Abstract
Chronic inflammatory disorders, such as rheumatoid arthritis, are often accompanied by systemic bone loss, which is thought to occur through inflammatory cytokine-mediated stimulation of osteoclast resorption and inhibition of osteoblast function. However, the mechanisms involved in osteoblast inhibition remain poorly understood. Here we test the hypothesis that increased Smad ubiquitin regulatory factor 1 (Smurf1)-mediated degradation of the bone morphogenetic protein pathway signaling proteins mediates reduced bone formation in inflammatory disorders. Osteoblasts derived from bone marrow or long bone samples of adult tumor necrosis factor (TNF) transgenic (TNF-Tg) mice were used in this study. TNF decreased the steady-state levels of Smad1 and Runx2 protein similarly to those in long bones of TNF-Tg mice. In the presence of the proteasome inhibitor MG132, TNF increased accumulation of ubiquitinated Smad1 protein. TNF administration over calvarial bones caused decreases in Smad1 and Runx2 protein levels and mRNA expression of osteoblast marker genes in wild-type, but not in Smurf1-/- mice. Vertebral bone volume and strength of TNF-Tg/Smurf1-/- mice were examined by a combination of micro-CT, bone histomorphometry, and biomechanical testing and compared with those from TNF-Tg littermates. TNF-Tg mice had significantly decreased bone volume and biomechanical properties, which were partially rescued in TNF-Tg/Smurf1-/- mice. We conclude that in chronic inflammatory disorders where TNF is increased, TNF induces the expression of ubiquitin ligase Smurf1 and promotes ubiquitination and proteasomal degradation of Smad1 and Runx2, leading to systemic bone loss. Inhibition of ubiquitin-mediated Smad1 and Runx2 degradation in osteoblasts could help to treat inflammation-induced osteoporosis.
Osteoporosis and fragility fractures are common and preventable complications of rheumatoid arthritis (RA).2 For example, one study has reported that 53.3% of RA patients had osteoporosis and 19.3% had vertebral fractures, rates that are much higher than those of the general population (1). These complications are thought to occur through inflammatory stimulation of osteoclast bone resorption and inhibition of osteoblast function (2) mediated by cytokines, such as TNF (3–5). TNF and other cytokines are overproduced by various cells in the inflamed joints of RA patients, which lead to severe local erosion of cartilage and bone, as well as periarticular osteopenia and systemic osteoporosis.
TNF is a strong inhibitor of osteoblast functions in vitro, but the molecular mechanisms that mediate the inhibitory effects of TNF on osteoblasts have not been fully investigated. TNF inhibits the recruitment of osteoblast progenitors, reduces expression of genes produced by mature osteoblasts, and promotes osteoblast apoptosis through the NF-κB pathway (6–10). It decreases the expression and DNA binding activity of runt-related transcription factor 2 (Runx2), which is partially through suppression of Runx2 gene transcription and destabilization of Runx2 mRNA through the TNF receptor 1 signaling pathway (11–13). Recently, we demonstrated that TNF increases the expression of Smad ubiquitin regulatory factor-1 (Smurf1) in osteoblasts and promotes the proteasomal degradation of Runx2 protein in osteoblast cell lines (14), suggesting that the proteasomal regulation of key transcription factors in osteoblasts may play an important role in inflammation-induced bone loss.
Smurf1 was identified as an interacting protein with high homology to the homologous to E6-AP C terminus subclass of E3 ubiquitin ligases using Xenopus Smad1 as bait (15). It negatively regulates osteoblast functions by promoting ubiquitination and proteasomal degradation of bone morphogenetic protein (BMP) signaling molecules, including Runx2 and Smads 1 and 5 in vitro (16–18). Small peptides that block Smurf1 from binding to Smads increase osteoblast functions (19). We have used genetic approaches to demonstrate that both overexpression of Smurf1 in osteoblasts by type I collagen promoter or deletion of Smurf1 cause bone phenotypes. Smurf1 transgenic mice have decreased bone volume and reduced bone formation rates at 3–4-month of age (20). In contrast, Smurf1-/- mice have an age-dependent increase in bone mass. Thus, Smurf1 appears to play important roles in regulating bone mass in adult mice under normal physiologic conditions. However, the role of Smurf1 in pathologic bone loss has not been studied.
Recently, we observed a significant reduction in bone volume in long bones of TNF transgenic (TNF-Tg) mice with established arthritis and a decrease in mineralized bone nodule formation from cells derived from bone marrow of these mice compared with wild-type (WT) littermates (14). Along with our previous finding that TNF increases Smurf1 expression in osteoblast cell lines (14), we hypothesize that in TNF-Tg mice, Smurf1-mediated degradation of BMP signaling proteins is increased in osteoblasts, leading to reduced bone formation. Deletion of Smurf1 prevents bone loss in TNF-Tg mice. In the present studies, we demonstrated that Smurf1 protein expression is significantly increased in bones from TNF-Tg mice, which is associated with decreased Runx2 and Smad1 protein levels. The mice have reduced bone volumes and biomechanical parameters of bone strength. Smurf1 deletion prevents systemic bone loss and improves bone strength in TNF-Tg mice. Thus, our findings provide a novel molecular mechanism of TNF-induced inhibition in osteoblast function and BMP signaling, which involves post-transcriptional regulation of protein levels through Smurf1 E3 ligase-mediated proteasomal degradation.
MATERIALS AND METHODS
Animals—We used 4–8-month-old animals in this study because alteration of bone volume occurs in adult genetic-modified Smurf1 mice and TNF-Tg mice (20). TNF-Tg mice (3647 TNF-Tg line) were originally obtained from Dr. G. Kollias and characterized by us (21). TNF-Tg mice have elevated serum TNF levels when they reach 2 months of age or older (mean concentration is 100 pg/ml, range 48–120) (21), which is compatible to published (22) serum TNF concentrations in RA patients (mean concentration was 71.2 pg/ml, range 10.7–556.5), making this model a close mimic of adult RA clinical presentations. They were crossed with C57/BL6 mice for 8 generations. Smurf1-/- mice are in a C57/BL6 background, as we described previously (23). TNF-Tg/Smurf1-/- mice were generated by inter-crossing TNF-Tg and Smurf1+/- mice to generate the TNF-Tg/Smurf1+/- F1 generation; then these F1 mice were crossed with Smurf1+/- mice to generate TNF-Tg/Smurf1-/- mice. The Institutional Animal Care and Use Committee approved all studies.
