Abstract
Topoisomerase I (Top1) is known to relax DNA supercoiling generated by transcription, replication, and chromatin remodeling. However, it can be trapped on DNA as cleavage complexes (Top1cc) by oxidative and carcinogenic DNA lesions, base damage, and camptothecin treatment. We show here that Top1 is also functionally involved in death receptor-induced programmed cell death. In cells exposed to TRAIL or Fas ligand, Top1cc form at the onset of apoptosis. Those apoptotic Top1cc are prevented by caspase inhibition and Bax inactivation, indicating that both caspases and the mitochondrial death pathway are required for their formation. Accordingly, direct activation of the mitochondrial pathway by BH3 mimetic molecules induces apoptotic Top1cc. We also show that TRAIL-induced apoptotic Top1cc are preferentially formed by caspase-3-cleaved Top1 at sites of oxidative DNA lesions with an average of one apoptotic Top1cc/100 kbp. Examination of Top1 knock-down cells treated with TRAIL revealed similar DNA fragmentation but a marked decrease in apoptotic nuclear fission with reduced formation of nuclear bodies. Thus, we propose that Top1 contributes to the full apoptotic responses induced by TRAIL.
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)2 is a promising therapeutic agent because it induces apoptosis in a wide variety of cancer cells without affecting normal tissues (1, 2). TRAIL belongs to the TNF family of cytokines, including TNFα and Fas ligand, which induce apoptosis by binding to their cognate plasma membrane receptors (3). The binding of TRAIL to the DR4 or DR5 receptors and the binding of Fas ligand to Fas receptor cause the intracellular death domains of those receptors to trimerize, which leads to the recruitment of FADD and the activation of caspase-8. Caspase-8 then cleaves and thereby activates caspase-3 either directly (type I cells) or/and indirectly (type II cells) (4) by activating the mitochondrial death pathway through the cleavage of Bid (5). Cleaved Bid binds to and activates the pro-apoptotic Bcl-2 relatives Bax and Bak proteins, causing the release of mitochondrial cytochrome c and the activation of caspase-9 and caspase-3 (6). Activated caspase-3 (and other downstream caspases) cleaves a broad array of intracellular targets including DNA topoisomerase I (Top1) (7). It is also required to induce the controlled rearrangement and degradation of nuclear structures with chromatin condensation, DNA fragmentation, nuclear fission, and release of apoptotic nuclear bodies in the extracellular space (8). TRAIL-induced apoptosis also involves an accumulation of intracellular reactive oxygen species (ROS) (9).
Top1 removes DNA superhelical tensions generated during transcription, replication, and chromatin remodeling (10) and is essential in higher eukaryotes (11). It relaxes DNA by forming transient DNA single-strand breaks that are produced as Top1 forms a covalent bond between its active site tyrosine (Tyr723) and a 3′-DNA phosphate. These Top1 cleavage complexes (Top1cc) allow controlled rotation of the broken DNA around the intact strand (10). Immediately after DNA relaxation, Top1 religates the break in the absence of added cofactor, such as ATP. Under normal conditions, the Top1cc are constitutively transient and almost undetectable because the DNA religation (closing) step is much faster than the DNA cleavage (nicking) step. The rapid resealing of Top1cc is inhibited by many common DNA base alterations (oxidation, alkylation, base mismatch, base loss), carcinogenic DNA adducts, and DNA backbone nicks (12). Top1cc can also be trapped with exquisite selectivity by camptothecin (CPT), a plant alkaloid (13) whose semi-synthetic derivatives topotecan and irinotecan are used to treat human cancers (14).
Top1cc have also been detected in cells undergoing apoptosis in response to a wide range of stimuli, including arsenic trioxide (15), staurosporine (16), tubulin and topoisomerase II (Top2) inhibitors (17, 18), TNFα (17), and UV-C radiation (19). In the present study, we demonstrated that apoptotic Top1cc also form in response to the physiological ligands TRAIL and Fas. We also investigated the mechanism of their production and provide evidence for their functional relevance during TRAIL-induced apoptosis.
EXPERIMENTAL PROCEDURES
Cells, Drugs, and Chemical Reagents—The human leukemia (Jurkat) and colon carcinoma (HCT116) cell lines (American Type Culture Collection, Manassas, VA) were cultured as described (16). HCT116 cells of each genotype (Bax+/-, Bax-/-, p53+/+, and p53-/-) (20, 21) were kind gifts from Dr. Bert Vogelstein (John Hopkins Oncology Center, Baltimore, MD). Recombinant human soluble TRAIL (KillerTRAIL™) was obtained from Alexis (Axxora, San Diego, CA), and agonistic anti-Fas receptor antibody (clone CH11) was from Upstate Biotechnology (Lake Placid, NY). Etoposide, antimycin A, and N-acetyl-l-cysteine (NAC) were obtained from Sigma. CPT was obtained from the Drug Synthesis and Chemistry Branch, Division of Cancer Treatment, NCI, National Institutes of Health. BH3I-2′ was purchased from Calbiochem (EMD Biosciences, La Jolla, CA), and the caspase peptide inhibitor benzyloxycarbonyl-Val-Ala-dl-Asp(OMe)-fluoromethylketone (Z-VAD-fmk) was from Bachem (Torrance, CA).
Detection of Cellular Topoisomerase Cleavage Complexes—Topoisomerase cleavage complexes were detected using the in vivo complex of enzyme bioassay as described (22). Briefly, the cells were lysed in 1% Sarkosyl and homogenized. The cell lysates were centrifuged on cesium chloride step gradients at 165,000 × g for 20 h at 20 °C. Twenty 0.5-ml fractions were collected and diluted (v/v) into 25 mm potassium phosphate buffer, pH 6.6. The DNA-containing fractions (fractions 7–11) were pooled (except in Fig. 1A) and applied to polyvinylidene difluoride membranes (Immobilon-P, Millipore, MA) using a slot blot vacuum manifold. Topoisomerase cleavage complexes were detected by immunoblotting using the C21 Top1 mouse monoclonal antibody (1:1,000 dilution) (a kind gift from Dr. Yung-Chi Cheng, Yale University, New Haven, CT) or a Top2α mouse monoclonal antibody (clone Ki-S1, 1:1,000 dilution) from Chemicon International (Temecula, CA).
FIGURE 1.
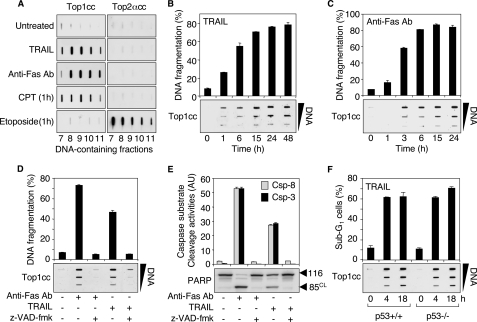
TRAIL and Fas induce apoptotic Top1cc. A, TRAIL (0.1 μg/ml, 48 h) and anti-Fas Ab (0.1 μg/ml, 48 h) induce Top1cc but fail to form Top2cc in Jurkat cells. CPT (1 μm, 1 h) and etoposide (100 μm, 1 h) were used as positive controls for Top1cc and Top2cc, respectively. The DNA-containing fractions were probed by immunoblotting with antibodies against Top1 and Top2α as indicated. B and C, the kinetics of formation of Top1cc parallel TRAIL- and Fas-induced DNA fragmentation. Jurkat cells were treated with 0.1 μg/ml TRAIL (B) or 0.1 μg/ml anti-Fas Ab (C) for the indicated times. Top panels, apoptotic DNA fragmentation (means ± S.D. of triplicate samples). Bottom panels, detection of Top1cc. The DNA-containing fractions were pooled and probed at three concentrations (10, 3, and 1 μg) with an antibody against Top1. D and E, the pan-caspase inhibitor Z-VAD-fmk inhibits TRAIL- and Fas-induced apoptotic Top1cc. Jurkat cells were treated with Z-VAD-fmk (100 μm, 30 min) before the addition of anti-Fas Ab (0.1 μg/ml) or TRAIL (0.1 μg/ml) for 6 h. D, top panel, apoptotic DNA fragmentation (means ± S.D. of triplicate samples). Bottom panel, detection of Top1cc as for B and C. E, top panel, IETD-AFC, (caspase-8 (Csp-8), gray bars) and DEVD-AFC (caspase-3 (Csp-3), black bars) peptide cleavage activity (means ± S.D. of triplicate samples). Bottom panel, whole cell extracts were examined for PARP cleavage by Western blotting. Molecular mass of PARP polypeptide and its cleaved product (CL) is indicated at right. F, TRAIL induces apoptotic Top1cc independently of p53. HCT116 cells of each genotype (p53+/+ and p53-/-) were treated with 0.1 μg/ml TRAIL for the indicated times. Top panel, percentages of cells with apoptotic sub-G1 DNA (means ± S.D. of triplicate samples). Bottom panel, detection of Top1cc as for B and C.
