Abstract
Hypoxia induces oxidative damage in skeletal muscle. Uncoupling protein 3 (UCP3) is the skeletal muscle enriched uncoupling protein and has previously been shown to confer resistance against oxidative stress. We show that hypoxia robustly up-regulates skeletal muscle UCP3 and that the absence of UCP3 in primary skeletal myocytes exacerbates hypoxia-induced reactive oxygen species generation. In this context, we reasoned that the investigation of the regulation of UCP3 may identify novel hypoxia-responsive regulatory pathways that modulate intrinsic anti-oxidant defenses. By screening a transcription factor array of 704 full-length cDNAs in murine C2C12 myoblasts following cotransfection of a murine UCP3 promoter-luciferase construct and myoD we identified numerous candidate regulatory factors that up-regulate UCP3. Active transcription factor-1 (ATF-1) was identified, and as this transcription factor is a known component of a multiprotein hypoxia-induced regulatory complex, we explored its role in hypoxia-mediated UCP3 up-regulation. Site-directed mutagenesis and chromatin immunoprecipitation assays identify a 10-bp region required for ATF-1 induction of UCP3 promoter activity. Hypoxia promotes the phosphorylation of ATF-1, and the knockdown of ATF-1 by shRNA prevents hypoxia-mediated up-regulation of UCP3. Pharmacologic inhibition of p38 MAP kinase prevents both hypoxia-mediated ATF-1 phosphorylation and UCP3 up-regulation. PKA signaling does not modulate hypoxia-induced UCP3 up-regulation and neither does HIF-1α activation by cobalt chloride. In conclusion, ATF-1, via p38 MAP kinase activation, functions as a novel regulatory pathway driving UCP3 expression. These data reinforce the role of ATF-1 as a hypoxia-responsive trans-activator and identifies a novel regulatory program that may modulate cellular responses to oxygen-deficit.
The skeletal and cardiac-enriched uncoupling protein 3 (UCP3)2 is proposed to function by modulating mitochondrial inner membrane proton conductance (1) resulting in the alteration in reactive oxygen species (ROS) production (2, 3) and to augment mitochondrial fatty acid metabolism (4). Consistent with its uncoupling effect, UCP3 attenuates skeletal and cardiac muscle ROS production (5-7). In line with the role of UCP3 in promoting fatty acid oxidation, the genetic depletion of UCP3 enhances the accumulation of triglyceride in skeletal muscle following a high fat diet (8), and skeletal muscle overexpression of UCP3 attenuates high fat diet-induced insulin resistance and diabetes (9). Taken together, these data suggest that the modest induction of this protein may have ameliorative therapeutic potential to enhance tolerance against oxidative stress and/or in the treatment of insulin resistance and diabetes.
Interestingly, hypoxemia in patients with chronic obstructive pulmonary disease results in excess skeletal muscle oxidative stress compared with similar patients who do not exhibit hypoxia (10). Evidence of oxidative stress is also shown in skeletal muscle of mice exposed to severe non-lethal hypoxia for 48 h (11). Despite these findings, the role of hypoxia in ROS production in skeletal muscle remains controversial (reviewed in Ref. 12). This controversy probably reflects, in part, the dynamic balance between ROS generation and the concordant activation of endogenous antioxidant systems as well as the relative lack of sensitivity of current technologies for ROS detection. Nevertheless, whether UCP3 functions to modulate the homeostatic balance of reactive oxygen species production in skeletal muscle in response to hypoxia has not been explored although an acute (<4 h) exposure to hypoxia does up-regulate skeletal muscle UCP3 levels (13).
The initial observation in our study confirms that UCP3 is induced by hypoxia in skeletal muscle and we show that this induction is sustained for up to 48 h. Furthermore, we demonstrate that the genetic deficiency of UCP3 in primary skeletal myocytes results in excess ROS levels under normoxic and hypoxic conditions. We exploited this regulation to identify novel hypoxia-responsive regulatory programs. Transcriptional array analysis identified ATF-1 as an activator of UCP3, and we show that ATF-1 is phosphorylated by hypoxia. The partial depletion of ATF-1 blunts hypoxia-mediated induction of UCP3. Finally, the UCP3 promoter region required for this induction was identified, and p38 MAP kinase signaling is implicated in this hypoxia-induced ATF-1-mediated up-regulation of UCP3.
EXPERIMENTAL PROCEDURES
Mouse in Vivo and Primary Cell Hypoxic Studies—The UCP3-/- mice and their wild-type littermate controls were obtained from T. Finkel (National Institutes of Health). Adult mice were studied at age of 8-12 weeks of age. Wild-type mice were exposed to normobaric hypoxia (10%, COY Laboratory Products Inc., Grass Lake, MI) prior to extraction of gastrocnemius muscle.
Primary skeletal muscle cultures were generated from 1-5-day-old neonatal UCP3 (+/+ and -/-) mice using hindlimb muscles as previously described (14). Differentiated myotubes were exposed to normobaric hypoxia using a hypoxic incubator (Sanyo Biomedical, Bensenville, IL). Animal experiments were approved by the NHLBI Animal Care and Use Committee.
Cell Line Hypoxia Studies—The mouse myoblast C2C12 and rat myoblast L6 cells were purchased from the American Type Culture Collection (Manassas, VA). Cells were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum as growth medium. Myoblast differentiation was initiated by transferring to Dulbecco's modified Eagle's medium with 2% horse serum once cells reached 90% confluence. Hypoxia was introduced following 4 days of differentiation by placing cells into a hypoxic incubator at 5% O2, 5% CO2, and 37 °C in differentiation medium. Normoxic control cells were maintained in 5% CO2 at 37 °C.