Antibodies—Monoclonal antibodies specific for c-Myc and β-Actin were purchased from Sigma; anti-Runx2 monoclonal antibody was from MBL (Woburn, MA); anti-Smad1/5 and anti-Ubiquitin monoclonal antibody was from Santa Cruz (Santa Cruz, CA); anti-pSmad1/5 polyclonal antibody was from Cell Signaling (Danvers, MA); and anti-Smurf1 polyclonal antibody was obtained from Abgent (San Diego, CA).
Cell Culture and Transfection Conditions—2T3 osteoblast precursors were cultured in α-minimal essential medium supplemented with 10% fetal bovine serum (JRH Biosciences, Lenexa, KS). When cells reached 90% confluence, the cDNA expression plasmid, pCMV-Myc-tagged Smad1 (M-Smad1), and/or a pCMV-FLAG-tagged Smurf1 (F-Smurf1) were transiently transfected into the cells using Lipofectamine™ 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions. Total amounts of transfected plasmids in each group were equalized by the addition of an empty vector. After transfection, the cells were further cultured in the presence and absence of TNF + the proteasome inhibitor, MG132 (Calbiochem, La Jolla, CA), and subjected to Western blot analysis.
Primary Bone Marrow Osteoblast Cultures—To generate bone marrow-derived osteoblasts from mice, bone marrow cells were flushed from femoral and tibial bones and cultured in α-medium plus 20% fetal bovine serum for 7 days to generate mesenchymal stromal cells. The cells were then cultured in osteoblast inducing medium (α-minimal essential medium containing 10% fetal bovine serum with 50 μg/ml ascorbic acid, 10 mm β-glycerophosphate) and treated with phosphate-buffered saline (PBS) or TNF (7.5 ng/ml) for various times. For human bone marrow osteoblast cultures, human mesenchymal stromal cells were isolated, as we described previously (24). Briefly, bone marrow aspirates were obtained from the iliac crests of patients during spinal surgery after informed consent under a protocol approved by the Institutional Review Board at the University of Rochester Medical Center. These were mixed with equal volumes of PBS. One volume of lymphocyte separation medium (Mediatech, Inc., Herndon, VA) was then slowly layered underneath the aspirate/PBS mixture to isolate mononuclear cells. The mononuclear cells were cultured in α-minimal essential medium plus 10% fetal bovine serum for 24 h and adherent cells were incubated for further expansion. Second to third passages of these human mesenchymal stromal cells were used in subsequent experiments. Cells were cultured in osteoblast inducing medium in the presence or absence of TNF, as described above.
Real-time Quantitative Reverse Transcriptase-PCR—Total RNA was extracted from cell cultures or bone samples using TRIzol reagent (Invitrogen). Bone samples were cut into small pieces and homogenized in TRIzol reagent. Expression levels of Smurf1, Smurf2, alkaline phosphatase (ALP), osteocalcin, RANKL, osteoprotegrin, and β-actin mRNA were examined using quantitative real-time PCR amplification and sequence-specific primers, as described previously (14, 25).
Western Blot Analysis—Cells were lysed with mammalian protein extraction reagent (Pierce) containing a protease inhibitor mixture (Roche Applied Science). Long bone samples were cut into small pieces and homogenized in tissue protein extraction reagent (Pierce). Samples were resolved by 12% SDS-PAGE gel, transferred to a nitrocellulose membrane, and immunoblotted with individual antibodies. Membranes were then washed and incubated with a horseradish peroxidase-conjugated secondary antibody (Bio-Rad) and visualized by an enhanced chemiluminesence system (Amersham Biosciences) according to the manufacturer's instructions.
Ubiquitination Assay—2T3 cells were transfected with M-Smad1 and treated with TNF for 72 h + MG132 for the last 4 h of TNF treatment. Cell lysates were incubated with anti-Myc antibody and protein G-agarose (Sigma) overnight at 4 °C. In some experiments, calvarial pre-osteoblasts were isolated from 3–5-day-old pups from Smurf1-/- and WT mice, as published previously (26). Cells were treated with TNF for 72 h plus MG132 for the last 4 h. Total Smad1 protein was immunoprecipitated with anti-Smad1 antibody. The immunoprecipitates were washed with 50 mm Tris-HCl buffer (pH 8.0), containing 150 mm NaCl, 1% Nonidet P-40, 0.05% deoxycholate, and 0.1% SDS, resuspended in 2× reducing sample buffer, and subjected to Western blot analysis with an anti-ubiquitin antibody, as described previously (14, 27).
Microcomputed Tomography (Micro-CT)—The fourth lumbar vertebra (L4) was used for micro-CT and mechanical testing because our preliminary study demonstrated that L4 gave the smallest inter-sample variation in mechanical testing perhaps due to its large size and ease of dissection compared with L5–7. L4 vertebrae were scanned at high resolution (10.5 μm) on a VivaCT40 micro-CT scanner (Scanco Medical, Basserdorf, Switzerland) using an integration time of 300 ms, energy of 55 kVp, and intensity of 145 μA. A volume of interest for quantitative analysis of trabecular bone was defined, extending from the proximal to the distal end of the vertebrae. The three-dimensional images were generated using a constant threshold of 275 for all samples. For each sample, bone volume fraction, trabecular number, trabecular thickness, trabecular separation, and connectivity density were measured.
Mechanical Testing—The L4 vertebra was used and mechanical testing was performed according to a published method with a minor modification (28). In brief, the L4 was dissected of soft tissue and the vertebral body was isolated from the posterior elements by cutting the pedicles with a 10-mm diameter diamond saw. The end plates of the vertebral body were cemented onto small plastic plates using bone cement (DePuy Orthopaedics, Inc., Warsaw, IN), to ensure axial alignment of the bodies and to distribute the load evenly on the surface of the cemented end plates. The specimens were then hydrated in PBS for 2 h at room temperature. The vertebral bodies were tested in compression at a rate of 1 mm/min until failure using an Instron DynaMight 8841 servo-hydraulic materials testing machine (Instron, Norwood, MA). Maximum compressive load at failure (N), maximum deformation at maximum force (mm), stiffness (N/mm), and energy to failure (area under the curve) (N.mm) were measured from the recorded load-deformation curves using MATLAB software (The Mathworks, Natick, MA).
Histology—Paraffin sections were stained with H&E or for TRAP activity. Histomorphometric analyses were performed using an OsteoMeasure program (Osteometrics, Atlanta, GA) (21). Peri-articular inflammation (mm2) and eroded articular surface (%) were measured around knee joints and the number of osteoclast per millimeter bone surface was countered in the proximal tibiae region.
Statistics Analysis—Data are presented as mean ± S.D., and all experiments were performed at least twice with similar results. Statistical analyses were performed with StatView statistical software (SAS, Cary, NC). Differences between the two groups were compared using un-paired Student's t test and more than two groups were compared using one-way analysis of variance, followed by a Bonferroni/Dunnet test. p values less than 0.05 were considered to be statistically significant.
