FtsH contributes to degradation of a CI-SsrA fusion protein in vivo (13). We observed a slight decrease in degradation rate of endogenous SsrA-tagged proteins in the clpP ftsH mutant compared with the clpP mutant during the first 10 min of the chase (supplemental Fig. S4), suggesting that FtsH makes a small contribution to the degradation (kdeg ≤ 0.015 min–1) (Table 2). Our measurements were complicated by the slow growth rate exhibited by this mutant, and the reduced degradation could simply reflect a decrease in metabolic activities in this strain. It should be emphasized that no cross-reactive bands accumulated in lon, ftsH, or clpQ mutants when ClpXP was present (Fig. 1A, lanes 6–8).
Degradation of SsrA-tagged Proteins during Stationary Phase—Protein degradation rates change in a complex manner during stationary phase depending on the specific substrates, the proteases responsible for the degradation, and the nature of the stationary phase. For example, some stable proteins are degraded in carbon- or nitrogen-starved cells (42), whereas the highly unstable RpoS protein is stabilized under starvation conditions (43, 44). GFP-SsrA was reported to be degraded in stationary phase cells after growth in a defined rich medium, and ClpAP plays an increased role in its degradation under those conditions (36). To investigate how accumulation and degradation of endogenous SsrA-tagged proteins change in cells in stationary phase after growth on LB, we first monitored the steady-state levels of tagged proteins throughout growth and into stationary phase. No tagged proteins were detectable at any time in wild type cells indicating that degradation keeps pace with synthesis of SsrA-tagged proteins in both growth phases (see supplemental Fig. S6). The same was true in a clpA mutant and in a clpA lon double mutant, indicating that ClpXP activity is sufficient to degrade all new SsrA-tagged proteins. Although total protein synthesis decreases in stationary phase, SsrA continues to be incorporated into newly synthesized proteins during stationary phase (see below). As expected, high levels of tagged proteins were seen in cells mutated in clpX or clpP, and the levels were 2–3-fold higher when clpX or clpP mutants also carried mutations in clpA or lon or both (supplemental Fig. S6). To directly measure the rate of degradation by ClpAP or Lon during stationary phase, we allowed cells to grow to an A600 ∼ 2.5, blocked protein synthesis with chloramphenicol, and measured the rate at which tagged proteins decreased (Fig. 9, A–D). Degradation was slow in the absence of ClpX, and mutating clpA or lon in addition to clpX had little or no effect on degradation (Table 2). Under these conditions, the clpX clpA double mutant had a slightly higher rate of degradation; however, as we note below, when an energy source was added to the cultures, overall degradation was faster and mutating clpA resulted in a 2-fold reduction in the degradation rate (Fig. 9, E and F and supplemental Fig. S5, A and B). Introducing the clpA and lon mutations together into the clpX strain led to only a slight reduction in the degradation rate during stationary phase (Table 2). We considered that the lower degradation rates measured in stationary phase could reflect a change in the SsrA-tagged proteins when they were allowed to accumulate for several hours, which could render them less susceptible to degradation.
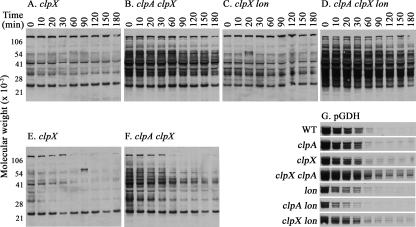
FIGURE 9.
