Abstract
Dickkopf (Dkk) proteins are antagonists of the canonical Wnt signaling pathway and are crucial for embryonic cell fate and bone formation. Wnt antagonism of Dkk requires the binding of the C-terminal cysteine-rich domain of Dkk to the Wnt coreceptor, LRP5/6. However, the structural basis of the interaction between Dkk and low density lipoprotein receptor-related protein (LRP) 5/6 is unknown. In this study, we examined the structure of the Dkk functional domain and elucidated its interactions with LRP5/6. Using NMR spectroscopy, we determined the solution structure of the C-terminal cysteine-rich domain of mouse Dkk2 (Dkk2C). Then, guided by mutagenesis studies, we docked Dkk2C to the YWTD β-propeller domains of LRP5/6 and showed that the ligand binding site of the third LRP5/6 β-propeller domain matches Dkk2C best, suggesting that this domain binds to Dkk2C with higher affinity. Such differential binding affinity is likely to play an essential role in Dkk function in the canonical Wnt pathway.
Dickkopf (Dkk) proteins are antagonists of the canonical Wnt signaling pathway and are crucial for embryonic cell fate and bone formation, and abnormal Dkk function has been implicated in cancers, bone diseases, and Alzheimer disease (1). Dkk is composed of two characteristic cysteine-rich domains, the N-terminal and C-terminal cysteine-rich domain, respectively, each containing 10 conserved cysteines, separated by a variable-length spacer region (2). Wnt antagonism by Dkk requires the binding of the C-terminal cysteine-rich domain of Dkk to the Wnt coreceptor, low density lipoprotein receptor-related protein (LRP)3 5 or 6 (LRP5/6) (3-6). The Dkk-LRP5/6 complex antagonizes canonical Wnt signaling by inhibiting LRP5/6 interaction with Wnt (4, 7, 8) and by forming a ternary complex with the transmembrane protein Kremen (9, 10) that promotes internalization of LRP5/6 (9). Despite the importance of the interaction between Dkk and LRP5/6, its structural basis is unknown.
The Dkk family has at least four members (2), and Dkk1 and Dkk2 share 50% identity in their N-terminal domains and 70% identity in their C-terminal cysteine-rich domains. We previously found that the C-terminal domain of human DKK1 and 2, which contains the second cysteine-rich region, is sufficient for antagonism of Wnt activity in mammalian cells (4). The same was also found to be true in Xenopus; the C-terminal domain of Dkk1 and 2 is both necessary and sufficient to inhibit Wnt-stimulated induction of secondary axis development and transcriptional activation of the Siamois promoter, cooperate with a dominant-negative BMP4 receptor to induce head structure development, and physically associate with LRP5/6 (5, 6). In this report, we defined the structure of the Dkk functional domain, the C-terminal cysteine-rich domain of mouse Dkk2 (amino acids Met172-Ile259) (Dkk2C), and elucidated its interactions with the extracellular β-propeller domains of LRP5/6. Our structural studies suggest that, comparing with other β-propeller domains, the third β-propeller domain of LRP5/6 binds to Dkk2C with greatest affinity. Such differential binding affinity is likely to play an essential role in Dkk function in the canonical Wnt pathway. Furthermore, this finding not only increases our understanding of the regulation of canonical Wnt signaling by Dkk but also may expand the range of options for innovative targeted therapies.
EXPERIMENTAL PROCEDURES
Dkk2C-mediated Inhibition of Wnt Activity—The recombinant protein Dkk2C (amino acids Met172-Ile259 of mouse Dkk2) was expressed and purified from an Escherichia coli system as described previously (11). The recombinant protein contained an N-terminal S tag and a thrombin cleavage site between the S tag and Dkk2C. The purified recombinant Dkk2C contained only one single band in SDS-PAGE. NIH3T3 cells were seeded in 24-well plates at 4 × 105 cells/well and transfected with a LEF-1 luciferase reporter plasmid, an enhanced green fluorescent protein plasmid, and LacZ plasmid (total 0.5 μg of DNA/well) by using Lipofectamine and Plus (Invitrogen), as suggested by the manufacturer. 24 h after transfection, cells were treated with Wnt3a conditioned medium and different dosages of purified Dkk2C for 6 h. Cells were treated with Wnt3a and vehicle as the control for 6 h. Then cells were lysed, and luciferase activity in the cell lysate was measured as described previously (12). Luminescence intensity, which represents Wnt activity, was normalized against the fluorescence intensity of enhanced green fluorescent protein.
S-protein Pulldown Experiments and Western Blotting Analysis—For preparation of the first β-propeller domain of mouse LRP5 with HA tag (LRP5-PD1-HA)-containing conditioned medium, HEK cells were seeded in 6-well plates at 4 × 105 cells/well and transfected with 1 μg of DNA/well. The conditioned medium was collected 30 h after transfection by centrifugation.