RESULTS
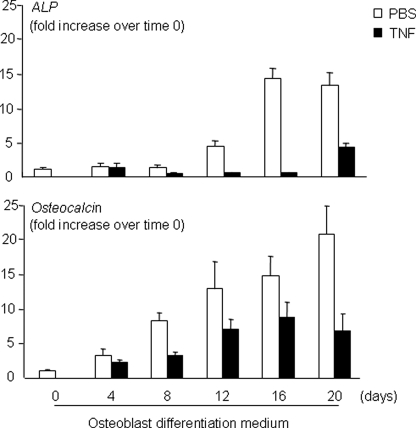
Chronic TNF Treatment of Mouse and Human Bone Marrow-derived Osteoblasts Increases Smurf1 and Reduces Basal Smad1 and Runx2 Protein Expression—Systemic osteoporosis occurs often in patients suffering from chronic inflammatory disorders and in TNF-Tg mice that have been chronically exposed to elevated blood TNF levels and have developed RA-like arthritis (14). Previously, we found that treatment of 2T3 pre-osteoblasts with TNF for 3 days increases Smurf1 expression and proteasomal degradation of Runx2 (14), leading to a hypothesis that elevated Smurf1-mediated BMP-signaling protein degradation may contribute to reduced osteoblast function in TNF-Tg mice. To test this hypothesis, we first attempted to establish a culture protocol that mimics the in vivo situation of TNF-Tg mice. Bone marrow mesenchymal stromal cells from 4–8-month-old WT mice were cultured in the osteoblast inducing medium and treated with TNF for 20 days. The expression levels of osteoblast marker genes were examined. TNF decreased the expression of ALP and osteocalcin starting from day 8, which became more significant thereafter, whereas ALP and osteocalcin expression increased markedly in PBS-treated cells (Fig. 1). In contrast, TNF-induced decreased ALP expression occurs usually much earlier in calvaria-derived or 2T3 pre-osteoblast cells (at about 24 h after TNF treatment, data not shown). These findings suggest that primary bone marrow-derived osteoblasts may respond to TNF differently and we decided to use 14-day TNF treatment as a protocol in our subsequent study.
FIGURE 1.
TNF reduces expression of osteoblast marker genes in bone marrow-derived osteoblasts from adult mice. Bone marrow cells from 4–8-month-old WT mice were cultured in osteoblast differentiation medium ± TNF (7.5 ng/ml) for various times. Expression of ALP, osteocalcin, and actin mRNA was determined by real time reverse transcriptase-PCR. -Fold changes were calculated by dividing the value of a given time point by the value obtained from day 0 as 1. Values are the mean ± S.D. of 3 loadings.
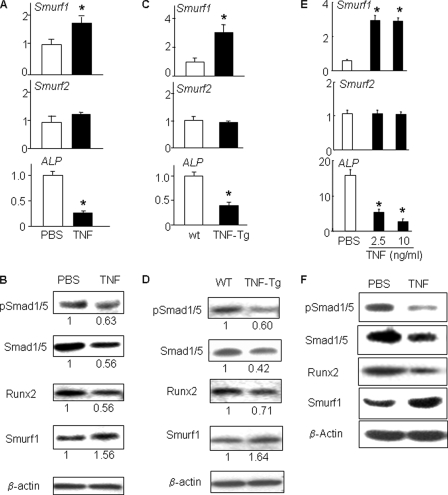
Under this culture condition, TNF decreased phosphorylated and total Smad1/5 as well as Runx2 protein levels and increased Smurf1 expression (Fig. 2, A and B). To determine whether this inhibitory effect of TNF on Smad1/5 and Runx2 protein expression also happened in vivo, we examined long bones of 4–5-month-old TNF-Tg mice, which had developed severe inflammatory arthritis and systemic bone loss. We found that Smurf1 protein expression was increased and phospho-Smad1/5, total Smad1/5, and Runx2 protein levels as well as ALP mRNA expression levels were decreased in bones from TNF-Tg mice compared with WT mice (Fig. 2, C and D).
FIGURE 2.
Chronic exposure to TNF decreases Smad1 and Runx2 protein levels in bone marrow-derived mature osteoblasts and in long bones from TNF-Tg mice. Murine (A and B) or human bone marrow cells (E and F) were cultured in osteoblast differentiation medium ± TNF (7.5 ng/ml) for 14 days, and total RNA and protein were harvested. Long bone samples (C and D) from 6-month-old TNF-Tg mice or WT littermates were harvested and pooled (n = 3/genotype). Expression of Smurf1, Smurf2, and ALP mRNA was determined by real time reverse transcriptase-PCR as in Fig. 1 (A, C, and E). Expression of phospho-Smad1/5, total Smad1/5, Runx2, and Smurf1 proteins was assessed by Western blot analysis (B, D, and F). The intensity of protein bands was measured in a densitometer. The -fold changes of a given protein over β-actin were calculated using PBS-treated samples as 1. *, p < 0.05 versus PBS-treated or WT samples.
To determine whether these inhibitory effects of TNF also occur in human osteoblasts, we treated osteoblasts derived from human bone marrow stromal cells with TNF. TNF increased Smurf1, decreased ALP, and had no effect on Smurf2 mRNA expression in human cells (Fig. 2E). The expression of phospho-Smad1/5, total Smad1/5, and Runx2 protein levels was significantly decreased in TNF-treated cells (Fig. 2F).
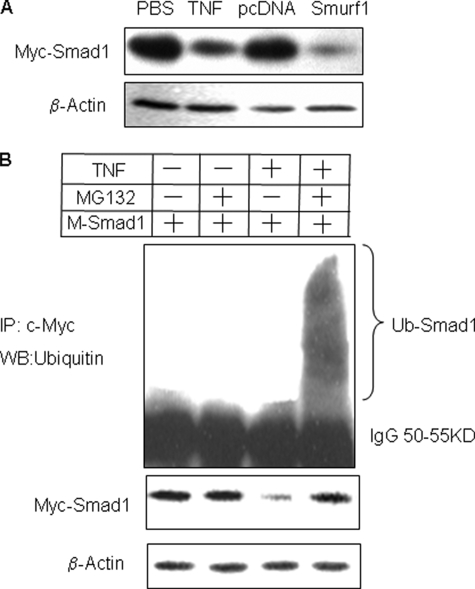
TNF Promotes Proteasomal Degradation of Smad1 through Smurf1-mediated Ubiquitination—We have demonstrated previously that Smurf1 mediates TNF-induced reduction in Runx2 protein levels in the 2T3 preosteoblast cell line (14). To determine whether TNF also affects Smad1 protein degradation through Smurf1, we transiently co-transfected 2T3 cells with M-Smad1 and F-Smurf1 expression vectors and treated them with TNF. TNF decreased exogenous Smad1 protein expression, assessed using an anti-c-Myc antibody, to a degree similar to that seen in cells overexpressing Smurf1 (Fig. 3A). To test if this was mediated by promoting ubiquitination and proteasomal degradation of Smad1, we treated cells with TNF + the proteasome inhibitor, MG132. Smad1 protein was immunoprecipitated with anti-c-Myc antibody and expression was assessed by Western blot using an anti-Ubiquitin antibody. In the presence of MG132, TNF increased the accumulation of ubiquitinated-Smad1 protein (Fig. 3B), indicating that TNF induces Smad1 ubiquitination and results in its rapid breakdown through proteasomal degradation.