The rate of degradation of endogenous SsrA-tagged proteins decreases in stationary phase. A–D, cultures were grown in LB to stationary phase (A600 ∼ 2.5); chloramphenicol (100 μg/ml) was added to stop new protein synthesis; samples were retrieved at regular intervals and probed for SsrA-tagged proteins by Western blotting. E and F, cultures were grown in LB to stationary phase (A600 ∼ 2.0), and arabinose was added to a final concentration of 0.2% (w/v). After 30 min, chloramphenicol (100 μg/ml) was added, and the cultures were processed as above. The strains used were as follows: A and E, clpX (ML30035); B and F, clpA clpX (ML30060); C, clpX lon (ML30064); D, clpA clpX lon (ML30067). G, strains bearing plasmid pBAD24-GDH were grown in LB supplemented with ampicillin (50 μg/ml) to stationary phase (A600 ∼ 2.0), and arabinose was added to a final concentration of 0.2% (w/v). After 30 min, chloramphenicol (100 μg/ml) was added to stop new protein synthesis, and samples were retrieved and probed for SsrA-tagged proteins as above. The images were trimmed to show only the major SsrA-tagged forms of GDH that are detected. To allow visual comparison of the decay rates, exposures were chosen to display approximately the same intensity at the first time point in each experiment. The strains were as follows: WT, wild type (MG1655); clpA (SG30069); clpX (ML30035); clpX clpA (ML30060); lon (ML30015); clpA lon (ML30065); clpX lon (ML30064).
To address the above possibility, we measured the degradation rate of newly synthesized GDH-SsrA in wild type and mutant cells by inducing GDH synthesis for 30 min during stationary phase, adding chloramphenicol to block synthesis, and measuring the remaining GDH-SsrA over time. Degradation in wild type cells was significantly slower than in exponential phase (Fig. 9G). The fastest degradation was observed in the clpA lon mutant, when only ClpXP is active, and the rate was about three times lower in the clpX lon mutant, when only ClpAP is active. Thus, ClpXP degrades SsrA-tagged proteins more efficiently than ClpAP in stationary phase, but the rate appears to be as much as 10 times slower than observed in exponential phase (Table 1). We do not know the cause for the lower activity of ClpXP in these cells, although reduced activity of ClpXP at the end of growth has been observed by others as well (45). A single mutation in clpX produced very little change in degradation rate, suggesting that ClpAP alone or in concert with other proteases can maintain the low rate of degradation in stationary phase. The degradation rate was ∼0.06 min–1, which is about two times the rate seen in exponential phase when SspB is present, but is lower than the maximum rate (0.14–0.2 min–1) seen in exponential phase when ClpX and SspB are both absent (Table 1). Paradoxically, when ClpX is present, degradation was faster in the clpA lon mutant, which was reminiscent of the increase in sequestration observed earlier with the clpA mutant, and we think it could have a similar explanation. Because degradation by ClpXP is faster than degradation by ClpAP, eliminating ClpA frees more ClpP for interaction with ClpX and allows faster degradation of SsrA-tagged proteins.
In a separate experiment, we measured the half-life of newly synthesized GDH-SsrA in stationary phase cultures of a clpX mutant and a double clpX sspB mutant and found that there was a very modest (∼30%) increase in degradation rate when SspB was absent (data not shown). When endogenous SsrA-tagged proteins were measured, we found that the levels declined less rapidly during stationary phase in the presence of SspB (see supplemental Fig. S6). This effect, however, was seen in the presence and absence of ClpA and therefore might not be due solely to competition between ClpA and SspB for SsrA-tagged proteins.
We noted that the rates of degradation of GDH-SsrA were higher than the rates of degradation of the pool of endogenous SsrA-tagged proteins in some mutant strains (as much as five times higher in clpX clpA, for example). One difference in these experiments was the addition of arabinose to the cultures to induce GDH synthesis. Although the cells did not show significantly new growth when arabinose was added, metabolism of the arabinose would be expected to partially restore ATP pools to these nutrient-deprived cells. To test whether addition of arabinose also affected degradation of the endogenous SsrA-tagged proteins, we performed a separate chloramphenicol chase with clpX and clpX clpA mutant cells after addition of arabinose in stationary phase. The results (Fig. 9, E and F) clearly showed that the degradation of the endogenous pool of SsrA-tagged proteins was faster in cells after arabinose addition. These data established that the accumulated SsrA-tagged proteins were not resistant to degradation and suggested that one cause of the reduced rate of degradation during stationary phase is depletion of the ATP needed to support ATP-dependent proteolytic activity. The results further suggest that ClpX activity is more substantially affected by the lower ATP pools in stationary phase cells, but additional studies will be required to support this conclusion.
