Abstract
The high aromatic content of the deep and narrow active-site gorge of acetylcholinesterase (AChE) is a remarkable feature of this enzyme. Here, we analyze conformational flexibility of the side chains of the 14 conserved aromatic residues in the active-site gorge of Torpedo californica AChE based on the 47 three-dimensional crystal structures available for the native enzyme, and for its complexes and conjugates, and on a 20-ns molecular dynamics (MD) trajectory of the native enzyme. The degree of flexibility of these 14 aromatic side chains is diverse. Although the side-chain conformations of F330 and W279 are both very flexible, the side-chain conformations of F120, W233, W432, Y70, Y121, F288, F290 and F331 appear to be fixed. Residues located on, or adjacent to, the Ω-loop (C67–C94), namely W84, Y130, Y442, and Y334, display different flexibilities in the MD simulations and in the crystal structures. An important outcome of our study is that the majority of the side-chain conformations observed in the 47 Torpedo californica AChE crystal structures are faithfully reproduced by the MD simulation on the native enzyme. Thus, the protein can assume these conformations even in the absence of the ligand that permitted their experimental detection. These observations are pertinent to structure-based drug design.
INTRODUCTION
The principal role of acetylcholinesterase (AChE) is termination of transmission at cholinergic synapses by rapid hydrolysis of the neurotransmitter acetylcholine (ACh) (1). Due to its crucial biologic role, AChE is the target of the first generation of anti-Alzheimer drugs, insecticides, and nerve agents (2). Solution of the three-dimensional (3D) structure of Torpedo californica AChE (TcAChE) revealed, unexpectedly, that its catalytic site is located near the bottom of a deep and narrow gorge, ∼20 Å deep (3). Its cross section at the narrowest point is substantially smaller than the diameter of the quaternary group of choline; thus, AChE must undergo substantial “breathing” motions to carry out its catalytic activity (3,4). A remarkable feature of the gorge is that it is lined by the rings of 14 conserved aromatic amino acids (F120, F288, F290, F330, F331, W84, W233, W279, W432, Y70, Y121, Y130, Y334, and Y442), which contribute ∼60% of its total surface area. These residues are highly conserved, and most of them can be assigned functional roles. Thus, W84 and F330 are involved in the so-called “anionic” subsite of the catalytic site (CAS), making π-cation interactions with the quaternary group of the substrate (3,5–8); W279 and Y70 contribute to the peripheral anionic site (PAS) at the mouth of the gorge, which serves as a relay station for the substrate en route to the active site (7–12); and W233, F288, and F290 contribute to the acyl pocket, which determines the specificity for ACh (6,13). Midway down the gorge is the narrow bottleneck mentioned above, to which both Y121 and F330 make important contributions (7,14,15). Crystallographic studies have also established the involvement of several of these conserved aromatic residues in interactions with anticholinesterase agents via both π-cation and π–π stacking interactions (5,16,17). A typical example is the drug E2020 (Aricept), which is used to treat Alzheimer disease. E2020 interacts with W84 at the bottom of the gorge and W279 at the entrance to the gorge through π–π stacking interactions, and it interacts with F330 in the middle of the gorge via a π-cation interaction (16). Steered molecular dynamics (MD) simulations of the dissociation of E2020 from AChE suggest a model in which the aromatic residues serve as a conveyer belt for E2020 entering and leaving the active-site gorge (18).
Since the solution of the initial crystal structure of TcAChE, more than 17 years ago (3), 3D structures of the native enzyme from several different species, as well as numerous complexes and conjugates with various ligands, inhibitors and substrate analogs, have been deposited in the Protein Data Bank (PDB) (www.pdb.org). Even though the enzyme has been crystallized under a number of very different conditions and in seven different space groups, the overall fold of the enzyme is remarkably well conserved in all these structures. The side chains of several aromatic residues, however, appear to be quite flexible. Thus, several crystal structures show that W279, at the PAS, can adopt several different conformations in various ligand/enzyme complexes (PDB codes 1q83 (19), 2gyu (20), 1odc (21), 2cek (22), 2ckm (23), 2gyw (20), and 2gyv (20)). F330 is one of two residues that form the bottleneck of the gorge, and it has been suggested that the orientation of its phenyl ring controls the opening/closing of the bottleneck (5,7,15,24,25). F330 has also been shown to adopt a spectrum of conformations in various ligand/AChE complexes (see the overlay representation in Fig. 7 in the work of Kryger, Silman, and Sussman (16)). Another aromatic residue that deserves attention is W84, which is the main contributor to the stabilization of the substrate in the active site. An early MD simulation of TcAChE suggested that a conformational change in the orientation of its indole ring could result in the transient opening of a short channel connecting the interior of the gorge with the bulk solvent (namely the backdoor) through which solvent, and perhaps substrate and products, might enter or leave the active site (4,26). Other possible back and side doors to the active-site gorge have also been suggested on the basis of theoretical studies (27–29). Reexamination of the Drosophila melanogaster AChE structure (30) by Nachon et al. (31) revealed that a flip in W83 (equivalent to W84 in TcAChE) had generated a channel ∼5 Å in diameter between W83, W472, and D482. Although various lines of experimental evidence have been produced both for (32,33) and against a functional backdoor (34,35), none of this evidence can be considered decisive.
FIGURE 7.
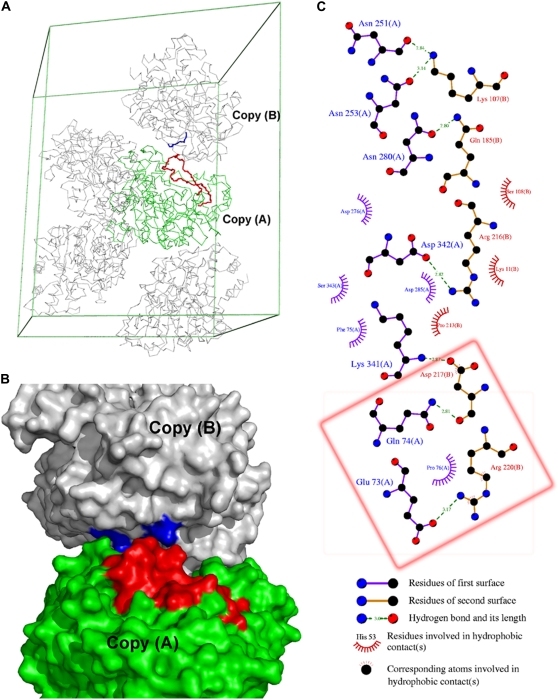
(A) Packing diagram of TcAChE (PDB code 1ea5). The backbones of the C67–C94 Ω-loop in copy A, and of the R216-R222 loop in copy B, are shown in red and blue, respectively. (B) Space-filling representation of two copies extracted from the packing diagram of TcAChE. The Ω-loop in copy A and residues R216–R222 in copy B are colored red and blue, respectively. (C) LIGPLOT (61) representation of the interactions between the two copies. The red box highlights the specific interactions between the Ω-loop in copy A and residues R216–R222 in copy B.
In this study, the side-chain flexibility of the 14 conserved aromatic residues in the active-site gorge of TcAChE is examined in the 47 available crystal structures of TcAChE and compared to those produced in a 20-ns MD simulation using the native enzyme as a starting model. The data presented emphasize the importance of taking into account side-chain flexibility in evaluating protein-ligand interactions. The data also suggest that MD simulation is an appropriate method for studying side-chain conformations, especially of proteins for which multiple crystal structures are not yet available (36).