To identify the covalent DNA binding of the 80-kDa cleaved form of Top1 (see Fig. 4D, bottom panel), the DNA-containing fractions isolated as described above were precipitated with isopropanol (v/v) following addition of 0.5 m ammonium and a 10-min incubation at -20 °C. After centrifugation (10,000 × g, 15 min, 4 °C), the pellets (DNA-protein complexes) were washed with 70% ethanol. Following resuspension in 5 mm CaCl2 buffer containing protease inhibitors (Complete; Roche Applied Science), the DNA was digested with 50 units of DNase I for 30 min at room temperature. The reactions were stopped by the addition of loading buffer (v/v) (125 mm Tris-HCl, pH 6.8, 10% β-mercaptoethanol, 4.6% SDS, 20% glycerol, and 0.003% bromphenol blue), boiled, and applied to 8% SDS-PAGE gels. Top1 was detected by immunoblotting with the C21 Top1 monoclonal antibody.
FIGURE 4.
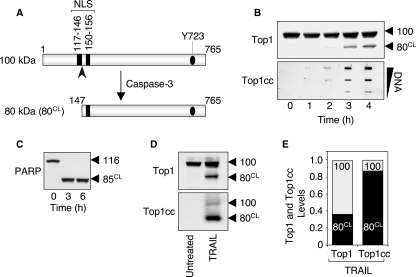
The caspase-3-cleaved Top1 catalytic polypeptide preferentially forms the apoptotic Top1cc induced by TRAIL. A, schematic representation of caspase-3-induced cleavage of Top1. Caspase-3 cleaves the 100-kDa native Top1 polypeptide after aspartate residue 146 (indicated by the arrowhead), and generates an 80-kDa C-terminal fragment (80CL) that still possesses one of the two nuclear localization signals (NLS) and the active site tyrosine (Tyr723, Y723) of the enzyme. B, time course of apoptotic Top1 cleavage and Top1cc in response to TRAIL. HCT116 cells were treated with 0.1 μg/ml TRAIL for the indicated times. Top panel, whole cell extracts were examined for caspase-mediated cleavage of Top1 by Western blotting. Molecular mass of Top1 polypeptide and its cleaved product (CL) are indicated at right. Bottom panel, detection of Top1cc. The DNA-containing fractions were pooled and probed at three concentrations (10, 3, and 1 μg) with an antibody against Top1. C, PARP is completely processed in response to TRAIL. HCT116 cells were treated with 0.1 μg/ml TRAIL for the indicated times. Whole cell extracts were examined for caspase-mediated cleavage of PARP by Western blotting. Molecular mass of PARP polypeptide and its cleaved product (CL) are indicated at right. D, the Top1 cleavage fragment is preferentially involved in the apoptotic Top1cc. HCT116 cells were treated with 0.1 μg/ml TRAIL for 18 h. Western blotting analyses of Top1 in whole cell extracts (top panel) and in the DNA-containing fractions isolated from CsCl gradient centrifugation (bottom panel) (see “Experimental Procedures”). E, quantification of the data shown in panel D. The 100-kDa native Top1 (100) is shown in gray, and the 80-kDa cleaved Top1 (80CL) is shown in black. Similar results were obtained following a 4-h TRAIL exposure (data not shown).
Western Blotting—Western blotting analyses on whole cell extracts were performed as described (16) using the C21 Top1 monoclonal antibody (1:1,000 dilution), the poly(ADP-ribose)-polymerase (PARP) polyclonal antibody (1:5,000 dilution) (Roche Applied Science), the γ-H2AX monoclonal antibody (1:2,000 dilution) (Upstate Biotechnology), and the tubulin monoclonal antibody (1:5,000 dilution) (clone Ab-4; NeoMarkers, Fremont, CA).
Identification of Apoptosis—Global DNA fragmentation was quantified by filter elution assay as described (23) and expressed as a percentage of fragmented DNA relative to total DNA. Oligonucleosomal DNA fragmentation was visualized by agarose gel electrophoresis using the quick apoptotic ladder kit from Biovision (Mountain View, CA). For sub-G1 analysis, DNA content was assessed by staining ethanol-fixed cells with propidium iodide and monitoring by FACScan (BD Biosciences, San Jose, CA). The number of cells with sub-G1 DNA content was determined with the CellQuest program (BD Biosciences). For electron microscopy, the cells were fixed in 0.1 m cacodylate buffer (pH 7.4) containing 2% glutaraldehyde and post-fixed in 1% osmium in same buffer. Then the cells were stained with 0.5% uranyl acetate, dehydrated in graded ethanol and propylene oxide, and infiltrated in equal volume of epoxy resin and propylene oxide overnight. The sample was embedded in epoxy resin and cured in 55 °C oven for 48 h. The cured block was thin sectioned and stained in uranyl acetate and lead citrate and examined by electron microscopy (Hitachi H700 microscope, Tokyo, Japan). Caspase activities were measured as described (18) using the fluorogenic peptide substrate Z-DEVD-AFC (caspase-3) and Z-IETD-AFC (caspase-8) from Calbiochem.
Confocal Microscopy Analyses of 8-Oxoguanine—After cytospin, Jurkat cells were fixed with 2% paraformaldehyde for 20 min and post-fixed/permeabilized with ice-cold 70% ethanol for 20 min. The cells were incubated with 5% bovine serum albumin for 1 h to block nonspecific binding before incubation with both mouse anti-8-oxoguanine (clone 483.15; Chemicon International) (1:250 dilution) and rabbit antihuman Top1 antibodies (raised against the 68-kDa C-terminal Top1 fragment; local source) (1:500 dilution) for 1.5 h. After washes, the cells were incubated with fluorescent secondary antibodies (Alexa-488 and Alexa-568; Molecular Probes, Eugene, OR) for 45 min. The slides were mounted using Vectashield mounting liquid (Vector Labs, Burlingame, CA), and fluorescences were visualized using a Nikon Eclipse TE-300 confocal laser microscope. Quantification of fluorescence intensity was determined using Adobe Photoshop 7.0 software and expressed as relative fluorescence intensity/cell.
Top1 Silencing by RNA Interference—Top1 expression was stably down-regulated in HCT116 cells by the transfection of U6 promoter-driven DNA vectors stably expressing small interfering RNA (siRNA) hairpins targeting human Top1 (HCT116siTop1; cDNA sequence: 5′-CTT GAC AGC CAA GGT ATT C-3′) or a negative control sequence (HCT116siCtrl; cDNA sequence, 5′-GCG TCC TTT CCA CAA GAT A-3′) as described (24). For transient down-regulation of Top1 expression, the cells were transfected with an siRNA duplex (Qiagen) against the sequence GGA CTC CAT CAG ATA CTA T from the Top1 mRNA. Transfections were performed using Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's protocol. A negative control siRNA duplex from Qiagen (target DNA sequence: TTC TCC GAA CGT GTC ACG T) was used. The Top1 knock-down experiments were carried out 72 h after transfection.