Quantitative Real-time PCR—Total RNA was isolated from cells using RNeasy mini kit (Qiagen, Valencia, CA). Reverse transcription of total RNA was performed using SuperScrip lll first-strand synthesis system (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Quantitative real-time PCR was performed using SYBR Green PCR master mix (Applied Biosystems, Foster City, CA) and run on Opticon2 DNA engine (Bio-Rad). All reactions were normalized using a Tbp endogenous control. All real-time PCR primers were purchased from predesigned primers of QuantiTect primer assays (Qiagen).
Western Blot Analyses—Total protein from myotubes and mice skeletal muscles were extracted using radioimmune precipitation assay buffer with Halt protease/phosphatase inhibitor mixture (Pierce). Nuclear protein was extracted using nuclear and cytoplasmic extraction reagents (Pierce). Samples were run on NuPAGE Bis-Tris gels (Invitrogen), transferred to nitrocellulose membranes, and incubated with various antibodies. The UCP3 (ab3477) and Tbp antibodies were purchased from Abcam (Cambridge, MA). ATF-1 (sc-243) and phospho-ATF-1 (Ser-133) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA) and Upstate (Billerica, MA), respectively. The actin antibody was from Sigma. Quantification of Western band intensities was performed with images on x-ray film using ImageJ software.
Analysis of Reactive Oxygen Species—Skeletal muscle primary cells from UCP3 wild-type and knock-out mice were grown in differentiation medium for 5 days. The cells exposed to hypoxia were incubated for 24 h at 5% O2, and were harvested in parallel with the normoxic control groups. 1 × 106 cells were incubated at 37 °C for 30 min with 5 μm H2DCFDA (Molecular Probes, Carlsbad, CA). They were then washed with phosphate-buffered saline and resuspended in medium at a concentration of 106 cells/100 μl. Fluorescence was measured using a GeniosPlus microplate reader at fluorescence excitation and emission of 495/525 nm (Tecan, Durham, NC). To measure the kinetics of H2O2 generation, we spectrophotometrically assayed the oxidation of amplex red to produce resofurin at 585 nm using the Amplex Red Hydrogen Peroxide/Peroxidase assay kit according to the manufacturer's protocol (Molecular Probes). A further measure to evaluate oxidative stress in these cells was performed by assaying the activity of aconitase (an oxidative stress-sensitive mitochondrial enzyme) by assessing spectrophotometric formation of NADPH at 340 nm using the Bioxytech Aconitase-340 kit (Oxis Research, Portland, OR).
UCP3 Promoter-reported Construct Studies—Fragments from -2050 to +63 of mouse UCP3 promoter were amplified by PCR from mouse genomic DNA and were cloned into pGL4.10 to generate luciferase 2k-luc. Ten deletion constructs from 1-luc to 10-luc with various lengths of UCP3 promoter region were generated from 2k-luc using unique internal restriction sites as shown in Fig. 2A. Deletion constructs from 21-luc to 28-luc were amplified by PCR and cloned into pGL4.10. Three point mutation constructs of 2m1-luc to 2m3-luc were generated from 2-luc using a QuikChange ll site-directed mutagenesis kit (Stratagene, La Jolla, CA) as shown in Fig. 3A. For all reporter constructs, numbers are based on +1 for the transcriptional start site. Cloning primers were listed in supplemental Table S1. Additional plasmids employed in this study include the mouse genes encoding for ATF-1 and MyoD. These were amplified by PCR and cloned into pcDNA3.1 (Invitrogen).
FIGURE 2.
Effects of myoD and ATF-1 on UCP3 murine promoter activity. A, luciferase reporter constructs of mouse UCP3 5′-end promoter region. Numerical annotation is relative to the transcriptional start site designation as +1. B, reporter constructs transfected into C2C12 myoblasts with MyoD or pcDNA empty control vector. Luciferase activities were normalized to the activity of 2k-luc co-transfected with the empty vector (value = 1) and represent mean ± S.D. of three independent experiments in this and all subsequent cotransfection experiments. C, similar study to B, with the exception that the luciferase activity in response to ATF-1 is assessed instead of that of myoD. D, cotransfection experiments to evaluate the combined ex vivo activation capacity of myoD and ATF-1 using the full-length and 2-luc UCP3 promoter-luciferase reporter constructs.
FIGURE 3.
Identification of the ATF-1-responsive region within UCP3 promoter. A, reporter constructs of deletion and point mutation of UCP3 promoter employed to identify region required for ATF-1 transactivation. The three mutation constructs of 2-luc are shown immediately below the index construct. Mutated nucleotides are underlined and deleted nucleotides shown as dashes. B, luciferase activity of deletion/mutation reporter constructs in response to the cotransfection of ATF-1 compared with the empty vector control in C2C12 cells. Results are normalized to 2k-luc activity co-transfected with empty vector (value = 1) and represent mean ± S.D. of three independent experiments. C, representative ChIP analysis of ATF1 binding to UCP3 promoter region corresponding to the region -1556 to -1342.
