Abstract
Induction of apoptosis by tensile forces is an important determinant of connective tissue destruction in osteoarthritis and periodontal diseases. We examined the role of molecular components of the unfolded protein response in force-induced apoptosis. Magnetic fields were used to apply tensile force through integrins to cultured fibroblasts bound with collagen-coated magnetite beads. Tensile force induced caspase 3 cleavage, DNA fragmentation, depolarization of mitochondria, and induction of CHOP10, all indicative of activation of apoptosis. Immunoblotting, immunocytochemistry, and release of Ca2+ from the endoplasmic reticulum showed evidence for both physical and functional associations between bound beads and the endoplasmic reticulum. Force-induced apoptosis was not detected in PERK null cells, but reconstitution of wild-type PERK in PERK null cells restored the apoptotic response. Force-induced apoptosis did not require PKR, GCN2, eIF2α, or CHOP10. Furthermore, force more than 24 h did not activate other initiators of the unfolded protein response including IRE-1 and ATF6. However, force-induced activation of caspase 3 was dependent on caspase 9 but was independent of mitochondria. We conclude that force-induced apoptosis depends on a novel function of PERK that occurs in addition to its canonical role in the unfolded protein response.
The ability of cells to sense and respond to physical stresses is required for tissue homeostasis and normal development. In muscle, bone, tendons, the periodontium, and the cardiovascular system, applied forces of physiological magnitude regulate cellular processes that are critical for normal tissue and organ function such as differentiation, proliferation, and migration (1). In contrast, pathophysiological forces can induce apoptosis (2). Indeed, force-induced death is an important mediator of connective tissue destruction in osteoarthritis (3) and periodontitis (4).
Cells in mechanically active environments constitutively exhibit protective responses that provide resistance to apoptosis in the face of either continuous or intermittent, high amplitude physical forces (5). As a result of these protective responses, cell viability and normal tissue function are maintained. For example, tensile and compressive forces applied to bone cells help to maintain bone mass by balancing bone apposition and resorption, in part by regulating cell proliferation and cell death (3). Cellular adaptations in high load environments include submembrane cytoskeletal buffering of high amplitude forces (5). In fibroblasts, tensile forces are delivered through collagen fibers and integrins to adhesion complexes; these forces are then transmitted intracellularly by actin filaments (6). Both focal adhesions and actin filaments are required for force-induced apoptosis (6, 7), but the intracellular mechanisms that initiate pro-apoptotic signals beyond the actin cytoskeleton are not defined.
Three pro-apoptotic pathways have been described and include the extrinsic pathway, involving activation of death receptors at the cell membrane, and two intrinsic pathways, involving induction of mitochondria-mediated death and activation of the unfolded protein response (UPR)4 in the endoplasmic reticulum (ER) (8). Notably, application of high amplitude tensile forces through collagen-coated magnetite beads in fibroblasts, which is a good model for the forces that are delivered to connective tissue cells in vivo (7), induces p38 and eIF2α (eukaryotic translation initiation factor 2α) phosphorylation (9) and cell death (10). eIF2α is regulated in the UPR as cells progress from physiological protein translation to compensatory responses to exogenous forces. These data suggest that the UPR pathway may translate force-induced signals into apoptotic responses.
The UPR is conventionally considered as a stress response that is activated by accumulation of misfolded proteins in the ER or by stressors such as hypoxia, changes in calcium concentration, and nutrient deprivation. The UPR is activated by three triggering molecules in the ER membrane: PERK (PKR (double-stranded RNA-activating protein kinase)-like ER kinase/pancreatic eIF2α kinase), IRE-1 (inositol requiring ER-to-nucleus signaling 1), and ATF6 (activating transcription factor 6) (11). Activation of these molecules initiates signaling cascades leading to the attenuation of general translation and the induction of chaperone proteins and protein degradation pathways. Ultimately, after prolonged application of force, apoptosis occurs (12).
Apoptosis arising from UPR stimulation involves the up-regulation of CHOP10 (CAATT enhancer binding homologous protein 10)/GADD153 (growth and DNA damage protein 153), which is a downstream target of PERK through eIF2α phosphorylation, and to a lesser extent ATF6 and IRE-1 (13). Although other mechanisms by which the UPR can initiate apoptosis have been studied, this particular pathway is well characterized and is best correlated with apoptotic signals originating from the ER. Based on these previous data, we determined whether force-induced cell death is mediated through signaling systems in the ER.
EXPERIMENTAL PROCEDURES
Reagents—JC-1 and mag-fura-2/AM were purchased from Molecular Probes (Eugene, OR). Magnetite beads, bovine serum albumin (BSA), poly-l-lysine, fibronectin, thapsigargin, tunicamycin, staurosporine, carbonyl cyanide 3-chlorophenylhydrazone, and camptothecin were obtained from Sigma. Acidified bovine type 1 collagen was from Angiotech (Palo Alto, CA). Swinholide A was purchased from Calbiochem. Rabbit polyclonal antibodies for phospho-eIF2α, total eIF2α, cleaved caspase 3, phospho-PERK, and caspase 9 were from Cell Signaling Technologies (Beverly, MA). Antibodies to CHOP10 (GADD153 (F-168)) were from Santa Cruz Biotechnology. Anti-GRP78 (glucose-regulated protein 78)/immunoglobulin heavy chain binding protein (BiP) mouse monoclonal, anti-calnexin mouse monoclonal, and anti-caspase 8 rabbit antibodies were from BD Transduction Laboratories. Anti-actin monoclonal antibody (clone AC-40) and anti-vinculin mouse monoclonal antibody (clone BM75.2) were from Sigma. Anti-ATF6 rabbit polyclonal antibody was from R. C. A. (14). Rabbit polyclonal antibody to caspase 12 was from Pharmingen. Anti-glyceraldehyde-3-phosphate dehydrogenase monoclonal antibody (MAB374) was obtained from Chemicon International (Temecula, CA).
Cell Culture—PERK wild-type, PERK null, GCN2 (general control amino acid 2) wild-type, and GCN2 null mouse embryonic fibroblasts (MEFs) were maintained according to Harding et al. (15, 16). Mouse embryonic fibroblasts expressing wild-type eIF2a (401-406 eIF2 Ser/Ser) or mutant, non-phosphorylatable eIF2a(S51A) (401-404 eIF2 Ala/Ala) were grown as described (17). PKR wild-type MEFs and PKR null MEFs were maintained as described (18). PKR cells were provided by Bryan Williams of the Cleveland Clinic Foundation. All rights, title, and interest in these materials are owned by the Cleveland Clinic Foundation. Human gingival fibroblasts (HGFs) were maintained according to Pender and McCulloch (19). HeLa cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen), supplemented with 10% fetal bovine serum, and 1% penicillin/streptomycin. CHOP10/GADD153 null MEFs were from R. C. A. Caspase 9 null MEFs were provided by Razq Hakem (Princess Margaret Hospital/Ontario Cancer Institute, Toronto, Ontario) (20), and APAF-1 (apoptotic protease-activating factor 1) null MEFs were obtained from Tak Mak (Princess Margaret Hospital/Ontario Cancer Institute) (20). In all experiments, control samples included untreated cells and thapsigargin-treated cells (1 μm for 0.5 or 1 h or 300 nm for 24 h).