METHODS
MD simulations
The monomer of native TcAChE, determined at 1.8 Å resolution (3), was used as the starting structure for the simulations (PDB code 1ea5). In brief, the protein, together with crystal water molecules, was inserted into a box of dimensions 10.6 × 10.6 × 10.6 nm3, with the minimal distance of the protein from its walls being 2.0 nm. This box was solvated by use of a simple point charge water model (37), and the resulting solvated box was then submitted to energy minimization. Counterions were subsequently added to provide a neutral simulation system. Energy minimization was then repeated on the entire system. After convergence had been reached, the solvent, the counterions, and the protein were coupled separately to a temperature bath at 300 K. The entire system was then equilibrated for 20 ns. To eliminate the influence of the main-chain movement on the side-chain conformational changes, a second 20-ns MD trajectory was run for the simulation system with a force restraint of 1000 kcal/mol imposed on all the main-chain atoms throughout the simulation time.
MD simulations were carried out with the GROMACS package (38,39), using NPT and periodic boundary conditions. The GROMOS96 force field (40) was applied to the protein. The pressure was kept constant at 1 bar by coupling to a Berendsen barostat with τp = 1.0 ps and a compressibility of 4.5 × 10−5 bar−1 (41). The temperature was maintained at 300 K by coupling to a Berendsen thermostat with a coupling time of τT = 0.1 ps (41). The LINCS method (42) was used to restrain bond lengths, allowing an integration step of 2 fs. The coordinates of the entire system were saved every 500 steps. Electrostatic interactions were calculated using the particle-mesh Ewald algorithm (43,44).
To assess the effect of packing restraints on the flexibility of the side chains of certain residues, MD simulations were also carried out on the physiologic dimer (PDB code 1ea5) using the same protocol.
Crystallographic data
All the side-chain conformations of the 14 aromatic residues included in the 47 3D structures of TcAChE available from the PDB were examined by calculating the χ1 (N-CA-CB-CG) and χ2 (CA-CB-CG-CD1) angles for each residue. The structures included native data sets (PDB codes 1ea5, 1qid, 1w75, and 2ace), complexes with reversibly bound ligands (PDB codes 1acj, 1acl, 1amn, 1ax9, 1dx6, 1e3q, 1e66, 1eve, 1fss, 1gpk, 1gpn, 1gqs, 1h22, 1h23, 1hbj, 1jjb, 1odc, 1qti, 1u65, 1ut6, 1vot, 1w4l, 1w6r, 1w76, 1zgb, 1zgc, 2ack, 2c5g, 2cek, 2ckm, 2cmf, and 2j4f), and covalent conjugates (PDB codes 1cfj, 1gqr, 1oce, 1som, 1vxo, 1vxr, 2bag, 2c4h, 2c5f, 2c58, and 2dfp). To avoid introducing bias via redundant entries, only one of the nine structures produced in the radiation damage study on TcAChE (45) was taken into account in the analysis, namely 1qid. Three crystal structures of recombinant mouse AChE (mAChE), 1q83, 2gyu, and 2gyv, were used to calculate the χ1 and χ2 angles of W286 (homologous to W279 in TcAChE).
RESULTS AND DISCUSSION
To our knowledge, 97 crystal structures of AChE are available from the PDB to date. The majority are for TcAChE (55 structures) and for mAChE (34 structures), as well as 2, 3, and 3 structures, respectively, for human, Electrophorus electricus, and D. melanogaster AChEs. Because the side-chain conformations observed for the 14 aromatic residues in the ensemble of TcAChE structures cover almost the entire range of conformations that occur in the structures of the AChEs of the other species and to eliminate any possible effects of differences in amino acid sequence, only the 47 crystal structures of TcAChE were used for the side-chain flexibility analysis of these residues.
The dihedral angles of an aromatic residue, χ1 and χ2, suffice to describe its side-chain conformation; hence, χ1/χ2 plots provide a simple representation of the shuffling between aromatic side-chain conformations along an MD trajectory. The χ1 and χ2 angles of the 14 conserved aromatic residues lining the active-site gorge of TcAChE were extracted from the MD simulation performed for the native structure, 1ea5, and plotted together with the experimental values obtained from the crystallographic data (Fig. 1). The black triangles represent the conformations observed in the crystal structures, whereas the gray dots are derived from the MD trajectory. Because our protocol involved sampling at 1-ps intervals, there are 20,000 such points in each representation. For most of the residues, though not for all, the side-chain conformational space explored by the MD simulation encompasses the space covered by the crystallographic data. Conversely, much of the conformational space explored by the simulation is devoid of experimental points.
FIGURE 1.
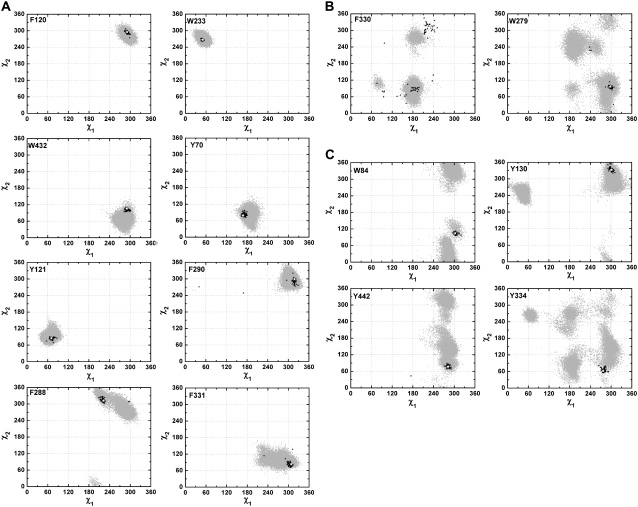
χ1/χ2 plots for the 14 aromatic residues lining the active-site gorge of TcAChE. (A) Group I: F120, W233, W432, Y70, Y121, F290, F288, and F331; (B) Group II: F330 and W279; (C) Group III: W84, Y130, Y442, and Y334. The gray dots are derived from a 20-ns MD trajectory of native TcAChE. Each one represents a pair of χ1 and χ2 angles calculated based on snapshot structures of TcAChE extracted from the MD trajectory at 1-ps intervals. Thus, each plot contains 20,000 such dots. The white pentacle is the crystal structure (PDB code 1ea5) used for the MD simulation. The dark triangles present pairs of χ1 and χ2 angles calculated based on 47 crystal structures of TcAChE deposited in the PDB.
The principal conclusions to be drawn from the data displayed in Fig. 1 are as follows. For most of the residues, the distribution of the black triangles corresponding to the experimentally determined angles is quite narrow; thus, their side-chain conformations in the various crystal structures, whether of native TcAChE or of its complexes and conjugates, are very similar. Considering that ∼80% of the crystal structures are of complexes or conjugates, this result indicates that the binding of a ligand to the enzyme has only subtle effects on the side-chain conformations of most of the aromatic residues lining the active-site gorge
With respect to the flexibility of their side chains, the 14 aromatic residues can be divided into three groups. Fig. 1 A shows that residues F120, W233, W432, Y70, Y121, F331, F288, and F290 (group I) fall into a category for which both the crystallographic data and MD simulation trajectory indicate very low flexibility. In contrast, for F330 and W279 (Fig. 1 B, group II), the MD trajectory and the crystallographic data indicate high flexibility of the side chains. The third category (Fig. 1 C, group III) includes W84, Y130, Y442, and Y334, whose side chains appear being quite flexible in the MD simulations, even though no substantial flexibility is evident in the crystal structures.