S-protein-agarose was obtained from Novagen, and the pulldown experiments were conducted in accordance with a standard protocol provided by the manufacturer. Briefly, for each pulldown experiment, 300 μl of S-tagged Dkk2C (5 μm) was incubated with 200 μl of S-protein-agarose at room temperature for 2 h with gentle agitation to allow the binding of S-tagged Dkk2C to S-protein-agarose, followed by washing four times to remove the unbound Dkk2C by centrifugation. Then, the S-tagged Dkk2C-charged agarose was incubated with the LRP5-PD1-HA conditioned medium in indicated concentrations at 4 °C for 4 h with gentle agitation. After the agarose was washed three times with buffer, it underwent SDS-PAGE and then Western blotting analysis. Western blotting analysis was performed following a standard protocol by using mouse monoclonal IgG3 against an HA tag (from Millipore) as the primary antibody and goat anti-mouse horseradish peroxidase-conjugated IgG (from Cell Signaling Technology) as the secondary antibody. Finally, the membrane was incubated in SuperSignal West Femto maximum sensitivity substrate (Pierce) at room temperature for 5 min, and the results were developed on the film (Eastman Kodak Co.).
Structural Determination of the Solution Structure of Dkk2C—The method used to determine the solution structure of Dkk2C is similar to those described previously (11, 13). Briefly, the 13C/15N double-labeled protein was produced in an E. coli system, and the N-terminal S tag was removed by thrombin. Typical NMR samples consisted of 1 mm 15N/13C-Dkk2C in 5 mm D4-acetic acid (pH 5.0) buffer with 10% (v/v) D2O. All NMR experiments were performed with Bruker 600- and 800-MHz NMR spectrometers at 25 °C. NMR spectra were processed and displayed by the NMRPipe (14) software package. The program XEASY (15) was used for data analysis and structure assignment. Backbone assignment was based mainly on HNCA, HN(CO)CA, HNCACB, and CBCA(CO)NH experiments. Side chain proton resonance was assigned by using 15N HSQC-TOCSY and HCCH-TOCSY. Aromatic side chain proton resonance was assigned with CB(CGCD)HD and CB(CGC-DCE)HE experiments. NOE distance constraints were obtained from NOE peaks in two-dimensional 1H-1H NOE spectroscopy, three-dimensional 15N HSQC-NOE spectroscopy, and three-dimensional 13C HSQC-NOE spectroscopy experiments.
Intensities of NOE peaks were calibrated and converted to distance constraints by the program CALIBA (16). CYANA2.1 (17) software was used for structure calculations, which were based on 1,879 proton-proton distance constraints and 112 dihedral angle restraints. Through the space proximity, the five disulfide bridges within the structure could be clearly identified in the earlier structural calculations. Based on such information, 30 disulfide distance constraints (three upper limits and three lower limits for each disulfide bridge) were added in the final structural calculation. The superimposition of backbone atoms of 20 conformers with smallest target function values among 200 calculated structures yielded a root mean square deviation of 0.36 ± 0.11 Å relative to the average structure, and the average target function value of ensemble structures was 1.57 ± 0.15 Å2 with no distance violations >0.2 Å or dihedral angle violations >5°. The statistical characteristics of these 20 best conformers are described in Table 1.
TABLE 1.
Statistical characteristics of the 20 conformers of the solution structure of Dkk2C
| Parameter | |
|---|---|
| No. of NOE distance restraints | |
| Intraresidue | 448 |
| Interresidue | |
| Sequential | 561 |
| Medium range | 243 |
| Long range | 627 |
| Total | 1879 |
| No. of disulfides restraints | 30 |
| No. of Talos dihedral angle restraints | |
| π | 56 |
| ϰ | 56 |
| r.m.s. deviations from the mean (Å)a | |
| Overall structure,b backbone | 0.36 ± 0.11 |
| Gly177-Gln217 and Glu226-Ile259, backbone | 0.18 ± 0.04 |
| Overall structure,b heavy atoms | 0.82 ± 0.10 |
| Gly177-Gln217 and Glu226-Ile259, heavy atoms | 0.68 ± 0.10 |
| Residuesbin Ramachandran plot (%)c | |
| Most favorable regions | 84.7 |
| Additionally allowed regions | 13.4 |
| Generously allowed regions | 1.9 |
| Disallowed regions | 0.0 |
The average root mean square deviation (r.m.s.d.) between the 20 structures with the lowest target functions and the mean coordinates of the protein.
Residues Gly177-Ile259.
Excluding glycines and prolines and calculated using the Ramachandran macro in CYANA software.
Mutagenesis Studies—To determine the effect of representative point mutants of Dkk1 on the inhibition of Wnt activity (10), NIH3T3 cells were transfected with LEF-1 luciferase reporter plasmids and 1 day later were treated with Wnt3a conditioned medium and Dkk1 or Dkk1 mutants conditioned medium prepared from HEK cells for 6 h. Then luciferase activity was measured as described above. To measure the binding of Dkk1 and its mutants to LRP6, HEK cells were transfected with LRP6 plasmids and 1 day later were incubated on ice with wild-type or mutant Dkk1-alkaline phosphatase (AP) fusion protein conditioned medium for 2 h (10). Then the cells were washed and lysed, and AP activity in cell lysate was measured by using a Tropix luminescence AP assay kit as described previously (10).