C2C12 and L6 myoblasts cells were seeded in 12-well plates. Transfection was carried out at 80% confluence using Lipofectamine 2000 (Invitrogen). Each well contained 1 μg of luciferase reporter construct, 0.6 μg of pcDNA-ATF-1 or pcDNA vector, and 6 ng of hRluc/SV40 used as an internal transfection control. For ATF-1 and MyoD co-transfection experiments, each well contained 1 μg of luciferase reporter construct, 0.3 μg of pcDNA-ATF-1, and/or 0.3 μg of pcDNA-MyoD, 6 ng of hRluc/SV40. Cells were harvested 48 h after transfection. Luciferase activities were measured in a GeniosPlus microplate reader using Dual-Glo luciferase assay system (Promega, Madison, WI).
Full-length cDNA Transcriptional Array Analysis—GFC-Array assays (Origene, Rockville, MD) was performed according to the manufacturer's protocol. Briefly, construct DNA, FuGene6 and C2C12 myoblast cells were added to 384-well plates that were preplated with different lyophilized transcription factor plasmids. The final added content to each well contained 75 ng of our full-length UCP3 promoter-luciferase construct, 10 ng of pcDNA-MyoD, 1.5 ng of hRluc/SV40, 0.26 μl of FuGENE6. Forty-eight hours after transfection, luciferase activities were measured in a Victor3 microplate reader (PerkinElmer Life Sciences) using the Dual-Glo luciferase assay system.
Chromatin Immunoprecipitation Assay—ChIP analysis was performed according to the manufacturer's EZ-ChIP kit protocol (Upstate). In briefly, C2C12 myotubes of both normoxia groups and hypoxia groups at 5% O2 for 2 days were treated with formaldehyde, and the cross-linked chromatin was sonicated to lengths spanning 200-1000 bp. The samples were precleared with 60 μl of protein A-agarose and subsequently incubated while gently rotating at 4 °C overnight with either 5 μg of mouse anti-ATF-1 or with 5 μg of normal mouse IgA as a negative control, to assess nonspecific antibody binding. Immunocomplexes were precipitated using protein A-agarose. Mouse IgA was from Bethyl (Montgomery, TX). After washing, elution, and reverse cross-linking, DNA fragments were isolated and purified with columns. PCR was performed using primers spanning the promoter region implicated in the promoter deletion and mutation transfection-reporter assays using the primers UCP3IP-F: 5′-ACAGACGACATGCCCAATTT-3′ and UCP3IP-R: 5′-GTCAAACCTCTGCTGGGAGT-3′. Primers spanning other regions were tested as negative controls.
shRNA Studies—The ATF-1 shRNA construct pLKO-shATF-1 was purchased from Open Biosystems (Huntsville, AL). For ATF-1 shRNA experiments, 1 × 106 C2C12 myoblast with 2 μg of pLKO-ATF-1 or vector were suspended in 100 μl of Nucleofector solution V and electroporated with Nucleofector II (Amaxa Biosystems, Gaithersburg, MD). Cells were then transferred to 6-well plates and grown in differentiation medium for 3 days prior to exposure to hypoxia versus normoxia for an additional 48 h. Myotubes were then harvested for protein analysis.
Pharmacologic Inhibitors and Chemicals—KT5720 and H89 were from Sigma-Aldrich. SB203580 was from Calbiochem (Gibbstown, NJ). All other chemicals were from Sigma-Aldrich unless stated otherwise.
Data Analysis—Data are expressed as mean ± S.D. Differences between groups were analyzed by Student's t test. Statistical significance was set at p < 0.05. Each experiment was performed at least three times in duplicate.
RESULTS
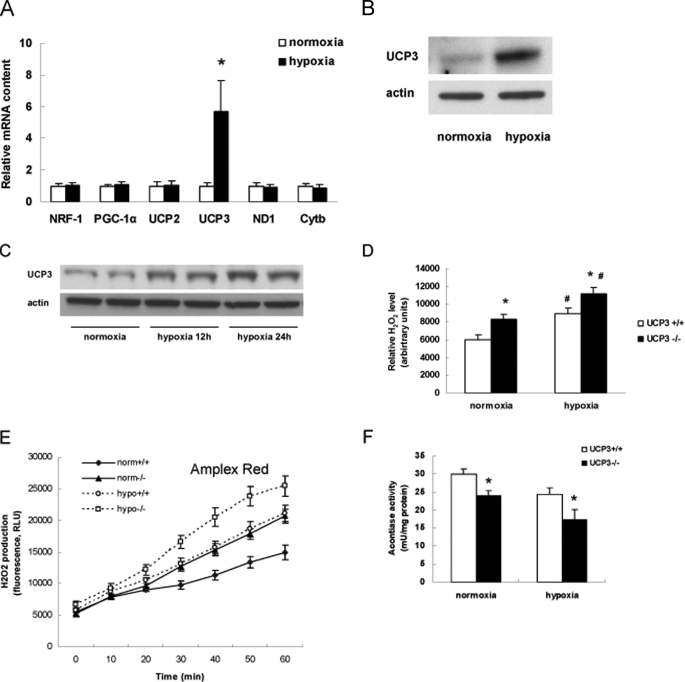
Regulation of UCP3 by Hypoxia and the Generation of Reactive Oxygen Species—To confirm that UCP3 is up-regulated by hypoxia, we assessed the gene expression and protein levels of UCP3 in C2C12 myotubes. Following 24 h of exposure to 5% oxygen UCP3 was induced in excess of 5-fold compared with the transcript levels in normoxic control cells (Fig. 1A). Of note this was not due to a generalized induction of mitochondrial biogenesis regulatory gene transcripts as nuclear respiratory factor 1 (NRF-1) and the peroxisome proliferator activated receptor γ coactivator 1 α (PGC-1α) were not modulated by hypoxia. Transcripts encoding electron transfer chain proteins and UCP2 were also unaltered, suggesting a distinct hypoxia-mediated regulation of UCP3 (Fig. 1A). This mRNA regulation was paralleled by the increase in UCP3 protein expression in both the C2C12 myotubes and in mouse gastrocnemius muscle exposed to 12 and 24 h of hypoxia (10% O2, Fig. 1, B and C). The increase in steady-state protein levels is sustained over this time period. Interestingly, this induction of UCP3 was only evident in differentiated tissue, as this regulatory pattern was not evident in C2C12 myoblasts (data not shown). As UCP3 is proposed to attenuate ROS generation in skeletal muscle, we compared the reactive oxygen species levels and production rates under normoxic and hypoxic conditions comparing primary skeletal myotubes from wild-type and UCP3 knock-out mice. At baseline (normoxia) and in response to 5% hypoxia, the UCP3 knock-out myotubes showed increased levels and generation of reactive oxygen species levels (Fig. 1, D and E). As a further measure of oxidative stress injury we find that the activity of the oxidative stress sensitive aconitase activity is diminished under normoxia and hypoxia in UCP3 knock-out myocytes compared with wild-type controls (Fig. 1F).