Group I
Fig. 1 A clearly shows that the side-chain conformations of the aromatic residues in group I are, in general, quite fixed. With the exception of F288, F290, and F331, all the experimental and MD points are concentrated in a specific region. For each of these latter residues, there are two aberrant experimental data sets. The two aberrant points in the plots for F288 and F290 correspond to PDB entries 2dfp and 2cmf, whereas those in the plot for F331 correspond to PDB entries 2dfp and 2cek. In both the 2dfp and 2cmf structures, the W279–S291 loop undergoes a substantial conformational rearrangement, due either to the modification of the catalytic S200 by an organophosphate with a bulky alkyl leaving group (2dfp) or to the binding of a bis-tacrine ligand with a short linker between the two tacrines (2cmf). In both cases, F288 and F290 rearrange and adopt side-chain conformations very different from their native ones (23,46). The conformational change observed for F288 in the 2dfp structure disrupts the π–π stacking interaction between the phenyl rings of F288 and F331. As a consequence, it increases the flexibility of the side chain of F331, which thus adopts an orientation different from its native one. In the 2cmf structure, the side-chain conformation of F331 is similar to its native one, yet a backbone movement is observed. Finally, there are two conformations for F330 and F331 in the 2cek structure (22). In one conformation, the F330 ring is stacked between the aromatic moiety of the ligand and F331, just as in the 2ckm structure (23). In the second conformation, F330 has moved away from the aromatic moiety of the ligand, and the aromatic ring of F331 assumes an unusual conformation.
Group II
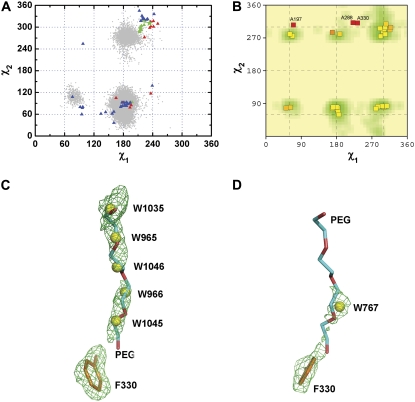
This group includes the two mobile residues W279 and F330. Figs. 1 B and 2 A show that the distribution of the experimental χ1/χ2 data points for F330 is very broad, clearly demonstrating an inherent flexibility of its side chain. Use of the protein structure validation program PROCHECK (47,48) to examine the allowed χ1/χ2 conformational angles reveals that a number of the data sets are clustered in one favored region. However, in many others, including all the native structures except 2ace, F330 adopts various unfavored side-chain conformations (Fig. 2, A and B). The unfavored side-chain conformations of F330 in the complex and conjugate structures can be ascribed to steric restrictions imposed by the respective ligands or covalently attached groups. But the cause of F330 adopting unfavored side-chain orientations in the three native structures, 1qid, 1ea5, and 1w75, was not immediately apparent. We earlier reported that continuous electron density observed within the active-site gorge in certain data sets collected for native TcAChE (49) and for complexes (50) could be ascribed to the polyethylene glycol 200 (PEG200), which had been used as a precipitant. Careful reexamination of the electron density maps of the four native structures revealed that the three structures in which F330 adopts an unfavored side-chain conformation all contain a PEG200 molecule bridging the PAS and the CAS, with the proximal tip of the bound PEG200 pushing the side chain of F330 into the unfavored position (Fig. 2 C). In the 2ace structure (Fig. 2 D), no PEG200 molecule can be seen, allowing the F330 side chain to assume a favored orientation. The 2ace data set was collected at an earlier time than the others, using a different batch of PEG200. Because PEG200 is a heterogeneous polymer, a difference in composition may explain the apparent discrepancy with the other native data sets.
FIGURE 2.
(A) χ1/χ2 plots for F330. The gray dots are derived from a 20-ns MD trajectory of native TcAChE as in Fig. 1. The green pentacle is the crystal structure (PDB code 1ea5) used for the MD simulation. The green, red, and blue triangles represent, respectively, the native structures, covalent conjugates, and reversible complexes obtained from the crystallographic data. (B) The χ1/χ2 coordinates of all Phe residues in TcAChE (PDB code 1ea5) overlaid on the favored regions for Phe derived using the program PROCHECK (47,48). Each square represents one residue. The color coding of the squares from yellow to red reflects the degree to which the orientation is favored; thus, yellow and red indicate that the residue is in a favored or an unfavored region, respectively. The residue numbers adjacent to the red squares, namely A197, A288, and A330 indicate that these three Phe residues reside in highly unfavored regions. The intensity of the green shading indicates how favored each region on the plot is—the darker the shading, the more favored the region. These assignments are based on a data set of 163 nonhomologous, high-resolution protein chains chosen from structures solved by x-ray crystallography to a resolution of 2.0 Å or better, and an R-factor no greater than 20% (47,48). (C and D) Difference electron density maps contoured at 3.0 σ, of the side chain of F330 and of neighboring water molecules, based on the 1ea5 (C) and 2ace (D) crystal structures of native TcAChE. The side chain of F330 and PEG200 are displayed as stick models; oxygen atoms of water molecules are displayed as yellow balls.
The MD simulation predicts three principal conformational states for the F330 side chain, also indicating that it is flexible. However, even though the starting side-chain conformation of F330 is unfavored, most of the points sampled by the MD simulation are located in favored regions. Because no bound PEG 200 is included in the simulation system, F330 can easily switch its side chain from an unfavored to a favored conformation. Because many of the experimental data points are located in unfavored regions, whereas most of the simulation points are in favored regions, the side-chain conformations observed for F330 in the MD simulation, unlike for other residues, do not encompass all the conformational possibilities revealed by the crystal structures.
F330 is situated on an α-helix in the middle of the active-site gorge and is one of two residues that form the bottleneck of the gorge. This location permits it to interact with ligands bound at the CAS and/or at the PAS. The crystal structures of the various complexes reveal that F330 always adopts an appropriate side-chain conformation that allows it to make π–π stacking and/or π-cation interactions with the bound ligand, resulting in the diverse repertoire of side-chain conformations observed. F330 and Y121 form the bottleneck of the gorge; therefore, changes in the conformations of their side chains influence its opening and closing. Fig. 1 A shows that the side-chain conformation of Y121 is quite fixed in all the crystal structures. This observation implies that it is the flexibility of the side chain of F330 that permits ligands to enter or leave the CAS. MD simulations of TcAChE in its native and complexed/conjugated states do, indeed, show that F330 is flexible, that the conformational switch of its side chain can control the opening of the bottleneck, and that it thus has the capacity to play a key role in both ligand binding and release (7,15,18,25). Moreover, as discussed above, a conformational change in F330 may affect the side-chain orientation of the adjacent residue F331. The side-chain reorientation of F331 may, in turn, affect the conformation of F288, as is seen in the structure of TcAChE complexed with a bis-tacrine containing a shorter linker (PDB code 2cmf) (23), also referred to above. Hence, the flexibility of F330 may induce other residues along the active-site gorge to change their conformations.