Elucidation of the Complex of LRP5 Bound with Dkk—The software package ICM (Molsoft) was used to build the structures of the first three β-propeller domains of LRP5 (termed as LRP5-PD1, PRP5-PD2, and LRP5-PD3) using the crystal structure of the low density lipoprotein receptor YWTD β-propeller domain (Protein Data Bank code 1IJQ (18)) as the template. The initial homology model structures were refined and evaluated by using software package AMBER8 (19). In this step, a 5-ns molecular dynamic simulation with 2 femtoseconds/step was performed by placing the individual propeller domain in a TIP3P water box.
The docking studies used the HADDOCK (20) program. First, the homology model of the third β-propeller domain of LRP5 (LRP5-PD3) and the solution structure of Dkk2C were used as starting structures. Mutation of the Tyr719, Glu721, Arg764, Trp780, Asp887, and Phe888 residues of LRP5-PD3 had an effect of >10% on Dkk-mediated inhibition of Wnt activity. These residues were defined as active residues, and the neighboring surface residues (Arg652, Ala653, Val694, Lys697, Asp718, Gln737, Gly738, Asn762, Gly781, Pro784, Arg805, Trp863, His866, and Met890) were defined as passive residues. In Dkk2C, mutation of His198 (His210 in Dkk1), Lys205 (Lys217), Arg230 (Arg242), and His254 (His267) strongly disrupted both Dkk-mediated Wnt inhibition and LRP6 binding; these residues were defined as active residues. Passive residues were neighboring surface residues Glu179, Phe199, Trp200, Thr201, Leu203, Pro206, Glu212, Val213, Lys216, Gln217, Glu226, Ile227, Gln229, Val241, Thr246, Ser249, Arg252, and Leu253. The flexible interface was defined as active and passive residues ±2 sequential residues for the purpose of docking. Ambiguous interaction restraints used in the docking process were defined as an ambiguous distance between all active and passive residues shown to be involved at the interaction interface.
The docking calculation was initiated with two proteins separated by 150 Å with random starting orientations. Three stages of docking solutions (rigid-body docking, semi-flexible simulated annealing, and a final refinement in water) were executed sequentially by energy minimization. Complex structures were sorted according to the intermolecular interaction energy (the sum of intermolecular van der Waals and electrostatic energies and restraint energies). In the last water refinement stage, the 100 docking structures with the lowest intermolecular interaction energies were generated and clustered on the basis of a 1.0-Å backbone root mean square deviation tolerance at the binding interface. The final docking complex structure was the structure that had the lowest intermolecular interaction energy within the cluster with the lowest average intermolecular interaction energy.
RESULTS
Using an E. coli expression system, we expressed and purified the C-terminal cysteine-rich domain of mouse Dkk2 (Dkk2C) (residues Met172-Ile259). The purified recombinant Dkk2C contained only one single band in SDS-PAGE (Fig. 1A). The Dkk2C possessed a significant inhibitory activity on canonical Wnt signaling; it inhibited Wnt3a activities with an IC50 value around 8 nm in the Wnt reporter gene assay (Fig. 1B), indicating the protein we produced is fully functional. Furthermore, in vitro pulldown experiments showed that Dkk2C directly bound to the first propeller domain of LRP5 (LRP5-PD1), further confirming that the protein we produced is well folded and functional (Fig. 1C).
FIGURE 1.
Inhibition of Wnt3a activities mediated by recombinant Dkk2C. A, SDS-PAGE of recombinant Dkk2C expressed and purified from an E. coli system. B, effects of Dkk2C on canonical Wnt signaling activity. NIH3T3 cells were transfected with a LEF-1 luciferase reporter plasmid and a green fluorescent protein expression plasmid. The next day, cells were treated with Dkk2C solution with the indicated concentrations and Wnt3a conditioned medium (50 ng/ml) for 6 h. The cells were then lysed, and luciferase activities were determined and normalized against the green fluorescent protein levels after 6 h. The activity from cells treated with Wnt3a only was taken as 100%. Two experiments were done individually, and the average values were taken as the results. C, in vitro binding of Dkk2C to LRP5-PD1 detected by pulldown experiments and Western blotting assay. LRP5-PD1-HA-containing conditioned medium (CM) in the indicated volume was incubated with purified S-tagged Dkk2C-coupled S-protein-agarose (lanes 2-4) or with S-protein-agarose only (lanes 5-7), respectively. After extensive washing, LRP5-PD1 bound to Dkk2C were detected using an HA antibody specific to an HA tag carried by LRP5-PD1 in Western blotting assay. Detailed experimental method is described under “Experimental Procedures.” Lane 1, expression of LRP5-PD1 in conditioned medium was detected by HA antibody.
