Abstract
The translation of the cyclin D1 and c-myc mRNAs occurs via internal ribosome entry site (IRES)-mediated initiation under conditions of reduced eIF-4F complex formation and Akt activity. Here we identify hnRNP A1 as an IRES trans-acting factor that regulates cyclin D1 and c-myc IRES activity, depending on the Akt status of the cell. hnRNP A1 binds both IRESs in vitro and in intact cells and enhances in vitro IRES-dependent reporter expression. Akt regulates this IRES activity by inducing phosphorylation of hnRNP A1 on serine 199. Serine 199-phosphorylated hnRNP A1 binds to the IRESs normally but is unable to support IRES activity in vitro. Reducing expression levels of hnRNP A1 or overexpressing a dominant negative version of the protein markedly inhibits rapamycin-stimulated IRES activity in cells and correlated with redistribution of cyclin D1 and c-myc transcripts from heavy polysomes to monosomes. Importantly, knockdown of hnRNP A1 also renders quiescent Akt-containing cells sensitive to rapamycin-induced G1 arrest. These results support a role for hnRNP A1 in mediating rapamycin-induced alterations of cyclin D1 and c-myc IRES activity in an Akt-dependent manner and provide the first direct link between Akt and the regulation of IRES activity.
A majority of eukaryotic mRNAs contain 5′-UTRs2 that are relatively unstructured and typically less than 100 nucleotides in length, which allows for efficient cap-dependent translation initiation. However, the leaders of some cellular mRNAs are relatively long and highly structured and can contain multiple upstream AUG or CUG codons such that scanning ribosomes are unlikely to efficiently initiate translation. In a number of these mRNAs, translation initiation is mediated by cap-independent mechanisms via an internal ribosome entry site (1). IRES-mediated translation initiation can occur during a variety of physiological conditions and has been reported to promote initiation for several mRNAs during cell cycle progression, differentiation, and apoptosis and during stress responses (2–6). IRESs are thought to directly recruit the ribosome to within close proximity to the start codon, thus bypassing the need for cap binding and ribosome scanning (7). Our previous data have demonstrated that both the cyclin D1 and c-myc mRNAs contain IRESs whose function is markedly enhanced following the inhibition of cap-dependent initiation by rapamycin in a manner dependent on Akt activity (8). In cells containing quiescent Akt, the IRESs of the cyclin D1 and c-myc mRNAs are constitutively active and are stimulated following rapamycin treatment; however, in cells containing active Akt cyclin D1 and c-myc, IRES activity is repressed and is not induced following rapamycin exposure.
Several proteins that regulate IRES activity, collectively termed IRES trans-acting factors (ITAFs), have been described (7). These ITAFs function by associating with the IRES and either facilitate direct ribosome binding with the mRNA or alter the structure of the IRES. For instance, the ITAFs PTB, Unr, and hnRNP C1/C2 have been shown to stimulate several IRESs, whereas it has been demonstrated that the ITAF HuR inhibits p27Kip1 IRES activity (9–12). Although several ITAFs have been described that affect the c-myc IRES (4), the factors that regulate the cyclin D1 IRES are unknown. Moreover, which ITAF(s) regulate both of these IRESs in an Akt-dependent manner coordinately is also unknown.
hnRNP A1 is a well studied ubiquitously expressed protein that has important roles in pre-mRNA and mRNA metabolism (13). hnRNP A1 binds nascent pre-mRNA in a sequence-specific manner and promotes the annealing of cRNA strands (14–16). hnRNP A1 also has demonstrated roles in nuclear export of mature mRNAs, mRNA turnover, and both cap-dependent and IRES-mediated translation initiation (17–20). Although primarily nuclear, hnRNP A1 continually shuttles between the nucleus and cytoplasm, and this nucleocytoplasmic shuttling activity depends on ongoing RNA polymerase II transcription and the integrity of the M9 domain, a 38-amino acid region that regulates its localization (21).
The Akt kinase is known to signal to the translational machinery via mTORC1, and activation of cap-dependent protein synthesis by Akt is associated with tumor formation and sensitivity to various chemotherapeutics (22). The activation of cap-dependent protein synthesis by Akt is phylogenetically conserved and Akt is known to directly activate a number of canonical translation initiation factors and stimulate ribosome biogenesis (23). The role of Akt signaling in IRES-mediated translation initiation is not clear. However, recently, the 14-3-3 proteins have been identified as critical factors that are required for IRES-mediated expression of the cyclin-dependent kinase Cdk11 (p58PITSLRE) during mitotic translation (24). It is known that specific 14-3-3 isoforms are regulated by Akt signaling (25), suggesting a link between IRES-mediated initiation and Akt signaling events. Thus, based on our previous studies, which implicated Akt signaling in the regulation of constitutive and rapamycin-stimulated cyclin D1 and c-myc IRES activity (8), we have investigated the mechanism by which this control takes place for these IRESs.
We report here that hnRNP A1 constitutively binds to both the cyclin D1 and c-myc IRESs and is a direct substrate of Akt. Knockdown of hnRNP A1 levels via RNA interference or overexpression of a dominant negative mutant markedly inhibits cyclin D1 and c-myc IRES activity in response to rapamycin in an Akt-dependent manner. Our data also suggest that Akt negatively regulates hnRNP A1-mediated IRES activity via phosphorylation at Ser199.
MATERIALS AND METHODS
Cell Lines, Transductions, and Transfections—The U87, U87PTEN, PTEN-/- and PTEN+/+ MEF cell lines have been described previously (8) and exhibit markedly different Akt activities as a result of their PTEN status. The LAPC-4puro and LAPC-4myrAKT lines have also been previously described (8) and differ in that LAPC-4myrAKT overexpresses a constitutively activated allele of Akt1, whereas LAPC-4puro is the matched control line transduced with the empty retroviral vector. All other cell lines were obtained from ATCC (Manassas, VA). Viral transduction of U87 and U87PTEN cells was performed as previously described (26). Briefly, infectious supernatants from transiently transfected Phoenix cells transfected with the LXSP or LXSP-NLS-A1 (26) (kind gift from Dr. Danilo Perrotti (Department of Molecular Virology, Immunology, and Medical Genetics, Ohio State University)) retroviral constructs were used to transduce U87 and U87PTEN cells. After infection, cells were split at varying dilutions and plated in the presence of puromycin (2.5 μg/ml), and resistant colonies were analyzed after 10 days. For DNA transfections, the indicated dicistronic reporter constructs were transfected into cells using X-treme GENE Q2 transfection reagent (Roche Applied Science) and normalized for transfection efficiency by co-transfection with pSVβGal (Promega, Madison, WI). Cells were harvested 18 h following transfection, and Renilla, firefly, and β-galactosidase activities were determined (Dual-Glo luciferase and β-galactosidase assay systems; Promega). Cell cycle distributions were determined by propidium iodide staining of cells, followed by flow cytometry. Statistical analysis was performed by using Student's t test and analysis of variance models using SigmaStat (Systat Software, San Jose, CA).
Constructs, Recombinant Proteins, and Antibodies—The parental dicistronic reporter constructs pRCD1F and pRmycF have been previously described (8) and were used to amplify the indicated cyclin D1 or c-myc 5′-UTR sequences, which were subsequently inserted into pRF for the deletion analysis. All constructs used for the yeast three-hybrid analysis were based on the RNA-Protein Hybrid Hunter System (Invitrogen). cDNAs derived from mRNA isolated from U87PTEN cells were inserted into either pYesTrp3 (VP16 activation domain fusions) or pYesTrp2 (B42 activation domain fusions) and were screened for positive interactors using the 165 nucleotide cyclin D1 or 233 nucleotide c-myc IRESs, which were inserted into the hybrid RNA expressing plasmid pRH5′ in either the sense or antisense orientations. Only clones that displayed IRES sequence-specific and -dependent growth were analyzed further. pRH3′-IRE and pYesTrp2-IRP were from Invitrogen. pGEX-2T/hnRNP A1 (full-length hnRNP A1-GST fusion) was kindly provided by Dr. Ronald Hay (Institute of Biomolecular Sciences, University of St. Andrews) (27). The hnRNP A1-GST fusion S199A (A1-S199A) was constructed by mutating pGEX-2T/hnRNP A1 using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The full-length hnRNP A1-GST and S199A-GST were amplified and cloned into pcDNA3.1 (Invitrogen) for mammalian expression. pLNCX HA Akt1 and pLNCX HA Akt1 K179M (Dr. William Sellers, Dana-Farber Cancer Institute, Boston, MA) (28, 29) were used as templates to amplify the HA-tagged Akt1 and HA-tagged Akt1 K179M coding sequences, which were subsequently cloned into pcDNA3.1 to generate pcDNA3 HA Akt1 (HA-Akt) and pcDNA3 HA Akt1 K179M (HA-AktKD), respectively. All plasmids were verified by sequencing, and primer information is available upon request. Purified glyceraldehyde-3-phosphate dehydrogenase was obtained from Sigma. Antibodies were from the following sources. Anti-GST was from MBL International Corp. (Woburn, MA); anti-hnRNP A1 was from Abcam (Cambridge, MA); anti-phospho-Akt-substrate, anti-Akt, anti-phospho-(Thr308)-Akt, and anti-phospho-(Ser473)-Akt were from Cell Signaling (Danvers, MA); anti-HA antibodies 3F10 and 12CA5 were from Roche Applied Science; cyclin D1 was from BD Pharmingen; c-myc (clone 9E11) was from Upstate Biotechnology; and anti-actin antibody was from Sigma. Serine 199 hnRNP A1 phospho-specific antibodies were generated in rabbits immunized with the phosphorylated peptide SQRGRSG-pSGNFGGGR (where pS represents phosphoserine) and subsequently affinity-purified. Rapamycin, LY294002, and wortmannin were from Calbiochem.