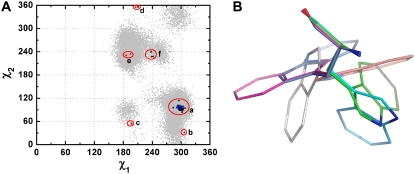
The side-chain conformations of W286 (corresponding to W279 in TcAChE) in the mAChE structures 1q83, 2gyu and 2gyv were not included in Fig. 1. The χ1 and χ2 angles of W286 in those three mAChE structures are, however, included with those for W279 in the χ1/χ2 plot shown in Fig. 3 A. As for F330, the experimental data points for W279 (and for mouse W286) are quite dispersed. To analyze these conformations in more detail, the χ1 and χ2 values obtained from the crystallographic data were grouped into six categories on the basis of their orientations (Fig. 3 A) (51). Group a includes the majority of the crystal structures, including that of native TcAChE. Group b is the single tacrine/TcAChE data set (PDB code 1acj) (5). Group c represents the complex of syn1 TZ2PA6/mAChE (PDB code 1q83) (19). There are two data points, one for each of the monomers in the dimer in 1q83. Groups d and e include the structures of mAChE complexed with the oximes HI-6 (PDB code 2gyu) (20) and Ortho-7 (PDB code 2gyv) (20), respectively. In both these cases, too, there are separate data points for each monomer in the dimer, as was the case for 1q83. Group f includes complexes of TcAChE with bis-tacrine derivatives (PDB codes 1odc (23), 2cek (22), and 2ckm (23)). If one side-chain conformation of W279 is selected from each group and fitted onto the backbone atoms of the residue, their superposition (Fig. 3 B) reveals that the six side-chain conformations span almost all the entire space (0–360°) around the Cα-Cβ bond. Thus, the flexibility of the side chain of W279 is very high. It is noteworthy that a standard χ1,χ2 rotamer plot (47,48) of tryptophan would miss several of these groups, in particular group f (51).
FIGURE 3.
(A) χ1/χ2 plot for TcAChE W279 and mAChE W286. The gray dots are derived from a 20-ns MD trajectory of native TcAChE as in Fig. 1. The blue pentacle corresponds to the conformation of W279 in the crystal structure (PDB code 1ea5) used as a starting model for the MD simulations. The dark triangles represent all the crystal structures of TcAChE; the red triangles represent those of mAChE (PDB codes 1q83, 2gyu, and 2gyv). (B) The snapshots of W279 (286) (fitted onto the backbone of the residue) corresponding to the six groups highlighted in A: (a) 1ea5 (green); (b) 1acj (cyan); (c) 1q83 (magenta); (d) 2gyu (gray); (e) 2gyv (blue); and (f) 2ckm (orange).
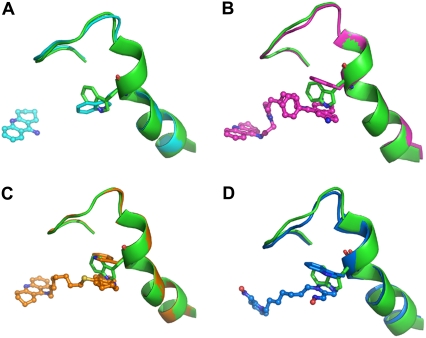
We selected several representative structures and aligned each of them with native TcAChE to illustrate how the conformation of the protein around W279, at the PAS, changes as a result of ligand binding (Fig. 4). Fig. 4 A shows the monofunctional inhibitor tacrine bound at the CAS of TcAChE. The orientation of the side chain of W279 is different from its position in the native data set. Fig. 4 B presents the inhibitor that had been shown to be synthesized within the active-site gorge by “click chemistry” (52); as might be expected, the inhibitor spans the CAS and the PAS of mAChE. Fig. 4 C shows a bis-tacrine derivative, NF595, with an 8-carbon spacer, which spans the CAS and the PAS of TcAChE. Fig. 4 D shows an example of a bifunctional oxime, Ortho-7, which also spans the two anionic sites of mAChE. Both the three structures with the bifunctional ligands and the one with tacrine only bound at the CAS show that only the side-chain conformation of W279/286 changes relative to the native structure, whereas the main chain maintains its position. Furthermore, in all three bifunctional complexes, the bound inhibitor makes a π–π stacking and/or a π-cation interaction with W279/286. It may be asked whether the conformations of the side chain of W279/286 observed in these complexes is generated de novo by ligand binding or represents a preexisting conformation present at low occupancy in the native enzyme and stabilized by interaction with the ligand. We consider the second possibility to be the plausible one for the following reasons. In the tacrine complex, the inhibitor does not interact with W279, but, nevertheless, its side-chain conformation changes (Fig. 4 A). This indicates that the side chain of this residue is flexible even in the absence of a directly interacting ligand. In the complexes with the inhibitors that span the CAS and PAS, the aromatic rings of the inhibitors always occupy a position occupied by the side chain of W279 in the native structure (Fig. 4, B–D). Because no large conformational change occurs in the vicinity of W279, its side chain must move away from its native position for the inhibitor to occupy the corresponding space. As shown in Fig. 3 A, the areas generated by the gray dots derived from the MD simulation of native TcAChE encompass all but one group (group d) of the experimental data points. This also suggests that the side-chain conformations of W279 observed in the various crystal structures preexist in dynamic equilibrium at differing occupancies in the native enzyme. Therefore, conformational changes of W279 upon ligand-binding involve preexisting equilibrium dynamics (51). The specific side-chain conformation of W279 preexists in the enzyme, and the inhibitor binds to the enzyme and stabilizes this specific conformation of W279 by π–π stacking and/or π-cation interactions. W279 is the main contributor to the stabilization of substrates and products at the PAS; hence, its preequilibrium dynamics are relevant to trafficking of substrate and product along the active-site gorge. The fact that F330, at the bottleneck of the gorge, and W279, at its rim, both display high conformational flexibility suggests that they play a dynamical role in the processes of attracting substrate to, and ejecting products from, the active site; their aromatic side chains may serve as relay stations interacting with the substrate/product via cation-π interactions.
FIGURE 4.
Structural alignments of 1acj (A, cyan), 1q83 (B, magenta), 2cek (C, orange), and 2gyv (D, blue) with 1ea5 (green). Only the loop corresponding to residues 270–290 is displayed. W279 is shown as a stick model; the inhibitors are shown as ball-and-stick models.