RESULTS
TRAIL and Fas Induce Apoptotic Top1cc—Trapped Top1cc can be detected in genomic DNA after cesium chloride gradient centrifugation and immunoblotting with Top1 antibody (16). In human leukemia Jurkat cells exposed to TRAIL or to an agonistic anti-Fas antibody (anti-Fas Ab) that mimics Fas ligand, we detected high levels of Top1cc (Fig. 1A). By contrast, topoisomerase IIα cleavage complexes (Top2αcc) were not detectable under conditions where etoposide (positive control) produced a clear signal (Fig. 1A). Similarly, Top2βcc were not induced by TRAIL (data not shown). Those experiments indicate that the binding of TRAIL and anti-Fas Ab to their plasma membrane receptors DR4/DR5 and Fas, respectively (3), effectively induce the formation of Top1cc.
TRAIL and Fas ligand are potent inducers of apoptosis in tumor cells (1, 2), suggesting that the observed Top1cc were apoptotic Top1cc resulting from the activation of cell death pathways. In line with this possibility, TRAIL and anti-Fas Ab induced Top1cc with kinetics that coincided with DNA fragmentation (Fig. 1, B and C). Top1cc and apoptotic DNA fragmentation were both detected after 1 h of exposure to TRAIL (or 3 h for anti-Fas Ab). Top1cc persisted throughout the apoptotic process as the DNA became extensively fragmented (Fig. 1, B and C).
To demonstrate that those Top1cc were due to apoptosis, we tested whether inhibition of the apoptotic pathways induced by TRAIL and anti-Fas Ab would prevent the formation of Top1cc. Pretreatment of cells with the pancaspase inhibitor Z-VAD-fmk inhibited TRAIL- and anti-Fas Ab-induced apoptosis, as well as the formation of Top1cc (Fig. 1D). As expected, Z-VAD-fmk also inhibited the activities of caspases 8 and 3 and the cleavage of PARP (Fig. 1E).
Because the formation of apoptotic Top1cc by UV-C radiation and colcemid was recently reported to involve p53 (17), we examined Top1cc in p53-/- HCT116 cells exposed to TRAIL. We found that p53+/+ and p53-/- HCT116 cells both displayed similar induction of Top1cc in response to TRAIL (Fig. 1F, bottom panel). Both cell lines also exhibited similar apoptotic response to TRAIL (Fig. 1F, top panel), which further demonstrates that TRAIL-induced apoptosis is p53-independent (25). Collectively, our experiments demonstrate a close association between the formation of Top1cc and the induction of p53-independent apoptosis in cells exposed to TRAIL.
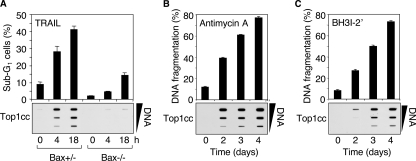
TRAIL-induced Top1cc Require Activation of the Mitochondrial Death Pathway—In most cancer cells, TRAIL induces apoptosis by increasing the permeability of mitochondrial membranes (3). Schematically, the proapoptotic Bax protein induces the formation of mitochondrial pores in the outer membrane by antagonizing the antiapoptotic Bcl-xL protein. Because the loss of Bax has been shown to protect HCT116 cells against TRAIL-induced apoptosis (25), we examined the formation of Top1cc in Bax+/- and Bax-/- HCT116 cells (21). Fig. 2A shows that Bax-/- cells failed to induce Top1cc (bottom panel). They were also defective in apoptosis (top panel). These experiments demonstrate that Bax plays a key role in TRAIL-induced apoptotic Top1cc.
FIGURE 2.
Activation of the mitochondrial pathway is required for the induction of apoptotic Top1cc by TRAIL. A, HCT116 cells of each genotype (Bax+/- and Bax-/-) were treated with 0.1 μg/ml TRAIL for the indicated times. Top panel, percentages of cells with apoptotic sub-G1 DNA (means ± S.D. of triplicate samples). Bottom panel, detection of Top1cc. The DNA-containing fractions were pooled and probed at three concentrations (10, 3, and 1 μg) with an antibody against Top1. B and C, antimycin A (10 μm) and BH3I-2′ (30 μm) induce apoptotic Top1cc in Jurkat cells. Top panels, apoptotic DNA fragmentation (means ± S.D. of triplicate samples). Bottom panels, detection of Top1cc as for A.
Next, we investigated whether direct activation of the apoptotic mitochondrial pathway (26) was sufficient to induce apoptotic Top1cc. Both the Bcl-2 homology domain 3 (BH3) mimetics antimycin A (27) (Fig. 2B) and BH3I-2′ (28) (Fig. 2C), which bind to and inhibit Bcl-xL, led to the induction of apoptotic Top1cc. Altogether, our results indicate a pivotal role of the mitochondrial death pathway for the induction of apoptotic Top1cc by TRAIL.
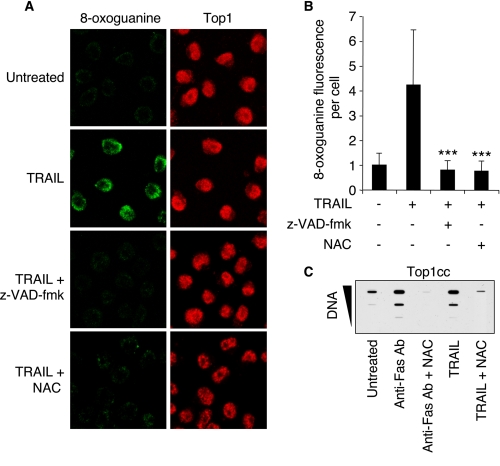
TRAIL-induced Apoptotic Top1cc Are Linked to the Generation of Oxidative DNA Lesions—Because Top1cc are readily induced by oxidative base damages (e.g. 8-oxoguanine (8-oxoG)), DNA strand breaks, and ROS (12, 29, 30), we searched for the presence of 8-oxoG in TRAIL-treated cells (9). Immunofluorescence confocal microscopy (Fig. 3, A and B) showed 8-oxoG nuclear staining in TRAIL-treated cells, indicative of TRAIL-induced oxidative damage (3, 31, 32). Moreover, caspases inhibition by Z-VAD-fmk (Fig. 1E) prevented TRAIL-induced 8-oxoG (Fig. 3, A and B), which is in agreement with recent studies showing ROS production by caspase activation (33, 34). Our results and these publications suggest that TRAIL-induced caspase activation leads to the generation of oxidative DNA lesions and apoptotic Top1cc.
FIGURE 3.
Relationship between oxidative DNA lesions and Top1cc in cell treated with TRAIL. A and B, Jurkat cells were preincubated for 30 min with Z-VAD-fmk (100 μm) or NAC (30 mm) before the addition of 0.1 μg/ml TRAIL for 6 h. A, confocal microscopy analysis of 8-oxoguanine (green) and Top1 (red). B, quantification of 8-oxoguanine fluorescence/cell (means ± S.D. of 100 cells). *** denotes statistically significant difference from TRAIL-treated cells (p < 0.001; unpaired t test). C, detection of Top1cc in Jurkat cells preincubated with 30 mm NAC for 30 min before the addition of anti-Fas Ab (0.1 μg/ml) or TRAIL (0.1 μg/ml) for 6 h. The DNA-containing fractions were pooled and probed at three concentrations (10, 3, and 1 μg) with an antibody against Top1.