Yeast Methods and Screening—Standard methods were employed for screening and yeast growth (30). The yeast three-hybrid analysis was performed in strain L40uraMS2 (MATa, ura3–52, leu2–3112, his3Δ200, trp1Δ1, ade2, LYS2::(LexA op)4-HIS3, ura3::(LexA-op)8-LacZ) carrying pLexA/MS2/Zeo. This strain is a derivative of L40-coat, described previously (31). All plasmids used were amplified in Escherichia coli strain DH5α. The indicated hybrid-RNA expression constructs and the constructs expressing the activation domain fusions or the activation domain construct only were cotransformed into L40uraMS2, as described (32). Transformants were grown on selective media containing 5 mm 3-aminotriazole.
RNA and Protein Analysis—For RNA-EMSA, either cyclin D1 or c-myc IRES RNAs were in vitro transcribed (mMessage T7 transcription kit; Ambion, Austin, TX) as previously described (8) and 5′-end-labeled with [γ-32P]ATP (GE Healthcare) using T4 polynucleotide kinase. Radiolabeled RNA was incubated with purified GST-hnRNP A1 in binding buffer containing 10 mm HEPES (pH 7.5), 90 mm potassium acetate, 1.5 mm magnesium acetate, 2.5 mm dithiothreitol, and 40 units of SUPERase-IN (Ambion) for 30 min at 30 °C. For supershift analysis, anti-GST antibody was added, and all subsequent procedures were as described in Ref. 33. Immunoprecipitation and RT-PCR were performed as described in Ref. 34. For the RNA pull-down assay (35), cytoplasmic lysates were prepared by hypotonic lysis in buffer containing 10 mm HEPES (pH 7.5), 10 mm potassium acetate, 1.5 mm magnesium acetate, 2.5 mm dithiothreitol, 0.05% Nonidet P-40, 10 mm NaF, 1 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, and 1.5% aprotinin using a Dounce homogenizer. Extracts were precleared by centrifugation, and SUPERase-IN (0.025 units/ml) and yeast tRNA (15 μg/ml) were added and subsequently applied to an equilibrated heparin-agarose column (Bio-Rad). Eluates were further cleared with 100 μl of streptavidin-Sepharose (Sigma) for 1 h at 4 °C. Following centrifugation, 10 μg of in vitro transcribed biotinylated RNA was added to the supernatant and incubated at 4 °C for 1 h. The protein and biotinylated RNA complexes were recovered by the addition of 30 μl of streptavidin-Sepharose, which was incubated for 2 h at 4°C. The streptavidin-Sepharose RNA-protein complexes were then washed five times in binding buffer (10 mm HEPES (pH 7.5), 90 mm potassium acetate, 1.5 mm magnesium acetate, 2.5 mm dithiothreitol, 0.05% Nonidet P-40, 10 mm NaF, 1 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, and 1.5% aprotinin) and then boiled in SDS and resolved by gel electrophoresis. For endogenous coimmunoprecipitations, treated cells were washed with ice-cold PBS and lysed in 0.2% Nonidet P-40 lysis buffer containing 1 mm EDTA, 10 mm NaF, 1 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, and 1.5% aprotinin. Clarified lysates were precleared with protein G-Sepharose (GE Healthcare), and Akt or hnRNP A1 was immunoprecipitated, followed by protein G-Sepharose. Immunoprecipitation of Akt was also performed with or without preincubation with an Akt peptide (Cell Signaling). Polysome analysis was performed as described previously (8). Briefly, cells were lysed in buffer containing 100 μg/ml cycloheximide at 4 °C. Following removal of mitochondria and nuclei, supernatants were layered onto 15–50% sucrose gradients and centrifuged at 38,000 rpm for 2 h at 4 °C in a SW 40 rotor (Beckman Instruments). Gradients were fractionated into eleven 1-ml fractions using a density gradient fractionator system (Brandel Instruments). The profiles of the gradients were monitored at 260 nm, and RNAs from individual fractions were pooled into a nonribosomal/monosomal pool and a polysomal pool. These RNAs (100 ng) were subsequently used in real time quantitative RT-PCR analysis for the indicated transcripts using amplicons located within the coding region. Real time amplifications were carried out on a Eppendorf Mastercycler equipped with a Realplex2 optical module (Eppendorf AG, Germany) and normalized to actin mRNA levels, as previously described (36). Primers for the amplicons are available upon request. For immunostaining, U87 and U87PTEN cells were grown on coverslips and fixed with 3.7% paraformaldehyde for 15 min at room temperature and permeabilized with 0.5% Triton X-100 plus phosphate-buffered saline for 5 min at 4 °C. Cells were then stained with anti-HA tag or anti-hnRNP A1 antibodies in phosphate-buffered saline containing 0.5% gelatin and 0.25% bovine serum albumin for 1.5 h. Samples were washed three times in phosphate-buffered saline containing 0.25% gelatin and incubated with an appropriate fluorochrome-labeled secondary antibody. Samples were washed three times in phosphate-buffered saline with 0.25% gelatin and mounted for analysis on an Olympus IX70 fluorescent microscope.
Filter Binding Assay—The indicated amounts of GST-hnRNP A1 were added to in vitro transcribed 32P-labeled RNAs corresponding to either the cyclin D1 or c-myc IRESs in separate reactions in a volume of 10 μl in buffer containing 5 mm HEPES (pH 7.6), 30 mm KCl, 2 mm MgCl2, 200 mm dithiothreitol, 4% glycerol, and 10 ng of yeast tRNA for 10 min at room temperature (20). For competition experiments, the indicated amounts of unlabeled competitor RNA were added to the reaction. The p27Kip1 IRES RNA was prepared as described previously (8). 8 μl of each binding reaction was applied to nitrocellulose membranes on a slot blot apparatus (Minifold II; Schleicher & Schuell). Membranes were washed and dried, and signals were quantified using a PhosphorImager. Binding curves of three independent experiments were fitted by using SigmaPlot to determine the apparent dissociation constants.
[32P]Orthophosphate Labeling—Transfected 293 cells were washed twice and incubated in phosphate-free Dulbecco's modified Eagle's medium for 1 h and then incubated with 100 μCi of [32P]orthophosphate/ml for 2 h in the presence or absence of dialyzed fetal bovine serum (Omega Scientific, Tarzana, CA). Following the indicated treatments, cells were washed twice with ice-cold Tris-buffered saline and lysed in 1% Nonidet P-40 lysis buffer (20 mm Tris (pH 8.0), 200 mm NaCl, 10% glycerol, 1 mm EDTA, 12 mm β-glycerophosphate, 10 mm NaF, 1 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, and 1.5% aprotinin). After lysates were clarified, GST-hnRNP A1 was immunoprecipitated with anti-GST antibody, followed by protein G-Sepharose (GE Healthcare). 32P incorporation into hnRNP A1 was visualized after SDS-PAGE and transfer to polyvinylidene difluoride (Bio-Rad) using a PhosphorImager. Quantitative densitometric analysis was performed with ImageQuant (Molecular Dynamics).
In Vitro Phosphorylation—GST fusion proteins were isolated using GST Purification Modules as recommended by the manufacturer (GE Healthcare). In vitro phosphorylation was performed using an Akt kinase kit (Cell Signaling Technology) using 200 ng of activated Akt (Upstate Biotechnology) and 500 ng of the indicated GST fusion protein in each reaction. Immunoblotting was performed using polyclonal anti-phospho-Akt substrate antibody (Cell Signaling Technology).
RNA Interference Analysis—siRNA transfections targeting human hnRNP A1 were performed using synthetic oligonucleotides (ON-TARGETplus SMARTpool™, Dharmacon, Lafayette, CO) directed at sequences within the coding region and 3′-UTR. An siRNA with a scrambled sequence was used as a negative targeting control.
In Vitro Translation of Dicistronic mRNA Reporters—The dicistronic plasmids were linearized using BamHI and capped RNA transcribed in vitro (mMessage T7 transcription kit; Ambion). Capped RNA transcripts were used to program extracts of U87 or U87PTEN cell lines as described previously (8).