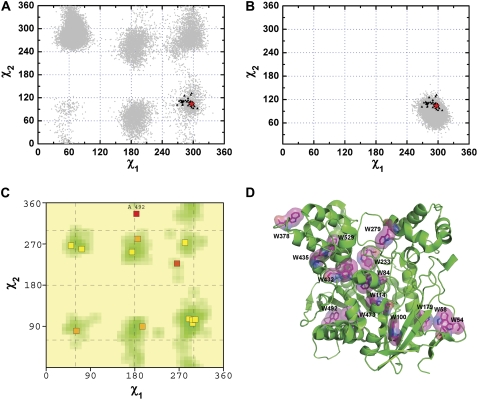
The side-chain flexibilities of all the Trp residues in TcAChE, not only those within the active-site gorge, were also analyzed on the basis of the MD trajectory. For most of them, the side-chain conformations are stable, and the χ1 and χ2 values obtained from the MD trajectory are in the vicinity of the starting crystal structure (data not shown). The residue with the most flexible side chain is W378 (Fig. 5 A). The data derived from the MD trajectory of the TcAChE monomer for this residue cover almost all six favorable regions for the Trp side chain yielded by PROCHECK, but the experimental data points are confined to only one of these regions (Fig. 5, A and C). W378 is located on a flexible loop with its side chain exposed to the solvent (Fig. 5 D). However, examination of the structure of the physiologic dimer shows that it is at the interface of the two monomers. Consequently, an MD simulation of the TcAChE dimer was performed. As Fig. 5 B shows, the theoretical points derived from the 20-ns MD trajectory of the dimer encompass the experimental points derived from the crystal structures. Thus, the MD simulation performed for the dimer accurately represents the true repertoire of conformations that the side chain of W378 can assume, whereas the MD simulation performed for the monomer does not.
FIGURE 5.
(A) χ1/χ2 plots for W378. The gray dots are derived from a 20-ns MD trajectory for the TcAChE monomer as in Fig. 1. The red pentacle is the crystal structure (PDB code 1ea5) used for MD simulation. The dark triangles present pairs of χ1 and χ2 angles calculated based on 47 crystal structures of TcAChE deposited in the PDB. (B) χ1/χ2 plots for W378. Details are all the same as in A except that the gray dots are derived from a 20-ns MD trajectory for the TcAChE dimer. (C) The χ1/χ2 positions of all Trp residues of TcAChE (PDB code 1ea5) overlaid on the favored regions for Trp derived using PROCHECK (47,48); details are the same as in Fig. 2 B. (D) The positions of the 14 Trp residues in native TcAChE (PDB code 1ea5). All Trp residues are shown as stick models enveloped by their van der Waals surfaces.
Group III
This group includes four residues: W84, Y130, Y442, and Y334. In this group, the side chains in the crystal structures occupy fixed positions, whereas the MD simulation exhibits diverse side-chain conformations (Fig. 1 C). Because W84 is the key component of the CAS and changes in the conformation of its side chain have been invoked to control the opening of the backdoor at the bottom of the active-site gorge (4,26), we focused our attention on this residue. As already noted, the χ1/χ2 plot for W84 shown in Fig. 1 C reveals that its side-chain conformation is constant in all the crystal structures, with no opening of the putative backdoor being observed. However, in the 20-ns MD trajectory, the side chain of W84 displays considerable flexibility. Whereas the χ1 angles are very similar and do not deviate much from those in the crystal structures, large fluctuations in the χ2 angles are observed. This large fluctuation in the χ2 angles makes possible a movement of the indole ring that would result in partial opening of the backdoor.
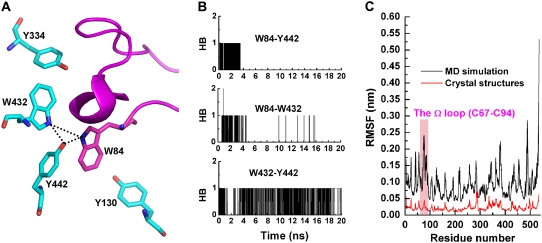
How can the flexibility of the side chain of W84 in the MD trajectory be reconciled with its fixed conformation in the crystal structures? Fig. 6 A shows that the indole is hydrogen-bonded to W432 and Y442 in the starting structure for the MD simulation. These two hydrogen bonds are only maintained during the first ∼4 ns of the 20-ns MD trajectory, whereas the hydrogen bond between W432 and Y442 is maintained throughout almost the entire trajectory (Fig. 6 B). Breaking its hydrogen bonds with W432 and Y442 would obviously give W84 more freedom to change its side-chain orientation. Breakage of these bonds might result from a conformational rearrangement of the C67–C94 Ω-loop. The root mean-square fluctuation (RMSF) of the Cα atoms in this loop during the 20-ns MD simulation was ∼2.5 Å, compared to an average RMSF value of 0.9 Å for the Cα atoms of the entire subunit (Fig. 6 C), indicating that it is relatively mobile. Such large movements of the Ω-loop would not only cause W84 to adopt different side-chain orientations but might affect the side-chain conformations of the other four aromatic residues with which it is interacting, namely Y130, Y334, W432, and Y442. However, in all the crystal structures, not only the side-chain of W84, but also the conformation of the entire Ω-loop is quite fixed. This is because the RMSF of its Cα atoms for the 47 TcAChE crystal structures is 0.2 Å, the same value as obtained for the whole polypeptide (Fig. 6 C). We considered the possibility that this might be due to restraints imposed by crystal packing. Native TcAChE crystallizes in three space groups; there are actually 42 data sets in P3121, 3 data sets in P212121, and a single data set in P3221. In all three, the Ω-loop is in contact with a symmetry-related copy of the enzyme. In the complex of TcAChE with the snake venom toxin fasciculin II (PDB code 1fss), which crystallizes in space group P21212, the Ω-loop interacts with the toxin. Fig. 7 shows how, in the 1ea5 crystal structure, which is in the P3121 space group, the Ω-loop in one copy interacts with the R216-R220 loop in one of the symmetry-related copies. E73 in copy A forms a salt bridge with R220 in copy B; Q74 in copy A forms a hydrogen bond to D217 in copy B; and P76 in copy A makes hydrophobic interactions with R220 in copy B (Fig. 7 C). These interactions, all of which help stabilize the conformation of the Ω-loop in the crystal structures, were not taken into account in the MD simulations because only one copy of the enzyme was used. Thus, in solution, the Ω-loop may be quite flexible, as may be the side chains of W84 and those of Y130, Y334, W432, and Y442. Indeed, using site-directed labeling in conjunction with time-resolved fluorescence anisotropy, Shi et al. (53) reported that the Ω-loop residues are significantly more mobile than nonloop residues facing the interior of the gorge, and so they are likely to contribute to transient gorge enlargements in the nonliganded enzyme.
FIGURE 6.
(A) Aromatic residues, shown as stick models, located in and around the C67–C94 Ω-loop. Hydrogen bonds are shown as dashed lines. The 1ea5 structure was used. (B) H-bond formation and cleavage between W84 and Y442, W84 and W432, and W432 and Y442. (C) The RMSF of Cα atoms of TcAChE calculated based on the 20-ns MD trajectory (black) and 47 crystal structures (red). The region of the Ω-loop (C67–C94) is highlighted in magenta.
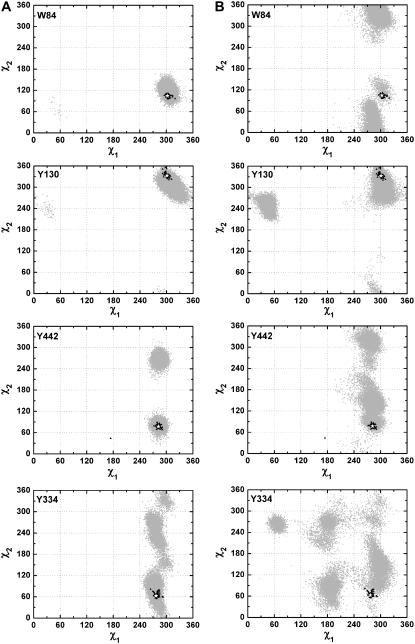
To evaluate whether overall movements of the Ω-loop affect the mobility of the side chains of W84, Y130, Y442, and Y334, a second 20-ns MD simulation of the monomer was performed. This time, the simulation was performed with the application of force restraints to the main-chain atoms. The χ1/χ2 plots for these four residues obtained using this trajectory are shown in Fig. 8 A. Clearly, the observed side-chain conformations of W84 are more restricted than when the main chain is not restrained (Figs. 1 and 8 B), and they agree well with the crystallographic data. The side-chain flexibility of the other three residues is also substantially reduced under the new simulation conditions (Fig. 8). Therefore, the discrepancies in side-chain flexibility of the residues in group III revealed by the MD simulation and by the crystal structures can be principally ascribed to the restricted mobility of the Ω-loop in the crystal structures due to the effects of crystal packing.