To further demonstrate that TRAIL-induced apoptotic Top1cc were related to oxidative DNA lesions, we tested the effect of the potent antioxidant N-acetyl-l-cysteine (NAC). Fig. 3 shows that quenching ROS with NAC prevented both TRAIL-induced 8-oxoG (Fig. 3, A and B) and Top1cc (Fig. 3C).
Fas-induced apoptosis has also been associated with the intracellular accumulation of ROS (35) and with the generation of oxidative DNA lesions (36). Accordingly, NAC prevented the induction of Top1cc by anti-Fas Ab (Fig. 3C). Altogether, our findings indicate that both caspase activation and oxidative DNA damage are involved in the production of apoptotic Top1cc induced by death receptor activation.
Caspase-3-cleaved Top1 Is Preferentially Associated with the Apoptotic Top1cc Induced by TRAIL—We next investigated whether modifications of Top1 itself might also contribute to the formation of apoptotic Top1cc induced by TRAIL. The 100-kDa native Top1 protein is indeed a known substrate of caspase-3 (7). Caspase-3 cleaves Top1 after aspartate residue 146 and generates an 80-kDa C-terminal fragment (Fig. 4A) that remains capable of forming Top1cc (7, 37) and still possesses one functional nuclear localization signal (38) (Fig. 4A).
Fig. 4B (upper panel) shows that TRAIL induced the appearance of the proteolytic Top1 form (80CL) (7). Compared with the other caspase substrate, PARP (Fig. 4C), Top1 cleavage remained incomplete (Fig. 4B, upper panel) (7). This partial cleavage of Top1 coincided with the generation of apoptotic Top1cc in response to TRAIL (Fig. 4B, lower panel).
We next investigated which of the two forms of Top1, the 100-kDa native Top1 or the 80-kDa caspase-processed Top1, formed the apoptotic Top1cc. To address this question, TRAIL-induced Top1cc were isolated from the DNA-containing cesium chloride fractions. Following DNA digestion with DNase I, Western blotting was performed to determine the molecular mass of the Top1 molecules involved in the TRAIL-induced Top1cc. Approximately 90% of the Top1cc consisted of the truncated 80-kDa Top1 fragment (Fig. 4, D, bottom panel, and E). By contrast, the 80-kDa Top1 form corresponded to only ∼40% of total Top1 in whole cell extracts (Fig. 4, D, top panel, and E). These experiments demonstrate that the apoptotic Top1cc generated during TRAIL-induced apoptosis correspond primarily to the 80-kDa C-terminal fragment of Top1 generated by caspase-3.
Functional Relevance of TRAIL-induced Apoptotic Top1cc—First, to determine whether apoptotic Top1cc could directly contribute to the apoptotic DNA fragmentation, we determined the frequency of TRAIL-induced Top1cc. Using CPT as a calibrator (39), we estimated that TRAIL induced approximately one Top1cc/100 kbp in cells treated with TRAIL for 4 h (Fig. 5). That number scores relatively low compared with the observed DNA double-strand break frequency produced by apoptotic endonucleases (8) (Fig. 6, G and H). Thus, it is unlikely that apoptotic Top1cc contribute to the direct breakdown of genomic DNA.
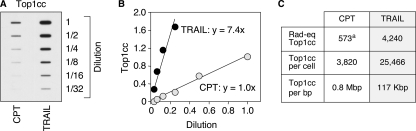
FIGURE 5.
Quantification of the apoptotic Top1cc induced by TRAIL. A, HCT116 cells were treated with 0.1 μg/ml TRAIL for 4 h. Top1cc were quantitated by pooling the DNA-containing fractions and probing with an antibody against Top1. The pools containing 10 μg of DNA were assayed by sequential 2-fold dilutions. HCT116 cells treated by CPT (0.5 μm, 1 h), for which the number of Top1cc/cell has been determined (39), were used to determine the genomic frequency of apoptotic Top1cc. B, quantification of the data shown in A. The curves displaying the levels of Top1cc as a function of the dilution of the samples show a 7.4-fold increased of Top1cc induced by TRAIL as compared with CPT (0.5 μm). C, table expressing the amount of rad equivalent (Rad-eq) Top1cc, and the number of Top1cc per cell and per bp in response to TRAIL. a, CPT (0.5 μm, 1 h) induces 573 rad equivalent Top1cc in HCT116 cells (39), and 1 rad equivalent Top1cc corresponds to ∼1 Top1cc/0.9 × 109 nucleotides (64). The values for the TRAIL-treated cells were obtained by multiplying by 7.4 (see B) those of the standard CPT.
FIGURE 6.
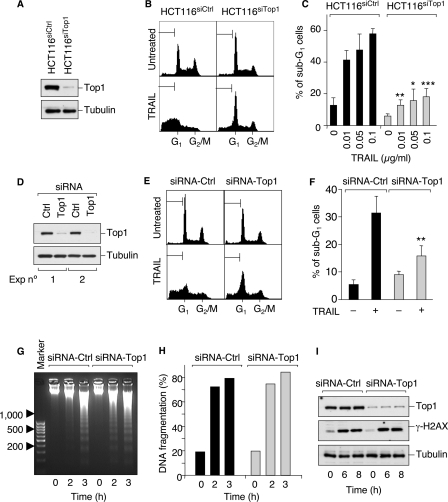
Apoptotic Top1cc are involved in TRAIL-induced apoptotic nuclear modifications. A–C, Top1 was silenced in HCT116 cells using U6 promoter-driven DNA vectors stably expressing siRNA hairpins targeting human Top1 (HCT116siTop1) or a negative control sequence (HCT116siCtrl). A, Top1 expression was examined by Western blotting. Tubulin was used as a loading control. B, cells were treated with 0.1 μg/ml TRAIL for 18 h. Apoptotic sub-G1 DNA contents are indicated by the brackets. C, percentage of cells with sub-G1 DNA (means ± S.D. of three independent experiments). The cells were treated with the indicated concentration of TRAIL for 18 h. Asterisks denote statistically significant difference from the TRAIL-treated HCT116siCtr cells (*, p < 0.05; **, p < 0.01; ***, p < 0.001; unpaired t test). D–I, HCT116 cells were transiently transfected with a duplex siRNA against Top1 (siRNA-Top1) or a negative control sequence (siRNA-Ctrl). D, Top1 expression was examined by Western blotting. Tubulin was used as a loading control. E, cells were treated with 0.1 μg/ml TRAIL for 3.5 h. Apoptotic sub-G1 DNA contents are indicated by the brackets. F, percentage of cells with sub-G1 DNA (means ± S.D. of four independent experiments). The cells were treated as in E. ** denotes statistically significant difference from the TRAIL-treated siRNA-Ctrl cells (p < 0.01; unpaired t test). G and H, cells were treated with 0.1 μg/ml TRAIL for the indicated times and DNA fragmentation was visualized by agarose gel electrophoresis (G) and quantified by filter elution assay (H). I, Western blotting analysis of Top1 and γ-H2AX in cells treated with 0.01 μg/ml TRAIL for the indicated times.
To address more directly the functional role of the apoptotic Top1cc, we wished to examine the impact of Top1 down-regulation on TRAIL-induced apoptosis. However, the lethality of knocking out Top1 in higher eukaryotes (11) precluded such an approach. Thus, we took advantage of HCT116 cells stably expressing siRNA targeting human Top1 (HCT116siTop1), which expressed low levels of Top1 as compared with cells expressing siRNA control sequence (HCT116siCtrl) (Fig. 6A) (24). Then we compared the apoptotic response of such HCT116siTop1 cells with that of HCT116siCtrl cells using flow cytometry. Typically, apoptotic cells are comprised in the sub-G1 fraction as they release apoptotic nuclear bodies. Fig. 6 (B and C) shows that Top1 down-regulation significantly reduced the number of nuclei with sub-G1 DNA content at the three concentrations of TRAIL tested, whereas the siRNA control cells remained sensitive to TRAIL. Similar results were obtained after transient transfections of siRNA duplex against Top1 (siRNA-Top1) versus control sequence (siRNA-Ctrl) in HCT116 cells (Fig. 6, D–F).