RESULTS
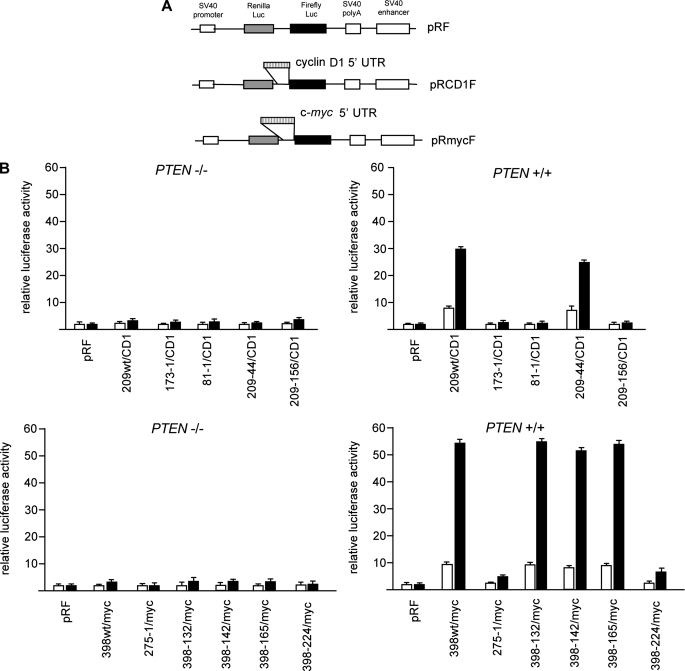
Identification of hnRNP A1 as a Cyclin D1 and c-myc 5′-UTR-binding Protein—Since we had previously demonstrated that the 5′-UTRs of the cyclin D1 and c-myc mRNAs possessed Akt-dependent IRES activity, we initially sought to identify which parts of the leader sequences were responsible for these effects. Thus, we generated deletion constructs of both the human cyclin D1 and c-myc 5′-UTRs and tested whether the dicistronic reporter mRNAs demonstrated Akt-dependent IRES activity following rapamycin exposure by transient transfection of PTEN-/- or PTEN+/+ MEFs. These constructs contained either cyclin D1 or c-myc mRNA leader sequences inserted within the intercistronic region of the plasmid pRF, and translation of the downstream firefly luciferase cistron occurred via internal ribosome entry (Fig. 1A) (8). The smallest construct containing sequences from the cyclin D1 leader that retained full Akt-dependent IRES activity was a segment spanning -209 to -44 upstream of the translation initiation codon (Fig. 1B). Similar dicistronic reporter mRNAs containing sequences from the c-myc 5′-UTR demonstrated that sequences spanning -398 to -165 upstream of the c-myc initiation codon were sufficient to confer Akt-dependent IRES activity following rapamycin exposure as effectively as the full-length 5′-UTR (Fig. 1B). Notably, this region did not include domain 2 of the c-myc IRES, which has been previously identified in chemical probing and secondary structure modeling studies (38). This region of the IRES has been hypothesized to be involved in the assembly of a competent translation initiation complex and has been shown to be required for maximal IRES activity (see “Discussion”). In addition, performing the dicistronic assays with reporters containing these minimal cyclin D1 and c-myc IRES elements and also containing a hairpin structure inserted upstream of the Renilla ORF showed reduced cap-dependent translation by 90–95%, whereas translation from either IRES persisted in an Akt-dependent manner (supplemental Fig. 1, A and B).
FIGURE 1.
The 5′-UTRs of the human cyclin D1 and c-myc mRNAs contain short regions that exhibit Akt-dependent IRES activity following rapamycin exposure. A, schematic diagram showing dicistronic constructs. B, DNA segments corresponding to the indicated regions of the 5′-UTRs of cyclin D1 and c-myc were inserted into the intercistronic region of the dicistronic plasmid pRF. PTEN-/- and PTEN+/+ MEFs were transfected with the indicated plasmids (2 μg each) and treated with or without rapamycin (rapa) (10 nm), and luciferase activities were determined. Relative firefly luciferase (IRES-mediated initiation) activity is shown in the absence (open bars) or presence (shaded bars) of rapamycin and normalized to values obtained for pRF in each cell line. The mean and S.D. are shown for three independent experiments.
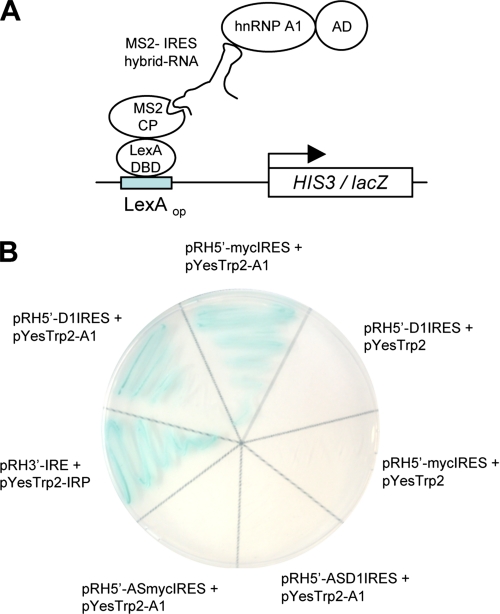
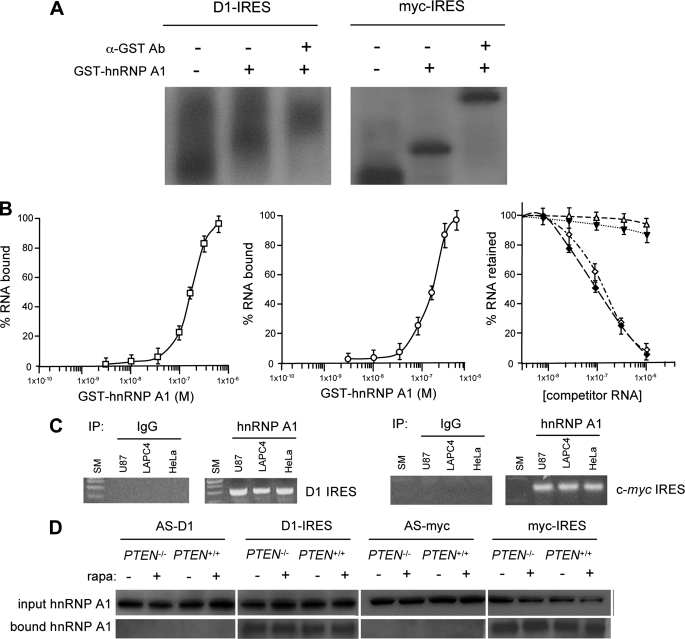
Since subsequent immunoblot assays for known c-myc ITAFs did not demonstrate any Akt-dependent differences in terms of expression levels or electrophoretic mobility changes, which may be indicative of possible regulation via post-translational mechanisms (data not shown), we attempted to identify Akt-regulated proteins that might function as novel cyclin D1 or c-myc ITAFs. We utilized a yeast three-hybrid screen (Fig. 2A) to identify proteins that specifically bound to the above described minimal Akt-dependent 5′-UTR sequences of the cyclin D1 and c-myc transcripts. From this screen, we identified several clones encoding hnRNP A1 that specifically bound to both the cyclin D1 and c-myc IRES sequences and was dependent on both the IRES sequences and the hnRNP A1 activation domain fusion for reporter expression (Fig. 2B). EMSA analysis confirmed the interaction between hnRNP A1 and these 5′-UTR sequences, which conferred Akt-dependent IRES activity. Recombinant GST-tagged hnRNP A1 was incubated with 32P-labeled cyclin D1 or c-myc IRES sequences and, as shown in Fig. 3A, inhibited electrophoretic mobility of these RNAs. Additionally, the cyclin D1 or c-myc IRES-hnRNP A1 complexes were supershifted by the addition of anti-GST antibody (Fig. 3A). To confirm that the cyclin D1 and c-myc IRESs contain high affinity hnRNP A1 binding sites, we determined the equilibrium dissociation constants (Kd) of GST-hnRNP A1 for both the cyclin D1 (Fig. 3B, left) and c-myc (Fig. 3B, center) minimal IRES RNAs in a filter binding assay. The Kd for both the GST-hnRNP A1 interaction with either the cyclin D1 or c-myc IRES RNAs was similar and was estimated to be ∼190 and 200 nm, respectively. These values were consistent with apparent Kd measurements obtained for other hnRNP A1-IRES RNA interactions (20). Our previous experiments had shown that the p27Kip1 IRES did not demonstrate Akt-dependent IRES activity (8), and thus we determined whether binding of hnRNP A1 was specific to the cyclin D1 and c-myc IRESs as compared with the p27Kip1 IRES RNA. We challenged complexes of hnRNP A1-cyclin D1 IRES RNA or hnRNP A1-c-myc IRES RNA with unlabeled cyclin D1 or c-myc IRES RNAs, respectively. These unlabeled RNAs effectively competed for binding of both hnRNP A1-IRES RNA complexes (Fig. 3B, right). However, unlabeled p27Kip1 IRES RNA did not significantly alter binding of hnRNP A1 to either the cyclin D1 or c-myc IRES RNAs. To further examine if endogenous hnRNP A1 would interact with cyclin D1 or c-myc mRNAs in vivo, we immunoprecipitated hnRNP A1 from several cell lines and analyzed the immunoprecipitates for the presence of either cyclin D1 or c-myc mRNAs by RT-PCR. As shown in Fig. 3C, both mRNAs were detected in these immunoprecipitates. Furthermore, in an RNA pull-down assay, extracts from either PTEN-/- or PTEN+/+ MEFs were mixed with biotinylated cyclin D1 or c-myc IRES sequences in the sense or antisense orientation. As shown in Fig. 3D, hnRNP A1 was preferentially precipitated by either of the IRES RNAs in the sense orientation, but no hnRNP A1 was detected in precipitates of the IRES RNAs in the antisense orientation. These data demonstrated that hnRNP A1 binds constitutively to both the cyclin D1 and c-myc IRESs. Furthermore, binding of hnRNP A1 to the IRESs was not affected by the degree of Akt activation or rapamycin exposure (Fig. 3D).
FIGURE 2.