FIGURE 8.
(A) χ1/χ2 plots for W84, Y130, Y442, and Y334 generated from a 20-ns MD simulation of the native TcAChE monomer, applying force restraints to the main chain atoms. (B) χ1/χ2 plots for W84, Y130, Y442, and Y334, extracted from Fig. 1 C, generated from the 20-ns MD trajectory performed without any force restraints. In both cases, the native data set used was PDB code 1ea5.
We were curious to know if there were significant correlations between conformational switches from one side-chain conformation to another. Accordingly, we performed principal component analysis (54) based on the coordinates of all 14 aromatic side chains, which were generated by the MD simulation with 1-ps intervals. Analysis of these results showed no evident correlation between side-chain flexibilities.
CONCLUSIONS
The χ1 and χ2 analysis of side-chain conformations, based on the ensemble of crystal structures and on the MD trajectories, permits classification of the 14 conserved aromatic residues in the active-site gorge of TcAChE into three groups. F120, W233, W432, Y70, Y121, F331, F288, and F290 fall into group I, which is the category of residues with fixed side-chain conformations. In contrast, the experimental and the MD data show that the side chains of F330 and W279 (group II) are very flexible. For F330, certain alternate conformations can be ascribed to interactions with the ligands. However, the alternative conformations observed in some native data sets appear to be due to the presence of the precipitant PEG200 in the active-site gorge. The side chains of W84, Y130, Y442, and Y334 (group III) display fixed side-chain conformations in the crystal structures, but they appear to be quite mobile in the MD simulations. This apparent discrepancy can be ascribed to crystal packing restraints.
The mobility of W279 revealed by the MD simulations, taken together with the crystallographic data, suggest that it may occur in a repertoire of conformations, with varying degrees of occupancies in the native enzyme, and that a given ligand may select one of these preexisting conformations preferentially. AChE has been shown to accelerate aggregation of the Aβ peptide, a process thought to be intimately involved with the progression of Alzheimer disease (55,56); this acceleration appears to involve the PAS, of which W279 is the key element (57,58). Many second-generation cholinesterase inhibitors for the treatment of Alzheimer disease are thus compounds that span the CAS and PAS (23,59,60). Structure-based drug design in this context, therefore, should explore the landscape of conformations revealed by MD simulations and not be confined to docking to a single average native structure. This concept should be of general applicability to structure-based drug design.
Acknowledgments
This work was supported by the Commissariat à l'Énergie Atomique, Centre National de la Recherche Scientifique, Université Joseph Fourier, National Institutes of Health CounterACT program, U.S. Army Defense Threat Reduction Agency, the European Commission Sixth Framework Research and Technological Development Program ‘SPINE2-COMPLEXES’ Project, under contract No. LSHG-CT-2006-031220, and ‘Teach-SG’ Project, under contract No. ISSG-CT-2007-037198, Nalvyco Foundation, and Kimmelman Center for Biomolecular Structure and Assembly. M.W. was supported by a grant from the Agence Nationale de la Recherche (project number JC05_45685). J.P.C. was supported by an EMBO short-term fellowship (ASTF230-2006). I.S. was supported by the Benoziyo Center for Neuroscience. J.L.S. is the Pickman Professor of Structural Biology.
Yechun Xu and Jacques-Philippe Colletier contributed equally to this work.
Editor: Ron Elber.
References
- 1.Rosenberry, T. L. 1975. Acetylcholinesterase. Adv. Enzymol. Relat. Areas Mol. Biol. 43:103–218. [DOI] [PubMed] [Google Scholar]
- 2.Silman, I., and J. L. Sussman. 2005. Acetylcholinesterase: ‘classical’ and ‘non-classical’ functions and pharmacology. Curr. Opin. Pharmacol. 5:293–302. [DOI] [PubMed] [Google Scholar]
- 3.Sussman, J. L., M. Harel, F. Frolow, C. Oefner, A. Goldman, L. Toker, and I. Silman. 1991. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science. 253:872–879. [DOI] [PubMed] [Google Scholar]
- 4.Gilson, M. K., T. P. Straatsma, J. A. McCammon, D. R. Ripoll, C. H. Faerman, P. H. Axelsen, I. Silman, and J. L. Sussman. 1994. Open “back door” in a molecular dynamics simulation of acetylcholinesterase. Science. 263:1276–1278. [DOI] [PubMed] [Google Scholar]
- 5.Harel, M., I. Schalk, L. Ehret-Sabatier, F. Bouet, M. Goeldner, C. Hirth, P. H. Axelsen, I. Silman, and J. L. Sussman. 1993. Quaternary ligand binding to aromatic residues in the active-site gorge of acetylcholinesterase. Proc. Natl. Acad. Sci. USA. 90:9031–9035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harel, M., D. M. Quinn, H. K. Nair, I. Silman, and J. L. Sussman. 1996. The X-ray structure of a transition state analog complex reveals the molecular origins of the catalytic power and substrate specificity of acetylcholinesterase. J. Am. Chem. Soc. 118:2340–2346. [Google Scholar]
- 7.Colletier, J. P., D. Fournier, H. M. Greenblatt, J. Stojan, J. L. Sussman, G. Zaccai, I. Silman, and M. Weik. 2006. Structural insights into substrate traffic and inhibition in acetylcholinesterase. EMBO J. 25:2746–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourne, Y., Z. Radic, G. Sulzenbacher, E. Kim, P. Taylor, and P. Marchot. 2006. Substrate and product trafficking through the active center gorge of acetylcholinesterase analyzed by crystallography and equilibrium binding. J. Biol. Chem. 281:29256–29267. [DOI] [PubMed] [Google Scholar]
- 9.Szegletes, T., W. D. Mallender, and T. L. Rosenberry. 1998. Nonequilibrium analysis alters the mechanistic interpretation of inhibition of acetylcholinesterase by peripheral site ligands. Biochemistry. 37:4206–4216. [DOI] [PubMed] [Google Scholar]
- 10.Szegletes, T., W. D. Mallender, P. J. Thomas, and T. L. Rosenberry. 1999. Substrate binding to the peripheral site of acetylcholinesterase initiates enzymatic catalysis. Substrate inhibition arises as a secondary effect. Biochemistry. 38:122–133. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, J. L., B. Cusack, M. P. Davies, A. Fauq, and T. L. Rosenberry. 2003. Unmasking tandem site interaction in human acetylcholinesterase. Substrate activation with a cationic acetanilide substrate. Biochemistry. 42:5438–5452. [DOI] [PubMed] [Google Scholar]
- 12.Morel, N., S. Bon, H. M. Greenblatt, D. Van Belle, S. J. Wodak, J. L. Sussman, J. Massoulié, and I. Silman. 1999. Effect of mutations within the peripheral anionic site on the stability of acetylcholinesterase. Mol. Pharmacol. 55:982–992. [DOI] [PubMed] [Google Scholar]