FIGURE 1.
Effects of hypoxia on UCP3 expression and on reactive oxygen species. A, mitochondria-related genes mRNA expression level in C2C12 myotubes exposed to 5% O2 for 24 h. Results are relative to normoxia content and represent the mean ± S.D. of three experiments. B, representative Western blot of UCP3 steady-state protein in C2C12 myotubes exposed to 5% O2 for 48 h. C, representative Western blot of UCP3 protein levels in gastrocnemius tissues from C57bl/6 mice subjected to 10% O2 versus normoxia for 12 and 24 h respectively. D, skeletal muscle primary cells from UCP3 wild-type and knock-out mice were incubated in condition of normoxia or hypoxia (5% O2) for 48 h. H2O2 level was detected by H2DCFDA. E, the rate of H2O2 production following 24 h of hypoxia was determined by measuring amplex red oxidation rates in wild-type and UCP3 knock-out primary myocytes compared with normoxic controls. F, aconitase activity, as an indirect measure of oxidative damage induced inhibition of enzyme activity was quantified under the same experimental conditions described in E. *, within normoxia or hypoxia group, p < 0.05. #, between groups, p < 0.05. PGC-1α, peroxisome proliferator-activated receptor γ co-activator 1α; NRF-1, nuclear respiratory factor-1; ND1-NADPH dehydrogenase subunit 1; Cytb, cytochrome b; UCP2, uncoupling protein 2; and UCP3, uncoupling protein 3.
Mouse UCP3 Promoter Analysis—To identify hypoxia-responsive transcription factors that up-regulate murine UCP3, we investigated the regulation of the murine UCP3 promoter. We cloned a 2.1-kb genomic fragment from +63 bp 3′ of exon 1 to-2010 bp 5′ of the exon 1. This 5′-flanking region was subcloned in-frame, into a luciferase reporter gene. Restriction digestion was employed to generate subsequent deletion promoter constructs (Fig. 2A). In light of UCP3 being induced in differentiated muscle (15), we evaluated the activity of the UCP3 promoter constructs following cotransfection with the muscle differentiation regulator myoD. In these cotransfection studies we found that the 1.5-kb UCP3 promoter construct was the most highly induced (Fig. 2B). This construct was termed the 2-luc construct and was employed as the index construct for all subsequent promoter analysis studies.
Identification of Potential Novel Transcriptional Activators of UCP3—Using C2C12 myoblasts we employed the 2-luc construct and myoD in cotransfection analysis to screen a transcription factor array incorporating 704 full-length cDNAs to identify putative novel regulatory proteins that could induce UCP3. The ten transcription factors that resulted in the highest induction of the UCP3 promoter are shown in Table 1. ATF-1 resulted in a 4-fold activation of the UCP3 promoter. As this transcription factor has previously been identified as a component of a hypoxia-mediated multiprotein regulatory complex, we focused on studying whether this transcription factor is a hypoxia-sensing transactivator of UCP3. ATF-1 co-transfection studies demonstrated robust activation of full-length 2k-luc and 2-luc UCP3 promoter-luciferase reporter constructs in both murine C2C12 (Fig. 2C) and rat L6 myotubes (data not shown). In parallel with the array data, the cotransfection of ATF-1 with myoD evoked exponential activation of the full-length and 2-luc UCP3 promoter constructs (Fig. 2D). The negative regulatory elements of the UCP3 promoter upstream of -1433 bp from the transcription start site, as shown in Fig. 2B, have not been investigated in this study.
TABLE 1.
Genome-wide full-length cDNA arrays results
| Transcription factor | Relative folda | Major function |
|---|---|---|
| DEAF1 | 5.19 | Embryonic development |
| PITX2 | 4.55 | Development of eye, tooth, abdominal organs |
| HSF1 | 4.18 | Temperature stress |
| ATF-1 | 4.1 | CREB family, cAMP signaling, cell survival |
| GABPB2 | 3.81 | Cytochrome oxidase expression, control of mitochondrial function |
| EBF | 3.73 | Regulation of B cell lineage commitment |
| SOX9 | 3.71 | Chondrocyte differentiation |
| NKX2-3 | 3.24 | Cell type specification, maintenance of differentiated tissue |
| TP53 | 2.87 | Regulation of cell cycle |
| SPDEF | 2.82 | Prostate-specific antigen expression |
Values are normalized to control group coated with blank vector and represent the mean of three independent experiments.