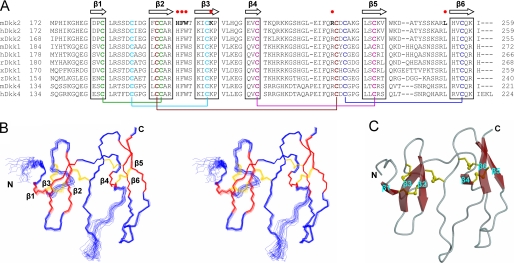
Using NMR spectroscopy, we determined the solution structure of Dkk2C (Fig. 2A). The structure was well defined except for one loop region and the N-terminal region (Fig. 2B and Table 1), and the five disulfide bonds were clearly identified. Within the structure of Dkk2C are two subdomains sharing very similar topology; each has a central anti-parallel β-sheet region consisting of three β strands (β1-β3 in subdomain 1 and β4-β6 in subdomain 2) and two finger-shaped loops linking the three β strands (Fig. 2C). The second subdomain has longer and flexible “finger loops” and is thus much larger than the first one. The flexibilities of the two finger loops in the second subdomain are clear in the relaxation data. The steady-state heteronuclear 15N[1H] NOE values versus the residue number of Dkk2C are shown in Fig. 3. Because the lengths of the N-H bonds are fixed, the 15N[1H] NOE values report information about the dynamics of N-H bonds and are used to determine the motion of a particular residue (21). Typically, the value for the heteronuclear 15N[1H] NOE of folded residues is ∼1-0.7, and the NOE for a flexible loop is <0.5. The dynamic study showed that the first finger loop in the second subdomain (loop β4-β5) is most flexible. The study also showed that the folded Dkk2C should start at Gly177.
FIGURE 2.
Solution structure of Dkk2C. A, amino acid sequence alignment of C-terminal cysteine-rich domains of Dkks in mouse (m), human (h), Xenopus (x), rabbit (r), and zebrafish (z).β strand elements identified in the three-dimensional structure of Dkk2C are indicated at the top. Ten conserved cysteines are in bold type, and pairs of cysteines forming disulfide bridges are colored identically and linked by lines. Amino acids that contact the third β-propeller domain of LRP5 in the docked model are in bold and indicated by the red dots. B, stereo view of the peptide backbone (N, C-α, C′) determined by superimposition of 20 conformers of Dkk2C with the lowest target function values. The figure was generated by using MOLMOL (39). β strands are red; disulfide bridges are yellow. C, ribbon diagram of Dkk2C with the lowest target function values, generated by using MOLSCRIPT (40).
FIGURE 3.
Plot of backbone amide heteronuclear 15N[1H] NOE values versus residue number for the Dkk2C. The steady-state heteronuclear 15N[1H] NOE value is plotted versus the residue number measured with a Bruker 600-MHz spectrometer at 25 °C. The secondary structure elements and disulfide brides in the Dkk2C are indicated at the top; amino acids that contact the third β-propeller domain of LRP5 in the docked model are indicated by the red dots. Because the lengths of the N-H bonds are fixed, the 15N[1H] NOE provides information about the dynamics of N-H bonds that can be used to determine whether a particular amide is in a well folded or a flexible region of a protein. The arrow indicates the starting residue, Gly177, of the folded Dkk2C.
Within each subdomain are two disulfide bonds to stabilize the β-core region; one connects the first and second β-sheets (Cys183-Cys195 connecting β1 and β2 in the subdomain 1 and Cys214-Cys239 connecting β4 and β5 in subdomain 2), and another (Cys189-Cys204 and Cys233-Cys256) connects the third β sheet (β3 and β6, respectively) to the first finger loop (loop β1-β2 and loop β4-β5, respectively). The fifth disulfide bond (Cys194-Cys231) links the two subdomains together. The five dihedral angles in each of five disulfide bonds in the solution structure of Dkk2C are well within the range of the established stereochemical preferences of a single disulfide bridge (22). Among the five disulfide bonds in the Dkk2C structure, only the first one (Cys183-Cys195) has the right-handed conformation; the rest of the four disulfide bonds are all in the left-handed conformation (23).
Analysis with DALI (24) software showed that the Dkk2C structure shares some features with those of colipase (Protein Data Bank code 1PCN) (25) and MIT1 (mamba intestinal toxin 1) (Protein Data Bank code 1IMT) (26), which belong to a family of proteins lacking extensive secondary structures and stabilized by abundant disulfide bridges (27). All three can be described as an assembly of protruding fingers, held together at one end by a network of five disulfide bridges. However, only the two central β-core regions of Dkk2C, colipase, and MIT1 are similar, and the connectivity patterns of disulfide bridges among the three proteins share a highly conserved feature (23). Indeed, the sequence identity shared by Dkk2C with the two proteins (24% with colipase and 29% with MIT1) is concentrated in the two central β-core regions, all of the finger-loop regions of Dkk2C are unique, and each has a different length and conformation.
The C-terminal cysteine-rich domains of Dkk1/2 interact with LRP5/6 (5, 10), whose extracellular region contains four well defined YWTD repeat domains that have a typical symmetrical six-bladed β-propeller fold (28, 29). Although each of the first three β-propeller domains can interact with Dkk1/2, only the third is required for Dkk1/2-mediated inhibition of Wnt signaling, presumably because it binds favorably to Dkk1/2 (10). Furthermore, the residues involved in Dkk-mediated Wnt inhibition in the third β-propeller domain of LRP5 (LRP5-PD3) were found by alanine substitution mapping (10) to be clustered on a concave, amphitheatre-shaped surface centered on the pseudo-6-fold axis atop the β-propeller (10). This amphitheatre-shaped ligand binding site is likely to be the common feature among these β-propeller domains (supplemental Fig. 1) (30, 31). Because of the network of hydrogen bonds within the β-propeller domains and the nature of the concave surface, the ligand binding sites of the β-propeller domains are rigid (30). Typical ligands of these the β-propeller domains are rigid as well (30, 31). The rigidity of the ligands and receptors minimizes loss of entropy upon binding and promotes a high affinity (30). The rigidity also makes it possible to model the complex by docking ligands to β-propeller domains. For example, at CAPRI, different groups successfully docked laminin to the β-propeller of nidogen successfully (32, 33). We therefore conducted a docking study to examine the interaction between Dkk2C and LRP5-PD3. To maximize the accuracy of this study, we used the program HADDOCK (20) to incorporate data from mutagenesis studies of the binding of Dkk1/2 to LRP5/6.