Collagen Bead Preparation and Mechanical Force Application—Collagen-coated magnetite beads (9) were bound to cells at 37 °C for 30 min, washed with phosphate-buffered saline (PBS), and incubated in medium containing 10% serum prior to force exposure (0.65 piconewtons/μm2) at 37 °C. After force induction, cells were washed with PBS, and lysates were collected in Laemmli sample buffer for determination of protein concentration and analysis. Tensile forces were applied by placing the magnet above the cells. Compressive forces were applied by placing the magnet below cells, and twisting forces were applied by rotating the magnet placed above cells by 90° after half the total length of exposure time.
Isolation of Focal Adhesions—After addition of collagen- or BSA-coated magnetite beads, bead-associated proteins were isolated from cells as described (9). Briefly, cells were washed with cold PBS to remove unbound beads, scraped into cold cytoskeleton extraction buffer (9), and sonicated for 10 s. The beads were isolated from the lysate with a magnet, resuspended in cold cytoskeleton extraction buffer, homogenized, and reisolated magnetically. Bead-associated proteins were removed by boiling in Laemmli sample buffer. In control experiments, cell cultures were treated with swinholide A (50 nm, 25 °C, 20 min) prior to bead binding to dissipate focal adhesion formation.
Ectopic Expression of ER Stress Proteins—A wild-type mouse PERK expression vector was transfected into PERK null MEFs using FuGENE 6 transfection reagent (Roche Applied Science). After force application, cell lysates were collected for protein concentration determination and analysis. Hemagglutinin-tagged mouse PERK constructs in pcDNA3 were kindly provided by T. Rutkowski and R. J. K.
Western Blotting—Adherent cells and the culture medium containing detached cells were combined for preparation of samples. Cell lysates were collected in Laemmli sample buffer, and protein concentrations were measured with the BCA protein assay. Equal amounts of protein were separated by SDS-PAGE, transferred to membranes, and blotted with antibodies. Quantitation of band intensity was performed using the NIH ImageJ 1.34n program. The values were normalized to actin loading controls within the same lane and expressed as a ratio relative to the zero time point for comparison between lanes.
Colocalization of ER Resident Proteins with Beads—Chamber slides (8-well, Labtek) were coated with poly-l-lysine (100 μg/ml in PBS). Cells were plated for 6 h prior to incubation with collagen-coated or BSA-coated latex microbeads for 30 min at 37 °C. Prior to immunostaining, cells were fixed (3.7% paraformaldehyde in PBS for 10 min at room temperature), blocked, and permeabilized (PBS with 0.2% Triton X-100 and 0.2% BSA) for 15 min at room temperature. Antibodies were diluted in PBS with 0.2% Triton X-100 and 0.2% BSA. Immunofluorescence staining was performed with mouse calnexin antibody for 1 h at room temperature. Slides were washed with PBS, incubated with goat anti-mouse fluorescein isothiocyanate-conjugated antibody for 1 h, washed, and sealed with coverslips. The slides were viewed with a Nikon 300 inverted fluorescence microscope equipped with differential interference contrast optics. Images were captured with a charge-coupled device camera (Imagepoint, Photometrics, Phoenix, AZ).
ER Calcium Release—Estimation of [Ca2+]ER was performed as described (21). Briefly, cells were incubated with mag-fura-2/AM (4 μm) for 150 min. The medium was replaced with nominally calcium-free HEPES buffer (9) for bead binding and then force application. Visual inspection of mag-fura-2/AM-loaded cells showed fluorescent labeling of intracellular organelles as shown earlier (22), which correspond spatially with the ER. [Ca2+]ER measurements based on the mag-fura-2 fluorescence ratio were obtained with alternating excitation wavelengths of 340 and 380 nm and an emission wavelength of 520 nm.
DNA Fragmentation—Staining for DNA fragmentation (TdT-mediated X-dUTP nick end labeling, TUNEL) was performed using an in situ cell death detection kit (Roche Diagnostics) as outlined in the manufacturer's instructions. Briefly, after force application, paraformaldehyde-fixed cells were permeabilized with Citrate buffer (0.1% Triton X-100 in 0.1% sodium citrate) for 15 min at room temperature, washed twice with PBS, and then stained with the TUNEL reagent mixture for at least 1 h at 37 °C. They were washed with PBS and counterstained with 4′,6-diamidino-2-phenylindole. Positive control cells were treated with 300 nm thapsigargin for 24 h and stained in the same manner. Stained cells were visualized on a Leica inverted fluorescence microscope, and digital images were captured using a Nikon CoolPix camera. For estimation of the percent of TUNEL-positive cells, three fields were photographed from each of three separate cultures, and the mean percent of TUNEL-positive cells was estimated from the total number of 4′,6-diamidino-2-phenylindole-stained cells.
Mitochondrial Membrane Potential (ψm)—JC-1 is a mitochondrial membrane potential-sensitive dye that exists largely as a green fluorescent monomer at low membrane potential in dead or dying cells. At higher (physiological) mitochondrial membrane potentials, JC-1 forms red fluorescent “J-aggregates.” JC-1 staining followed by analysis of JC-1 aggregates and monomers by two-color flow cytometry was conducted as described (7). Cells were loaded with collagen beads and treated with or without force. Floating (presumptive dead) cells were washed away and removed. Attached cells were trypsinized, washed, and suspended in PBS containing 10% serum and stained with JC-1 at 0.32 μg/ml for 1 h at 37 °C in the dark. Cells were washed and resuspended in PBS for analysis as described (23). Cells denoted as positively stained for aggregates were determined from signal thresholds of untreated controls and for monomers cells treated with carbonyl cyanide 3-chlorophenylhydrazone for 2 h to dissipate ψm.
Reverse Transcription (RT)-PCR Detection of XBP-1 Cleavage—RT-PCR detection of XBP-1 (XhoI site-binding protein 1) cleavage was performed as described by Ref. 24 with modifications to primers for human XBP-1. Preparation of cDNA from RNA was performed using the Qiagen Omniscript RT kit (Qiagen, Mississauga, Ontario) by diluting RNA in RNase-free water, denaturing at 65 °C for 5 min, and incubating with 5 mm dNTPs, 10× RT buffer, 10 μm oligo(dT)20, and Omni RT mixture at 42 °C for 2 h for cDNA synthesis. PCR was performed using the Qiagen HotStarTaq master mixture kit by mixing cDNA with HotStarTaq mixture and 25 μm forward and reverse primers for PCR amplification of XBP-1 products. The PCR products were resolved on a 3% NuSieve 3:1 agarose gel (Cambrex, Rockland, ME). The primers used for hXBP-1 were forward, 5′-GAGAACCAGGAGTTAAGACA-3′, and reverse, 5′-TAAGACTAGGGGCTTGGTA-3′.
Animal Model—For assessment of force-induced cell death in vivo, latex rubber separators were placed between the first and second mandibular molar teeth of male Wistar rats (250 g; n = 3) (25). This maneuver applies tensile forces to rat periodontal ligament cells in vivo and initiates apoptosis in periodontal ligament cell populations (7). Controls (n = 3) consisted of animals without separators. Rats were killed at 2 days after stimulation. Jaws were fixed in paraformaldehyde, decalcified, and prepared for paraffin sections. Cells in the periodontal ligament were stained with an antibody to phosphorylated PERK at 1:100 as described (26). The tissue sections were not counterstained. The experiments were conducted according to an experimental protocol approved by the Animal Care Committee at the University of Toronto.