DISCUSSION
We have measured accumulation and degradation of the global pool of endogenous proteins tagged with SsrA by the tmRNA system in E. coli and determined the contributions of the various ATP-dependent proteases to their degradation. Using an antibody that recognizes the SsrA peptide allowed us to detect and quantitate SsrA-tagged proteins without perturbing the tmRNA tagging system and to measure their degradation rates (kdeg) without changing the sequence or structural context of the endogenously added tag. We found that ClpXP is responsible for ≥90% of the degradation of the SsrA-tagged proteins made. No SsrA-tagged proteins were detected in cells missing other ATP-dependent proteases, ClpAP, Lon, HslUV, or FtsH, as long as active ClpXP was present. We estimate that the kdeg by ClpXP must be ≥1.5 min–1 (t½ < 0.5 min). The kdeg measured here for endogenous SsrA-tagged proteins is in line with that found for small plasmid-expressed proteins either engineered with an SsrA tag (32) or tagged by the tmRNA system because of a lack of an in-frame stop codon in its mRNA (12), but it is significantly faster than found for other constructs (36, 37). Although the degradation rates observed for engineered SsrA constructs vary, we found that all the SsrA-tagged proteins were degraded at similar rates, indicating that the endogenous SsrA tags in different contexts are recognized very efficiently by the proteolytic systems, particularly ClpXP and SspB.
SsrA tagging occurs with high frequency. In mutants deficient in multiple proteases, SsrA-tagged proteins made up >0.5% of the total protein, indicating that 1 in 200 or more nascent polypeptides acquires an SsrA tag. Although we did not rule out that the absence of multiple ATP-dependent proteases might affect the rate of tagging, our measurements are in agreement with those obtained by Moore and Sauer (25), who used a method not requiring protease mutants and estimated that tagging occurs on 0.4% of translated polypeptides. The implication from that study and ours is that stalling and other interruptions in translation are common in rapidly growing cells, and that the tmRNA system efficiently recycles blocked ribosomes and promotes elimination of the incomplete polypeptides. An uncertainty that remains is whether the multitude of SsrA-tagged proteins derive from all products of translation or only a limited set of open reading frames.
If ≥0.5% of proteins are made with an SsrA tag, then ∼12–15,000 SsrA-tagged proteins are produced each generation, or about 400–500 per min. Estimates of the number of ClpXP complexes in growing cells range from ∼50 to ∼200 (36, 39). Each ClpXP complex must degrade between two and five molecules/min, which is within the capacity of ClpXP based on in vitro studies (29). In wild type cells only 2% of SsrA-tagged proteins or 200–300 molecules are present in the steady state, equivalent to an intracellular concentration5 of 300–500 nm, which is lower than the Km for SsrA-tagged proteins and close to the Km for the SspB-mediated reaction (32). SspB would be expected to stimulate degradation ∼3-fold under these conditions, consistent with the effect we see on GDH-SsrA accumulation when SspB is mutated (Fig. 6) and with the small effects seen on degradation of other SsrA-tagged proteins in vivo (35, 36).
A puzzling finding of this study and others (12, 35–37) is the low rate of degradation of SsrA-tagged proteins in vivo by ClpAP compared with ClpXP. Low activity of ClpAP is not because of sequestration of SsrA-tagged proteins by SspB. The clpX mutant contains ∼5000 molecules of SsrA-tagged protein, far in excess of the SspB levels, which are only sufficient to sequester 200–400 molecules (36). Overexpressed SspB does compete with SsrA-tagged proteins (Fig. 4B) as has been found for GFP-SsrA (36). Under wild type conditions, SspB might indeed direct SsrA-tagged proteins away from ClpA, but other factors or conditions are mostly responsible for the low activity of ClpAP.