FIGURE 3.
TNF increases Smad1 degradation by promoting its ubiquitination. A, 2T3 preosteoblasts were either transfected with M-Smad1 expression vector ± TNF or co-transfected with F-Smurf1 expression vectors for 72 h. The expression of M-Smad1 was examined by Western blot analysis with anti-c-Myc antibody. B, cells were transfected with M-Smad1 ± TNF for 72 h. MG132 (10 μm) was added for the last 4 h. M-Smad1 was immunoprecipitated (IP) by anti-c-Myc antibody, and ubiquitinated Smad1 protein ladders were detected by anti-ubiquitin antibody (upper panel). Total Smad1 and β-actin protein levels were determined by Western blot (WB) using anti-c-Myc or anti-actin antibody, respectively (lower panel).
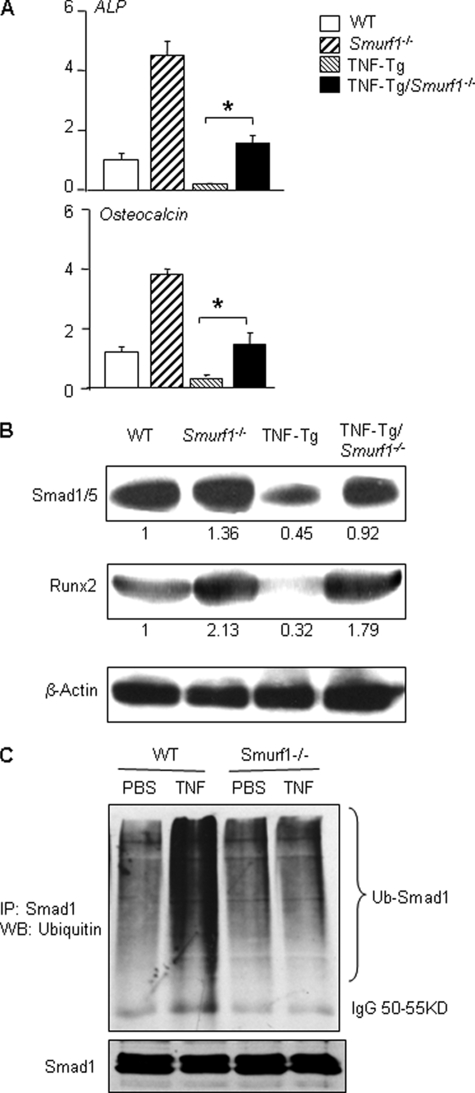
TNF-decreased Smad1 Protein Levels Are Attenuated in Smurf1-/- Mice—If Smurf1 mediates TNF-induced inhibition of osteoblast function, the effect should be attenuated or abolished in the absence of Smurf1. To examine this possibility, we generated TNF-Tg/Smurf1-/- mice and examined their bones when they were 7–8 months old. ALP and osteocalcin mRNA and Smads and Runx2 protein levels were examined. ALP and osteocalcin mRNA expression and Smad1/5 and Runx2 protein levels were decreased in TNF-Tg mice and these were restored to the WT levels in TNF-Tg/Smurf1-/- mice (Fig. 4, A and B). To confirm that TNF-induced decrease Smad1 protein levels is through Smurf1-mediated Smad1 ubiquitination in vivo, we treated calvarial pre-osteoblasts from Smurf1-/- and WT mice with TNF and subjected them to a ubiquitination assay. As shown in the 2T3 preosteoblast cell line, TNF increased the ubiquitination of endogenous Smad1 protein, which was prevented in cells from Smurf1-/- mice (Fig. 4C).
FIGURE 4.
TNF-induced osteoblast inhibition is reduced in Smurf1-/- mice. Long bone samples were harvested and pooled from 7.5-month-old WT, Smurf1-/-, TNF-Tg, and TNF-Tg/Smurf1-/- mice (n = 3/genotype). Total RNA and protein were extracted. The expression of ALP and osteocalcin mRNA (A), and Smad1/5 and Runx2 proteins (B) were determined by real time reverse transcriptase-PCR and Western blot (WB) analysis, respectively, as in Fig. 2. The intensity of protein bands was measured in a densitometer. The -fold changes of a given protein over β-actin were calculated using WT samples as 1. *, p < 0.05 versus TNF-Tg mice. Calvarial pre-osteoblasts were isolated from 3–5-day-old newborn pups of Smurf1-/- and WT mice and treated with TNF for 72 h. MG132 (10 μm) was added for the last 4 h. Endogenous Smad1 was immunoprecipitated (IP) by anti-Smad1 antibody, and ubiquitinated (upper panel) and total Smad1 (lower panels) proteins were detected by anti-ubiquitin or anti-Smad1 antibody, respectively (C).