We then tested whether Top1 down-regulation affected TRAIL-induced oligonucleosomal DNA fragmentation or global DNA fragmentation. Fig. 6 (G and H) shows that this was not the case. Accordingly, phosphorylation of histone H2AX at Ser139 (γ-H2AX), a landmark marker for DNA double-strand breaks (40), was similar in siRNA-Top1 and siRNA-Ctrl cells (Fig. 6I). These results indicate that Top1 acts independently from the apoptotic endonucleases in promoting apoptotic bodies.
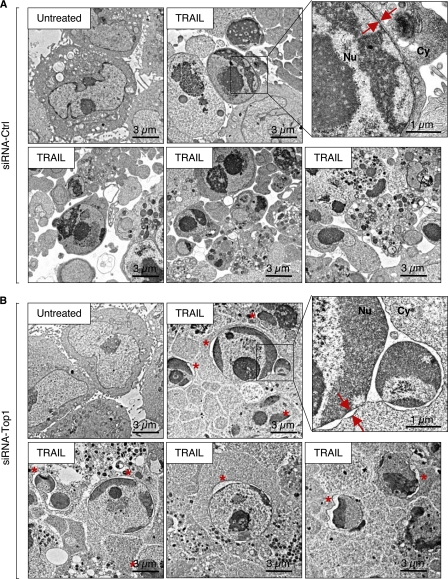
To further characterize how Top1 affected the formation of apoptotic nuclear bodies (see Fig. 6, B, C, E, and F), we performed electron microscopy analyses in TRAIL-treated siRNA-Top1 and siRNA-Ctrl cells. Representative examples of those analyses are shown in Fig. 7. Comparison of panels A (siRNA-Ctrl) and B (siRNA-Top1) suggests a yet undescribed role of Top1 in promoting apoptotic nuclear rearrangements. Indeed, nuclei remained bigger with less apoptotic nuclear bodies in the Top1-deficient cells than in the Top1-proficient cells (Fig. 7). In the Top1-deficient cells (Fig. 7B), chromatin formed a condensed area in close contact with long segments of the inner nuclear envelope. At those sites, the two layers of the nuclear envelope tended to separate, resulting in nuclear envelope distension (see asterisks in Fig. 7B). Such alterations, which concerned 44% ± 6 (means ± S.E.) of siRNA Top1 cells exposed to TRAIL (Fig. 7B), were not observed in the siRNA control cells (Fig. 7A). Indeed, the siRNA control cells formed well defined apoptotic bodies surrounded by plasma membrane, and chromatin was condensed at the center of well defined micronuclei (Fig. 7A).
FIGURE 7.
Nuclear morphology of Top1-deficient cells in response to TRAIL. Electron microcopy analysis of HCT116 cells transiently transfected with a duplex siRNA against Top1 (siRNA-Top1) (B) or a negative control sequence (siRNA-Ctrl) (A) before treatment with 0.1 μg/ml TRAIL for 2.5 h. A, TRAIL-treated siRNA-Ctrl cells show typical apoptotic features including cell shrinkage, highly condensed chromatin, numerous vesicles in the cytoplasm, and apoptotic bodies in the extracellular space. B, in TRAIL-treated siRNA-Top1 cells, chromatin is condensed mostly around the envelope of lobulated or fragmented nuclei and much less apoptotic bodies are observed. Condensed chromatin areas are in close contact with the inner nuclear envelope and are associated with nuclear envelope blebbing. The double arrows indicate inner and outer nuclear membranes. An asterisk indicates nuclear envelope blebbing in Top1-deficient TRAIL-treated cells. Nu, nucleus; Cy, cytoplasm.
Altogether, the Top1 down-regulation experiments suggest a role for Top1 in nuclear apoptosis, independent of apoptotic DNA fragmentation. Top1 appears to promote nuclear fission leading to the release of apoptotic bodies surrounded by plasma membrane.
DISCUSSION
The formation of Top1cc in apoptotic cells has been previously reported in response to cytotoxic drugs (15–18, 41, 42) as well as TNFα (17) and UV-C (19). It has been proposed that these Top1cc contribute to DNA fragmentation (15, 16) and histone release (41) during apoptosis. In the present study, we demonstrate for the first time that apoptotic Top1cc are induced by the endogenous ligands, TRAIL and Fas, downstream from mitochondrial activation and elevated ROS production. We also show that the apoptotic Top1cc induced by death receptor engagement preferentially consist of the caspase-3-cleaved Top1 fragment. Finally, our data provide evidence for a novel and unexpected function of Top1 in promoting nuclear fission and the release of apoptotic bodies, which are essential steps for the recognition and elimination of apoptotic cells by neighboring cells.
The Death Receptor Ligands TRAIL and Fas Induce Apoptotic Top1cc—This study demonstrates the formation of Top1cc by TRAIL and Fas ligand, two physiological cytokines that induce apoptosis by binding to their plasma membrane receptors DR4/DR5 and Fas, respectively (3). We have proposed to refer to these Top1cc as apoptotic Top1cc (15, 16, 18, 43) for the following reasons. First, they occur in the early phase of apoptosis and persist throughout the apoptotic process (Fig. 1, B and C). Second, inactivation of the apoptotic pathways by caspase inhibition (Fig. 1D) or by Bax inactivation (Fig. 2A) prevents the formation of Top1cc. Third, down-regulation of Top1 decreases the formation of apoptotic nuclear bodies (Figs. 6 and 7), and fourth, the formation of Top1cc appears to represent a general and conserved process during apoptosis. Indeed, besides TRAIL and Fas ligand (present study), Top1cc also form in apoptotic human cells in response to arsenic trioxide (15), staurosporine (16), tubulin and Top2 inhibitors (17, 18), TNFα (17), and UV-C radiation (19). Apoptotic Top1cc have also been reported in the parasite Leishmania donovani (42).
Although Top2 has been reported to participate in chromatin condensation (44) and in the excision of DNA loops (45) during apoptosis, we did not detect Top2αcc (Fig. 1A) or Top2βcc (data not shown) in apoptotic cells following exposure to TRAIL and anti-Fas Ab. Apoptotic Top1cc also formed in the absence of detectable Top2cc in prior studies with arsenic trioxide (15) or vinblastine (18). It is conceivable that the catalytic activity of Top2, rather than its stabilization on DNA as Top2cc, contributes to chromatin fragmentation. However, we cannot rule out the possibility that the levels of Top2cc formed during apoptosis are below the threshold of detection of our assays.
TRAIL-induced Apoptotic Top1cc Are Likely the Consequence of Oxidative DNA Lesions—Here, we show that oxidative DNA lesions (8-oxoG) are produced during TRAIL-induced apoptosis (Fig. 3, A and B) as a result of ROS production (9). Considering that oxidative DNA lesions are known to trap Top1cc (29, 30) and that quenching of ROS, which prevented TRAIL-induced 8-oxoG (Fig. 3, A and B), also prevented Top1cc (Fig. 3C), we propose that TRAIL-induced Top1cc form at sites of oxidative DNA lesions (Fig. 8). Besides oxidized nucleobases, it is possible that the DNA breaks produced by ROS and apoptotic nucleases (8) also contribute to the apoptotic Top1cc because Top1 can be directly trapped by DNA breaks (46). The ROS-dependent formation of oxidative DNA lesions appears to represent a common mechanism for the trapping of Top1cc during apoptosis as such a mechanism is also involved in response to arsenic trioxide (15), staurosporine (16), and etoposide (18). Recently, Sen et al. (42) reported the conservation of the ROS mechanism in L. donovani.