The mutation E721A on the amphitheatre surface of LRP5-PD3 that binds to Dkk had the strongest effect on Dkk1-mediated inhibition of Wnt1 activity (∼70% reduction) and abolished binding of LRP5-PD3 to Dkk1 (10). Because the corresponding residue in nidogen, Glu994, forms a salt bridge with a lysine residue in bound laminin (30), we speculated that Glu721 of LRP5 would form a salt bridge with a basic residue in the bound Dkk1/2. In mutagenesis studies, we substituted each positively charged amino acid in the C-terminal cysteine-rich domain of Dkk1 (including lysine, arginine, and histidine) with glutamic acid and investigated the effect on Dkk1-LRP6 binding. Among the four Dkk1 mutants, H210E, K217E, and R242E reduced the binding of Dkk1 to LRP6 by more than 50%, and H267E reduced the binding of Dkk1 to LRP6 by about 43% (Fig. 4A). We also examined the effects of these four mutants on Dkk1-mediated inhibition of Wnt3a activity and found that, indeed, these mutants attenuated Dkk1-mediated antagonism of Wnt signaling (Fig. 4B).
FIGURE 4.
Ability of Dkk1 and its mutants to bind to LRP6 and antagonize Wnt activity. A, binding of Dkk1 and its mutants to LRP6. HEK cells were transfected with the LRP6 plasmid and 1 day later were incubated on ice with conditioned medium containing wild-type and mutant mouse Dkk1-AP for 2 h. After cells were washed and lysed, the AP activities in cell lysates were determined. The activity resulted from the binding of wild-type Dkk1-AP to LRP6 was taken as 100%. B, effect of Dkk1 and its mutants on the inhibition of Wnt activity. NIH3T3 cells were transfected with a LEF-1 activity reporter plasmid and an enhanced green fluorescent protein plasmid. 24 h after transfection, cells were treated with Wnt3a conditioned medium and wild-type or mutant Dkk1 conditioned medium for 6 h. The luciferase activities in cell lysates were determined as described in the legend to Fig. 1.
On the basis of the mutagenesis studies, we first used the HADDOCK software package to build the complex structure of Dkk2C bound to LRP5-PD3. As mutation of the Tyr719, Glu721, Arg764, Trp780, Asp887, and Phe888 residues of LRP5-PD3 had an effect of >10% on Dkk-mediated inhibition of Wnt activity (10), we proposed that these residues are involved in the interaction between Dkk2C and LRP5-PD3. We thus defined these residues as active residues, and 14 neighboring residues as passive residues. Similarly, in Dkk2C, as mutation of His198 (His210 in Dkk1), Lys205 (Lys217), Arg230 (Arg242), and His254 (His267) strongly disrupted both Dkk-mediated inhibition and LRP6 binding, we defined these residues and 14 of their neighbor residues as active and passive residues, respectively. In the docking process, the flexible interfaces of both Dkk2C and LRP5-PD3 were defined as active and passive residues ±2 sequential residues and the ambiguous interaction restraints as the distances between all active and passive residues. The docking calculation was initiated with the two proteins separated by 150 Å with random starting orientations. Three stages of docking solutions (rigid-body docking, initial refinement with semi-flexible simulated annealing, and a final refinement in water) were executed sequentially by energy minimization. During the first-stage rigid-body docking, a total of 1,000 docking structures were generated, and the top 100 docking structures with the lowest intermolecular interaction energies were selected in the second and third refinement stages. After the docking, a cluster analysis was also carried out to further evaluate the final 100 structures. To our surprise, the 100 final docking structures could be clustered on the basis of a 1.0-Å backbone root mean square deviation tolerance at the binding interface between Dkk2C and LRP5-PD3, indicating that complex structure generated by our docking studies has very high accuracy (34).
DISCUSSION
Despite the extensive efforts, the structures of the β-propeller domains of LRP5/6 have not been experimentally elucidated. However, it was well documented that because of their unique folding properties, the models of these β-propeller domain can be built with reasonable accuracy (29, 35), and based on the well established protocol, we built the homology models of the β-propeller domains of LRP5/6. These structural models provided us an opportunity to exploit the interaction between Dkk and LRP5/6. To further enrich the precision of our studies, we utilized HADDOCK to carry out the docking studies. There were two advantages of using this approach. First, HADDOCK, which stands for high ambiguity driven protein-protein docking, is a method in which biochemical data are used directly to drive the docking (20); such a feature allowed us to take the advantage of the wealth information generated by the mutagenesis studies. Second, HADDOCK explicitly includes backbone flexibility in the docking process (36). Because we used homology models in our docking study, the feasible docking feature was particularly helpful in terms of addressing the issue of loop uncertainties associated with the structures of Dkk2C and LRP5 β-propeller domains. As HADDOCK has been proven a reliable docking tool and widely used to study protein-protein interactions (37, 38), we believe that, even if it is not perfect, our docking studies, which are driven by the data from the mutagenesis studies, can provide valuable biological insight.