Binding of hnRNP A1 to minimal cyclin D1 and c-myc IRESs in a yeast three-hybrid system. A, schematic diagram showing components of the three-hybrid assay. Clones encoding hnRNP A1 fused to a transcriptional activation domain bound a bifunctional hybrid RNA composed of either the cyclin D1 or c-myc IRES fused to MS2 coat protein RNA binding sites. The hybrid RNA also bound a LexA DNA-binding domain fusion to the bacteriophage MS2 coat protein. This ternary interaction reconstituted a functional transcription factor that interacted with LexA operator sites and resulted in activation of HIS3 and lacZ reporter gene transcription. B, L40uraMS2 cells carrying pHybLex/Zeo-MS2 (LexA DNA-binding domain/MS2 coat protein fusion-expressing plasmid) were transformed with the indicated constructs. Transformants were grown on selective media lacking tryptophan, uracil, and histidine and containing 5 mm 3-aminotriazole and analyzed via a colony color assay for lacZ expression. pRH5′-D1IRES, plasmid expressing MS2-cyclin D1 IRES hybrid RNA; pRH5′-mycIRES, plasmid expressing MS2-myc IRES hybrid RNA; pRH5′-ASD1IRES, plasmid expressing MS2-cyclin D1 IRES in antisense orientation; pRH5′-ASmycIRES, plasmid expressing MS2-myc IRES in antisense orientation; pYesTrp2, plasmid expressing activation domain only; pYesTrp2-A1, plasmid expressing hnRNP A1-activation domain fusion. As a positive control, the interaction between the iron-responsive protein (IRP) and its RNA binding sites (IRE) was detected using the hybrid RNA-expressing plasmid, pRH3′-IRE, and the IRP-activation domain fusion protein expressing plasmid pYesTrp2-IRP.
FIGURE 3.
Association of hnRNP A1 with the cyclin D1 and c-myc IRESs in vitro and in intact cells. A, RNA-EMSA of 32P-labeled cyclin D1 or c-myc minimal IRES sequences incubated with GST-control (-,-) or GST-hnRNP A1 (-,+). Supershift was assessed by the addition of anti-GST antibody (+,+). B, hnRNP A1 binding curves for the cyclin D1 (left) IRES RNA (165 nucleotides) and the c-myc (center) IRES RNA (233 nucleotides). Right, competition filter binding assays. hnRNP A1-cyclin D1 IRES RNA complexes competed for binding against unlabeled cyclin D1 IRES RNA (⋄) or unlabeled p27Kip1 IRES RNA (▵). Similarly, hnRNP A1-c-myc IRES RNA complexes competed for binding against unlabeled c-myc IRES RNA (♦) or unlabeled p27Kip1 IRES RNA (▾). C, hnRNP A1 binds cyclin D1 IRES and c-myc IRES in cells. Control IgG or anti-hnRNP A1 antibody was used to immunoprecipitate (IP) lysates from the indicated cell lines, and bound RNA was amplified by PCR of the cyclin D1 (left) or c-myc (right) IRES sequences. PCR products were visualized via ethidium bromide staining. SM, DNA size markers. D, identification of hnRNP A1 in RNA pull-down assays utilizing biotinylated cyclin D1 or c-myc IRES RNAs. Cytoplasmic extracts of PTEN-/- or PTEN+/+ MEFs, which had been treated with or without rapamycin (10 nm), were incubated with biotinylated cyclin D1 or c-myc IRES RNAs or control RNAs corresponding to the antisense cyclin D1 or c-myc IRES sequences and precipitated with streptavidin-Sepharose beads. Input and bound fractions were analyzed by immunoblotting for hnRNP A1.
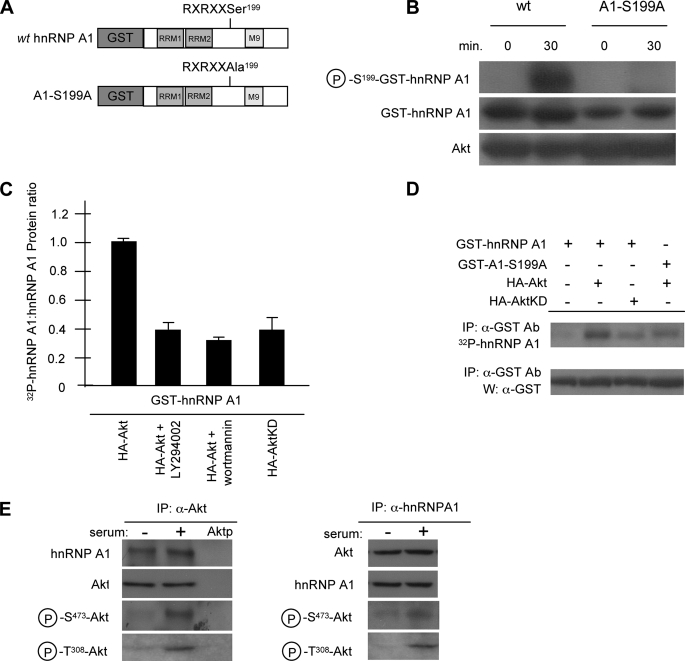
hnRNP A1 Is a Substrate of Akt and Is Phosphorylated on Serine 199—Sequence analysis of hnRNP A1 identified a putative Akt phosphorylation consensus site (RXRXX(S/T)) within the C-terminal half of the protein just proximal to the M9 export domain at serine 199 (39). To investigate whether Akt was able to phosphorylate this residue in vitro, we used an antibody that recognizes amino acids phosphorylated by Akt. For these experiments, we used two GST fusion proteins, which consisted of the full-length human hnRNP A1 with GST fused to the N terminus and the identical protein in which serine 199 had been mutated to alanine (Fig. 4A). Fig. 4B shows that Akt is able to specifically phosphorylate hnRNP A1 as detected by the anti-phospho-Akt substrate antibody but is unable to phosphorylate the alanine substitution mutant. This suggests that serine 199 on hnRNP A1 is accessible to Akt phosphorylation in vitro. We then examined whether this phosphorylation was also detectable in intact cells by assaying the extent of 32P-orthophosphate labeling of hnRNP A1 (GST-hnRNP A1) in transfected 293 cells (Fig. 4C). In cells stimulated with serum, cotransfection of Akt (HA-Akt) and hnRNP A1 induced a high level of hnRNP A1 phosphorylation, which was inhibited by ∼50% by either the phosphoinositide 3-kinase inhibitors LY294002 or wortmannin or by cotransfection of hnRNP A1 with a kinase-dead Akt construct (HA-AktKD). This suggests that Akt phosphorylates hnRNP A1 in cells. To determine whether serine 199 of hnRNP A1 was the critical residue in Akt-mediated phosphorylation in cells, we again employed the S199A hnRNP A1 mutant and performed additional 32P labeling experiments. As shown in Fig. 4D, a relatively low level of 32P-labeled hnRNP A1 was observed in cells transfected with hnRNP A1 alone, and introduction of native Akt markedly enhanced hnRNP A1 32P incorporation, whereas introduction of a kinase-inactive mutant of Akt yielded a level of hnRNP A1 phosphorylation approximately the same as that for hnRNP A1 alone. Mutation of hnRNP A1 serine 199 to alanine decreased Akt-dependent phosphorylation, suggesting that hnRNP A1 serine 199 represents a relevant phosphorylation site in intact cells.
FIGURE 4.
Akt phosphorylates hnRNP A1 in vitro and in cells. A, schematic diagram of GST fusion proteins used for in vitro and in vivo Akt phosphorylation experiments. B, 500 ng of either wild type (wt) hnRNP A1 or A1-S199A was incubated with 200 ng of activated Akt and immunoblotted with anti-phospho-Akt substrate antibody, anti-GST antibody, or anti-Akt antibody. Phosphorylation reactions were performed for 0 min (negative control) and 30 min. C, hnRNP A1 phosphorylation in cells stimulated by serum. 293 cells transiently transfected for 24 h with 1 mg of each of the indicated constructs were deprived of phosphate and serum for 1 h, incubated in the absence or presence of LY294002 (20 mm) or wortmannin (100 nm) for 1 h, and exposed to [32P]orthophosphate and dialyzed serum for 2 h. Immunoprecipitated hnRNP A1 was assessed for 32P-labeling and protein levels with anti-GST antibody. The data are shown as mean densitometric ratios (32P labeling/protein) from three independent experiments. D, Akt phosphorylates hnRNP A1 on serine 199 in serum-stimulated 293 cells. Cells were transfected with the indicated constructs and were deprived of phosphate and serum for 1 h and exposed to [32P]orthophosphate and dialyzed serum as in C. Data are representative of three independent experiments. IP, immunoprecipitation; W, Western blot. E, endogenous association of Akt and hnRNP A1 in 293 cells. Cells were serum-starved for 24 h and then treated with or without dialyzed serum for 2 h. Endogenous Akt (left) or hnRNP A1 (right) was immunoprecipitated (IP) from extracts and immunoblotted for the indicated proteins. Aktp, immunoprecipitation with anti-Akt antibody preincubated with an Akt-blocking peptide. Three independent experiments were performed with similar results.
To further confirm a direct association of Akt with hnRNP A1 in cells, we conducted co-immunoprecipitation experiments. As shown in Fig. 4E, endogenous hnRNP A1 from 293 cells was detectable in immunoprecipitates of Akt in the presence or absence of serum stimulation, suggesting that both inactive and active forms of Akt are able to associate with hnRNP A1. Similarly, both forms of Akt were also detected in immunoprecipitates of hnRNP A1 in the reciprocal experiment.