FIGURE 8.
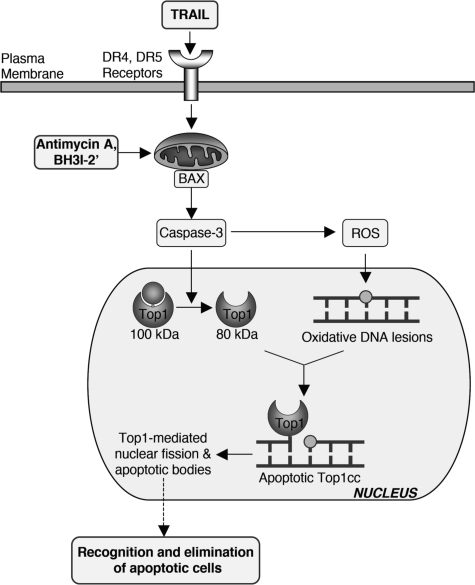
Proposed molecular pathways for the induction of apoptotic Top1cc by TRAIL. The binding of TRAIL to its DR4 and DR5 receptors activates the Bax-dependent mitochondrial death pathway, causing caspase-3 activation, and accumulation of ROS. The mitochondrial pathway can be activated directly by the BH3 mimetics antimycin A and BH3I-2′. Activated caspase-3 cleaves Top1 and generates ROS, which produce oxidative DNA lesions (8-oxoguanine, abasic sites, and strand breaks). Caspase-cleaved Top1 binds at the proximity of oxidative DNA lesions by forming apoptotic Top1cc (suicide complexes). Those complexes participate in apoptotic-related nuclear modifications including nuclear fission and the release of nuclear bodies in the extracellular space and may therefore contribute to the recognition and elimination of apoptotic cells.
Caspases have been involved in the generation of ROS (33, 34). Our study showing that caspase inhibition prevents TRAIL-induced oxidative DNA lesions (Fig. 3, A and B) and Top1cc (Fig. 1D) is consistent with the possibility that caspases serve, at least in part, to generate the ROS that lead to apoptotic Top1cc (Fig. 8).
Caspase-3-cleaved Top1 Is Preferentially Involved in the Apoptotic Top1cc Induced by TRAIL—Our study indicates that TRAIL-induced Top1cc consist preferentially in the 80-kDa C-terminal fragment of Top1 generated by caspase-3 (Figs. 4 and 8). It remains to be determined whether caspase-mediated cleavage of Top1 precedes Top1cc formation or whether Top1 is cleaved after Top1cc formation by the 100-kDa form, i.e. whether caspase-mediated cleavage of Top1 contributes to or results from Top1cc formation. Prior studies have shown that cleavage of Top1 to the 80-kDa form does not prevent its nuclear localization (38) and that 80-kDa Top1 retains its ability to form Top1cc in cells exposed to camptothecin (7). In vitro experiments have further demonstrated that truncated 68-kDa C-terminal recombinant Top1 is still catalytically active (37).
Although the N-terminal domain of Top1 is dispensable for Top1 catalytic activity (37), it binds to the E3 ubiquitin ligase Topors (47). Top1 ubiquitination and subsequent degradation by the 26 S proteasome (48) have been suggested as an early step in the repair of Top1cc prior to tyrosyl-DNA phosphodiesterase (Tdp1) action (49). PARP-1 also binds to the N-terminal of Top1 and facilitates the religation of Top1cc (50). It is therefore plausible that the caspase-3-dependent cleavage of Top1 serves to prevent the repair of Top1cc during apoptosis.
The cleavage of Top1 may also affect other cellular functions because the N-terminal domain of Top1 binds to proteins involved in the splicing of RNA (SF2/ASF (51)) and in the subnuclear localization of Top1 (nucleolin (52), SUMO-1 E2 enzyme UBC9 (53)) (for review, see Ref. 54).
Functional Involvement of Top1cc in TRAIL-induced Apoptosis—Our present study demonstrates the importance of Top1 for the apoptotic response. Indeed, knocking down Top1 by siRNA decreased TRAIL-induced nuclear fission and the release of apoptotic nuclear bodies (Figs. 6 and 7). The role of Top1 in promoting those nuclear events appears to represent a general phenomenon during apoptosis because siRNA-mediated Top1 down-regulation also reduces apoptosis-associated DNA content loss (sub-G1) in response to arsenic trioxide (15) and staurosporine (16). In addition, we found that Top1 down-regulation by siRNA had no detectable effect on apoptosis-associated DNA fragmentation in response to TRAIL (Fig. 6, G and H), suggesting that Top1 acts independently of apoptotic endonucleases in promoting apoptotic bodies. Prior studies have suggested that Top1 could also participate in DNA fragmentation based on the lack of oligonucleosomal and global DNA fragmentation in Top1-deficient cells (P388/CPT45) exposed to arsenic trioxide (15) and staurosporine (16). However, these results should be interpreted with caution, because P388/CPT45 cells were selected by continuous exposure to camptothecin (55), and other molecular changes within the apoptotic pathway may contribute to apoptotic defects in camptothecin-resistant cells (56). More recently, Ganguly et al. (41) reported that siRNA-mediated Top1 knock-down prevents apoptosis-associated cytoplasmic histone accumulation in response to staurosporine. Thus, besides nuclear fission (present study), Top1 can also play a role in chromatin breakdown during apoptosis. Although histone release seems to precede nuclear fission (57), it is still unclear whether these two events are connected or separated in two independent processes. Further studies will be required to determine how Top1 promotes these nuclear events during apoptosis induced by various agents in different cell lines.
In TRAIL-treated cells, Top1 deficiency is further associated with nuclear envelope separation that occurs principally in areas where chromatin is highly condensed (Fig. 7). Such separation of nuclear envelopes has also been reported in lamin-deficient Caenorhabditis elegans embryos (58). However, we did not find that Top1 down-regulation affects kinetics of TRAIL-induced lamin A/C degradation during apoptosis (data not shown). Nevertheless, one attractive possibility is that nuclear envelope blebbing prevents proper nuclear fission and/or the release of apoptotic nuclear bodies.
The Top1 siRNA knock-down experiments are consistent with the involvement of Top1cc in TRAIL-induced nuclear fission, apoptotic body release, and nuclear envelope blebbing. It is possible that these apoptotic events could depend on nonclassic functions of Top1, independently of its catalytic activity. Indeed, Top1 has been shown to regulate transcription initiation independently of its nicking closing activity (59, 60) and to promote phosphorylation of splicing factors (51, 61). Recently, nonclassical functions of Top1 have also been associated with DNA replication and genomic stability, nucleolar structure, and gene-specific transcription (in particular for asparagine synthetase expression) (24). It is possible that nonclassic functions of Top1 could be related to its N terminus, i.e. mediated by binding of Top1 with other nuclear proteins (54). Further studies will be required to determine which function(s) of Top1 is responsible for nuclear fission. However, such studies are not straightforward because there is presently no specific catalytic inhibitor of Top1 known, and the lethality of knocking out Top1 in eukaryotic cells (11) is a major obstacle for genetic approaches.
Based on the present experiments, we conclude that the physiological ligand TRAIL induces the formation of Top1cc during apoptosis in various TRAIL-sensitive human cancer cells: Jurkat cells, Bax+/-, and p53+/+ HCT116 cells. We propose that apoptotic Top1cc are preferentially produced by the 80-kDa C-terminal fragment of Top1 (generated by caspase-3) bound at the proximity of oxidative DNA lesions (8-oxoG) generated by TRAIL-induced ROS (Fig. 8). Moreover, we show that Top1 plays a role in apoptotic nuclear fission and the release of apoptotic nuclear bodies. Hence, Top1, which is ubiquitous and essential in mammals (11), may contribute to the proper recognition and elimination of apoptotic cells. At the organism level, the complete elimination of apoptotic cells is essential to prevent autoimmune disease (62), as well as chronic inflammation that may lead to cancers (63).