In the complex of Dkk2C bound to LRP5-PD3 generated by our HADDOCK studies, the side chains of those residues at the interface form an extensive network of contacts between the two molecules, and those contacts are consistent with the mutagenesis studies reported in this study as well as in the earlier studies (10). In particular, the second finger loop (loop β2-β3) of DkkC2 in the first subdomain lies atop LRP5-PD3 and the three aromatic residues, His198, Phe199, and Trp200, form a “roof” that covers the amphitheatre of LRP5-PD3 (Fig. 5A). At this interface, the side chain of His198 in Dkk2C (His210 in Dkk1) forms a hydrogen bond with the side chain of Glu721 in LRP5-PD3, and Trp200 in Dkk2C (W212 in Dkk1) also interacts with Glu721 of LRP5-PD3 (Fig. 5B). Furthermore, there are many hydrophobic interactions between the roof and the surface of the “amphitheatre” that also significantly contribute to the binding between the two molecules; among them, the benzoic ring contacts between Phe199 and Trp200 in Dkk2C and Tyr719 and Trp780 in LRP5-PD3 are most visible. Another interaction hot spot between Dkk2C and LRP5-PD3 involves the second subdomain of Dkk2C. In this binding site, the side chain of Arg230 (Arg242 in Dkk1) in Dkk2C forms hydrogen bonds with the side chain of Asp887 in LRP5-PD3. In addition, the side chain Leu253 (Leu266 in Dkk1) in Dkk2C and Phe888 in LRP5-PD3 form a hydrophobic network. Indeed, mutation of this hydrophobic residue in Dkk1, L266A, efficiently disrupted inhibition of Dkk1-mediated Wnt3a activity (about 30%; data not shown) and the binding of Dkk1 to LRP6 (about 40%; data not shown). Interestingly, although the second subdomain of Dkk2C has two mobile finger loops, in the complex, the residues that are involved in the interaction with LRP5-PD3 are all in a relatively rigid conformation (Fig. 3).
FIGURE 5.
Complex structure of the third β-propeller domain of LRP5 (LRP5-PD3) bound to Dkk2C. A, a ribbon diagram of the complex of LRP5-PD3 bound to Dkk2C. B, side chain interactions between Dkk2C and LRP5-PD3. Dashed lines represent hydrogen bonds. Residue numbers in brackets are the numbers in mouse Dkk1. Corresponding amino acids in LRP5-PD1 and LRP5-PD2 to those involved in LRP5-PD3 binding interface are listed in the right bottom panel. Figures were generated by using the Pymol program (DeLano Scientific).
In the mutagenesis studies, we also identified two other residues, Lys205 (Lys217 in Dkk1) and His254 (His267 in Dkk1) in Dkk2C, that also played a significant role in Dkk1-LRP6 binding and Dkk1-mediated inhibition of Wnt3a activity (Fig. 4). In the complex structure of Dkk2C bound to LRP5-PD3, Lys205 locates at the boundary of the two binding hot spot, forming a hydrogen bond with Asp887 of LRP5-PD3 (Fig. 5B). Interestingly, mutant K217E of Dkk1 significantly disrupted the binding of Dkk1 to both LRP6 and another Dkk receptor Kremen protein (as described in the accompanying article (41)); perhaps this residue plays a key role in coordinating the interactions between Dkk and its two binding partners, LRP5/6 and Kremen. On the other hand, in the structure of Dkk2C, His254 (His267 in Dkk1) is buried inside the folded structure of Dkk2C (Fig. 5B). Therefore the mutation H267E in Dkk1 would likely affect the overall stability of Dkk structure.