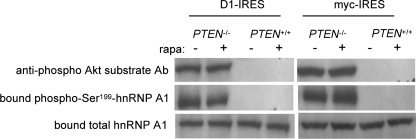
hnRNP A1 Serine 199 Is Phosphorylated in an Akt-dependent Manner, and Phosphorylated hnRNP A1 Is Inactivated as an ITAF—To identify endogenous Akt-mediated phosphorylation of hnRNP A1 and additionally determine whether Ser199-phosphorylated hnRNP A1 was associated with the IRESs, we again employed the RNA pull-down assay in isogenic lines differing by Akt activation. Using biotinylated cyclin D1 or c-myc IRES RNAs to isolate hnRNP A1, we then examined hnRNP A1 for serine 199 phosphorylation. As shown in Fig. 5, in PTEN null MEFs (with activated Akt) treated with rapamycin, hnRNP A1 serine 199 was phosphorylated to a much greater degree, as detected by the anti-phospho-Akt substrate antibody, as compared with PTEN+/+ MEFs following the same treatment. We subsequently generated phospho-specific hnRNP A1 antibodies in rabbits that specifically recognized serine 199 phosphorylation. The specificity of this antibody was confirmed in preabsorption experiments in which the immunogenic hnRNP A1 phosphopeptide used to generate the antisera effectively abrogated immunoblot recognition of recombinant serine 199-phosphorylated hnRNP A1, whereas an identical nonphosphorylated control peptide was unable to do so (not shown). As shown in Fig. 5, these antibodies also reacted strongly with IRES-bound hnRNP A1 isolated from PTEN null MEFs relative to PTEN+/+ MEFs following rapamycin exposure. These results indicated that hnRNP A1 is phosphorylated at serine 199 in cells containing elevated levels of Akt and remained mostly unphosphorylated in cells with quiescent Akt activity.
FIGURE 5.
Serine 199 is differentially phosphorylated in cyclin D1 or c-myc IRES bound hnRNP A1 in an Akt-dependent manner following rapamycin exposure. Biotinylated cyclin D1 or c-myc IRES RNAs were used to pull-down hnRNP A1 from cytoplasmic extracts of PTEN-/- or PTEN+/+ MEFs treated with or without rapamycin (10 nm) for 2.5 h. RNA-protein complexes were resolved by gel electrophoresis and immunoblotted with anti-hnRNP A1, anti-phospho-Akt substrate or serine 199 phospho-specific hnRNP A1 antibodies. Data are representative of three independent experiments.
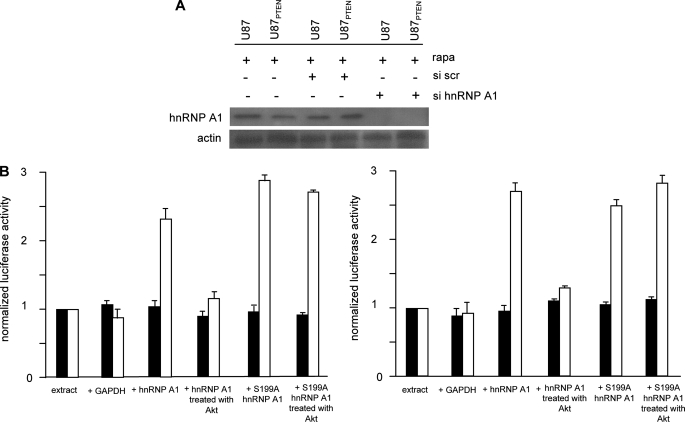
To investigate whether this phosphorylation event regulates hnRNP A1-mediated IRES activity, we first attempted to test whether native or serine 199-phosphorylated hnRNP A1 would support cyclin D1 or c-myc IRES activity in vitro. Translation-competent cell extracts were prepared from cells in which hnRNP A1 had been knocked down via RNA interference (Fig. 6A) and programmed with in vitro transcribed dicistronic mRNAs containing either the cyclin D1 or c-myc IRESs within the intercistronic region. As shown in Fig. 6A, siRNA-mediated knockdown of hnRNP A1 resulted in undetectable levels of the protein as determined by Western analysis, whereas a nontargeting scrambled sequence had no appreciable affect on expression. The reduction in hnRNP A1 levels did not significantly affect the kinetics of overall cap-dependent Renilla protein synthesis of exogenous mRNAs in these extracts (supplemental Fig. 2A). We then supplemented the in vitro extracts with either glyceraldehyde-3-phosphate dehydrogenase, which we utilized as a negative control and is an ITAF that specifically binds the HAV IRES (40, 41), native hnRNP A1, the serine-to-alanine S199A hnRNP A1 mutant, or serine 199-phosphorylated hnRNP A1. As shown in Fig. 6B, the addition of recombinant hnRNP A1 (GST-hnRNP A1) or S199A hnRNP A1 markedly stimulated either cyclin D1 or c-myc IRES activity in vitro, whereas the addition of glyceraldehyde-3-phosphate dehydrogenase (negative control) or hnRNP A1 phosphorylated in vitro with activated Akt (serine 199-phosphorylated hnRNP A1) was unable to support IRES function. The serine to alanine-mutated hnRNP A1 was also capable of stimulating IRES function even following exposure to activated Akt. We also confirmed that the in vitro phosphorylated hnRNP A1 remained Ser199-phosphorylated for the duration of the translation reactions (supplemental Fig. 2C). These data suggest that phosphorylation of serine 199 on hnRNP A1 negatively regulates IRES activity in vitro.
FIGURE 6.
Akt negatively regulates cyclin D1 and c-myc IRES activity in vitro. A, siRNA mediated knockdown of hnRNP A1. U87 and U87PTEN were transiently transfected with siRNAs targeting hnRNP A1 or a nontargeting scrambled (scr) sequence and exposed to rapamycin (10 nm) for 24 h, and extracts were prepared and immunoblotted for hnRNP A1 and actin as indicated. Similar results were found in two additional independent experiments. B, translation-competent extracts were prepared from U87PTEN cells in which hnRNP A1 expression was knocked down via siRNA treatment, and the indicated proteins were added to the extracts prior to programming the extracts with either in vitro transcribed dicistronic cyclin D1 (B, left) or dicistronic c-myc (B, right) reporter mRNAs. Translations were performed at 30 °C for 40 min. An irrelevant ITAF, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was added as a negative control. Renilla (shaded bars) and firefly (open bars) luciferase activities were determined and normalized to values obtained for extracts alone. The mean and S.D. are shown for three independent experiments.
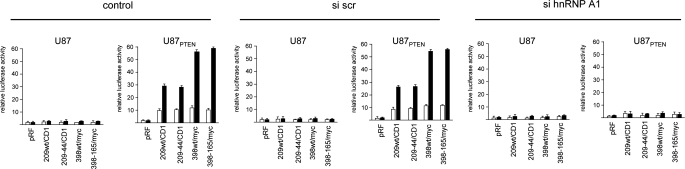
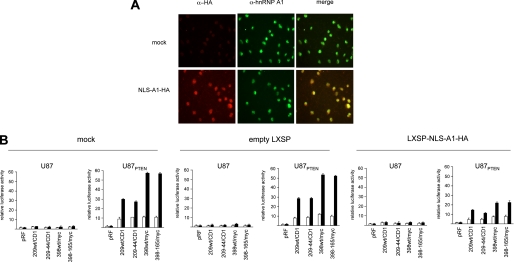
Knockdown or Overexpression of a Dominant Negative Mutant of hnRNP A1 Inhibits IRES Activity—To determine whether the effects on IRES-mediated translation we had observed for hnRNP A1 in vitro were also consistent in cells, we determined the effects on cyclin D1 and c-myc IRES activity following rapamycin exposure in cells knocked down for hnRNP A1. U87 and U87PTEN cells in which hnRNP A1 expression was inhibited (as in Fig. 6A) were transiently transfected with the indicated constructs as shown in Fig. 7 and treated with or without rapamycin, and luciferase activities were determined. As can be seen, in control or cells treated with a nontargeting scrambled siRNA, cyclin D1 and c-myc IRES-derived firefly luciferase activity was stimulated 5–6-fold by rapamycin in cells containing quiescent Akt as compared with those containing active Akt. However, in cells treated with the siRNA targeting hnRNP A1 rapamycin, treatment did not appreciably stimulate Akt-dependent cyclin D1 or c-myc IRES activity. Similarly, we tested whether overexpression of a dominant negative mutant of hnRNP A1 would affect rapamycin stimulated Akt-dependent cyclin D1 or c-myc IRES activity. The NLS-A1-HA construct contains the bipartite-basic type NLS of hnRNP K1 fused in frame with the N terminus of an HA-tagged hnRNP A1 mutant, which lacks both nuclear import and export activities and inhibits hnRNP A1-mediated mRNA export when micro-injected into nuclei of Xenopus oocytes (42, 43). This hnRNP A1 mutant, which retains hnRNP A1 nuclear localization, lacks nuclear export activity (26). As such, nuclear localized NLS-A1-HA competes with native hnRNP A1 for binding to mRNAs and for nuclear export (44). A retroviral vector LXSP-NLS-A1-HA was used to ectopically express this mutant in U87 and U87PTEN cells. Following viral transduction, immunostaining of the resistant population revealed that ∼80% of the cells (U87PTEN staining not shown) were positive for HA labeling (Fig. 8A). Dual immunostaining indicated that both endogenous hnRNP A1 (anti-hnRNP A1; green) and ectopically expressed NLS-A1-HA (anti-HA; red) displayed diffuse nuclear staining excluding the nucleoli. We then determined whether overexpression of this shuttling-deficient hnRNP A1 mutant was interfering with IRES activity. As shown in Fig. 8B, mock- and control empty vector (LXSP)-infected cells retained Akt-dependent cyclin D1 and c-myc IRES activity following rapamycin exposure. However, in cells overexpressing the NLS-A1-HA mutant, firefly luciferase activities were markedly reduced as compared with controls. We subsequently examined whether rapamycin exposure affected the predominantly nuclear distribution of the dominant negative hnRNP A1 mutant or possibly altered the export of the dicistronic reporter mRNAs. As shown in supplemental Fig. 3, rapamycin did not vary the nuclear localization of the hnRNP A1 mutant or result in inhibiting export of the reporter transcripts. These data suggested that hnRNP A1 was required for cyclin D1 and c-myc IRES activity.