UCP3 Promoter Deletion/Mutation Constructs and Chromatin Immunoprecipitation Identifies an ATF-1 Responsive Promoter Region—The ATF-1-induced UCP3 promoter-luciferase activity was markedly attenuated by the deletion of a 204-bp region from the 5′-flank of the 2-luc promoter construct as shown in Fig. 2C. Deletion and mutation constructs of this region were generated and tested to identify the functional activation region of the UCP3 promoter (Fig. 3A). The promoter construct-luciferase activity assays identified that the 10-bp region between positions -1426 and -1417 was required for ATF-1 activation of the murine UCP3 promoter construct (Fig. 3B). To date, this sequence CTCCACAGAT is not a recognized consensus sequence for transcription factor binding. Interesting a CREB consensus binding sequence is present in the murine UCP3 promoter at -767 to -775. We mutated this site, but did not show any modulation of UCP-luciferase promoter activity in response to ATF-1 cotransfection (data not shown). To confirm the necessity of this putative regulatory region for ATF-1-mediated activation, chromatin immunoprecipitation assays were performed using primer sets that spanned the region -1556 bp to -1342 bp of this 5′-flanking region. In parallel with the deletion/mutation analysis, we show that ATF-1 does bind to this region (Fig. 3C). ChIP analysis with regions 5′ and 3′ to this region did not show binding of ATF-1 (data not shown).
Signal Transduction Pathways and ATF-1-mediated Activation of UCP3—We next evaluated whether ATF-1 is activated by hypoxia in C2C12 myotubes. ATF-1 activation requires phosphorylation (Ser-133) and we show a modest increase in phosphorylation at 3 to 6 h with a maximal 3.8-fold increase in phosphorylation at 24 h following the onset of hypoxia (Fig. 4A). To establish whether this ATF-1 activation is directly linked to the induction of UCP3 we transfected C2C12 myotubes with a shRNA directed against ATF-1. This approach resulted in a 74% reduction in ATF-1 steady-state protein levels (Fig. 4B). Following ATF-1 depletion, UCP3 levels were assessed under normoxic and hypoxic conditions. The genetic knockdown of ATF-1 had no effect on UCP3 levels under normoxic conditions; however, under hypoxia the induction of UCP3 was completely blunted in the ATF-1-deficient myotubes (Fig. 4C).
FIGURE 4.
Effects of hypoxia on ATF-1 phosphorylation and of ATF-1 knockdown on hypoxia-mediated UCP3 up-regulation. A, representative Western blot of temporal levels of ATF-1 and phosphorylated ATF-1 levels in nuclear protein from C2C12 myotubes exposed to 5% O2 from 3 to 24 h compared with levels in normoxic control cells. B, representative Western blot showing knockdown of ATF-1 in C2C12 myotubes, 5 days after insertion into myoblasts and growth in differentiation media. Nuclear protein loading is normalized to TATA-binding protein (Tbp). Histogram shows mean and S.D. nuclear protein levels of ATF-1 comparing cells electroporated with the control vector construct versus the shATF-1 construct. C, representative Western blot showing expression of UCP3 protein levels in C2C12 myotubes transfected with pLKO-shATF-1 or control vector in response to 48 h of normoxia or hypoxia (5% O2), respectively. pLKO, control vector; pLKO-shATF-1, vector harboring the ATF-1 shRNA.
As ATF-1 belongs to a subfamily of transcription factors that are activated by cAMP we evaluated whether the pharmacologic inhibition of PKA using either KT5720 or H89, would attenuate hypoxia-mediated up-regulation of UCP3 protein levels. As shown in Fig. 5, A and B, neither of these PKA inhibitors blunted UCP3 induction by hypoxia. In contrast, the p38 MAP kinase inhibitor SB203580 blocked the phosphorylation of ATF-1 (Fig. 5, C and D) and partially attenuated the up-regulation of UCP3 in response to hypoxia (Fig. 5, C and E). Finally, as HIF-1α is another putative candidate transcription factor that may hypothetically up-regulate UCP3 under hypoxic conditions, we investigated whether cobalt chloride, which stabilizes HIF-1α (16), could modulate UCP3 protein levels under normoxic conditions. The steady-state protein levels of HIF-1α protein levels were increased in response to cobalt chloride administration, however, this did not result in an induction of UCP3 protein levels (Fig. 5F). In parallel there was no induction of the UCP3 transcript levels in C2C12 myotubes exposed to cobalt chloride (Fig. 5G).
FIGURE 5.
Signal transduction pathways of ATF-1-mediated UCP3 up-regulation. A, a histogram showing the transcript levels of UCP3 under a normoxic and hypoxic environment for 48 h under control conditions compared with myotubes treated with vehicle or the PKA inhibitors KT5720 (5uM) or H89 (5uM). B, representative Western blot comparing UCP3 levels under a normoxic and hypoxic environment for 48 h under control conditions compared with myotubes treated with vehicle or the PKA inhibitors KT5720 (5 μm) or H89 (5 μm). C, representative Western blot comparing phospho-ATF-1 and UCP3 levels under a normoxic and hypoxic environment for 48 h comparing myotubes treated with vehicle or the p38 MAP kinase inhibitor SB203580 (30 μm). Quantification of the protein levels of phospho-ATF-1 (D) and UCP3 (E) protein levels under the same conditions described in C. DMSO, dimethyl sulfoxide. F, representative Western blot of HIF-1α and UCP3 levels in response to cobalt chloride administration (0.5 mm) or vehicle control for 48 h. Equal protein loading is shown by immunoblot of actin levels. G, histogram showing UCP3 transcript levels in control and cobalt chloride-treated C2C12 myotubes under the same conditions described in F.*, p < 0.05.