- 13.Harel, M., J. L. Sussman, E. Krejci, S. Bon, P. Chanal, J. Massoulié, and I. Silman. 1992. Conversion of acetylcholinesterase to butyrylcholinesterase: modeling and mutagenesis. Proc. Natl. Acad. Sci. USA. 89:10827–10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Botti, S. A., C. E. Felder, S. Lifson, J. L. Sussman, and I. Silman. 1999. A modular treatment of molecular traffic through the active site of cholinesterase. Biophys. J. 77:2430–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu, Y., J. Shen, X. Luo, I. Silman, J. L. Sussman, K. Chen, and H. Jiang. 2003. How does huperzine A enter and leave the binding gorge of acetylcholinesterase? Steered molecular dynamics simulations. J. Am. Chem. Soc. 125:11340–11349. [DOI] [PubMed] [Google Scholar]
- 16.Kryger, G., I. Silman, and J. L. Sussman. 1999. Structure of acetylcholinesterase complexed with E2020 (Aricept): implications for the design of new anti-Alzheimer drugs. Structure. 7:297–307. [DOI] [PubMed] [Google Scholar]
- 17.Greenblatt, H. M., H. Dvir, I. Silman, and J. L. Sussman. 2003. Acetylcholinesterase: a multifaceted target for structure-based drug design of anticholinesterase agents for the treatment of Alzheimer's disease. J. Mol. Neurosci. 20:369–383. [DOI] [PubMed] [Google Scholar]
- 18.Niu, C., Y. Xu, X. Luo, W. Duan, I. Silman, J. L. Sussman, W. Zhu, K. Chen, J. Shen, and H. Jiang. 2005. Dynamic mechanism of E2020 binding to acetylcholinesterase: a steered molecular dynamics simulation. J. Phys. Chem. B. 109:23730–23738. [DOI] [PubMed] [Google Scholar]
- 19.Bourne, Y., H. C. Kolb, Z. Radic, K. B. Sharpless, P. Taylor, and P. Marchot. 2004. Freeze-frame inhibitor captures acetylcholinesterase in a unique conformation. Proc. Natl. Acad. Sci. USA. 101:1449–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ekstrom, F., Y. P. Pang, M. Boman, E. Artursson, C. Akfur, and S. Borjegren. 2006. Crystal structures of acetylcholinesterase in complex with HI-6, Ortho-7 and obidoxime: structural basis for differences in the ability to reactivate tabun conjugates. Biochem. Pharmacol. 72:597–607. [DOI] [PubMed] [Google Scholar]
- 21.Wong, D. M., H. M. Greenblatt, H. Dvir, P. R. Carlier, Y. F. Han, Y. P. Pang, I. Silman, and J. L. Sussman. 2003. Acetylcholinesterase complexed with bivalent ligands related to Huperzine A: experimental evidence for species-dependent protein-ligand complementarity. J. Am. Chem. Soc. 125:363–373. [DOI] [PubMed] [Google Scholar]
- 22.Colletier, J. P., B. Sanson, F. Nachon, E. Gabellieri, C. Fattorusso, G. Campiani, and M. Weik. 2006. Conformational flexibility in the peripheral site of Torpedo californica acetylcholinesterase revealed by the complex structure with a bifunctional inhibitor. J. Am. Chem. Soc. 128:4526–4527. [DOI] [PubMed] [Google Scholar]
- 23.Rydberg, E. H., B. Brumshtein, H. M. Greenblatt, D. M. Wong, D. Shaya, L. D. Williams, P. R. Carlier, Y. P. Pang, I. Silman, and J. L. Sussman. 2006. Complexes of alkylene-linked tacrine dimers with Torpedo californica acetylcholinesterase: binding of bis5-tacrine produces a dramatic rearrangement in the active-site gorge. J. Med. Chem. 49:5491–5500. [DOI] [PubMed] [Google Scholar]
- 24.Tai, K., T. Shen, U. Borjesson, M. Philippopoulos, and J. A. McCammon. 2001. Analysis of a 10-ns molecular dynamics simulation of mouse acetylcholinesterase. Biophys. J. 81:715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen, T., K. Tai, R. H. Henchman, and J. A. McCammon. 2002. Molecular dynamics of acetylcholinesterase. Acc. Chem. Res. 35:332–340. [DOI] [PubMed] [Google Scholar]
- 26.Axelsen, P. H., M. Harel, I. Silman, and J. L. Sussman. 1994. Structure and dynamics of the active site gorge of acetylcholinesterase: synergistic use of molecular dynamics simulation and X-ray crystallography. Protein Sci. 3:188–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovach, I. M., N. Qian, and A. Bencsura. 1994. Efficient product clearance through exit channels in substrate hydrolysis by acetylcholinesterase. FEBS Lett. 349:60–64. [DOI] [PubMed] [Google Scholar]
- 28.Wlodek, S. T., T. W. Clark, L. R. Scott, and J. A. McCammon. 1997. Molecular dynamics of acetylcholinesterase dimer complexed with tacrine. J. Am. Chem. Soc. 119:9513–9522. [Google Scholar]
- 29.Van Belle, D., L. De Maria, G. Iurcu, and S. J. Wodak. 2000. Pathways of ligand clearance in acetylcholinesterase by multiple copy sampling. J. Mol. Biol. 298:705–726. [DOI] [PubMed] [Google Scholar]
- 30.Harel, M., G. Kryger, T. L. Rosenberry, W. D. Mallender, T. Lewis, R. J. Fletcher, J. M. Guss, I. Silman, and J. L. Sussman. 2000. Three-dimensional structures of Drosophila melanogaster acetylcholinesterase and of its complexes with two potent inhibitors. Protein Sci. 9:1063–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nachon, F., Y. Nicolet, M. Harel, T. L. Rosenberry, P. Masson, I. Silman, and J. L. Sussman. 2007. A second look at the crystal structures of Drosophila melanogaster acetylcholinesterase: evidence for backdoor opening and stabilization of an enzyme/carboxylate complex. Abstract. IXth International Meeting on Cholinesterase, Suzhou, China. p 113.
- 32.Bartolucci, C., E. Perola, L. Cellai, M. Brufani, and D. Lamba. 1999. “Back door” opening implied by the crystal structure of a carbamoylated acetylcholinesterase. Biochemistry. 38:5714–5719. [DOI] [PubMed] [Google Scholar]
- 33.Colletier, J. P., A. Royant, A. Specht, B. Sanson, F. Nachon, P. Masson, G. Zaccai, J. L. Sussman, M. Goeldner, I. Silman, D. Bourgeois, and M. Weik. 2007. Use of a ‘caged’ analogue to study the traffic of choline within acetylcholinesterase by kinetic crystallography. Acta Crystallogr. D Biol. Crystallogr. 63:1115–1128. [DOI] [PubMed] [Google Scholar]
- 34.Kronman, C., A. Ordentlich, D. Barak, B. Velan, and A. Shafferman. 1994. The “back door” hypothesis for product clearance in acetylcholinesterase challenged by site-directed mutagenesis. J. Biol. Chem. 269:27819–27822. [PubMed] [Google Scholar]
- 35.Faerman, C., D. Ripoll, S. Bon, Y. Le Feuvre, N. Morel, J. Massoulie, J. L. Sussman, and I. Silman. 1996. Site-directed mutants designed to test back-door hypotheses of acetylcholinesterase function. FEBS Lett. 386:65–71. [DOI] [PubMed] [Google Scholar]
- 36.Dooley, A. J., N. Shindo, B. Taggart, J. G. Park, and Y. P. Pang. 2006. From genome to drug lead: identification of a small-molecule inhibitor of the SARS virus. Bioorg. Med. Chem. Lett. 16:830–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, and J. Hermans. 1981. Interaction models for water in relation to protein hydration. In Intermolecular Forces. Pullman B, editor. Reidel Publishing, Dordrecht (Holland). p 331–342.