FIGURE 7.
Knockdown of hnRNP A1 abrogates Akt-dependent cyclin D1 and c-myc IRES activity following rapamycin exposure. U87 and U87PTEN cells in which hnRNP A1 expression was inhibited via siRNA were transiently transfected with the indicated dicistronic reporter constructs as in Fig. 1. Relative firefly luciferase (IRES-mediated initiation) activity is shown in the absence (open bars) or presence (shaded bars) of rapamycin and normalized to values obtained for pRF in each cell line. The mean and S.D. are shown for three independent experiments.
FIGURE 8.
Effects of a dominant negative shuttling-deficient hnRNP A1 mutant on Akt-dependent cyclin D1 and c-myc IRES activity following rapamycin exposure. A, expression of the NLS-A1-HA mutant in U87 cells. Immunofluorescence microscopy of untransduced (mock-infected, top row) or NLS-A1-HA-transduced (bottom row) cells after dual immunofluorescence staining using anti-HA (red) and anti-hnRNP A1 (green) antibodies; the panels on the right shows the overlay composite images. B, U87 and U87PTEN cells were stably transduced with the indicated viral constructs and transiently transfected with the indicated dicistronic reporter constructs as before. Relative firefly luciferase (IRES-mediated initiation) activity is shown in the absence (open bars) or presence (shaded bars) of rapamycin and normalized to values obtained for pRF in each cell line. The mean and S.D. are shown for three independent experiments.
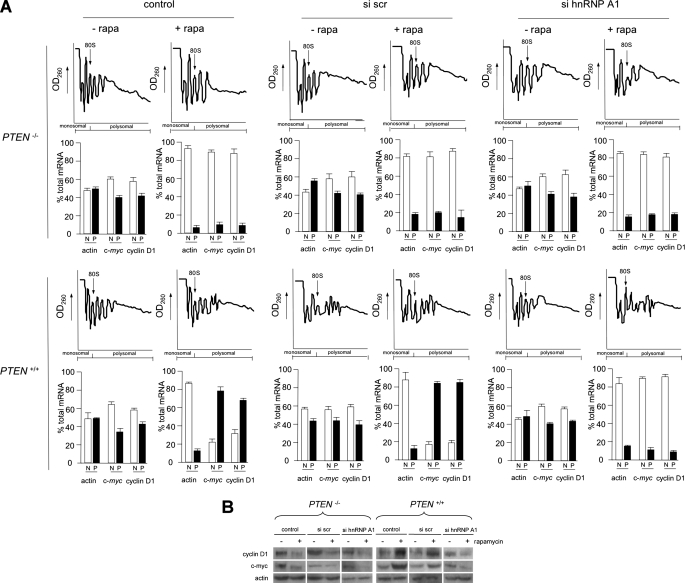
Knockdown of hnRNP A1 Affects Polysome Distribution of Cyclin D1 and c-myc mRNAs under Conditions Favoring IRES-dependent Initiation—The role of hnRNP A1 in cyclin D1 and c-myc mRNA translation was also assessed by analyzing the distribution of these transcripts in ribosome profiles in which either PTEN-/- or PTEN+/+ MEFs were knocked down for hnRNP A1 and treated with or without rapamycin. PTEN-/- or PTEN+/+ MEF extracts were subjected to sucrose density gradient fractionation, and individual fractions were pooled into a nonribosomal, monosomal fraction or a polysomal fraction and then subjected to real time quantitative RT-PCR analysis to quantify the indicated mRNA levels in these fractions. As shown in Fig. 9A, in PTEN-/- MEFs containing active Akt, the cyclin D1 and c-myc transcripts are present at ∼40–45% of total cyclin D1 and c-myc mRNAs in the polysomal fractions in control, scrambled siRNA, or hnRNP A1 targeting siRNA-treated cells. Upon inhibition of cap-dependent initiation following rapamycin treatment, the cyclin D1 and c-myc mRNA shifted into mainly the nonribosomal, monosomal fractions in all three groups. We also monitored actin polysome distribution, and, as shown, this mRNA, whose translation is mediated by eIF-4E mediated initiation, was redistributed to nonribosomal, monosomal fractions in each treatment group, demonstrating efficient inhibition of cap-dependent initiation by rapamycin. However, in PTEN+/+ MEFs containing quiescent Akt, there was a dramatic shift in cyclin D1 and c-myc mRNAs to mainly polysomal fractions following rapamycin treatment in control and nontargeting scrambled siRNA-treated groups, consistent with the enhancement of IRES activity. In hnRNP A1 siRNA-treated cells, we observed that this rapamycin-induced redistribution to polysomes was markedly inhibited and resulted in cyclin D1 and c-myc mRNAs present in mostly nonribosomal, monosomal fractions. We also assessed the levels of endogenous cyclin D1 and c-myc in hnRNP A1 knockdown PTEN-/- and PTEN+/+ MEFs treated with or without rapamycin as before. As shown in Fig. 9B, the steady-state accumulation of these proteins was reflective of the translational states of their mRNAs. In PTEN-/- MEFs, containing active Akt, rapamycin exposure resulted in marked reductions of cyclin D1 and c-myc in all three groups. In PTEN+/+ MEFs, containing quiescent Akt, rapamycin treatment resulted in significant increases in cyclin D1 and c-myc protein in control and nontargeting siRNA-treated groups, whereas in the hnRNP A1 knockdown group, these rapamycin-induced increases in cyclin D1 and c-myc were blunted. These data strongly suggested that hnRNP A1 plays an important role in the Akt-dependent translation of both cyclin D1 and c-myc mRNAs under conditions of reduced cap-dependent initiation.
FIGURE 9.
Knockdown of hnRNP A1 alters polysome distribution of cyclin D1 and c-myc mRNAs. A, polysome distributions of cyclin D1, c-myc, and actin mRNAs in hnRNP A1 knockdown PTEN-/- and PTEN+/+ MEFs treated with or without rapamycin. Extracts were prepared from PTEN-/- or PTEN+/+ MEFs transfected with the indicated siRNAs (control, nontargeting scrambled siRNA (scr) or hnRNP A1-targeting siRNA) and treated with or without rapamycin (10 nm) for 24 h. Extracts were subjected to sucrose density gradient centrifugation and then divided into 11 1-ml fractions, which were pooled into a nonribosomal, monosomal fraction (N, white bars) and a polysomal fraction (P, black bars). Purified RNAs were used in real time quantitative RT-PCR analysis to determine the distributions of actin, cyclin D1, and c-myc mRNAs across the gradients. Polysome tracings are shown above values obtained from the RT-PCR analyses, which are displayed graphically below. RT-PCR measurements were done in quadruplicate, and the mean and S.D. are shown. B, cyclin D1, c-myc, and actin protein levels in hnRNP A1 knockdown PTEN-/- or PTEN+/+ MEFs treated with or without rapamycin and transfected with the indicated siRNAs as in A. Experiments were repeated three times with similar results.
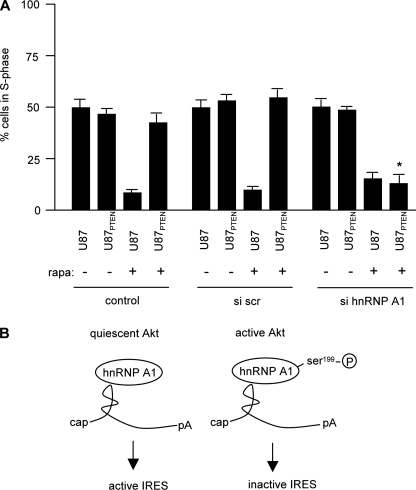
Inhibition of hnRNP A1 Expression Confers Sensitivity to Rapamycin of Quiescent Akt-containing Cells—To determine whether knockdown of hnRNP A1 would affect the sensitivity of cells containing elevated Akt levels as compared with those with relatively quiescent Akt, we treated U87 and U87PTEN cells with siRNA to inhibit hnRNP A1 expression and determined the cell cycle distributions of these cells following rapamycin exposure. As shown in Fig. 9, in control and nontargeting scrambled siRNA-treated cultures, U87 cells were very sensitive to rapamycin (∼10% of cells in S-phase) as compared with untreated cells (∼50% of cells in S-phase). U87PTEN cells were relatively resistant, with ∼50% of cells in S-phase prior to and following rapamycin exposure. However, in cells treated with the siRNA targeting hnRNP A1, both U87 and U87PTEN cells displayed reduced S-phase cell numbers relative to controls. These data demonstrated that knockdown of hnRNP A1 resulted in sensitivity of quiescent Akt containing U87PTEN cells to rapamycin.