Statistical Analysis—For continuous variables, means and S.E. were computed. Band intensities reported are averaged data from three separate measurements. Statistical significance was estimated by Student's t test using individual values from three different measurements. All experiments were performed at least three times in triplicate.
RESULTS
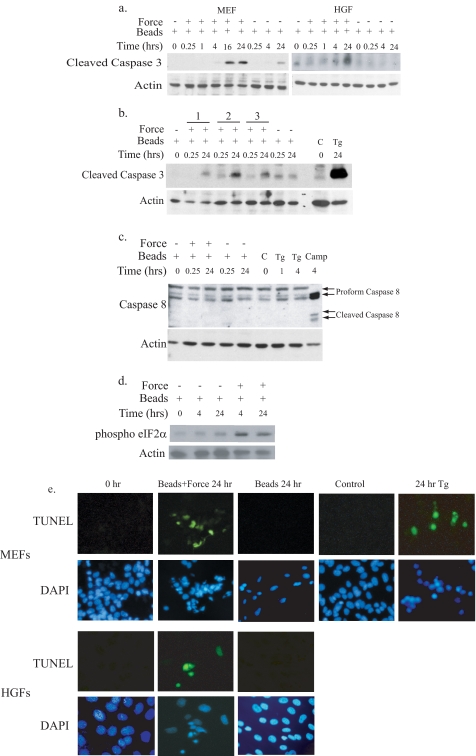
Mechanical Force Induces Apoptosis—Robust time-dependent increases in the active (cleaved) form of caspase 3 were observed in MEFs and in HGFs exposed to tensile force (Fig. 1a). This was consistently observed as early as 16 h after force application and was detected with application of stretching, compression, and twisting forces (Fig. 1b). To ensure complete and reliable detection of activation in all subsequent studies, cleavage of caspase 3 was monitored after 24 h of force exposure. Tensile force did not activate caspase 8, an indicator of death receptor-mediated apoptosis activation (Fig. 1c), but did increase phosphorylation of eIF2α (Fig. 1d). DNA fragmentation detected by TUNEL staining was observed in MEFs (45% TUNEL-positive at 24 h) and in human gingival fibroblasts (35% TUNEL-positive at 24 h) after exposure to mechanical forces (Fig. 1e) and was restricted to cells that bound collagen-coated beads. In contrast, cells with attached collagen-coated magnetite beads that were not exposed to tensile force exhibited minimal caspase 3 cleavage and no detectable DNA frag-mentation (Fig. 1, a-c and e). Together, these data indicate that the primary inducer of apoptosis is mechanical force and not collagen bead binding alone.
FIGURE 1.
Induction of apoptosis by mechanical stretching. a, collagen-coated magnetite beads were bound to MEFs or HGFs. Cells were either undisturbed or exposed to tensile force with magnets. Lysates were collected from cells at different durations of force exposure and resolved by immunoblotting to detect caspase 3 cleavage. Actin expression was used as a loading control. b, MEFs were exposed to stretching force (lane 1), compression force (lane 2), and twisting force (lane 3) for the time duration indicated and analyzed for induction of caspase 3 cleavage as described under “Experimental Procedures.” Untreated cells (C) and thapsigargin-treated cells (Tg; 300 nm, 24 h) were used as negative and positive controls, respectively. c, an immunoblot of lysates from HGFs exposed to force for detection of caspase 8 cleavage is shown. Proform 55/50-kDa (inactive) and cleaved form 40/36-kDa (active) caspase 8 were detected using a polyclonal antibody to caspase 8. Actin expression was used as a loading control. Negative control samples were treated with thapsigargin (1 μm, 1 or 4 h). Positive controls for active caspase 8 were from cells treated with camptothecin (Camp;1 μg/ml, 4 h). d, MEFs were treated with force as described above, and lysates were immunoblotted for phosphorylated eIF2α and for actin as the loading control. e, MEFs and HGFs grown on coverslips were stained by TUNEL and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Control samples with bound beads were not exposed to force (0 h) or not treated with beads (control). Cells treated with thapsigargin (300 nm, 24 h) were used as a positive control. Magnifications were MEFs, ×375, and HGFs, ×450.
Cells loaded with JC-1 to monitor mitochondrial membrane potential, incubated with collagen beads, and then exposed to force or no force showed that tensile force reduced the ratio of JC-1 aggregates to monomers (Table 1), an estimate of ψm. The aggregate-to-monomer ratio was reduced to less than one-third of that of cells not exposed to force, indicating that stretching forces cause dissipation of ψm. Cells treated with carbonyl cyanide 3-chlorophenylhydrazone, a positive control for dissipation of ψm, exhibited a ratio of aggregates to monomers that was 1/15 that of control ratios. Thus tensile force may be able to induce apoptosis in fibroblasts through intrinsic pathways associated with both the ER and mitochondria.
TABLE 1.
Mitochondrial membrane potential in attached cells
Cells were incubated with collagen-coated beads and treated as indicated. Attached cells were removed by trypsinization, stained with JC-1, and analyzed by two-color flow cytometry. JC-1 aggregates were estimated by red fluorescence and JC-1 monomers by green fluorescence. For each treatment group, at least 1000 cells were analyzed to generate an aggregate-to-monomer ratio as an estimate of mitochondrial membrane potential. This experiment was repeated three times. The data from a single representative experiment are shown below. The mean aggregate:monomer ratios for three experiments are given in parentheses after the single experimental data.
| Time (h)/treatment | No. of aggregates | No. of monomers | Aggregate:monomer |
|---|---|---|---|
| 0/Beads + no force | 1962 | 612 | 3.2 (3.4) |
| 24/Beads + no force | 7872 | 2190 | 3.6 (3.3) |
| 24/Beads + force | 1384 | 1429 | 1.0 (0.9) |
| 2/Carbonyl cyanide 3-chlorophenylhydrazone | 205 | 1144 | 0.2 (0.3) |
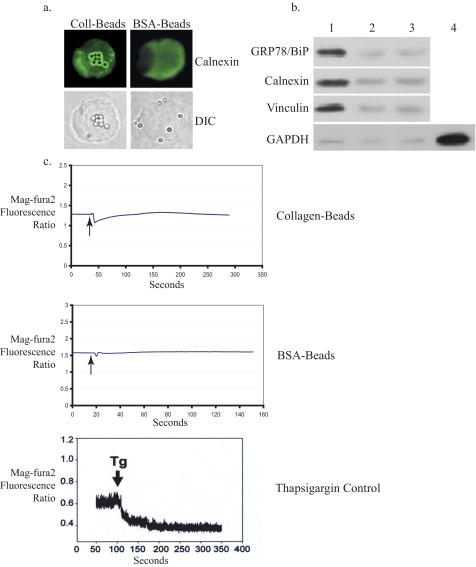
Plasma Membrane Interactions with the ER at Focal Adhesions—Increased eIF2α phosphorylation has been observed in response to force application (9), and our data above (Fig. 1d) indicated that the UPR is activated in this system. Accordingly, we determined if there was a mechanical continuity between focal adhesions and the ER. Colocalization and coisolation experiments were performed to assess whether focal adhesion proteins bound to magnetite beads can interact with the ER. In human fibroblasts, collagen or BSA-coated beads were bound to cells and stained for calnexin, an ER protein. Enrichment of calnexin was observed around collagen-coated beads but not around BSA-coated beads in permeabilized cells (Fig. 2a) or in unpermeabilized cells (data not shown). Second, focal adhesion-associated proteins were isolated from cells after binding to collagen-coated beads, and the bead-associated proteins were immunoblotted. Both calnexin and GRP78/BiP, another ER resident chaperone protein, were detected in immunoblots of the focal adhesion-associated proteins from collagen-coated bead-bound samples (Fig. 2b, lane 1) but not from BSA-coated bead-bound samples (Fig. 2b, lane 2). Vinculin, a focal adhesion-associated protein, was also detected in these samples, indicating that the adhesions formed around the beads were authentic focal adhesions. Inhibition of actin filament polymerization with swinholide A blocked focal adhesion formation and coaggregation of calnexin and GRP78/BiP (Fig. 2b, lane 3), indicating that the interaction between focal adhesions and ER-resident proteins depends on the integrity of actin filaments and the formation of focal adhesions.