Acknowledgments
We thank Carlos Almanza and Ling-hua Meng for technical help.
This work was supported, in whole or in part, by a National Institutes of Health grant from the National Cancer Institute, Center for Cancer Research Intramural Research Program. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TRAIL, TNF-related apoptosis-inducing ligand; TNF, tumor necrosis factor; BH3, Bcl-2 homology domain 3; CPT, camptothecin; NAC, N-acetyl-l-cysteine; PARP, poly(ADP-ribose)-polymerase; ROS, reactive oxygen species; Top1, topoisomerase I; Top1cc, topoisomerase I cleavage complexes; Z, benzyloxycarbonyl; fmk, fluoromethylketone; siRNA, small interfering RNA; Ab, antibody; 8-oxoG, 8-oxoguanine.
References
- 1.Ashkenazi, A., Pai, R. C., Fong, S., Leung, S., Lawrence, D. A., Marsters, S. A., Blackie, C., Chang, L., McMurtrey, A. E., Hebert, A., DeForge, L., Koumenis, I. L., Lewis, D., Harris, L., Bussiere, J., Koeppen, H., Shahrokh, Z., and Schwall, R. H. (1999) J. Clin. Investig. 104 155-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walczak, H., Miller, R. E., Ariail, K., Gliniak, B., Griffith, T. S., Kubin, M., Chin, W., Jones, J., Woodward, A., Le, T., Smith, C., Smolak, P., Goodwin, R. G., Rauch, C. T., Schuh, J. C., and Lynch, D. H. (1999) Nat. Med. 5 157-163 [DOI] [PubMed] [Google Scholar]
- 3.Ashkenazi, A. (2002) Nat. Rev. Cancer 2 420-430 [DOI] [PubMed] [Google Scholar]
- 4.Scaffidi, C., Fulda, S., Srinivasan, A., Friesen, C., Li, F., Tomaselli, K. J., Debatin, K. M., Krammer, P. H., and Peter, M. E. (1998) EMBO J. 17 1675-1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo, X., Budihardjo, I., Zou, H., Slaughter, C., and Wang, X. (1998) Cell 94 481-490 [DOI] [PubMed] [Google Scholar]
- 6.Wei, M. C., Lindsten, T., Mootha, V. K., Weiler, S., Gross, A., Ashiya, M., Thompson, C. B., and Korsmeyer, S. J. (2000) Genes Dev. 14 2060-2071 [PMC free article] [PubMed] [Google Scholar]
- 7.Samejima, K., Svingen, P. A., Basi, G. S., Kottke, T., Mesner, P. W., Jr., Stewart, L., Durrieu, F., Poirier, G. G., Alnemri, E. S., Champoux, J. J., Kaufmann, S. H., and Earnshaw, W. C. (1999) J. Biol. Chem. 274 4335-4340 [DOI] [PubMed] [Google Scholar]
- 8.Samejima, K., and Earnshaw, W. C. (2005) Nat. Rev. Mol. Cell Biol. 6 677-688 [DOI] [PubMed] [Google Scholar]
- 9.Lee, M. W., Park, S. C., Yang, Y. G., Yim, S. O., Chae, H. S., Bach, J. H., Lee, H. J., Kim, K. Y., Lee, W. B., and Kim, S. S. (2002) FEBS Lett. 512 313-318 [DOI] [PubMed] [Google Scholar]
- 10.Wang, J. C. (2002) Nat. Rev. Mol. Cell Biol. 3 430-440 [DOI] [PubMed] [Google Scholar]
- 11.Morham, S. G., Kluckman, K. D., Voulomanos, N., and Smithies, O. (1996) Mol. Cell Biol. 16 6804-6809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pourquier, P., and Pommier, Y. (2001) Adv. Cancer Res. 80 189-216 [DOI] [PubMed] [Google Scholar]
- 13.Sirikantaramas, S., Yamazaki, M., and Saito, K. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 6782-6786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pommier, Y. (2006) Nat. Rev. Cancer 6 789-802 [DOI] [PubMed] [Google Scholar]
- 15.Sordet, O., Liao, Z., Liu, H., Antony, S., Stevens, E. V., Kohlhagen, G., Fu, H., and Pommier, Y. (2004) J. Biol. Chem. 279 33968-33975 [DOI] [PubMed] [Google Scholar]
- 16.Sordet, O., Khan, Q. A., Plo, I., Pourquier, P., Urasaki, Y., Yoshida, A., Antony, S., Kohlhagen, G., Solary, E., Saparbaev, M., Laval, J., and Pommier, Y. (2004) J. Biol. Chem. 279 50499-50504 [DOI] [PubMed] [Google Scholar]
- 17.Rockstroh, A., Kleinert, A., Kramer, M., Grosse, F., and Soe, K. (2007) Oncogene 26 123-131 [DOI] [PubMed] [Google Scholar]
- 18.Sordet, O., Goldman, A., and Pommier, Y. (2006) Mol. Cancer Ther. 5 3139-3144 [DOI] [PubMed] [Google Scholar]
- 19.Soe, K., Rockstroh, A., Schache, P., and Grosse, F. (2004) DNA Repair. (Amst.) 3 387-393 [DOI] [PubMed] [Google Scholar]
- 20.Bunz, F., Dutriaux, A., Lengauer, C., Waldman, T., Zhou, S., Brown, J. P., Sedivy, J. M., Kinzler, K. W., and Vogelstein, B. (1998) Science 282 1497-1501 [DOI] [PubMed] [Google Scholar]
- 21.Zhang, L., Yu, J., Park, B. H., Kinzler, K. W., and Vogelstein, B. (2000) Science 290 989-992 [DOI] [PubMed] [Google Scholar]
- 22.Subramanian, D., Kraut, E., Staubus, A., Young, D. C., and Muller, M. T. (1995) Cancer Res. 55 2097-2103 [PubMed] [Google Scholar]
- 23.Bertrand, R., Kohn, K. W., Solary, E., and Pommier, Y. (1995) Drug Dev. 34 138-144 [Google Scholar]
- 24.Miao, Z. H., Player, A., Shankavaram, U., Wang, Y. H., Zimonjic, D. B., Lorenzi, P. L., Liao, Z. Y., Liu, H., Shimura, T., Zhang, H. L., Meng, L. H., Zhang, Y. W., Kawasaki, E. S., Popescu, N. C., Aladjem, M. I., Goldstein, D. J., Weinstein, J. N., and Pommier, Y. (2007) Cancer Res. 67 8752-8761 [DOI] [PubMed] [Google Scholar]
- 25.Ravi, R., and Bedi, A. (2002) Cancer Res. 62 1583-1587 [PubMed] [Google Scholar]
- 26.Grad, J. M., Cepero, E., and Boise, L. H. (2001) Drug Resist. Updates 4 85-91 [DOI] [PubMed] [Google Scholar]
- 27.Tzung, S. P., Kim, K. M., Basanez, G., Giedt, C. D., Simon, J., Zimmerberg, J., Zhang, K. Y., and Hockenbery, D. M. (2001) Nat. Cell Biol. 3 183-191 [DOI] [PubMed] [Google Scholar]
- 28.Degterev, A., Lugovskoy, A., Cardone, M., Mulley, B., Wagner, G., Mitchison, T., and Yuan, J. (2001) Nat. Cell Biol. 3 173-182 [DOI] [PubMed] [Google Scholar]
- 29.Daroui, P., Desai, S. D., Li, T. K., Liu, A. A., and Liu, L. F. (2004) J. Biol. Chem. 279 14587-14594 [DOI] [PubMed] [Google Scholar]