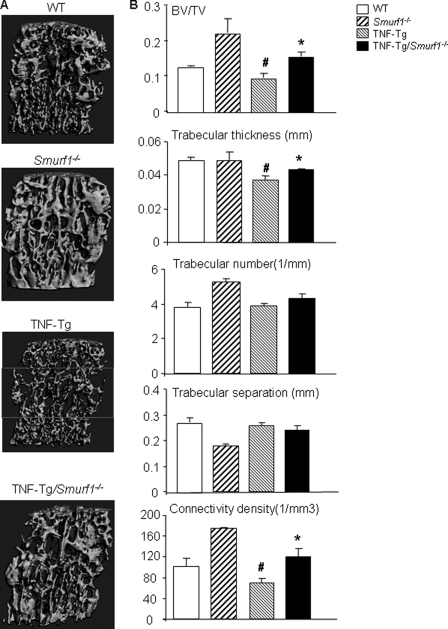
Smurf1 Deletion Attenuates Reduction in Bone Volume and Strength in TNF-Tg Mice—To determine whether Smurf1 deletion prevents the systemic bone loss in TNF-Tg mice, we analyzed vertebral bones of TNF-Tg/Smurf1-/- mice and compared them with WT, Smurf1-/-, and TNF-Tg mice from the same litters using micro-CT, histology, and mechanical testing. We used vertebral bones in these studies because persistent severe inflammation in the knee and ankle joints of TNF-Tg mice cause deformation and destruction of joints and, in some cases, damage to the epiphyses of femora and tibiae due to massive bone remodeling. This prohibited us from reliably examining the role of Smurf1 deletion in bone loss of the long bones of these mice. Micro-CT analyses of cancellous regions of the vertebrae (Fig. 5, A and B) show that the bone volume of Smurf1-/- mice was significantly increased as previously reported in long bones of these mice (23). TNF-Tg mice had a statistically significant decrease in bone volume, particularly in the middle portions of the vertebral bodies (see box in the Fig. 5A). Bone volume was restored to WT levels in TNF-Tg/Smurf1-/- mice (Fig. 5B). In contrast, micro-CT analysis of cortical regions in the same vertebrae found that both TNF-Tg and TNF-Tg/Smurf1-/- mice had significantly reduced cortical bone thickness compared with WT or Smurf1-/- mice (cortical thickness (mm): 0.236 ± 0.020 in WT, 0.211 ± 0.026 in Smurf1-/-, *0.190 + 0.016 in TNF-Tg, and *0.186 ± 0.012 in TNF-Tg/Smurf1-/- mice; *, p < 0.05 versus WT mice). The micro-CT findings were confirmed histologically (supplemental Fig. S2).
FIGURE 5.
TNF-induced systemic bone loss is prevented in TNF-Tg/Smurf1-/- mice. Bodies of 4th and 2nd lumber vertebrae were isolated from 7.5-month-old WT, Smurf1-/-, TNF-Tg, or TNF-Tg/Smurf1-/- mice and subjected to micro-CT analysis and histology examination, respectively. A, representative three-dimensional reconstructed image shows significantly reduced trabecular bone structure in TNF-Tg mice, particularly in the middle portion of the vertebral bodies (box). Reduced bone volume was not observed in TNF-Tg/Smurf1-/- mice. B, trabecular bone parameters were analyzed at the middle portion of the vertebral bodies. Values are the mean ± S.D. of 4–8 mice. *, p < 0.05 versus TNF-Tg mice; #, p < 0.05 versus WT mice.
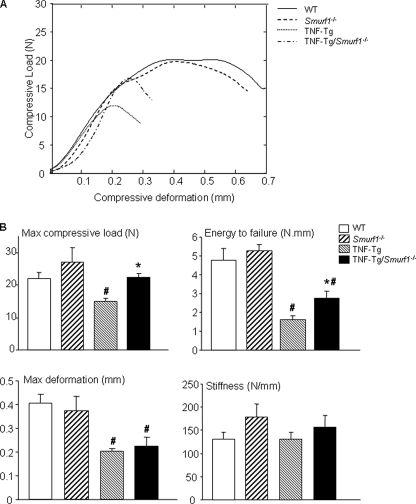
Osteoporosis is typically associated with reduced bone strength. To examine if TNF-Tg mice have reduced bone quality and if this can be rescued by Smurf1 deletion, we examined the compressive biomechanical properties of mouse vertebral bones using a modified Instron material testing system (28). The biomechanical properties measured include maximum deformation, maximum compressive load at failure, energy to failure, and stiffness. Compared with WT littermates, TNF-Tg mice have significantly reduced biomechanical properties except stiffness (Fig. 6, A and B). TNF-Tg/Smurf1-/- mice have improved mechanical strength in certain degrees. The maximum compressive load in TNF-Tg/Smurf1-/- mice was higher than in TNF-Tg mice and restored to the WT levels. Their energy to failure was improved compared with TNF-Tg mice but still lower than in WT mice (Fig. 6B). The maximum deformation was unchanged between TNF-Tg/Smurf1-/- and TNF-Tg mice, both of which were significantly lower than WT mice (Fig. 6B). In addition, there were no differences in Smurf1-/- and WT mice vertebral biomechanical properties except stiffness.
FIGURE 6.
Chronic exposure to TNF reduces bone mechanical strength, which is partially restored in TNF-Tg/Smurf1-/- mice. Lumbar 4 vertebral bodies from 7.5-month-old WT, Smurf1-/-, TNF-Tg, and TNF-Tg/Smurf1-/- mice were tested in compression at a rate of 1 mm/min until failure. The representative load-deformation curves from WT, Smurf1-/-, TNF-Tg, and TNF-Tg/Smurf1-/- mice show that TNF-Tg mice have reduced maximal load and work to failure but TNF-Tg/Smurf1-/- mice have a similar compressive strength compared with WT mice (A). Biomechanical properties including maximum deformation, maximum compressive load to failure, energy to failure, and stiffness are shown in B. Values are the mean ± S.D. of 4–9 mice. *, p < 0.05 versus TNF-Tg mice; #, p < 0.05 versus WT mice.
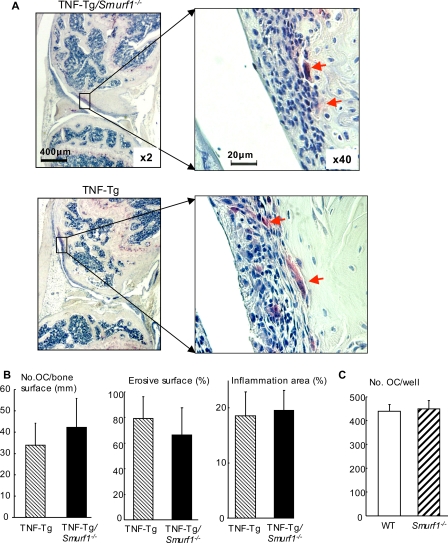
TNF is known to stimulate osteoclast formation and bone erosion in joints of RA patients and animals (29). To determine whether partially restored bone mass in TNF-Tg/Smurf1-/- mice is due to potent influence of Smurf1 on osteoclast function, we examined osteoclast number and local bone erosion in knee joints of TNF-Tg/Smurf1-/- and TNF-Tg mice. No difference in osteoclast numbers, bone erosion, and synovial area was observed between TNF-Tg/Smurf1-/- and TNF-Tg mice (Fig. 7, A and B). In addition, the same number of osteoclasts was formed when spleen cells from Smurf1-/- and WT mice were cultured with RANKL and macrophage colony-stimulating factors (Fig. 7C).
FIGURE 7.