FIGURE 2.
Association between plasma membrane and the endoplasmic reticulum. a, human gingival fibroblasts plated on poly-l-lysine were incubated with collagen (Coll)- or BSA-coated latex microbeads for 30 min at 37 °C and immunostained for calnexin followed by fluorescein isothiocyanate-conjugated second antibody. The microscope was focused on the dorsal surface of the cell to image bead-associated proteins. DIC, differential interference contrast. Magnification was ×800. b, immunoblots of proteins from focal adhesion complexes show coisolation of calnexin and GRP78/BiP with complexes prepared from cells bound with collagen-coated beads (lane 1) but not with BSA beads (lane 2). Disruption of the actin cytoskeleton with swinholide A blocked formation of focal adhesion complexes and the association of calnexin and GRP78/BiP (lane 3). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), used as a control for FAC purity, was detected only in whole cell lysates (lane 4). c, mag-fura-2 ratio data are shown for estimation of [Ca2+]ER. Cells plated on poly-l-lysine were loaded with mag-fura-2/AM and preincubated with collagen- or BSA-coated beads. Traces are representative of three independent experiments, each with n = 3-5 cells. The bottom panel shows composite traces of five cells incubated with thapsigargin (Tg;1 μm) at the indicated time as a positive control for release of calcium from the ER.
To determine whether applied forces can generate an ER-specific response, [Ca2+]ER was estimated from measurements of the mag-fura-2 excitation ratio in single cells. Dye-loaded fibroblasts were switched to Ca2+-free medium (to prevent entry of Ca2+ through stretch-activated plasma membrane channels (27)), and within 1 min, force was applied (for a 1-s duration). Under these conditions, cells exhibited rapid reduction of the mag-fura-2 ratio when forces were applied through collagen-coated beads but not BSA-coated beads (Fig. 2c), indicating a close physical association between focal adhesions (the site of force application in this model) and the ER. As a control, thapsigargin-treated cells exhibited rapid reduction of the magfura-2 ratio (Fig. 1c).
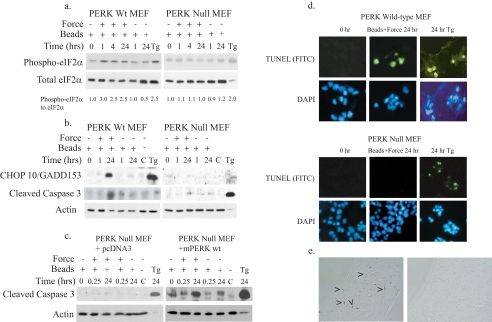
PERK Mediates Force-induced Apoptosis—The data above indicated that mechanical forces could be transmitted to the ER and thereby activate apoptosis, possibly through the UPR. Because tensile forces can increase eIF2α phosphorylation in fibroblasts (9), and as shown also by our data above, force induction of apoptosis was examined by focusing on the PERK signaling pathway. Detection of eIF2α phosphorylation and induction of CHOP10/GADD153 by immunoblotting were used as downstream indicators of PERK activity and UPR activation, whereas cleavage of caspase 3 was used to estimate apoptosis. In PERK wild-type MEFs, force application enhanced eIF2α phosphorylation at 1 h, slightly higher than levels seen with thapsigargin treatment (Fig. 3a). There was also marked induction of CHOP10/GADD153 and caspase 3 cleavage after prolonged exposure to sustained force (Fig. 3b). When PERK null MEFs were examined under the same conditions, eIF2α phosphorylation, induction of CHOP10/GADD153, and caspase 3 cleavage were not observed above background levels (Fig. 3, a and b). Ectopic expression of hemagglutinin-tagged wild-type mouse PERK into PERK null MEFs, confirmed by hemagglutinin expression (data not shown), restored force-induced caspase 3 cleavage (Fig. 3c). Relative to cells at zero time, caspase 3 activation was induced 2-3-fold after force. Notably, in some experiments, thapsigargin-treated cells showed little or no detectable actin, possibly because of cell death and degradation of cytoskeletal proteins by caspases. Furthermore, in some experiments, cells binding large numbers of beads showed caspase 3 cleavage even when not exposed to force; however, the levels of induction were below that of force-treated cells (Fig. 3, b and c, see values below blots and Fig. 1a). For all experiments, bead-bound cells exposed to prolonged force consistently exhibited higher caspase 3 activation compared with bead-bound cells in the absence of force.
FIGURE 3.
Force-induced apoptosis is mediated through PERK. Tensile forces were applied to PERK wild-type (Wt) or PERK null MEFs. a and b, immunoblots of cell lysates show enhanced eIF2α phosphorylation, up-regulation of CHOP10/GADD153, and caspase 3 cleavage in PERK wild-type MEFs but not PERK null MEFs. Actin was used as a loading control; thapsigargin was used as a positive control (Tg; 300 nm, 24 h). Band intensity for phosphorylated eIF2α was measured and expressed as a ratio relative to the 0-h time point as described under “Experimental Procedures.” Values comparing the statistical significance of force-treated and untreated samples at 24 h were analyzed using Student's t-test. The reported average values at 1-24 h after force were significantly higher (p < 0.05). C, untreated cells. c, immunoblots of cell lysates show no induction of caspase 3 cleavage in PERK null cells, whereas in PERK null cells transfected with wild-type mouse PERK there was abundant cleaved caspase 3 at 24 h. Cleaved caspase 3 band intensities were measured and expressed as a ratio relative to the 0-h time point as described under “Experimental Procedures.” Student's t test showed significantly higher (p < 0.002) cleaved caspase 3 at 24 h compared with 0 h. Control samples include untreated cells and thapsigargin-treated samples (1 μm for 1 h or 300 nm for 24 h). d, TUNEL staining in PERK wild-type (top panel) and PERK null (bottom panel) cells after force application is shown. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Magnification was ×350. Cells treated with thapsigargin (300 nm, 24 h) were used as positive controls. FITC, fluorescein isothiocyanate. e, immunohistochemical staining for phospho-PERK in rat periodontal ligament subjected to orthodontic forces (left panel) or to no force (right panel) is shown. Magnification was ×225.