The low activity of ClpAP is not because of inhibition by endogenous ClpS (Fig. 3), which we and others (36) have found to be present at low intracellular levels. However, overexpression of ClpS also had only a slight effect on degradation of endogenous SsrA-tagged proteins by ClpAP. This result differs from reported effects on GFP-SsrA levels, which increase significantly after overexpression of ClpS, especially in stationary phase cells (36). Our estimate of the concentration of SsrA-tagged proteins in a clpX clpA mutant (∼40% of the total or 6000–8000 molecules) is similar to the number of GFP-SsrA molecules measured (36). Insensitivity of the degradation to ClpS might reflect higher affinity of ClpA for endogenous SsrA-tagged proteins. ClpA is known to interact with end motifs and so-called body motifs in proteins, and endogenous tagged substrates might have more body motifs exposed. Tighter binding of substrates would not necessarily promote faster turnover, and in other studies we have found that binding of unfolded proteins by the ClpA N-domains can impede degradation (42).
The trapping experiments reveal that ClpA is much less efficient than ClpX in delivering SsrA-tagged proteins to ClpPin. Trapping by ClpX actually increased when ClpA was absent. As ClpPin was present in excess in the cells, slow translocation of SsrA-tagged proteins by ClpA should not have prevented ClpX from translocating SsrA-tagged proteins to most of the ClpPin. We propose that ClpA competes with ClpX by translocating its own substrates to ClpPin, thus decreasing the amount of ClpPin available for ClpX-promoted translocation of SsrA-tagged proteins. This model implies that specific substrates for ClpA must be present in cells and have higher binding affinity than SsrA-tagged proteins. The competing substrates have not been identified. ClpA degrades proteins bearing N-degrons (46), and this degradation is mediated by ClpS (40). The low activity of ClpA against SsrA-tagged proteins would not appear to be due to competition from N-end rule substrates, because eliminating ClpS should have prevented this competition. However, ClpA can degrade some proteins with N-degrons in the absence of ClpS (47) and in general can recognize proteins with N-terminal peptides containing an array of hydrophobic residues (48). Degradation of SsrA-tagged proteins by the very slightly elevated levels of ClpA also argues that endogenous ClpA is blocked from acting on SsrA-tagged proteins, most likely because it is saturated with its substrates.
We found that Lon can degrade endogenous SsrA-tagged proteins. GFP-SsrA was reported to be either slowly degraded (37) or not degraded (36) by Lon. Lon degrades several specific substrates, such as SulA and RcsA, at high rates (kdeg 0.2–0.3 min–1) (49, 50). The degradation rate for SsrA-tagged proteins is lower (kdeg ∼ 0.05 min–1), and Lon is obviously able to recognize and process SsrA-tagged proteins efficiently. However, the detection method does not allow us to determine whether Lon recognizes the tag or sees these proteins because they are unfolded. If Lon recognizes the SsrA tag, it might have lower activity on stably folded proteins like GFP and degrade less stable SsrA-tagged nascent chains more rapidly. Lon degraded all tagged proteins at similar rates, and there were no obvious Lon-resistant tagged proteins, which might have been expected if at least some of the SsrA-tagged proteins were well folded proteins. Lon activity appeared to be higher in cells with ClpX and ClpA (Table 2), perhaps indicating that these chaperones helped to maintain the tagged proteins in a form susceptible to Lon.