Smurf1 deletion does not affect osteoclast function. Knee sections from 7.5-month-old TNF-Tg and TNF-Tg/Smurf1-/- mice were stained for TRAP activity for identifying osteoclasts (arrows). The number of TRAP+ osteoclasts and inflammatory area were assessed. Representative TRAP-stained sections indicate a similar degree of osteoclastogenesis, bone erosion, and inflammation between TNF-Tg and TNF-Tg/Smurf1-/- mice (A). Histomorphometric analyses of the number of osteoclasts and percentage of erosion and inflammation are shown in B. Values are the mean ± S.D. of 6 mice per group. Spleen cells from WT and Smurf1-/- mice were cultured with RANKL and macrophage colony-stimulating factors for 7 days to form osteoclasts. The cells were fixed in formalin and stained for TRAP activity. The number of TRAP+ cells was counted. Values are the mean ± S.D. of 4 wells (C). A similar result was repeated using another pair of WT and Smurf1-/- mice.
DISCUSSION
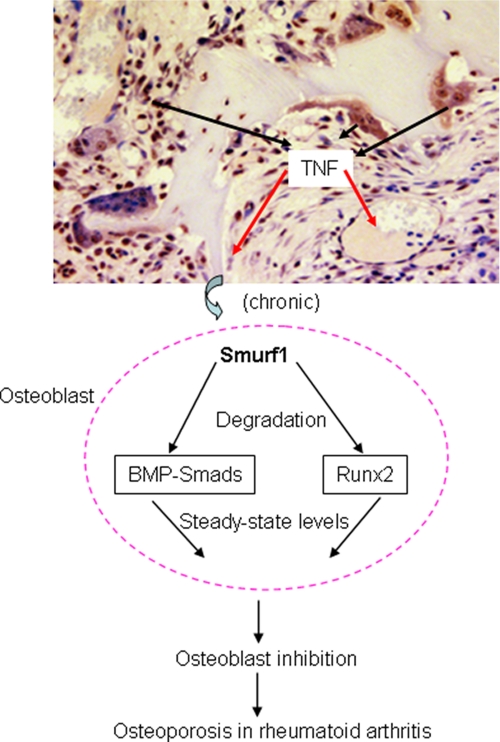
In the present study, we demonstrate that expression of the E3 ubiquitin ligase, Smurf1, is increased in TNF-treated osteoblasts in vitro and in bones of TNF-Tg mice with established arthritis. Deletion of Smurf1 prevented the TNF-induced reduction in Smad1/5 and Runx2 protein levels in osteoblasts and partially rescued systemic bone loss. Smurf1 deletion does not affect inflammation and osteoclastic bone erosion in joints of TNF-Tg mice. Thus, Smurf1 is the primary mediator of TNF-induced degradation of Smad1 and Runx2 proteins, and it is likely responsible for inhibition of bone formation. Based on these findings, we propose a model to explain the involvement of Smurf1 in inhibition of bone formation in patients with chronic inflammatory disorders (Fig. 8). In arthritic joints, secretion of pro-inflammatory cytokines, such as TNF, is elevated. TNF can affect osteoblasts locally or at distant sites to increase their expression of Smurf1. Smurf1 promotes the ubiquitination and proteasomal degradation of Smad1 and Runx2 proteins, leading to consistently decreased steady-state levels of these key positive regulators in osteoblasts. Consequently, bone formation cannot match the increased bone resorption, which is also induced by pro-inflammatory cytokines, resulting in systemic bone loss.
FIGURE 8.
A model of Smurf1-induced BMP signal protein degradation in TNF-induced osteoblast inhibition in inflammatory arthritis. In inflamed joints, elevated TNF increases Smurf1 expression in osteoblasts locally and systemically. Increased Smurf1 constitutively increases proteasomal degradation of BMP signal proteins Runx2 and Smad1 and decreases steady-state levels of these osteoblast positive regulators, leading to osteoblast inhibition and bone loss.
We and others have reported previously that overexpression of Smurf1 in osteoblastic cell lines promotes ubiquitination and degradation of Smad1 and Runx2, and that Smurf1 transgenic mice have decreased bone volume (20, 30). These in vitro and in vivo findings indicate that Smurf1-mediated protein degradation plays a role in bone remodeling. Here we provide in vivo evidence that Smurf1 mediates TNF-induced pathologic bone loss. We believe that this is the first demonstration showing up-regulation of an E3 ligase in an in vivo model of a common bone disorder, which implies a role for increased protein degradation in inflammatory bone loss.
Increased degradation of muscle protein has been reported in patients suffering from stress and sepsis and in animals receiving large amounts of lipopolysaccharide (26). For example, lipopolysaccharide or cytokines promote muscle protein degradation through NF-κB-induced up-regulated expression of the E3 ubiquitin ligase, Muscle RING-finger (MuRF) 1 (31, 32). The degree of muscle atrophy in this model can be reduced by deletion of MuRF1 (31).
The mechanisms that mediate TNF-induced increased Smurf1 expression in osteoblasts or in bones of TNF-Tg mice are not clear. We identified a putative NF-κB binding sequence at -1131/-1127 of the mouse Smurf1 promoter using Genomatix software and generated a luciferase reporter construct containing 1.8 kb of the Smurf1 promoter. However, we did not detect any effect of TNF or IL-1 on reporter activity (supplemental Fig. S1). This finding suggests that the TNF-regulatory region is not within this 1.8-kb promoter fragment.
Ubiquitin protein ligases have largely been considered to be activated constitutively and regulated only at the level of target binding. However, it has become evident recently that homologous E6-AP C terminus-type E3 ligases can be regulated by other mechanisms, including phosphorylation of the ligase or substrate, utilization of adaptor proteins or intra- and inter-molecular interactions (33–35). Whether or not these regulatory mechanisms contribute to increased degradation of Smad1/Runx2 proteins in a TNF overexpression condition is not known. Because of multiple functions of the TNF signaling pathway in cells, it is very likely that TNF increases protein degradation through several mechanisms. For instance, TNF can increase phosphorylation stage proteins and/or increase the production of adaptor proteins to facilitate protein degradation. These possibilities need to be investigated in the future.
In the original study characterizing Smurf1-/- mice using osteoblasts derived from calvariae of 3–5-day-old neonatal pups, canonical Smad-mediated BMP signaling appears unchanged but Smurf1-/- cells have an enhanced JNK-MAPK cascade. Ubiquitination and degradation of mitogen-activated protein kinase kinase 2 (MEKK2), an upstream kinase of JNK, was impaired in the absence of Smurf1 (23). We do not know currently if MEKK2 is required for an inhibitory effect of TNF on osteoblasts because the bone phenotype of MEKK2-/- mice has not been studied (36). Because MEKK2 up-regulates TNF promoter activity and works upstream of TNF (37), it is difficult using elevated steady-state levels of MEKK2 protein to explain rescued osteoblast phenotypes in TNF-Tg/Smurf1-/- mice.