As a second approach for evaluating apoptosis, TUNEL staining was performed and was observed in PERK wild-type MEFs after 24 h exposure to force but not in PERK null MEFs (Fig. 3, d and e). Only PERK wild-type MEFs with attached collagen-coated magnetite beads were TUNEL-positive (45% after 24 h). In contrast, PERK null MEFs did not stain positively for TUNEL regardless of whether they had bound collagen-coated beads after force application (Fig. 3d). These observations cannot be attributed to differential binding capacities of the two cell lines because collagen bead binding by PERK wild-type and PERK null MEFs were comparable (data not shown). Staining for TUNEL and caspase 3 cleavage after treatment with thapsigargin (a chemical activator of the UPR) indicated that activation of apoptosis by the UPR was intact in PERK null MEFs (Fig. 3, a and b).
We asked whether PERK may be involved in force-dependent signaling events in vivo. Accordingly, cells in rat periodontal ligament exposed to mechanical forces or not were examined by immunohistochemistry using previously described methods (7). Phosphorylated PERK was detected in force-loaded tissues but not in unloaded tissues (Fig. 3e).
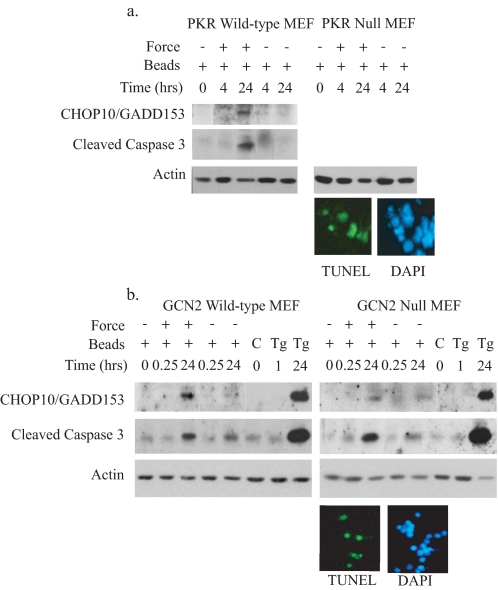
Force Induction of Apoptosis Is Not Dependent on eIF2a Phosphorylation—Previous reports have shown that eIF2α phosphorylation in the UPR is reliant on PERK expression (28). As force-induced eIF2α phosphorylation and caspase 3 cleavage were both ablated in the absence of PERK, we examined the contribution of other eIF2α kinases (PKR and GCN2) to force induction of apoptosis. In PKR null MEFs (Fig. 4a) and GCN2 null MEFs (Fig. 4b), force induction of both CHOP10/GADD153 (used as a downstream indicator of eIF2α phosphorylation) and caspase 3 cleavage remained intact, indicating that neither PKR or GCN2 is required for force-induced apoptosis. Immunoblots showed that force application activated caspase 3 above levels seen with bead binding alone (Fig. 4, a and b), similar to previous findings in the other cell lines. Notably, in both the GCN2 and PKR null cell lines, force induced TUNEL-positive cells (Fig. 4, a and b; ∼40% for both cell types).
FIGURE 4.
Force-induced apoptosis is not dependent on PKR or GCN2. Tensile force was applied to cells. Lysates were collected for immunoblotting to detect CHOP10/GADD153 and caspase 3 cleavage. Actin was used as a loading control. a, PKR wild-type or PKR null MEFs. CHOP10/GADD153 and cleaved caspase 3 was detected by immunoblotting. TUNEL staining was done on PKR null cells after 24 h of force application. Band intensity was measured and expressed as a ratio relative to the 0-h time point as described under “Experimental Procedures.” DAPI, 4′,6-diamidino-2-phenylindole. b, GCN2 wild-type or GCN2 null MEFs. Control samples include untreated cells (C) as negative controls and cells treated with thapsigargin (Tg;1 μm for 1 h or 300 nm for 24 h) as positive controls. TUNEL staining was done on GCN2 null cells 24 h after force application. Magnification for TUNEL staining was ×250.
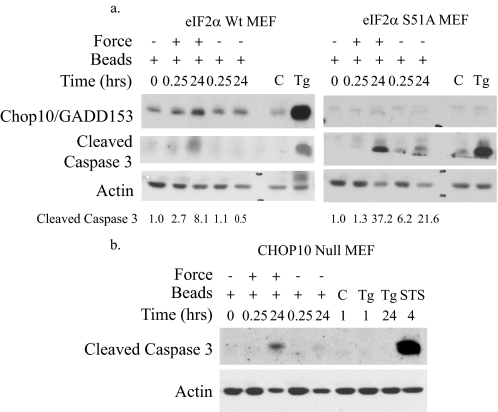
We determined whether phosphorylation of eIF2α is required for force-induced apoptosis because it is important for apoptosis in classical UPR mechanisms (17). We examined MEFs expressing wild-type eIF2α or eIF2α harboring a point mutation at Ser-51, the site of PERK-induced phosphorylation. Because eIF2α phosphorylation cannot be detected in the S51A mutant cell line, CHOP10/GADD153 elevation was used as a downstream functional readout for eIF2α phosphorylation. Increased expression of both CHOP10/GADD153 and caspase 3 cleavage were observed in cells expressing wild-type eIF2α after force application. In eIF2α mutant cells, as anticipated, CHOP10/GADD153 expression was not observed, but caspase 3 cleavage was still detected, indicating that caspase 3 activation by force does not require eIF2α phosphorylation or CHOP10/GADD153 (Fig. 5a). This notion was confirmed with CHOP10/GADD153 null MEFs that showed induction of caspase 3 cleavage by force and staurosporine (Fig. 5b). CHOP10/GADD153 expression was not detected by immunoblotting (data not shown), and this result was confirmed by absence of an UPR-mediated apoptotic response (caspase 3 cleavage) to thapsigargin. Quantification of cleaved caspase 3 bands from both eIF2α cell lines indicates that activation of caspase 3 was enhanced after prolonged force induction (Fig. 5a), and quantification of TUNEL staining showed ∼35% after 24 h of force.
FIGURE 5.
Phosphorylation of eIF2a is not required for force-induced apoptosis. a, force was applied to MEFs expressing wild-type (Wt) eIF2α (Ser-51, Ser/Ser) or mutant eIF2α (S51A, Ala/Ala). Lysates were immunoblotted for CHOP10/GADD153 and caspase 3 cleavage. Cleaved caspase 3 band intensity was measured as described under “Experimental Procedures.” Values comparing the statistical significance of force-treated and untreated samples at 24 h were analyzed using Student's t test. The reported average values were significant with p < 0.05. Control samples include untreated cells (C) as negative controls and cells treated with thapsigargin (Tg; 300 nm, 24 h) as positive controls. b, immunoblot of lysates from CHOP10/GADD153 null MEFs shows cleavage of caspase 3 with force. Cells treated with thapsigargin (1 μm for 1 h, 300 μm for 24 h) or staurosporine (STS; 200 μm, 4 h) were used as positive controls.