DISCUSSION
The current study finds that ATF-1 functions as a hypoxia-sensing transcription factor to up-regulate UCP3 in skeletal muscle. This functions through the activation of p38 MAP kinase, which in turn phosphorylates and activates ATF-1. Moreover, the absence of UCP3 enhances skeletal muscle ROS production. Collectively, these data suggest that this regulatory program may play an adaptive role to protect skeletal muscle in response to exposure to hypoxia.
Numerous environmental modulators, signaling intermediates, gene transcription factors, and post-translational modifications have been identified that regulate UCP3 functioning. The environmental triggers include hypoxia (13), hyperoxia (17), transient ischemia (6), high dietary fat intake, and acute exercise (13). The regulatory mediators identified to date include the activation of AMPK (13), thyroid hormone (18), retinoic acid, and activation of peroxisome proliferator-activated receptor γ (PPARγ) (19). At the post-translational level UCP3 is activated by fatty acids and free-radical-derived alkenals (20, 21). This study shows that the ATF-1 transcription factor also up-regulates skeletal muscle UCP3, and that this regulatory program is hypoxia-sensing and is operational in the context of oxygen-deficit.
The ATF-1 transcription along with cyclic AMP-response element (CRE)-binding protein (CREB) and CRE modulator (CREM) constitute a subfamily of β-Zip transcription factors. CREB being the most extensively studied (reviewed in Ref. 22) is activated by phosphorylation in response to, among other signals, cAMP. Interestingly, in response to mitogen or stress signaling target gene activation by CREB requires additional promoter bound transcription factors (23). Furthermore, under certain conditions phosphorylated CREB appears to function as a facilitator of transcription without the direct occupancy of a CRE (24). Interestingly, CREB has been shown to be markedly induced by hypoxia (25). Although the transcription factor ATF-1 has been less well characterized, it has been shown to function during development, during tumorigenesis and in regulating the immune system (26-28). Although a robust phenotype following the genetic depletion of ATF-1 has, to date not been identified (29), the combined knock-out of ATF-1 and CREB shows that these transcription factors have overlapping functions and are important in early embryogenesis (29). Moreover, ATF-1 has been shown to have tumorigenic properties (30), a biological process known to be operational in a milieu of relative hypoxia. Furthermore, ATF-1 does interact with the HIF-1 DNA-recognition site (31) and it has been identified as a component of a hypoxia-inducible multiprotein complex that activates oxygen responsive genes (32). The activation of ATF-1 in hypoxia is supported by the data from this study which shows that ATF-1 is phosphorylated by hypoxia and that its induction can drive UCP3 expression and in parallel, that the genetic knockdown of ATF-1 abrogates hypoxia-mediated UCP3 induction. In this study we did not test whether the phosphorylation of ATF-1 modifies its direct or indirect interaction with the UCP3 promoter. Moreover, it is of interest to note that time to ATF-1 phosphorylation in response to hypoxia requires numerous hours compared with the more rapid phosphorylation of proteins more commonly seen in response to cell stimulation. Whether this reflects the activation of intermediate effector/s in this signaling pathway is unknown.
The most characterized hypoxia-mediated signaling cascade involves the regulation of HIF-1α (reviewed in Ref. 33), although multiple signaling kinases are also activated by hypoxia and these include the AMPK, p38 MAPK, and cAMP (34-36). Despite the known capacity of ATF-1 to bind to HIF-1α (32) and to its cognate binding site (31), the direct induction of HIF-1α by cobalt chloride in this study did not result in the induction of UCP3. This shows that the induction of HIF-1α is insufficient to up-regulate UCP3, but does not preclude an interaction between ATF-1 with HIF-1α in this regulation. This potential interaction was not explored but cannot be discounted. We show that the phosphorylation of ATF-1 during hypoxia is dependent on p38 MAP kinase signaling in that the pharmacologic inhibition of this kinase precluded hypoxia-mediated ATF-1 phosphorylation and partially but significantly blunts UCP3 induction. The incomplete attenuation of hypoxia-mediated UCP3 up-regulation by the prevention of ATF-1 phosphorylation suggests that additional transcriptional mediators are operation in this regulation of UCP3. Although not explored additional candidates, identified in the transcriptional array screen are shown in Table 1. Furthermore, the classic CREB cAMP pathway does not appear to be required as the PKA inhibitors H-89 and KT5720 did not disrupt UCP3 induction under hypoxic conditions.
It has been well established that UCP3 is up-regulated during myoblast differentiation (37) and its differentiation induction is dependent on myoD (38). Interestingly, we show that UCP3 expression was not up-regulated by hypoxia in immature C2C12 myoblasts (data not shown), in contrast to the hypoxia-mediated induction in mature myotubes. This shows that the up-regulation of UCP3 is dependent on myoblast differentiation and could therefore infer that a threshold level of myoD may be required to mediate this hypoxia-induced regulation. To account for this we included myoD in the transcriptional factor array analysis where we identified ATF-1. The co-transfection of ATF-1 and myoD also shows additive UCP3 promoter activation in our study. However, the transactivator function of ATF-1 does not require myoD in ex vivo promoter-reporter activation studies. The human UCP3 promoter has been more extensively studied than the mouse promoter. However, our promoter deletion-construct reporter activity studies show remarkably similarly required regions conferring activator and repressor promoter functions compared with the human UCP3 promoter (39). These include the requirement of the ∼251bp 5′ of the transcription start site for transcriptional activation and a repressor promoter region between -2050 bp and -1433 bp upstream of the transcription start site. The 10-bp promoter region show to be necessary for ATF-1-mediated promoter activation is not a recognized DNA binding sequence for transcriptional activation. Whether this region is important from a structural perspective to maintain the integrity of a regulatory complex may be possible, but has not been established. The ChIP assay does show that ATF-1 does interact with the UCP3 promoter in a region including this 10-bp region. Of note, the direct mutation of the CRE region 3′ to this putative regulatory region did not diminish AFT-1-mediated activation of the UCP3 promoter (data not shown).