- 38.Berendsen, H. J. C., D. van der Spoel, and R. van Drunen. 1995. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 91:43–56. [Google Scholar]
- 39.Lindahl, E., B. Hess, and D. van der Spoel. 2001. Gromacs 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Model. 7:306–317. [Google Scholar]
- 40.van Gunsteren, W. F., S. R. Billeter, A. A. Eising, P. H. Hunenberger, P. Kruger, A. E. Mark, W. R. P. Scott, and I. G. Tironi. 1996. Biomolecular Simulations: The GROMOS96 Manual and User Guide. vdf Hochschulverlag, Zürich.
- 41.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, A. DiNola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- 42.Hess, B., B. Bekker, H. J. C. Berendsen, and J. G. E. M. Fraaije. 1997. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463–1472. [Google Scholar]
- 43.Darden, T., D. York, and L. Pedersen. 1993. Particle mesh Ewald: an Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. [Google Scholar]
- 44.Essmann, U., L. Perera, M. L. Berkowitz, T. Darden, H. Lee, and L. G. Pedersen. 1995. A smooth particle mesh Ewald potential. J. Chem. Phys. 103:8577–8592. [Google Scholar]
- 45.Weik, M., R. B. Ravelli, G. Kryger, S. McSweeney, M. L. Raves, M. Harel, P. Gros, I. Silman, J. Kroon, and J. L. Sussman. 2000. Specific chemical and structural damage to proteins produced by synchrotron radiation. Proc. Natl. Acad. Sci. USA. 97:623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Millard, C. B., G. Kryger, A. Ordentlich, H. M. Greenblatt, M. Harel, M. L. Raves, Y. Segall, D. Barak, A. Shafferman, I. Silman, and J. L. Sussman. 1999. Crystal structures of aged phosphonylated acetylcholinesterase: nerve agent reaction products at the atomic level. Biochemistry. 38:7032–7039. [DOI] [PubMed] [Google Scholar]
- 47.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26:283–291. [Google Scholar]
- 48.Morris, A. L., M. W. MacArthur, E. G. Hutchinson, and J. M. Thornton. 1992. Stereochemical quality of protein structure coordinates. Proteins. 12:345–364. [DOI] [PubMed] [Google Scholar]
- 49.Koellner, G., T. Steiner, C. B. Millard, I. Silman, and J. L. Sussman. 2002. A neutral molecule in a cation-binding site: specific binding of a PEG-SH to acetylcholinesterase from Torpedo californica. J. Mol. Biol. 320:721–725. [DOI] [PubMed] [Google Scholar]
- 50.Greenblatt, H. M., G. Kryger, T. Lewis, I. Silman, and J. L. Sussman. 1999. Structure of acetylcholinesterase complexed with (-)-galanthamine at 2.3 Å resolution. FEBS Lett. 463:321–326. [DOI] [PubMed] [Google Scholar]
- 51.Xu, Y., J. P. Colletier, H. Jiang, I. Silman, J. L. Sussman, and M. Weik. 2008. Induced-fit or preexisting equilibrium dynamics? Lessons from protein crystallography and MD simulations on acetylcholinesterase and implications for structure-based drug design. Protein Sci. 17:601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lewis, W. G., L. G. Green, F. Grynszpan, Z. Radic, P. R. Carlier, P. Taylor, M. G. Finn, and K. B. Sharpless. 2002. Click chemistry in situ: acetylcholinesterase as a reaction vessel for the selective assembly of a femtomolar inhibitor from an array of building blocks. Angew. Chem. Int. Ed. Engl. 41:1053–1057. [DOI] [PubMed] [Google Scholar]
- 53.Shi, J., K. Tai, J. A. McCammon, P. Taylor, and D. A. Johnson. 2003. Nanosecond dynamics of the mouse acetylcholinesterase cys69-cys96 omega loop. J. Biol. Chem. 278:30905–30911. [DOI] [PubMed] [Google Scholar]
- 54.Amadei, A., A. B. Linssen, and H. J. Berendsen. 1993. Essential dynamics of proteins. Proteins. 17:412–425. [DOI] [PubMed] [Google Scholar]
- 55.Inestrosa, N. C., A. Alvarez, C. A. Perez, R. D. Moreno, M. Vicente, C. Linker, O. I. Casanueva, C. Soto, and J. Garrido. 1996. Acetylcholinesterase accelerates assembly of amyloid-beta-peptides into Alzheimer's fibrils: possible role of the peripheral site of the enzyme. Neuron. 16:881–891. [DOI] [PubMed] [Google Scholar]
- 56.Bartolini, M., C. Bertucci, V. Cavrini, and V. Andrisano. 2003. β-Amyloid aggregation induced by human acetylcholinesterase: inhibition studies. Biochem. Pharmacol. 65:407–416. [DOI] [PubMed] [Google Scholar]
- 57.Eichler, J., A. Anselmet, J. L. Sussman, J. Massoulié, and I. Silman. 1994. Differential effects of “peripheral” site ligands on Torpedo and chicken acetylcholinesterase. Mol. Pharmacol. 45:335–340. [PubMed] [Google Scholar]
- 58.Radic, Z., R. Duran, D. C. Vellom, Y. Li, C. Cervenansky, and P. Taylor. 1994. Site of fasciculin interaction with acetylcholinesterase. J. Biol. Chem. 269:11233–11239. [PubMed] [Google Scholar]
- 59.Bolognesi, M. L., R. Banzi, M. Bartolini, A. Cavalli, A. Tarozzi, V. Andrisano, A. Minarini, M. Rosini, V. Tumiatti, C. Bergamini, R. Fato, G. Lenaz, P. Hrelia, A. Cattaneo, M. Recanatini, and C. Melchiorre. 2007. Novel class of quinone-bearing polyamines as multi-target-directed ligands to combat Alzheimer's disease. J. Med. Chem. 50:4882–4897. [DOI] [PubMed] [Google Scholar]
- 60.He, X. C., S. Feng, Z. F. Wang, Y. Shi, S. Zheng, Y. Xia, H. Jiang, X. C. Tang, and D. Bai. 2007. Study on dual-site inhibitors of acetylcholinesterase: Highly potent derivatives of bis- and bifunctional huperzine B. Bioorg. Med. Chem. 15:1394–1408. [DOI] [PubMed] [Google Scholar]
- 61.Wallace, A. C., R. A. Laskowski, and J. M. Thornton. 1995. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8:127–134. [DOI] [PubMed] [Google Scholar]