As shown in this work as well as in the earlier studies, the first and second β-propeller domains of LRP5/6 also interact with Dkk1/2 but do not play an essential role in Dkk1-mediated inhibition of Wnt activity (10). Because of the structural similarity of the YWTD β-propeller domains, it is likely that Dkk molecules can fit into the ligand binding sites of all β-propeller domains of LRP5/6 but can fit perfectly into only that of the third β-propeller domain. Furthermore, in the complex of LRP5-PD3 bound to Dkk2C, the residues of LRP5-PD3 at the interface are relatively conserved in the first three propeller domains of LRP5/6 (Fig. 5B). To further elucidate why the interactions of Dkk2C with different β-propeller domains of LRP5 lead to different effects on regulating Wnt signaling, we built the complexes of Dkk2C bound with LRP5-PD1 and Dkk2C with LRP5-PD2 by using the complex model of Dkk2C bound with LRP5-PD3 as the template. In the studies, LRP5-PD3 in the complex template was first replaced with LRP5-PD1 and LRP5-PD2, respectively, by superimposition of the propeller domains; then the new complexes of Dkk2C bound with LRP5-PD1 and LRP5-PD2 were refined by the water refinement algorithm in the HADDOCK program. As expected, the Dkk2C fits well to the ligand binding sites of LRP5-PD1 and LRR5-PD2. Comparing the three complexes in detail, we observed that although Dkk2C has similar interactions with the first two β-propeller domains as it does with the third one, not all of the intermolecular interactions found in the complex of Dkk2C bound to the third β-propeller domain are preserved in the complexes of Dkk2C bound to the first two propeller domains. The most pronounced one is the amino acid Glu721 in LRP5-PD3, which plays an important role in the interaction; this amino acid is replaced by aspartic acid in the first two propeller domains of LRP5/6 (supplemental Fig. 1). Because of the rigidity of both the ligand and receptor, the aspartic acids in the first two β-propeller domains of LRP5 are unable to interact with the bound Dkk2C (data not shown). Indeed, the calculated intermolecular interaction energies (mainly intermolecular van der Waals and electrostatic energies and restraint energies) between Dkk2C and the first two propellers were about 30% weaker than that between Dkk2C and LRP5-PD3 (-402.263 kcal/mol and -374.747 kcal/mol versus -558.5 kcal/mol), suggesting that Dkk2C binds to the first two β-propeller domains of LRP5 with lower affinities. Therefore, binding affinity is likely to be the factor that determines the selection of Dkk binding partners in the Wnt signaling pathway.
Supplementary Material
Acknowledgments
We thank the Hartwell Center for Bioinformatics and Biotechnology at St. Jude for providing computational time, Scott Malone and Dr. Charles Ross for providing computer-related technical support, Dr. Weixing Zhang for assistance with the NMR experiments, and Sharon Naron for editing the manuscript.
The atomic coordinates and structure factors (code 2JTK) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grants GM081492 (to J. J. Z.) and AR051476 and CA132317 to D. W.). This work was also supported by the American Lebanese Syrian Associated Charities. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains a supplemental Fig. 1.
Footnotes
The abbreviations used are: LRP, low density lipoprotein receptor-related protein; Dkk2C, the C-terminal cysteine-rich domain of mouse Dkk2; LRP5-PD1, the first β-propeller domain of mouse LRP5; LRP5-PD2, the second β-propeller domain of mouse LRP5; LRP5-PD3, the third β-propeller domain of mouse LRP5; HA, hemagglutinin; NOE, nuclear Overhauser effect; AP, alkaline phosphatase; HEK, human embryonic kidney.
References
- 1.Niehrs, C. (2006) Oncogene 25 7469-7481 [DOI] [PubMed] [Google Scholar]
- 2.Krupnik, V. E., Sharp, J. D., Jiang, C., Robison, K., Chickering, T. W., Amaravadi, L., Brown, D. E., Guyot, D., Mays, G., Leiby, K., Chang, B., Duong, T., Goodearl, A. D., Gearing, D. P., Sokol, S. Y., and McCarthy, S. A. (1999) Gene 238 301-313 [DOI] [PubMed] [Google Scholar]
- 3.Semenov, M. V., Tamai, K., Brott, B. K., Kuhl, M., Sokol, S., and He, X. (2001) Curr. Biol. 11 951-961 [DOI] [PubMed] [Google Scholar]
- 4.Li, L., Mao, J. H., Sun, L., Liu, W. Z., and Wu, D. Q. (2002) J. Biol. Chem. 277 5977-5981 [DOI] [PubMed] [Google Scholar]
- 5.Brott, B. K., and Sokol, S. Y. (2002) Mol. Cell. Biol. 22 6100-6110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao, B., and Niehrs, C. (2003) Gene 302 179-183 [DOI] [PubMed] [Google Scholar]
- 7.Bafico, A., Liu, G., Yaniv, A., Gazit, A., and Aaronson, S. A. (2001) Nat. Cell Biol. 3 683-686 [DOI] [PubMed] [Google Scholar]