In mice deficient in UCP3, the amount of ROS generated in skeletal muscle mitochondria is increased (1). This observation was confirmed in our study where we show that the lack of UCP3, results in an additional increase in ROS generation under normoxic and hypoxic conditions. It is important to note, that the change in UCP3 levels is not the exclusive modulator of ROS production and that even with the induction of UCP3 in wild-type muscle ROS generation is increased in response to hypoxia, albeit to a lesser extent than in the UCP3 knock-out muscle. Thus the up-regulation of UCP3 appears to be an important component of skeletal muscle adaptation to hypoxia. Additionally, the genetic induction of UCP3 has been shown to be of benefit in the prevention of diabetes (9) and exercise-mediated elevation of endogenous UCP3 reduces mitochondrial ROS production (7). Taken together, these data support targeting the activation of this protein for therapeutic purposes. Although it is known that the thiazolidinedione class of compounds up-regulates UCP3 the therapeutic potential of the thiazolidinediones has recently been assuaged by evidence linking them to systemic adverse effects (40, 41). Hence, identifying additional regulatory programs that up-regulate UCP3 may have clinical utility. ATF-1 is a novel target to explore in this context and the additional transcription factors identified in our transcriptional array screen, shown in Table 1, also need to be characterized as putative activators of UCP3.
In conclusion this study has identified a novel regulatory program that up-regulates UCP3 in the context of hypoxia. Moreover, this study has identified ATF-1, through the activation of p38 MAP kinase as a novel hypoxia-responsive transcription factor.
Supplementary Material
Acknowledgments
We thank Dr. Marc L. Reitman for his gift of the UCP3 knockout mice to the NHLBI and the Laboratory of Animal Medicine and Surgery for their assistance with the murine hypoxia studies.
This work was supported, in whole or in part, by the National Institutes of Health NHLBI, Division of Intramural Research. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
Footnotes
The abbreviations used are: UCP, uncoupling protein; ROS, reactive oxygen species; MAP, mitogen-activated protein; PKA, cAMP-dependent kinase; ATF, active transcription factor; ChIP, chromatin immunoprecipitation assay.
References
- 1.Vidal-Puig, A. J., Grujic, D., Zhang, C. Y., Hagen, T., Boss, O., Ido, Y., Szczepanik, A., Wade, J., Mootha, V., Cortright, R., Muoio, D. M., and Lowell, B. B. (2000) J. Biol. Chem. 275 16258-16266 [DOI] [PubMed] [Google Scholar]
- 2.Brand, M. D., and Esteves, T. C. (2005) Cell Metab. 2 85-93 [DOI] [PubMed] [Google Scholar]
- 3.Sack, M. N. (2006) Cardiovasc Res. 72 210-219 [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Martinez, C., Sibille, B., Solanes, G., Darimont, C., Mace, K., Villarroya, F., and Gomez-Foix, A. M. (2001) Faseb. J. 15 2033-2035 [DOI] [PubMed] [Google Scholar]
- 5.Talbot, D. A., and Brand, M. D. (2005) Biochim. Biophys. Acta 1709 150-156 [DOI] [PubMed] [Google Scholar]
- 6.McLeod, C. J., Aziz, A., Hoyt, R. F., Jr., McCoy, J. P., Jr., and Sack, M. N. (2005) J. Biol. Chem. 280 33470-33476 [DOI] [PubMed] [Google Scholar]
- 7.Anderson, E. J., Yamazaki, H., and Neufer, P. D. (2007) J. Biol. Chem. 282 31257-31266 [DOI] [PubMed] [Google Scholar]
- 8.Costford, S. R., Chaudhry, S. N., Salkhordeh, M., and Harper, M. E. (2006) Am. J. Physiol. Endocrinol. Metab. 290 E1304-E1312 [DOI] [PubMed] [Google Scholar]
- 9.Choi, C. S., Fillmore, J. J., Kim, J. K., Liu, Z. X., Kim, S., Collier, E. F., Kulkarni, A., Distefano, A., Hwang, Y. J., Kahn, M., Chen, Y., Yu, C., Moore, I. K., Reznick, R. M., Higashimori, T., and Shulman, G. I. (2007) J. Clin. Investig. 117 1995-2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koechlin, C., Maltais, F., Saey, D., Michaud, A., LeBlanc, P., Hayot, M., and Prefaut, C. (2005) Thorax 60 834-841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magalhaes, J., Ascensao, A., Soares, J. M., Ferreira, R., Neuparth, M. J., Marques, F., and Duarte, J. A. (2005) J. Appl. Physiol. 99 1247-1253 [DOI] [PubMed] [Google Scholar]