DISCUSSION
Our previous studies implicated IRES-mediated translation initiation of cyclin D1 and c-myc mRNAs in regulating Akt-dependent sensitivity of cells to mTOR inhibitors (8). In this report, we have identified an ITAF that specifically interacts with both the IRESs of cyclin D1 and c-myc and regulates IRES activity in an Akt-dependent manner. Moreover, we demonstrate that Akt directly regulates the ability of hnRNP A1 to promote cyclin D1 and c-myc IRES activity via phosphorylation. Our data suggest that serine 199 phosphorylation of hnRNP A1 inactivates its ITAF function for the cyclin D1 and c-myc IRESs. We also demonstrate by two different approaches that the lack of hnRNP A1 results in the inability of the cell to activate IRES-mediated translation initiation of cyclin D1 and c-myc following rapamycin exposure. Finally, we show that knockdown of hnRNP A1 results in redistribution of cyclin D1 and c-myc mRNAs from polysomes to monosomes under conditions of reduced cap-dependent translation and confers rapamycin sensitivity to quiescent Akt-containing cells. Our data are consistent with a working model (Fig. 10B) in which cells containing quiescent Akt have constitutively associated with the cyclin D1, and c-myc IRESs bound hnRNP A1, which is required for IRES-dependent translation initiation and permits synthesis of these critical cell cycle proteins following the inhibition of cap-dependent initiation by mTOR inhibitors. However, in cells with active Akt, hnRNP A1 is phosphorylated at serine 199, which inactivates its ability to promote cyclin D1 and c-myc IRES activity and results in the blockade of protein synthesis of these determinants following mTOR inhibitor exposure.
FIGURE 10.
Knockdown of hnRNP A1 confers sensitivity to quiescent Akt containing rapamycin resistant cells. A, U87 or U87PTEN cells were transfected with the indicated siRNAs and treated with or without rapamycin (100 nm) for 48 h. S-phase cell cycle distribution was subsequently determined on propidium iodide-stained cells by flow cytometry. The mean and S.D. are shown for three independent experiments (*, p < 0.05). B, in cells containing active Akt, hnRNP A1 is differentially phosphorylated at serine 199, which inactivates IRES-mediated translation of cyclin D1 and c-myc mRNAs. The absence of this phosphorylation event results in hnRNP A1-mediated IRES activity in the face of rapamycin exposure.
hnRNP A1 is a nuclear-cytoplasmic shuttling protein that has been implicated as an ITAF for FGF-2 (20) and more recently XIAP (45). Our identification of hnRNP A1 as an ITAF for both the cyclin D1 and c-myc IRESs in addition to those above suggests an important role for this protein in IRES-mediated initiation. However, how hnRNP A1 may regulate IRES activity is unclear. hnRNP A1 was reported to be required for FGF-2 IRES activity and stimulated IRES-dependent initiation of FGF-2 in vitro but, interestingly, negatively regulated XIAP IRES activity in response to cellular stress. A reduction in XIAP IRES-dependent translation correlated with the cytoplasmic accumulation of hnRNP A1. Our results support a role for the phosphorylation of serine 199 of hnRNP A1 in regulating IRES activity and may partially explain the differences in hnRNP A1-mediated ITAF activity observed for the FGF-2 and XIAP IRESs. Akt activity has been associated with cell survival and reported to be stimulated by certain stress stimuli, including sorbitol-induced osmotic shock (46, 47). Elevated Akt activity following sorbitol exposure may result in the phosphorylation of hnRNP A1 serine 199 and inhibit XIAP IRES activity, whereas Akt activity may be relatively low under the conditions in which hnRNP A1-mediated FGF-2 IRES activity was examined, thus permitting IRES function.
The observation that hnRNP A1 is mostly a nuclear localized protein (48) begs the question of how hnRNP A1-mediated IRES initiation of cyclin D1 and c-myc is maintained in the translationally active cytoplasmic compartment of quiescent Akt-containing cells following rapamycin exposure. It is possible that localization of hnRNP A1 may also participate in the regulation of cyclin D1 and c-myc IRES activity. Our previous data suggested that, following rapamycin exposure, p38 mitogen-activated protein kinase is markedly stimulated in quiescent Akt-containing cells as compared with cells with elevated Akt activity (8). Several studies have documented the affects of p38 mitogen-activated protein kinase/Mnk signaling on hnRNP A1 phosphorylation and subcellular distribution (49–51). We are currently investigating whether p38 mitogen-activated protein kinase/Mnk signaling regulates hnRNP A1-mediated cyclin D1 and c-myc IRES activity.
Several studies have suggested that biologically relevant substrates of Akt are localized in the nucleus (52–54). Additionally, activated Akt has been found in the nucleus in several cancers and has been reported to translocate to the nucleus following insulin stimulation (37, 55, 56). It is tempting to speculate that hnRNP A1 is a nuclear target of Akt phosphorylation activity and that this event affects nucleo-cytoplasmic transport of the mRNA-hnRNP A1 complex as well as ITAF activity. In fact, Akt-mediated phosphorylation of hnRNP A1 may effectively serve to partition the bound mRNA from the cytoplasm and subsequent translation initiation.
We noted that domain 2 of the predicted c-myc IRES secondary structure model (38) was not required for full Akt-dependent c-myc IRES activity in our deletion analysis. The c-myc IRES contains two structurally defined domains. Domain 1 is predicted to have an elaborate and complex structure containing two overlapping pseudoknots and a large internal loop, whereas domain 2 is much less structured, containing a relatively smaller AUUU tetraloop. Deletion analysis demonstrated that the removal of either domain reduces c-myc IRES activity to ∼60% of the native IRES, suggesting that neither domain is crucial for maximal IRES activity in HeLa cells and points to the modular nature of the IRES whereby each domain may contribute singly to IRES function (4). Our analysis suggests that, under the conditions of rapamycin-induced stimulation of the c-myc IRES, domain 1 and sequences corresponding to the downstream ribosome landing site are sufficient for full IRES function.
In summary, our data identify hnRNP A1 as an ITAF for the cyclin D1 and c-myc IRESs and demonstrate that the protein is phosphorylated in vitro and in intact cells directly by Akt. In vitro studies also suggest that phosphorylation of hnRNP A1 at serine 199 by Akt inactivates its ITAF function. We also show that knockdown or overexpression of a dominant negative mutant of hnRNP A1 inhibits rapamycin-induced hnRNP A1-mediated cyclin D1 and c-myc IRES activity. Finally, we demonstrate that inhibiting hnRNP A1 expression sensitizes quiescent AKT-containing cells to rapamycin. These data implicate the hnRNP A1-cyclin D1 or c-myc IRES interactions as potential targets for intervention and suggest that the serine 199 phosphorylation state of hnRNP A1 may be an effective indicator of tumor cell responses to mTOR inhibitors.
Supplementary Material
Acknowledgments
We thank Drs. Danilo Perrotti, Robert Hay, and William Sellers for providing reagents and Megan Marderosian for assistance with the yeast screening. Flow cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center and Center for AIDS Research, Janis Giorgi Flow Cytometry Core Facility (supported by National Institutes of Health Grants CA-16042 and AI-28697 and by the Jonsson Comprehensive Cancer Center, the UCLA AIDS Institute, and the David Geffen School of Medicine at UCLA). We thank Drs. Robert Nishimura and Richard Weisbart for comments on the manuscript and Ardella Sherwood for excellent administrative assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01CA111448 (to A. L.) and R01CA109312 (to J. G.). This work was also supported in part by funds from the Department of Veterans Affairs. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
Footnotes
The abbreviations used are: UTR, untranslated region; IRES, internal ribosome entry site; ITAF, IRES trans-acting factor; hnRNP, heterogeneous nuclear ribonucleoprotein; GST, glutathione S-transferase; HA, hemagglutinin; RT, reverse transcription; siRNA, small interfering RNA; NLS, nuclear localization sequence; MEF, mouse embryo fibroblast.