SsrA-tagged proteins are degraded at reduced rates in stationary phase cells after growth in LB. Although ClpXP activity is sufficient to degrade the low levels of SsrA-tagged proteins produced as cell growth slows, degradation by ClpXP occurs at only one-tenth the rate seen during exponential phase. The most likely explanation for slower degradation is that intracellular levels of ATP drop below the levels needed for optimal activity of ClpX. In support of this proposal, we observed that activity in the absence of ClpX, mostly by ClpAP and Lon, is also impaired in stationary phase cells and can be partly restored by addition of a metabolizable carbon source. Our results would imply that reduced ATP levels affect ClpX to a greater degree than Lon and ClpA. Lon has a lower Km value for ATP than do ClpX and ClpA, and it is possible that Lon remains relatively more active as ATP levels decline in nongrowing cells. ClpA but not ClpX levels increase during stationary phase, but the 2-fold increase under our conditions (data not shown) should not be sufficient to offset the drop in ATP. Another possible explanation for the low ClpXP activity is that specific inhibitors or new substrates appear in stationary phase to compete with SsrA-tagged proteins for ClpXP. Competition between different classes of substrates has been proposed to explain stabilization of RpoS under specific carbon starvation conditions (44). In addition, specific inhibitors that block degradation of RpoS have been found under several starvation conditions (45), although none has been shown to interact directly with ClpX. Farrell et al. (36) reported that ClpAP activity against GFP-SsrA increased about 2-fold during stationary phase, in part because ClpA levels increased 3–4-fold in the same period. The stationary phase conditions in the two studies are not comparable, because the medium used in those studies included high amounts of glucose, making it possible that the cells were not energy-starved to the same degree.
In conclusion, our study shows that SsrA-tagged proteins are made at high frequency during protein translation, and the tagged proteins are degraded extremely rapidly. ClpXP, together with SspB, efficiently degrades ≥90% of the SsrA-tagged proteins during all growth phases. Degradation by ClpXP is extremely fast, with a half-life of less than a minute in actively growing cells. ClpAP and Lon protease limits the amounts of SsrA-tagged proteins that can accumulate in a clpX mutant but contribute to only 5–10% of the degradation in wild type cells. All three proteases target the global pool of SsrA-tagged proteins without any obvious discrimination.
Supplementary Material
Acknowledgments
We thank former members of our laboratory, S. K. Singh and S. G. Kang, for the original preparation of anti-SsrA antibody, and Jan Rozycki, for synthesis of SsrA peptide derivatives. We also thank Susan Gottesman, NCI, National Institutes of Health, for suggestions and comments on the manuscript.
This work was authored, in whole or in part, by National Institutes of Health staff. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods, additional references, Figs. S1–S7, and Tables S1 and S2.
Footnotes
The abbreviations used are: BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; MES, 4-morpholineethanesulfonic acid; GDH, glutamate dehydrogenase.
In exponentially growing cells, the fraction of a newly synthesized unstable protein that accumulates is given by the ratio of its synthesis rate to the sum of its synthesis and degradation rates, ksyn/(ksyn + kdeg) (51). The synthesis rate is approximately equal to the instantaneous rate of growth, which is ∼0.030 min–1 for all the strains except the ftsH mutant. For the clpP mutant with a kd of 0.081 min–1 (Table 2), the calculated fractional recovery would be 0.030/(0.030 + 0.081) or 0.27.
The overall degradation rate constant is the sum of the rate constants for the individual proteases. Because the accumulated SsrA-tagged proteins in the absence of ClpX is 10–20 times the amount observed in the presence of ClpX (the limit of detection by our antibody), the rate constant for ClpX-dependent degradation is between 10 and 20 times that of all other proteases combined or at least 1.0 min–1.
The volume of an E. coli cell is on the order of 1 μm3 (52); 100 molecules per cell is ∼150 nm.