- 12.Clanton, T. L. (2007) J. Appl. Physiol. 102 2379-2388 [DOI] [PubMed] [Google Scholar]
- 13.Zhou, M., Lin, B. Z., Coughlin, S., Vallega, G., and Pilch, P. F. (2000) Am. J. Physiol. Endocrinol. Metab. 279 E622-E629 [DOI] [PubMed] [Google Scholar]
- 14.Rando, T. A., and Blau, H. M. (1994) J. Cell Biol. 125 1275-1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, D., Jitrapakdee, S., and Thompson, M. (2007) J. Biochem. Mol. Biol. 40 921-927 [DOI] [PubMed] [Google Scholar]
- 16.Wang, G. L., and Semenza, G. L. (1995) J. Biol. Chem. 270 1230-1237 [DOI] [PubMed] [Google Scholar]
- 17.Flandin, P., Donati, Y., Barazzone-Argiroffo, C., and Muzzin, P. (2005) FEBS Lett. 579 3411-3415 [DOI] [PubMed] [Google Scholar]
- 18.Solanes, G., Pedraza, N., Calvo, V., Vidal-Puig, A., Lowell, B. B., and Villarroya, F. (2005) Biochem. J. 386 505-513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solanes, G., Pedraza, N., Iglesias, R., Giralt, M., and Villarroya, F. (2003) Mol. Endocrinol. 17 1944-1958 [DOI] [PubMed] [Google Scholar]
- 20.Echtay, K. S., Roussel, D., St-Pierre, J., Jekabsons, M. B., Cadenas, S., Stuart, J. A., Harper, J. A., Roebuck, S. J., Morrison, A., Pickering, S., Clapham, J. C., and Brand, M. D. (2002) Nature 415 96-99 [DOI] [PubMed] [Google Scholar]
- 21.Echtay, K. S., Esteves, T. C., Pakay, J. L., Jekabsons, M. B., Lambert, A. J., Portero-Otin, M., Pamplona, R., Vidal-Puig, A. J., Wang, S., Roebuck, S. J., and Brand, M. D. (2003) EMBO J. 22 4103-4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayr, B., and Montminy, M. (2001) Nat. Rev. Mol. Cell. Biol. 2 599-609 [DOI] [PubMed] [Google Scholar]
- 23.Bonni, A., Ginty, D. D., Dudek, H., and Greenberg, M. E. (1995) Mol. Cell Neurosci. 6 168-183 [DOI] [PubMed] [Google Scholar]
- 24.Brindle, P., Nakajima, T., and Montminy, M. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 10521-10525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beitner-Johnson, D., Rust, R. T., Hsieh, T. C., and Millhorn, D. E. (2001) Cell Signal. 13 23-27 [DOI] [PubMed] [Google Scholar]
- 26.Rosenberg, D., Groussin, L., Jullian, E., Perlemoine, K., Bertagna, X., and Bertherat, J. (2002) Ann. N. Y. Acad. Sci. 968 65-74 [DOI] [PubMed] [Google Scholar]
- 27.Fujimura, Y., Siddique, H., Lee, L., Rao, V. N., and Reddy, E. S. (2001) Oncogene 20 6653-6659 [DOI] [PubMed] [Google Scholar]
- 28.Pongubala, J. M., and Atchison, M. L. (1995) J. Biol. Chem. 270 10304-10313 [DOI] [PubMed] [Google Scholar]
- 29.Bleckmann, S. C., Blendy, J. A., Rudolph, D., Monaghan, A. P., Schmid, W., and Schutz, G. (2002) Mol. Cell. Biol. 22 1919-1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jean, D., Tellez, C., Huang, S., Davis, D. W., Bruns, C. J., McConkey, D. J., Hinrichs, S. H., and Bar-Eli, M. (2000) Oncogene 19 2721-2730 [DOI] [PubMed] [Google Scholar]
- 31.Kvietikova, I., Wenger, R. H., Marti, H. H., and Gassmann, M. (1995) Nucleic Acids Res. 23 4542-4550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ebert, B. L., and Bunn, H. F. (1998) Mol. Cell. Biol. 18 4089-4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Semenza, G. L. (2007) Biochem. J. 405 1-9 [DOI] [PubMed] [Google Scholar]
- 34.Taylor, C. T. (2008) Biochem. J. 409 19-26 [DOI] [PubMed] [Google Scholar]
- 35.Sen, C. K., Khanna, S., and Roy, S. (2006) Cardiovasc Res. 71 280-288 [DOI] [PubMed] [Google Scholar]
- 36.Kitagawa, K. (2007) Febs. J. 274 3210-3217 [DOI] [PubMed] [Google Scholar]
- 37.Shimokawa, T., Kato, M., Ezaki, O., and Hashimoto, S. (1998) Biochem. Biophys. Res. Commun. 246 287-292 [DOI] [PubMed] [Google Scholar]
- 38.Solanes, G., Pedraza, N., Iglesias, R., Giralt, M., and Villarroya, F. (2000) Faseb. J. 14 2141-2143 [DOI] [PubMed] [Google Scholar]
- 39.Tu, N., Chen, H., Winnikes, U., Reinert, I., Pirke, K. M., and Lentes, K. U. (2000) Life Sci. 67 2267-2279 [DOI] [PubMed] [Google Scholar]
- 40.Nissen, S. E., and Wolski, K. (2007) N. Engl. J. Med. 356 2457-2471 [DOI] [PubMed] [Google Scholar]
- 41.Wan, Y., Chong, L. W., and Evans, R. M. (2007) Nat. Med 13 1496-1503 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.