References
- 1.Hellen, C. U. T., and Sarnow, P. (2001) Genes Dev. 15 1593-1612 [DOI] [PubMed] [Google Scholar]
- 2.Spriggs, K. A., Bushell, M., Mitchell, S. A., and Willis, A. E. (2005) Cell Death Differ. 12 585-591 [DOI] [PubMed] [Google Scholar]
- 3.Komar, A. A., and Hatzoglou, M. (2005) J. Biol. Chem. 280 23425-23428 [DOI] [PubMed] [Google Scholar]
- 4.Stoneley, M., and Willis, A. E. (2004) Oncogene 23 3200-3207 [DOI] [PubMed] [Google Scholar]
- 5.Vagner, S., Galy, B., and Pyronnet, S. (2001) EMBO Rep. 2 893-898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holcik, M., and Sonenberg, N. (2005) Nat. Rev. Mol. Cell Biol. 6 318-327 [DOI] [PubMed] [Google Scholar]
- 7.Baird, S. D., Turcotte, M., Korneluk, R. G., and Holcik, M. (2006) RNA 12 1755-1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi, Y., Sharma, A., Wu, H., Lichtenstein, A., and Gera, J. (2005) J. Biol. Chem. 280 10964-10973 [DOI] [PubMed] [Google Scholar]
- 9.Mitchell, S. A., Brown, E. C., Coldwell, M. J., Jackson, R. J., and Willis, A. E. (2001) Mol. Cell Biol. 21 3364-3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holcik, M., Gordon, B. W., and Korneluk, R. G. (2003) Mol. Cell Biol. 23 280-288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell, S. A., Spriggs, K. A., Bushell, M., Evans, J. R., Stoneley, M., Le Quesne, J. P., Spriggs, R. V., and Willis, A. E. (2005) Genes Dev. 19 1556-1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kullmann, M., Gopfert, U., Siewe, B., and Hengst, L. (2002) Genes Dev. 16 3087-3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreyfuss, G., Kim, V. N., and Kataoka, N. (2002) Nat. Rev. Mol. Cell Biol. 3 195-205 [DOI] [PubMed] [Google Scholar]
- 14.Burd, C., and Dreyfuss, G. (1994) EMBO J. 13 1197-1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cobianchi, F., Calvio, C., Stoppini, M., Buvoli, M., and Riva, S. (1993) Nucleic Acids Res. 21 949-955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Idriss, H., Kumar, A., Casas-Finet, J. R., Gou, H., Damuni, Z., and Wilson, S. H. (1994) Biochemistry 33 11382-11390 [DOI] [PubMed] [Google Scholar]
- 17.Hamilton, B. J., Nagy, E., Malter, J. S., Arrick, B. A., and Rigby, W. F. (1993) J. Biol. Chem. 268 8881-8887 [PubMed] [Google Scholar]
- 18.Henics, T., Sanfridson, A., Hamilton, B. J., Nagy, E., and Rigby, W. F. (1994) J. Biol. Chem. 269 5377-5383 [PubMed] [Google Scholar]
- 19.Svitkin, Y., Ovchinnikov, L., Dreyfuss, G., and Sonenberg, N. (1996) EMBO J. 15 7147-7155 [PMC free article] [PubMed] [Google Scholar]
- 20.Bonnal, S., Pileur, F., Orsini, C., Parker, F., Pujol, F., Prats, A. C., and Vagner, S. (2005) J. Biol. Chem. 280 4144-4153 [DOI] [PubMed] [Google Scholar]
- 21.Pinol-Roma, S., and Dreyfuss, G. (1991) Science 253 312-314 [DOI] [PubMed] [Google Scholar]
- 22.Mamane, Y., Petroulakis, E., LeBacquer, O., and Sonenberg, N. (2006) Oncogene 25 6416-6422 [DOI] [PubMed] [Google Scholar]
- 23.Ruggero, D., and Sonenberg, N. Oncogene 24 7426-7434 [DOI] [PubMed]
- 24.Wilker, E. W., van Vugt, M. A. T. M., Artim, S. C., Huang, P. H., Petersen, C. P., Reinhardt, H. C., Feng, Y., Sharp, P. A., Sonenberg, N., White, F. M., and Yaffe, M. B. (2007) Nature 446 329-332 [DOI] [PubMed] [Google Scholar]
- 25.Powell, D. W., Rane, M. J., Chen, Q., Singh, S., and McLeish, K. R. (2002) J. Biol. Chem. 277 21639-21642 [DOI] [PubMed] [Google Scholar]
- 26.Iervolino, A., Santilli, G., Trotta, R., Guerzoni, C., Cesi, V., Bergamaschi, A., Gambacorti-Passerini, C., Calabretta, B., and Perrotti, D. (2002) Mol. Cell Biol. 22 2255-2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hay, D. C., Kemp, G. D., Dargemont, C., and Hay, R. T. (2001) Mol. Cell Biol. 21 3482-3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh, A. C., Bo, R., Manola, J., Vazquez, F., Bare, O., Khvorova, A., Scaringe, S., and Sellers, W. R. (2004) Nucleic Acids Res. 32 893-901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramaswamy, S., Nakamura, N., Vazquez, F., Batt, D. B., Perera, S., Roberts, T. M., and Sellers, W. R. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 2110-2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherman, F. (2002) Methods Enzymol. 350 3-41 [DOI] [PubMed] [Google Scholar]
- 31.SenGupta, D. J., Zhang, B., Kraemer, B., Pochart, P., Fields, S., and Wickens, M. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 8496-8501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gera, J. F., Hazbun, T. R., and Fields, S. (2002) Methods Enzymol. 350 499-512 [DOI] [PubMed] [Google Scholar]
- 33.Marderosian, M., Sharma, A., Funk, A. P., Vartanian, R., Masri, J., Jo, O. D., and Gera, J. F. (2006) Oncogene 25 6277-6290 [DOI] [PubMed] [Google Scholar]
- 34.Tenenbaum, S. A., Lager, P. J., Carson, C. C., and Keene, J. D. (2002) Methods 26 191-198 [DOI] [PubMed] [Google Scholar]
- 35.Takagi, M., Absalon, M. J., McLure, K. G., and Kastan, M. B. (2005) Cell 123 49-63 [DOI] [PubMed] [Google Scholar]
- 36.Sharma, A., Masri, J., Jo, O. D., Bernath, A., Martin, J., Funk, A., and Gera, J. (2007) J. Biol. Chem. 282 9505-9516 [DOI] [PubMed] [Google Scholar]
- 37.Andjelkovic, M., Alessi, D. R., Meier, R., Fernandez, A., Lamb, N. J. C., Frech, M., Cron, P., Cohen, P., Lucocq, J. M., and Hemmings, B. A. (1997) J. Biol. Chem. 272 31515-31524 [DOI] [PubMed] [Google Scholar]
- 38.Le Quesne, J. P., Stoneley, M., Fraser, G. A., and Willis, A. E. (2001) J. Mol. Biol. 310 111-126 [DOI] [PubMed] [Google Scholar]
- 39.Yaffe, M. B., Leparc, G. G., Lai, J., Obata, T., Volinia, S., and Cantley, L. C. (2001) Nat. Biotechnol. 19 348-353 [DOI] [PubMed] [Google Scholar]
- 40.Schultz, D. E., Hardin, C. C., and Lemon, S. M. (1996) J. Biol. Chem. 271 14134-14142 [DOI] [PubMed] [Google Scholar]
- 41.Yi, M., Schultz, D. E., and Lemon, S. M. (2000) J. Virol. 74 6459-6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Izaurralde, E., Jarmolowski, A., Beisel, C., Mattaj, I. W., Dreyfuss, G., and Fischer, U. (1997) J. Cell Biol. 137 27-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michael, W. M., Choi, M., and Dreyfuss, G. (1995) Cell 83 415-422 [DOI] [PubMed] [Google Scholar]
- 44.Kress, E., Baydoun, H., Bex, F., Gazzolo, L., and Duc Dodon, M. (2005) Retrovirology 2 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lewis, S. M., Veyrier, A., Hosszu Ungureanu, N., Bonnal, S., Vagner, S., and Holcik, M. (2007) Mol. Biol. Cell 18 1302-1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Konishi, H., Matsuzaki, H., Tanaka, M., Ono, Y., Tokunaga, C., Kuroda, S. i., and Kikkawa, U. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 7639-7643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, X., Finegan, K. G., Robinson, A. C., Knowles, L., Khosravi-Far, R., Hinchliffe, K. A., Boot-Handford, R. P., and Tournier, C. (2006) Cell Death Differ. 13 2099-2108 [DOI] [PubMed] [Google Scholar]
- 48.Pinol-Roma, S., and Dreyfuss, G. (1992) Nature 355 730-732 [DOI] [PubMed] [Google Scholar]
- 49.van der Houven van Oordt, W., Diaz-Meco, M. T., Lozano, J., Krainer, A. R., Moscat, J., and Caceres, J. F. (2000) J. Cell Biol. 149 307-316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buxade, M., Parra, J. L., Rousseau, S., Shpiro, N., Marquez, R., Morrice, N., Bain, J., Espel, E., and Proud, C. G. (2005) Immunity 23 177-189 [DOI] [PubMed] [Google Scholar]
- 51.Guil, S., Long, J. C., and Caceres, J. F. (2006) Mol. Cell Biol. 26 5744-5758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brownawell, A. M., Kops, G. J. P. L., Macara, I. G., and Burgering, B. M. T. (2001) Mol. Cell Biol. 21 3534-3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pekarsky, Y., Koval, A., Hallas, C., Bichi, R., Tresini, M., Malstrom, S., Russo, G., Tsichlis, P., and Croce, C. M. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 3028-3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brunet, A., Bonni, A., Zigmond, M., Lin, M., Juo, P., Hu, L., Anderson, M., Arden, K., Blenis, J., and Greenberg, M. (1999) Cell 96 857-868 [DOI] [PubMed] [Google Scholar]
- 55.Wang, R., and Brattain, M. (2006) Cell. Signal. 18 1722-1731 [DOI] [PubMed] [Google Scholar]
- 56.Hsu, J., Shi, Y., Krajewski, S., Renner, S., Fisher, M., Reed, J. C., Franke, T. F., and Lichtenstein, A. (2001) Blood 98 2853-2855 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.