We found that the -fold increase in Smad1 and Runx2 protein levels in Smurf1-/- osteoblasts compared with WT cells (1.36 for Smad1 and 2.1 for Runx2, Fig. 4B) is lower than the -fold increase in cells from TNF-Tg/Smurf1-/- mice compared with TNF-Tg mice (2.04 for Smad1 and 5.6 for Runx2, Fig. 4B). This observation raises a possibility that Smurf1 may play a more important role in mediating the degradation of Smad1 and Runx2 proteins under the activated condition. A recent article (38) reported that phosphorylation of Smad1 increases Smurf1-induced degradation. Whether this is the case in TNF chronic exposure-induced osteoblast inhibition needs to be studied further.
Although our data indicate that Smurf1 may be the primary mediator of TNF-induced degradation of Smad1 and Runx2, it does not mean that Smurf1 is the only mechanism by which TNF inhibits osteoblast function. For example, TNF decreases BMP-induced osteoblast differentiation by activating stress-activated protein kinase/JNK signaling (39). It reduces maximal peak bone mass and inhibits activation of the Smad-Luciferase reporter construct in MC3T3-E1 osteoblasts (40). TNF also negatively regulates bone formation through activation of p38 MAPK (41) and induction of apoptosis (42). Thus, TNF-mediated osteoblast inhibition likely works through multiple mechanisms.
One of our important findings in this study is the remarkable reductions in bone strength and toughness of bones from TNF-Tg mice (Fig. 6). Increased fragility fractures are common in RA patients (1), but it has not been formally demonstrated in a mouse model of RA. Our finding is the first demonstration of reduced bone strength in TNF-Tg mice. This is important because although Smurf1 deletion restored bone volume of TNF-Tg mice to WT levels (Fig. 5), it only partially rescued the biomechanical properties, such as maximum load and energy to failure, and had no effect on maximum deformation (Fig. 6). Furthermore, although the algorithm to assess overall trabecular numbers, thickness, and connectivity for the TNF-Tg/Smurf1-/- vertebrae are “globally” restored in number value (Fig. 5B), the micro-CT images show that the TNF-Tg/Smurf1-/- mice exhibit an abnormal hypertrophic cancellous bone microarchitecture as compared with WT (Fig. 5A). These findings suggest that deletion of Smurf1 only partially improves abnormal bone microarchitectures and bone strength of TNF-Tg mice. Other factors must also contribute to the bone phenotypes of these animals. Smurf1 deletion does not affect the increased osteoclastic activity in TNF-Tg mice (Fig. 7), which may result in increased resorption pits that act as stress riser sites and undermine the restoration of biomechanical strength even with increased bone volume. Another underlying mechanism is that Smurf1 deletion improves the trabecular but not cortical bone defect of TNF-Tg mice, whereas biomechanical testing is a function of combinations of cortical and trabecular bones. All of the above factors likely take into account the partial restoration of the mechanical properties of TNF-Tg/Smurf1-/- mice.
Increased osteoclast function in TNF-Tg mice may also explain identical stiffness between the WT and TNF-Tg vertebrae (Fig. 6). Increased osteoclastic resorption pits may act as stress concentration sites that influence the failure properties such as maximum compressive load and the energy to failure, but will have no significant effects on subfailure properties such as the stiffness. On the other hand, Smurf1 deletion enhances osteoblast function in WT and TNF-Tg mice, which effects partial rescue of bone volume, leading only to minute and statistically insignificant increases in the subfailure stiffness property.
In conclusion, our study has provided evidence that TNF inhibits BMP signaling through Smurf1-mediated proteasomal degradation of Smad1/5 and Runx2 proteins, leading to impaired osteoblast function in vivo. Deletion of Smurf1 partially prevents systemic bone loss and improves the bone quality of TNF-Tg mice. Thus, the regulation of protein degradation alone, or in combination with osteoclast inhibition, may represent a new strategy to treat periarticular osteopenia and systemic osteoporosis in patients with chronic inflammatory arthritis.
Supplementary Material
Acknowledgments
We thank Xiaoyun Zhang for technical assistance with histology and Dr. Paul Rubery for providing human bone marrow cells.
This work was supported, in whole or in part, by National Institutes of Health Grants AR48697 and AR53586 (to L. X.) and AR43510 (to B. F. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: RA, rheumatoid arthritis; Smurf1, Smad ubiquitin regulatory factor 1; BMP, bone morphogenetic protein; Tg, transgenic; TNF, tumor necrosis factor; WT, wild type; PBS, phosphate-buffered saline; CT, computed tomography; MEKK2, mitogen-activated protein kinase kinase 2; JNK, c-Jun NH2-terminal kinase; MAPK, mitogen-activated protein kinase; ALP, alkaline phosphatase.