- 30.Pourquier, P., Ueng, L.-M., Fertala, J., Wang, D., Park, H.-J., Essigman, J. M., Bjornsti, M.-A., and Pommier, Y. (1999) J. Biol. Chem. 274 8516-8523 [DOI] [PubMed] [Google Scholar]
- 31.Izeradjene, K., Douglas, L., Tillman, D. M., Delaney, A. B., and Houghton, J. A. (2005) Cancer Res. 65 7436-7445 [DOI] [PubMed] [Google Scholar]
- 32.Mohr, A., Buneker, C., Gough, R. P., and Zwacka, R. M. (2007) Oncogene [DOI] [PubMed]
- 33.Ricci, J. E., Gottlieb, R. A., and Green, D. R. (2003) J. Cell Biol. 160 65-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ricci, J. E., MunoZ-Pinedo, C., Fitzgerald, P., Bailly-Maitre, B., Perkins, G. A., Yadava, N., Scheffler, I. E., Ellisman, M. H., and Green, D. R. (2004) Cell 117 773-786 [DOI] [PubMed] [Google Scholar]
- 35.Shakibaei, M., Schulze-Tanzil, G., Takada, Y., and Aggarwal, B. B. (2005) Antioxid. Redox. Signal 7 482-496 [DOI] [PubMed] [Google Scholar]
- 36.Nathan, I., Dizdaroglu, M., Bernstein, L., Junker, U., Lee, C., Muegge, K., and Durum, S. K. (2000) Cytokine 12 881-887 [DOI] [PubMed] [Google Scholar]
- 37.Pommier, Y., Laco, G. S., Kohlhagen, G., Sayer, J. M., Kroth, H., and Jerina, D. M. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 10739-10744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mo, Y. Y., Wang, C., and Beck, W. T. (2000) J. Biol. Chem. 275 41107-41113 [DOI] [PubMed] [Google Scholar]
- 39.Goldwasser, F., Bae, I., Valenti, M., Torres, K., and Pommier, Y. (1995) Cancer Res. 55 2116-2121 [PubMed] [Google Scholar]
- 40.Rogakou, E. P., Boon, C., Redon, C., and Bonner, W. M. (1999) J. Cell Biol. 146 905-916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ganguly, A., Das, B., Roy, A., Sen, N., Dasgupta, S. B., Mukhopadhayay, S., and Majumder, H. K. (2007) Cancer Res. 67 11848-11858 [DOI] [PubMed] [Google Scholar]
- 42.Sen, N., Banerjee, B., Das, B. B., Ganguly, A., Sen, T., Pramanik, S., Mukhopadhyay, S., and Majumder, H. K. (2007) Cell Death Differ. 14 358-367 [DOI] [PubMed] [Google Scholar]
- 43.Sordet, O., Khan, Q. A., and Pommier, Y. (2004) Cell Cycle 3 1095-1097 [PubMed] [Google Scholar]
- 44.Durrieu, F., Samejima, K., Fortune, J. M., Kandels-Lewis, S., Osheroff, N., and Earnshaw, W. C. (2000) Curr. Biol. 10 923-926 [DOI] [PubMed] [Google Scholar]
- 45.Li, T. K., Chen, A. Y., Yu, C., Mao, Y., Wang, H., and Liu, L. F. (1999) Genes Dev. 13 1553-1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pourquier, P., Pilon, A. A., Kohlhagen, G., Mazumder, A., Sharma, A., and Pommier, Y. (1997) J. Biol. Chem. 272 26441-26447 [DOI] [PubMed] [Google Scholar]
- 47.Haluska, P., Jr., Saleem, A., Rasheed, Z., Ahmed, F., Su, E. W., Liu, L. F., and Rubin, E. H. (1999) Nucleic Acids Res. 27 2538-2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Desai, S. D., Li, T. K., RodrigueZ-Bauman, A., Rubin, E. H., and Liu, L. F. (2001) Cancer Res. 61 5926-5932 [PubMed] [Google Scholar]
- 49.Pommier, Y., Barcelo, J. M., Rao, V. A., Sordet, O., Jobson, A. G., Thibaut, L., Miao, Z. H., Seiler, J. A., Zhang, H., Marchand, C., Agama, K., Nitiss, J. L., and Redon, C. (2006) Prog. Nucleic Acids Res. Mol. Biol. 81 179-229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park, S. Y., and Cheng, Y. C. (2005) Cancer Res. 65 3894-3902 [DOI] [PubMed] [Google Scholar]
- 51.Rossi, F., Labourier, E., Forne, T., Divita, G., Derancourt, J., Riou, J. F., Antoine, E., Cathala, G., Brunel, C., and Tazi, J. (1996) Nature 381 80-82 [DOI] [PubMed] [Google Scholar]
- 52.Bharti, A. K., Olson, M. O., Kufe, D. W., and Rubin, E. H. (1996) J. Biol. Chem. 271 1993-1997 [DOI] [PubMed] [Google Scholar]
- 53.Rallabhandi, P., Hashimoto, K., Mo, Y. Y., Beck, W. T., Moitra, P. K., and D'Arpa, P. (2002) J. Biol. Chem. 277 40020-40026 [DOI] [PubMed] [Google Scholar]
- 54.Leppard, J. B., and Champoux, J. J. (2005) Chromosoma 114 75-85 [DOI] [PubMed] [Google Scholar]
- 55.Mattern, M. R., Hofmann, G. A., Polsky, R. M., Funk, L. R., McCabe, F. L., and Johnson, R. K. (1993) Oncol. Res. 5 467-474 [PubMed] [Google Scholar]
- 56.Reinhold, W. C., Kouros-Mehr, H., Kohn, K. W., Maunakea, A. K., Lababidi, S., Roschke, A., Stover, K., Alexander, J., Pantazis, P., Miller, L., Liu, E., Kirsch, I. R., Urasaki, Y., Pommier, Y., and Weinstein, J. N. (2003) Cancer Res. 63 1000-1011 [PubMed] [Google Scholar]
- 57.Gabler, C., Blank, N., Hieronymus, T., Schiller, M., Berden, J. H., Kalden, J. R., and Lorenz, H. M. (2004) Ann. Rheum. Dis. 63 1135-1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cohen, M., Tzur, Y. B., Neufeld, E., Feinstein, N., Delannoy, M. R., Wilson, K. L., and Gruenbaum, Y. (2002) J. Struct. Biol. 140 232-240 [DOI] [PubMed] [Google Scholar]
- 59.Merino, A., Madden, K. R., Lane, W. S., Champoux, J. J., and Reinberg, D. (1993) Nature 365 227-232 [DOI] [PubMed] [Google Scholar]
- 60.Shykind, B. M., Kim, J., Stewart, L., Champoux, J. J., and Sharp, P. A. (1997) Genes Dev. 11 397-407 [DOI] [PubMed] [Google Scholar]
- 61.Soret, J., Gabut, M., Dupon, C., Kohlhagen, G., Stevenin, J., Pommier, Y., and Tazi, J. (2003) Cancer Res. 63 8203-8211 [PubMed] [Google Scholar]
- 62.Napirei, M., Karsunky, H., Zevnik, B., Stephan, H., Mannherz, H. G., and Moroy, T. (2000) Nat. Genet. 25 177-181 [DOI] [PubMed] [Google Scholar]
- 63.Coussens, L. M., and Werb, Z. (2002) Nature 420 860-867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kohn, K. W., Ewig, R. A., Erickson, L. C., and Zwelling, L. A. (1981) DNA Repair: A Laboratory Manual of Research Techniques, pp. 379-401, Marcel Dekker, New York