- 8.Li, Y., Lu, W., He, X., Schwartz, A. L., and Bu, G. (2004) Oncogene 23 9129-9135 [DOI] [PubMed] [Google Scholar]
- 9.Mao, B., Wu, W., Davidson, G., Marhold, J., Li, M., Mechler, B. M., Delius, H., Hoppe, D., Stannek, P., Walter, C., Glinka, A., and Niehrs, C. (2002) Nature 417 664-667 [DOI] [PubMed] [Google Scholar]
- 10.Zhang, Y., Wang, Y., Li, X., Zhang, J., Mao, J., Li, Z., Zheng, J., Li, L., Harris, S., and Wu, D. (2004) Mol. Cell. Biol. 24 4677-4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong, H. C., Mao, J., Nguyen, J. T., Srinivas, S., Zhang, W., Liu, B., Li, L., Wu, D., and Zheng, J. (2000) Nat. Struct. Biol. 7 1178-1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, L., Yuan, H., Xie, W., Mao, J., Caruso, A. M., McMahon, A., Sussman, D. J., and Wu, D. (1999) J. Biol. Chem. 274 129-134 [DOI] [PubMed] [Google Scholar]
- 13.Wong, H. C., Bourdelas, A., Krauss, A., Lee, H.-J., Shao, Y.-M., Wu, D., Mlodzik, M., Shi, D. L., and Zheng, J. (2003) Mol. Cell 12 1251-1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax, A. (1995) J. Biomol. NMR 6 277-293 [DOI] [PubMed] [Google Scholar]
- 15.Eccles, C., Guntert, P., Billeter, M., and Wuthrich, K. (1991) J. Biomol. NMR 1 111-130 [DOI] [PubMed] [Google Scholar]
- 16.Guntert, P., Braun, W., and Wuthrich, K. (1991) J. Mol. Biol. 217 517-530 [DOI] [PubMed] [Google Scholar]
- 17.Guntert, P., Mumenthaler, C., and Wuthrich, K. (1997) J. Mol. Biol. 273 283-298 [DOI] [PubMed] [Google Scholar]
- 18.Jeon, H., Meng, W. Y., Takagi, J., Eck, M. J., Springer, T. A., and Blacklow, S. C. (2001) Nat. Struct. Biol. 8 499-504 [DOI] [PubMed] [Google Scholar]
- 19.Case, D. A., Cheatham, T. E., III, Darden, T., Gohlke, H., Luo, R., Merz, K. M., Jr., Onufriev, A., Simmerling, C., Wang, B., and Woods, R. J. (2005) J. Comput. Chem. 26 1668-1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominguez, C., Boelens, R., and Bonvin, A. M. (2003) J. Am. Chem. Soc. 125 1731-1737 [DOI] [PubMed] [Google Scholar]
- 21.Wong, H. C., Liu, G., Zhang, Y. M., Rock, C. O., and Zheng, J. (2002) J. Biol. Chem. 277 15874-15880 [DOI] [PubMed] [Google Scholar]
- 22.Sali, A., and Overington, J. P. (1994) Protein Sci. 3 1582-1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thornton, J. M. (1981) J. Mol. Biol. 151 261-287 [DOI] [PubMed] [Google Scholar]
- 24.Holm, L., and Sander, C. (1993) J. Mol. Biol. 233 123-138 [DOI] [PubMed] [Google Scholar]
- 25.van, T. H., Sarda, L., Verger, R., and Cambillau, C. (1992) Nature 359 159-162 [DOI] [PubMed] [Google Scholar]
- 26.Boisbouvier, J., Albrand, J. P., Blackledge, M., Jaquinod, M., Schweitz, H., Lazdunski, M., and Marion, D. (1998) J. Mol. Biol. 283 205-219 [DOI] [PubMed] [Google Scholar]
- 27.van, T. H., Bezzine, S., Cambillau, C., Verger, R., and Carriere, F. (1999) Biochim. Biophys. Acta 1441 173-184 [DOI] [PubMed] [Google Scholar]
- 28.Springer, T. A. (1998) J. Mol. Biol. 283 837-862 [DOI] [PubMed] [Google Scholar]
- 29.Springer, T. A. (2002) Curr. Opin. Struct. Biol. 12 802-813 [DOI] [PubMed] [Google Scholar]
- 30.Takagi, J., Yang, Y., Liu, J. H., Wang, J. H., and Springer, T. A. (2003) Nature 424 969-974 [DOI] [PubMed] [Google Scholar]
- 31.Rudenko, G., Henry, L., Henderson, K., Ichtchenko, K., Brown, M. S., Goldstein, J. L., and Deisenhofer, J. (2002) Science 298 2353-2358 [DOI] [PubMed] [Google Scholar]
- 32.Gray, J. J. (2006) Curr. Opin. Struct. Biol. 16 183-193 [DOI] [PubMed] [Google Scholar]
- 33.Mendez, R., Leplae, R., Lensink, M. F., and Wodak, S. J. (2005) Proteins 60 150-169 [DOI] [PubMed] [Google Scholar]
- 34.van Dijk, A. D., Boelens, R., and Bonvin, A. M. (2005) FEBS J. 272 293-312 [DOI] [PubMed] [Google Scholar]
- 35.Springer, T. A. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 65-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van, D. M., van Dijk, A. D., Hsu, V., Boelens, R., and Bonvin, A. M. (2006) Nucleic Acids Res. 34 3317-3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonvin, A. M. (2006) Curr. Opin. Struct. Biol. 16 194-200 [DOI] [PubMed] [Google Scholar]
- 38.de Vries, S. J., van Dijk, A. D., Krzeminski, M., van, D. M., Thureau, A., Hsu, V., Wassenaar, T., and Bonvin, A. M. (2007) Proteins 69 726-733 [DOI] [PubMed] [Google Scholar]
- 39.Koradi, R., Billeter, M., and Wuthrich, K. (1996) J. Mol. Graph. 14 29-32 [DOI] [PubMed] [Google Scholar]
- 40.Esnouf, R. M. (1997) J. Mol. Graph. Model. 15 132-133 [DOI] [PubMed] [Google Scholar]
- 41.Wang, K., Zhang, Y., Li, X., Chen, L., Wang, H., Wu, J., Zheng, J., and Wu, D. (2008) J. Biol. Chem. 283 23371-23375 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.