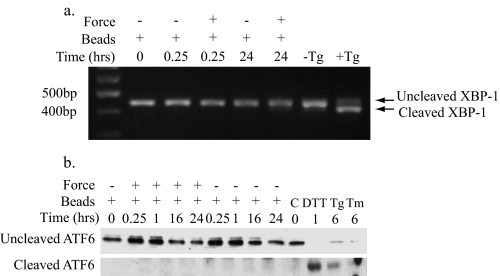
Force Application Does Not Activate Canonical ER Stress Responses—As phosphorylation of eIF2α and induction of CHOP10/GADD153 were neither necessary nor sufficient for force-induced apoptosis, the canonical UPR mechanism leading to cell death through PERK and eIF2α evidently does not mediate force-induced apoptosis. To determine whether other UPR mechanisms are involved, the activities of IRE-1 and ATF6 were examined. Cleavage of XBP-1 mRNA by IRE-1 was used to detect activation of IRE-1. RNA was isolated from cells after force treatment and analyzed with RT-PCR using primers specific for inactive (uncleaved) and active (cleaved) XBP-1. Cells exposed to force for 15 min or 24 h did not exhibit XBP-1 cleavage (Fig. 6a). In contrast, thapsigargin-treated controls showed typical XBP-1 cleavage (29). Therefore, the IRE-1 pathway of the UPR was not activated by mechanical force, as indicated by the absence of XBP-1 cleavage, and it is unlikely to mediate force-induced apoptosis.
FIGURE 6.
IRE-1 and ATF6 pathways are not induced by force. a, RNA from HGFs was isolated for RT-PCR using primers specific for uncleaved (inactive) and cleaved (active) XBP-1 to detect IRE-1-dependent RNase activity in response to force. Samples were resolved by agarose gel electrophoresis. Cells treated with thapsigargin (Tg; 1 mm, 1 h) were used as positive control for IRE-1 activity. Negative control cells were untreated. b, an immunoblot of lysates from HGFs exposed to force for detection of ATF6 cleavage is shown. Full-length (inactive) and cleaved (active) ATF6 were detected using a polyclonal antibody for ATF6. Positive control samples were treated with dithiothreitol (DTT; 1 mm, 1 h), thapsigargin (300 nm, 6 h), or tunicamycin (Tm;2 μg/ml, 6 h). C, untreated cells.
We also examined the ATF6 pathway of the UPR by immunoblotting for cleaved forms of ATF6 in cell lysates after exposure to force. In contrast to cells treated with dithiothreitol, thapsigargin, and tunicamycin, which were used as positive controls, activation of ATF6 was not detected in cells exposed to mechanical force (Fig. 6b), indicating that the ATF6 pathway of the UPR is not activated by force. Notably, it is possible that cleavage of XBP-1 or ATF6 in response to force occurs at very low levels that are below the detection threshold by RT-PCR and Western blotting, respectively. If this were the case, it is unlikely that the low level of activation of XBP-1 or ATF6 contributes significantly to the apoptotic response induced by mechanical forces because activation of caspase 3 by mechanical force is lost in PERK null cells where IRE-1 and ATF6 remain intact (Fig. 3b).
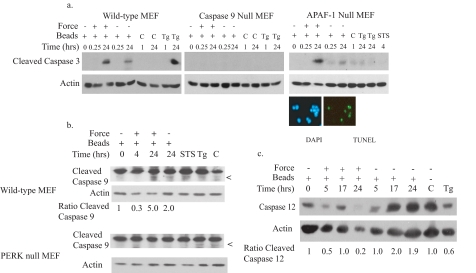
PERK-mediated Force Induction of Apoptosis Involves and Caspase 9 but Not APAF-1—As we detected mitochondrial depolarization after force application (Fig. 1f), mitochondria-mediated apoptotic signals involving the cytochrome C·APAF-1·caspase 9 apoptosome complex could be involved in force-induced cell death (30). To examine the role of mitochondrial systems in facilitating force-induced apoptosis, cells deficient in APAF-1 or caspase 9, proteins required for formation of the apoptosome, were exposed to force and examined for caspase 3 activation. In the absence of APAF-1 expression, caspase 3 cleavage was stimulated after force application, and TUNEL staining was still noted (30% TUNEL-positive after 24 h); however, the deficiency in caspase 9 expression abrogated caspase 3 activity (Fig. 7a). These data imply that force-induced apoptosis occurs through caspase 9 activity, independent of APAF-1 and possibly mitochondria. Although the JC-1 data above indicated mitochondrial depolarization, suggesting that mitochondria-mediated apoptotic signals are activated, these pathways may be not necessary for force induced apoptosis but instead function to enhance the PERK-mediated apoptotic signal (31).
FIGURE 7.
Force-induced apoptosis is dependent on caspase 9 but not APAF-1. a, force was applied to control MEFs, caspase 9 null MEFs, or APAF-1 null MEFs. Immunoblots of cell lysates show that force-induced caspase 3 cleavage is preserved in the absence of APAF-1 but not in the absence of caspase 9. Cells treated with thapsigargin (Tg; 1 mm for 1 h, 300 nm for 24 h) were used as positive control. Staurosporine (STS; 4 h) did not induce caspase 3 cleavage. Negative controls cells were untreated (C). APAF null cells were TUNEL-positive after force application. Magnification was ×300. DAPI, 4′,6-diamidino-2-phenylindole. b, an immunoblot of wild-type MEF lysates shows cleavage of caspase 9 after force. Actin was used as a loading control. Cells treated with staurosporine (200 μm, 4 h) or thapsigargin (1 μm for 1 h, 300 μm for 24 h) were used as positive controls. Untreated cells were used as negative controls. Band intensity for cleaved caspase 9 was measured as described under “Experimental Procedures.” Values comparing the statistical significance of force-treated and untreated samples at 24 h were analyzed using Student's t test. Reported average values were significantly different (p < 0.002). c, caspase 12 immunoblots of MEFs show disappearance of caspase 12 at 24 h after force because of force-induced processing.
We next examined whether force can activate caspase 9. In wild-type MEFs, caspase 9 cleavage was observed after force application in a time-dependent manner (Fig. 7b, arrows). Densitometry of the caspase 9 cleaved products showed increased detection of these proteins with prolonged exposure to force in the wild-type cells, but the cleaved products were not detected in the PERK null cells (Fig. 7b). We also examined caspase 12 cleavage, and these data showed loss of the caspase 12 band between 5 and 24 h after force treatment (Fig. 7c). Replicate analyses of caspase 12 showed that compared with 0 h, caspase 12 was reduced to 0.3 ± 0.3 (n = 3 separate experiments) of the density at 24 h after force application. Together with previously published work showing that apoptosis can be stimulated through caspase 9 activation in an ER-dependent manner (32), these results are consistent with our observation that force can elicit an apoptotic response through the ER.
DISCUSSION
In connective tissue cells, transmission of tensile forces from fibrillar extracellular matrix proteins through integrins to the actin cytoskeleton induces signals that can mediate cell proliferation or death (1). In tissues exposed to high amplitude forces such as bone, joint, or periodontium, inappropriate responses to physical forces contribute to the pathogenesis of several high prevalence diseases including osteoarthritis and periodontitis (3, 4). Here we explored the signaling mechanisms that mediate force-induced apoptosis and demonstrated that in this system, apoptosis is mediated by a novel function of PERK. Applied forces induced apoptosis as assessed by caspase 3 cleavage, DNA fragmentation, depolarization of mitochondria, and induction of CHOP10/GADD153. In some experiments in which cells bound high numbers of collagen-coated beads for prolonged periods, caspase 3 cleavage was observed also at low levels in the absence of applied magnetic forces. This result likely arose from bead-induced attractive interactions attributable to the beads themselves that induce small force effects. In situ analysis of DNA fragmentation by TUNEL showed that apoptosis was restricted to cells that bound collagen beads. Furthermore, the levels of detectable caspase 3 cleavage and TUNEL-positive staining were consistently higher after prolonged exposure to beads in different cell lines. We conclude that force-induced apoptosis was mediated largely by force-induced mechanical stretching.