References
- 1.Nakayama, H. (2007) Clin. Calcium 17 1607-1612 [PubMed] [Google Scholar]
- 2.Goldring, S. R. (2003) Calcif. Tissue Int. 73 97-100 [DOI] [PubMed] [Google Scholar]
- 3.Nair, S. P., Williams, R. J., and Henderson, B. (2000) Rheumatology 39 821-834 [DOI] [PubMed] [Google Scholar]
- 4.Srivastava, S., Weitzmann, M. N., Cenci, S., Ross, F. P., Adler, S., and Pacifici, R. (1999) J. Clin. Investig. 104 503-513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eigler, A., and Endres, S. (1997) Z. Gastroenterol. 35 899-901 [PubMed] [Google Scholar]
- 6.Li, Y. P., and Stashenko, P. (1992) J. Immunol. 148 788-794 [PubMed] [Google Scholar]
- 7.Taichman, R. S., and Hauschka, P. V. (1992) Inflammation 16 587-601 [DOI] [PubMed] [Google Scholar]
- 8.Kitajima, I., Soejima, Y., Takasaki, I., Beppu, H., Tokioka, T., and Maruyama, I. (1996) Bone 19 263-270 [DOI] [PubMed] [Google Scholar]
- 9.Jilka, R. L., Weinstein, R. S., Bellido, T., Parfitt, A. M., and Manolagas, S. C. (1998) J. Bone Miner. Res. 13 793-802 [DOI] [PubMed] [Google Scholar]
- 10.Gilbert, L., He, X., Farmer, P., Boden, S., Kozlowski, M., Rubin, J., and Nanes, M. S. (2000) Endocrinology 141 3956-3964 [DOI] [PubMed] [Google Scholar]
- 11.Gilbert, L., He, X., Farmer, P., Rubin, J., Drissi, H., van Wijnen, A. J., Lian, J. B., Stein, G. S., and Nanes, M. S. (2002) J. Biol. Chem. 277 2695-2701 [DOI] [PubMed] [Google Scholar]
- 12.Abbas, S., Zhang, Y. H., Clohisy, J. C., and Abu-Amer, Y. (2003) Cytokine 22 33-41 [DOI] [PubMed] [Google Scholar]
- 13.Gilbert, L. C., Rubin, J., and Nanes, M. S. (2005) Am. J. Physiol. 288 E1011-E1018 [DOI] [PubMed] [Google Scholar]
- 14.Kaneki, H., Guo, R., Chen, D., Yao, Z., Schwarz, E. M., Zhang, Y. E., Boyce, B. F., and Xing, L. (2006) J. Biol. Chem. 281 4326-4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu, H., Kavsak, P., Abdollah, S., Wrana, J. L., and Thomsen, G. H. (1999) Nature 400 687-693 [DOI] [PubMed] [Google Scholar]
- 16.Chen, D., Zhao, M., and Mundy, G. R. (2004) Growth Factors 22 233-241 [DOI] [PubMed] [Google Scholar]
- 17.Izzi, L., and Attisano, L. (2004) Oncogene 23 2071-2078 [DOI] [PubMed] [Google Scholar]
- 18.Datto, M., and Wang, X. F. (2005) Cell 121 2-4 [DOI] [PubMed] [Google Scholar]
- 19.Sangadala, S., Boden, S. D., Viggeswarapu, M., Liu, Y., and Titus, L. (2006) J. Biol. Chem. 281 17212-17219 [DOI] [PubMed] [Google Scholar]
- 20.Zhao, M., Qiao, M., Oyajobi, B. O., Mundy, G. R., and Chen, D. (2003) J. Biol. Chem. 278 27939-27944 [DOI] [PubMed] [Google Scholar]
- 21.Li, P., Schwarz, E. M., O'Keefe, R. J., Ma, L., Looney, R. J., Ritchlin, C. T., Boyce, B. F., and Xing, L. (2004) Arthritis Rheum. 50 265-276 [DOI] [PubMed] [Google Scholar]
- 22.Di Comite, G., Marinosci, A., Di Matteo, P., Manfredi, A., Rovere-Querini, P., Baldissera, E., Aiello, P., Corti, A., and Sabbadini, M. G. (2006) Ann. N. Y. Acad. Sci. 1069 428-437 [DOI] [PubMed] [Google Scholar]
- 23.Yamashita, M., Ying, S. X., Zhang, G. M., Li, C., Cheng, S. Y., Deng, C. X., and Zhang, Y. E. (2005) Cell 121 101-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito, H., Goater, J. J., Tiyapatanaputi, P., Rubery, P. T., O'Keefe, R. J., and Schwarz, E. M. (2004) Gene Ther. 11 34-41 [DOI] [PubMed] [Google Scholar]
- 25.Mimura, T., Amano, S., Usui, T., Kaji, Y., Oshika, T., and Ishii, Y. (2001) Exp. Eye Res. 72 71-78 [DOI] [PubMed] [Google Scholar]
- 26.Franzoso, G., Carlson, L., Xing, L., Poljak, L., Shores, E. W., Brown, K. D., Leonardi, A., Tran, T., Boyce, B. F., and Siebenlist, U. (1997) Genes Dev. 11 3482-3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen, R., Chen, M., Wang, Y. J., Kaneki, H., Xing, L., O'Keefe, R. J., and Chen, D. (2006) J. Biol. Chem. 281 3569-3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva, M. J., Brodt, M. D., and Uthgenannt, B. A. (2004) Bone 35 425-431 [DOI] [PubMed] [Google Scholar]
- 29.Goldring, S. R. (2003) Rheumatology (Oxford) 42 Suppl. 2, ii11-ii16 [DOI] [PubMed] [Google Scholar]
- 30.Zhao, M., Qiao, M., Harris, S. E., Oyajobi, B. O., Mundy, G. R., and Chen, D. (2004) J. Biol. Chem. 279 12854-12859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bodine, S. C., Latres, E., Baumhueter, S., Lai, V. K., Nunez, L., Clarke, B. A., Poueymirou, W. T., Panaro, F. J., Na, E., Dharmarajan, K., Pan, Z. Q., Valenzuela, D. M., DeChiara, T. M., Stitt, T. N., Yancopoulos, G. D., and Glass, D. J. (2001) Science 294 1704-1708 [DOI] [PubMed] [Google Scholar]
- 32.Glass, D. J. (2003) Nat. Cell Biol. 5 87-90 [DOI] [PubMed] [Google Scholar]
- 33.Wiesner, S., Ogunjimi, A. A., Wang, H. R., Rotin, D., Sicheri, F., Wrana, J. L., and Forman-Kay, J. D. (2007) Cell 130 651-662 [DOI] [PubMed] [Google Scholar]
- 34.Gao, M., and Karin, M. (2005) Mol. Cell 19 581-593 [DOI] [PubMed] [Google Scholar]
- 35.Gallagher, E., Gao, M., Liu, Y. C., and Karin, M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 1717-1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo, Z., Clydesdale, G., Cheng, J., Kim, K., Gan, L., McConkey, D. J., Ullrich, S. E., Zhuang, Y., and Su, B. (2002) Mol. Cell. Biol. 22 5761-5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cuevas, B. D., Uhlik, M. T., Garrington, T. P., and Johnson, G. L. (2005) Oncogene 24 801-809 [DOI] [PubMed] [Google Scholar]
- 38.Sapkota, G., Alarcon, C., Spagnoli, F. M., Brivanlou, A. H., and Massague, J. (2007) Mol. Cell 25 441-454 [DOI] [PubMed] [Google Scholar]
- 39.Mukai, T., Otsuka, F., Otani, H., Yamashita, M., Takasugi, K., Inagaki, K., Yamamura, M., and Makino, H. (2007) Biochem. Biophys. Res. Commun. 356 1004-1010 [DOI] [PubMed] [Google Scholar]
- 40.Li, Y., Li, A., Strait, K., Zhang, H., Nanes, M. S., and Weitzmann, M. N. (2007) J. Bone Miner. Res. 22 646-655 [DOI] [PubMed] [Google Scholar]
- 41.Yang, C., Zhou, W., Jeon, M. S., Demydenko, D., Harada, Y., Zhou, H., and Liu, Y. C. (2006) Mol. Cell 21 135-141 [DOI] [PubMed] [Google Scholar]
- 42.Kitajima, I., Nakajima, T., Imamura, T., Takasaki, I., Kawahara, K., Okano, T., Tokioka, T., Soejima, Y., Abeyama, K., and Maruyama, I. (1996) J. Bone Miner. Res. 11 200-210 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.