References
- 1.Kisselev, L. L., and Buckingham, R. H. (2000) Trends Biochem. Sci. 25 561–566 [DOI] [PubMed] [Google Scholar]
- 2.Muto, A., Sato, M., Tadaki, T., Fukushima, M., Ushida, C., and Himeno, H. (1996) Biochimie (Paris) 78 985–991 [DOI] [PubMed] [Google Scholar]
- 3.Keiler, K. C., Waller, P. R., and Sauer, R. T. (1996) Science 271 990–993 [DOI] [PubMed] [Google Scholar]
- 4.Ray, B. K., and Apirion, D. (1979) Mol. Gen. Genet. 174 25–32 [DOI] [PubMed] [Google Scholar]
- 5.Oh, B. K., Chauhan, A. K., Isono, K., and Apirion, D. (1990) J. Bacteriol. 172 4708–4709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komine, Y., Kitabatake, M., Yokogawa, T., Nishikawa, K., and Inokuchi, H. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 9223–9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karzai, A. W., Susskind, M. M., and Sauer, R. T. (1999) EMBO J. 18 3793–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saguy, M., Gillet, R., Metzinger, L., and Felden, B. (2005) Biochimie (Paris) 87 897–903 [DOI] [PubMed] [Google Scholar]
- 9.Withey, J. H., and Friedman, D. I. (2002) Curr. Opin. Microbiol. 5 154–159 [DOI] [PubMed] [Google Scholar]
- 10.Tu, G. F., Reid, G. E., Zhang, J. G., Moritz, R. L., and Simpson, R. J. (1995) J. Biol. Chem. 270 9322–9326 [DOI] [PubMed] [Google Scholar]
- 11.Williams, K. P., Martindale, K. A., and Bartel, D. P. (1999) EMBO J. 18 5423–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottesman, S., Roche, E., Zhou, Y., and Sauer, R. T. (1998) Genes Dev. 12 1338–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herman, C., Thevenet, D., Bouloc, P., Walker, G. C., and D'Ari, R. (1998) Genes Dev. 12 1348–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ueda, K., Yamamoto, Y., Ogawa, K., Abo, T., Inokuchi, H., and Aiba, H. (2002) Genes Cells 7 509–519 [DOI] [PubMed] [Google Scholar]
- 15.Roche, E. D., and Sauer, R. T. (1999) EMBO J. 18 4579–4589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sunohara, T., Abo, T., Inada, T., and Aiba, H. (2002) RNA (N. Y.) 8 1416–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayes, C. S., Bose, B., and Sauer, R. T. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 3440–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de la Cruz, J., and Vioque, A. (2001) RNA (N. Y.) 7 1708–1716 [PMC free article] [PubMed] [Google Scholar]
- 19.Abo, T., Ueda, K., Sunohara, T., Ogawa, K., and Aiba, H. (2002) Genes Cells 7 629–638 [DOI] [PubMed] [Google Scholar]
- 20.Vioque, A., and de la Cruz, J. (2003) FEMS Microbiol. Lett. 218 9–14 [DOI] [PubMed] [Google Scholar]
- 21.Abo, T., Inada, T., Ogawa, K., and Aiba, H. (2000) EMBO J. 19 3762–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ranquet, C., Geiselmann, J., and Toussaint, A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 10220–10225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Handley, D., and Nakai, H. (2002) J. Mol. Biol. 322 311–324 [DOI] [PubMed] [Google Scholar]
- 24.Defenbaugh, D. A., and Nakai, H. (2003) J. Biol. Chem. 278 52333–52339 [DOI] [PubMed] [Google Scholar]
- 25.Moore, S. D., and Sauer, R. T. (2005) Mol. Microbiol. 58 456–466 [DOI] [PubMed] [Google Scholar]
- 26.Roche, E. D., and Sauer, R. T. (2001) J. Biol. Chem. 276 28509–28515 [DOI] [PubMed] [Google Scholar]
- 27.Asano, K., Kurita, D., Takada, K., Konno, T., Muto, A., and Himeno, H. (2005) Nucleic Acids Res. 33 5544–5552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, Y. I., Burton, R. E., Burton, B. M., Sauer, R. T., and Baker, T. A. (2000) Mol. Cell 5 639–648 [DOI] [PubMed] [Google Scholar]