Binding of extracellular matrix proteins to integrins induces focal adhesion formation at sites of bead binding (33). Both focal adhesions and the associated actin filaments are required for force-induced apoptosis (6, 7). Notably, focal adhesions contain large numbers of proteins involved in apoptotic signaling pathways (9) and may provide a physical and functional connection to organelles such as the ER (9, 34) that can contribute to activation of apoptotic processes (12). We determined whether focal adhesions associate with the ER to mediate apoptosis through ER-specific pathways, possibly the UPR. By immunofluorescence and coisolation methods, we found a physical association between focal adhesions and ER proteins. This association facilitated force-induced Ca2+ efflux from the ER and suggested that the ER may be involved in force-induced apoptosis.
Apoptotic signals arising from the ER may be mediated by the UPR-associated initiator molecules PERK, IRE-1, and ATF6 (11). These molecules are generally thought to be involved in apoptosis stimulated by up-regulation of CHOP10/GADD153. Because previous studies showed that force promotes eIF2α phosphorylation (9), we focused on the role of the PERK pathway in ER-associated apoptosis. Our analysis of apoptosis in PERK wild-type and null MEFs, as well as ectopic expression of wild-type PERK in null MEFs, demonstrated that force-induced apoptosis requires PERK and suggested that force-induced cell death is mediated through the ER. PERK is generally considered to be anti-apoptotic, as PERK null MEFs are more susceptible to apoptosis after treatment with thapsigargin (15), a response that we also observed (Fig. 3, b and c). However, after prolonged ER stress, PERK activates apoptotic signals (13). Similarly, we found that after prolonged force application, PERK is required for force induction of apoptosis. Consequently, PERK may function as an environmental force sensor by regulating cell death and thereby contributing to tissue homeostasis.
PERK phosphorylation of eIF2α initiates a signaling cascade that leads to CHOP10/GADD153 induction (16). As expected, phosphorylated eIF2α and CHOP10/GADD153 up-regulation were increased by force, but caspase 3 cleavage was preserved in MEFs expressing mutant eIF2α (not phosphorylated at Ser-9) and in MEFs null for CHOP10/GADD153. These data indicate that force-enhanced eIF2α phosphorylation and CHOP10/GADD153 expression are not essential for force-induced apoptosis. Force application also induced dissipation of mitochondrial membrane potential, an event that initiates apoptotic signaling cascades from the mitochondria and involves the formation of the cytochrome C·APAF-1·caspase 9 apoptosome (30). These findings suggested that mitochondrial signaling mechanisms are involved in force-induced apoptosis, consistent with the observation that mitochondrial mediated apoptotic pathways can also be activated upon UPR stimulation (35). However, we found that tensile force induction of caspase 3 cleavage is maintained in the absence of APAF-1, indicating that although force-induced and mitochondria-mediated apoptotic pathways are both activated, they are separate and functionally independent.
In contrast, caspase 9 is essential for force-induced caspase 3 activation. Despite its association with APAF-1 in the apoptosome, caspase 9 activation can occur independent of APAF-1/cytochrome C release (32). Indeed, the mitochondrial-apoptosome mediated pathway is considered to amplify rather that initiate caspase cascades leading toward apoptosis (31). Because CHOP10/GADD153 is known to inhibit Bcl-2 expression and in turn can affect the mitochondrial membrane potential (13), force-induced up-regulation of CHOP10/GADD153 may serve to amplify a force-induced apoptotic signal by stimulating the mitochondria.
In addition to the pro-apoptotic properties attributed to CHOP10/GADD153 (13), it can also activate genes such as GADD34, which promotes recovery from UPR-induced suppression of translation (36). Depending on the type of stress conditions, CHOP10/GADD153 may not function strictly as a pro-apoptotic molecule but may instead maintain cellular homeostasis, a function that would be lost with its deletion. Conceivably, after application of tensile force, the induction of eIF2a phosphorylation and CHOP10/GADD153 may be a recovery response to stress. Furthermore, our observations that force did not induce CHOP10/GADD153 in both PERK null cells and eIF2α Ala/Ala mutant cells imply that ATF6 and IRE-1 pathways, which can also up-regulate CHOP10/GADD153, are not activated by force in both cell lines. This notion was confirmed by the absence of ATF6 and XBP-1 cleavage in stretched cells.
Activation of the UPR generally involves the integration of PERK, ATF6, and IRE-1 signaling pathways, but their selective activation has been observed previously in myoblast and B cell differentiation (37). Perhaps in connective tissues exposed to high tensile forces such as tendon, ligaments, and bone, PERK can function separately from ATF6 and IRE-1. Notably, skeletal abnormalities are associated with defects in the PERK/eIF2α signaling pathway in both PERK null and ATF4 null mice and in humans affected by the Wolcott-Rallison syndrome, a disease linked to mutations in the human PERK gene (38). These skeletal abnormalities have been attributed to the inefficient processing and secretion of extracellular matrix proteins such as collagen, and manifest as fragmented and distended ER in the osteoblasts of PERK null mice. Loss of PERK function may also dysregulate the expression of growth factors that direct bone development.
Our data indicate that PERK is essential for force-induced apoptosis and suggest that it may play a role in integrating force-related signals with other extracellular stimuli. In contrast, CHOP10/GADD153 expression and eIF2α phosphorylation are not essential, and the canonical UPR is not induced. The PERK signaling pathway also acts independently of mitochondrial signals leading to apoptosis but requires the activation of caspase 9 and evidently involves caspase 12, thereby linking PERK to the caspase pathway. The discovery that PERK expression is required for force-induced apoptosis in a system that is activated in addition to its effects in the UPR indicates that PERK may play a more universal role in cell and tissue homeostasis than was considered previously.
This work was supported, in whole or in part, by National Institutes of Health Grants DK47119 and ES08681 (to D. R. and H. P. H.) and HL052173 and DK42394 (to R. J. K.). This work was also supported by Canadian Institutes of Health Research Grant 418386 (to B. C. M., Q. W., C. L., W. L., and C. A. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: UPR, unfolded protein response; BiP, immunoglobulin heavy chain binding protein; BSA, bovine serum albumin; ER, endoplasmic reticulum; HGF, human gingival fibroblast; MEF, mouse embryonic fibroblast; PBS, phosphate buffered saline; RT, reverse transcription; TUNEL, TdT-mediated X-dUTP nick end labeling.