- 29.Singh, S. K., Grimaud, R., Hoskins, J. R., Wickner, S., and Maurizi, M. R. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 8898–8903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flynn, J. M., Levchenko, I., Seidel, M., Wickner, S. H., Sauer, R. T., and Baker, T. A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 10584–10589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGinness, K. E., Bolon, D. N., Kaganovich, M., Baker, T. A., and Sauer, R. T. (2007) J. Biol. Chem. 282 11465–11473 [DOI] [PubMed] [Google Scholar]
- 32.Levchenko, I., Seidel, M., Sauer, R. T., and Baker, T. A. (2000) Science 289 2354–2356 [DOI] [PubMed] [Google Scholar]
- 33.Levchenko, I., Grant, R. A., Flynn, J. M., Sauer, R. T., and Baker, T. A. (2005) Nat. Struct. Mol. Biol. 12 520–525 [DOI] [PubMed] [Google Scholar]
- 34.McGinness, K. E., Baker, T. A., and Sauer, R. T. (2006) Mol. Cell 22 701–707 [DOI] [PubMed] [Google Scholar]
- 35.Bohn, C., Binet, E., and Bouloc, P. (2002) Mol. Genet. Genomics 266 827–831 [DOI] [PubMed] [Google Scholar]
- 36.Farrell, C. M., Grossman, A. D., and Sauer, R. T. (2005) Mol. Microbiol. 57 1750–1761 [DOI] [PubMed] [Google Scholar]
- 37.Choy, J. S., Aung, L. L., and Karzai, A. W. (2007) J. Bacteriol., 189 6564–6571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levchenko, I., Smith, C. K., Walsh, N. P., Sauer, R. T., and Baker, T. A. (1997) Cell 91 939–947 [DOI] [PubMed] [Google Scholar]
- 39.Ortega, J., Lee, H. S., Maurizi, M. R., and Steven, A. C. (2004) J. Struct. Biol. 146 217–226 [DOI] [PubMed] [Google Scholar]
- 40.Erbse, A., Schmidt, R., Bornemann, T., Schneider-Mergener, J., Mogk, A., Zahn, R., Dougan, D. A., and Bukau, B. (2006) Nature 439 753–756 [DOI] [PubMed] [Google Scholar]
- 41.Dougan, D. A., Reid, B. G., Horwich, A. L., and Bukau, B. (2002) Mol. Cell 9 673–683 [DOI] [PubMed] [Google Scholar]
- 42.Maurizi, M. R., and Rasulova, F. (2002) Arch. Biochem. Biophys. 397 206–216 [DOI] [PubMed] [Google Scholar]
- 43.Schweder, T., Lee, K. H., Lomovskaya, O., and Matin, A. (1996) J. Bacteriol. 178 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bougdour, A., Wickner, S., and Gottesman, S. (2006) Genes Dev. 20 884–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fredriksson, A., Ballesteros, M., Peterson, C. N., Persson, O., Silhavy, T. J., and Nystrom, T. (2007) Genes Dev. 21 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tobias, J. W., Shrader, T. E., Rocap, G., and Varshavsky, A. (1991) Science 254 1374–1377 [DOI] [PubMed] [Google Scholar]
- 47.Wang, K. H., Sauer, R. T., and Baker, T. A. (2007) Genes Dev. 21 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoskins, J. R., and Wickner, S. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 909–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mizusawa, S., and Gottesman, S. (1983) Proc. Natl. Acad. Sci. U. S. A. 80 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stout, V., Torres-Cabassa, A., Maurizi, M. R., Gutnick, D., and Gottesman, S. (1991) J. Bacteriol. 173 1738–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moesteller, R. D., Goldstein, R. V., and Nishimoto, K. R. (1980) J. Biol. Chem. 255 2524–2532 [PubMed] [Google Scholar]
- 52.Neidhardt, F. C. (1987) in Escherichia coli and Salmonella typhimurium, Cellular and Molecular Biology (Ingraham, J. L., Brooks Low, K., Magasanik, B., Schaechter, M., and Umbarger, H. E., eds) Vol. 1, pp. 3–6, American Society for Microbiology, Washington, DC [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.