References
- 1.Wang, J. H., and Thampatty, B. P. (2006) Biomech. Model Mechanobiol. 5 1-16 [DOI] [PubMed] [Google Scholar]
- 2.Cheng, W., Li, B., Kajstura, J., Li, P., Wolin, M. S., Sonnenblick, E. H., Hintze, T. H., Olivetti, G., and Anversa, P. (1995) J. Clin. Investig. 96 2247-2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuhn, K., D'Lima, D. D., Hashimoto, S., and Lotz, M. (2004) Osteoarthritis Cartilage 12 1-16 [DOI] [PubMed] [Google Scholar]
- 4.Yamaguchi, M., and Kasai, K. (2005) Arch. Immunol. Ther. Exp. (Warsz) 53 388-398 [PubMed] [Google Scholar]
- 5.Malek, A. M., and Izumo, S. (1996) J. Cell Sci. 109 713-726 [DOI] [PubMed] [Google Scholar]
- 6.Glogauer, M., Arora, P., Yao, G., Sokholov, I., Ferrier, J., and McCulloch, C. A. (1997) J. Cell Sci. 110 11-21 [DOI] [PubMed] [Google Scholar]
- 7.Kainulainen, T., Pender, A., D'Addario, M., Feng, Y., Lekic, P., and McCulloch, C. A. (2002) J. Biol. Chem. 277 21998-22009 [DOI] [PubMed] [Google Scholar]
- 8.Holcik, M., and Sonenberg, N. (2005) Nat. Rev. Mol. Cell Biol. 6 318-327 [DOI] [PubMed] [Google Scholar]
- 9.Wang, J., Laschinger, C., Zhao, X. H., Mak, B., Seth, A., and McCulloch, C. A. (2005) Biochem. Biophys. Res. Commun. 330 123-130 [DOI] [PubMed] [Google Scholar]
- 10.Glogauer, M., Arora, P., Chou, D., Janmey, P. A., Downey, G. P., and McCulloch, C. A. (1998) J. Biol. Chem. 273 1689-1698 [DOI] [PubMed] [Google Scholar]
- 11.Wu, J., and Kaufman, R. J. (2006) Cell Death Differ. 13 374-384 [DOI] [PubMed] [Google Scholar]
- 12.Xu, C., Bailly-Maitre, B., and Reed, J. C. (2005) J. Clin. Investig. 115 2656-2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oyadomari, S., and Mori, M. (2004) Cell Death Differ. 11 381-389 [DOI] [PubMed] [Google Scholar]
- 14.Haze, K., Yoshida, H., Yanagi, H., Yura, T., and Mori, K. (1999) Mol. Biol. Cell 10 3787-3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harding, H. P., Zhang, Y., Zeng, H., Novoa, I., Lu, P. D., Calfon, M., Sadri, N., Yun, C., Popko, B., Paules, R., Stojdl, D. F., Bell, J. C., Hettmann, T., Leiden, J. M., and Ron, D. (2003) Mol. Cell 11 619-633 [DOI] [PubMed] [Google Scholar]
- 16.Harding, H. P., Zhang, Y., Bertolotti, A., Zeng, H., and Ron, D. (2000) Mol. Cell 5 897-904 [DOI] [PubMed] [Google Scholar]
- 17.Scheuner, D., Song, B., McEwen, E., Liu, C., Laybutt, R., Gillespie, P., Saunders, T., Bonner-Weir, S., and Kaufman, R. J. (2001) Mol. Cell 7 1165-1176 [DOI] [PubMed] [Google Scholar]
- 18.Kumar, A., Yang, Y. L., Flati, V., Der, S., Kadereit, S., Deb, A., Haque, J., Reis, L., Weissmann, C., and Williams, B. R. (1997) EMBO J. 16 406-416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pender, N., and McCulloch, C. A. (1991) J. Cell Sci. 100 187-193 [DOI] [PubMed] [Google Scholar]
- 20.Soengas, M. S., Alarcon, R. M., Yoshida, H., Giaccia, A. J., Hakem, R., Mak, T. W., and Lowe, S. W. (1999) Science 284 156-159 [DOI] [PubMed] [Google Scholar]
- 21.Golovina, V. A., and Blaustein, M. P. (1997) Science 275 1643-1648 [DOI] [PubMed] [Google Scholar]
- 22.Arora, P. D., Silvestri, L., Ganss, B., Sodek, J., and McCulloch, C. A. (2001) J. Biol. Chem. 276 14100-14109 [DOI] [PubMed] [Google Scholar]
- 23.Kulkarni, G. V., and McCulloch, C. A. (1995) J. Cell. Physiol. 165 119-133 [DOI] [PubMed] [Google Scholar]
- 24.Lee, K., Tirasophon, W., Shen, X., Michalak, M., Prywes, R., Okada, T., Yoshida, H., Mori, K., and Kaufman, R. J. (2002) Genes Dev. 16 452-466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts, W. E., Chase, D. C., and Jee, S. S. (1974) Arch. Oral Biol. 19 665-670 [DOI] [PubMed] [Google Scholar]
- 26.Zhou, J., Lhotak, S., Hilditch, B. A., and Austin, R. C. (2005) Circulation 111 1814-1821 [DOI] [PubMed] [Google Scholar]
- 27.Glogauer, M., Ferrier, J., and McCulloch, C. A. (1995) Am. J. Physiol. 269 C1093-C1104 [DOI] [PubMed] [Google Scholar]
- 28.Harding, H. P., Zhang, Y., and Ron, D. (1999) Nature 397 271-274 [DOI] [PubMed] [Google Scholar]
- 29.Yoshida, H., Matsui, T., Yamamoto, A., Okada, T., and Mori, K. (2001) Cell 107 881-891 [DOI] [PubMed] [Google Scholar]
- 30.Saleh, A., Srinivasula, S. M., Acharya, S., Fishel, R., and Alnemri, E. S. (1999) J. Biol. Chem. 274 17941-17945 [DOI] [PubMed] [Google Scholar]
- 31.Marsden, V. S., O'Connor, L., O'Reilly, L. A., Silke, J., Metcalf, D., Ekert, P. G., Huang, D. C., Cecconi, F., Kuida, K., Tomaselli, K. J., Roy, S., Nicholson, D. W., Vaux, D. L., Bouillet, P., Adams, J. M., and Strasser, A. (2002) Nature 419 634-637 [DOI] [PubMed] [Google Scholar]
- 32.Rao, R. V., Castro-Obregon, S., Frankowski, H., Schuler, M., Stoka, V., del Rio, G., Bredesen, D. E., and Ellerby, H. M. (2002) J. Biol. Chem. 277 21836-21842 [DOI] [PubMed] [Google Scholar]
- 33.Hughes-Fulford, M. (2004) Science's STKE 2004 RE12. [DOI] [PubMed] [Google Scholar]
- 34.Tran, H., Pankov, R., Tran, S. D., Hampton, B., Burgess, W. H., and Yamada, K. M. (2002) J. Cell Sci. 115 2031-2040 [DOI] [PubMed] [Google Scholar]
- 35.Nakamura, K., Bossy-Wetzel, E., Burns, K., Fadel, M. P., Lozyk, M., Goping, I. S., Opas, M., Bleackley, R. C., Green, D. R., and Michalak, M. (2000) J. Cell Biol. 150 731-740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marciniak, S. J., Yun, C. Y., Oyadomari, S., Novoa, I., Zhang, Y., Jungreis, R., Nagata, K., Harding, H. P., and Ron, D. (2004) Genes Dev. 18 3066-3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, K., Wong, H. N., Song, B., Miller, C. N., Scheuner, D., and Kaufman, R. J. (2005) J. Clin. Investig. 115 268-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, P., McGrath, B., Li, S., Frank, A., Zambito, F., Reinert, J., Gannon, M., Ma, K., McNaughton, K., and Cavener, D. R. (2002) Mol. Cell. Biol. 22 3864-3874 [DOI] [PMC free article] [PubMed] [Google Scholar]