Abstract
Background and Aims
The proteasome is a major cellular proteinase. Its activity is modulated by cellular oxidants. Hepatitis C core protein and ethanol exposure both cause enhanced oxidant generation. The aim was to investigate whether core protein, by its ability to generate oxidants, alters proteasome activity and whether these alterations are further affected byethanol exposure.
Methods
These interactions were examined in Huh-7 cell lines that expressed inducible HCV core protein and/or constitutive cytochrome P450 2E1 (CYP2E1) and as purified components in a cell free system. Chymotrypsin-like proteasome activity was measured fluorometrically.
Results
Proteasome activity in core-positive 191-20 cells was 20% higher than that in core-negative cells and was enhanced three-fold in CYP2E1-expressing L14 cells. Exposure of core-positive cells to glutathione ethyl ester, catalase, or the CYP2E1 inhibitor, DAS, partially reversed the elevation of proteasome activity in core-positive cells, while ethanol exposure suppressed proteasome activity. The results indicate that proteasome activity was up-regulated by low levels of core-induced oxidative stress and down-regulated by high levels of ethanol-elicited stress. These findings were partially mimicked in cell free system. Addition of core protein enhanced the peptidase activity of purified 20S proteasome containing the proteasome activator, PA28 and was further potentiated by addition of liver mitochondrial and/or microsome fractions. However, proteasome activation was significantly attenuated when fractions were obtained from ethanol-fed animals.
Conclusions
HCV core protein interacts with PA28, mitochondrial and ER proteins to cause low levels of oxidant stress and proteasome activation, which is dampened during ethanol metabolism when oxidant generation is higher.
INTRODUCTION
The proteasome is the major cellular protein-degrading enzyme that degrades 80% of intracellular protein. The enzyme is crucial for cell survival, as it plays a pivotal role in destroying not only normal, but also damaged proteins. By degrading short-lived signal transduction factors, the proteasome regulates signal transduction events and the inflammatory response 1, 2. In the immune response, the proteasome cleaves antigenic proteins to generate peptides for MHC class I-restricted antigen presentation 3.
Proteasome function is regulated by the levels of intracellular oxidants. In liver cells, the proteasome is continuously exposed to oxidants, because of mitochondrial electron transport as well as high expression levels of cytochrome P4502E1 (CYP2E1). Multiple agents, including viral proteins and ethanol enhance oxidant generation in the liver.
HCV core protein plays an important role in HCV infection pathogenesis. The viral protein is known to induce oxidative stress by its ability to associate with the outer membrane of the mitochondrion. This interaction increases Ca2+ entry, mitochondrial superoxide production, and subsequently elevates generation of reactive oxygen species (ROS) by mitochondrial electron transport complex I. This results in a decrease in mitochondrial GSH and mitochondrial depolarization which can be augmented by simultaneous ER oxidative stress 4, 5. The ability of core protein to increase oxidant production has been reported in isolated mitochondria, in cells expressing core protein, in full-length HCV replicon and in liver mitochondria derived from HCV transgenic mice 5, 6. These effects of core protein are enhanced in CYP2E1-expressing cells and are further potentiated by alcohol exposure 4-7. Because core protein induces oxidant formation and because the proteasome is sensitive to their levels in cells 8-10, we postulated that core protein regulates proteasome activity. These properties of core protein may be further potentiated by ethanol exposure, as ethanol metabolism suppresses proteasome activity 11, 12. Suppression of proteasome activity may enhance disease progression in HCV–infected alcohol-consuming patients, as it is known that ethanol consumption exacerbates the clinical course of HCV infection in these individuals 13, 14. To date, specific interactions between proteasome and HCV core and the effects of ethanol on these interactions have not been investigated. Hence, we sought to determine specific interactions between HCV core protein and proteasome activity in vitro and in cultured hepatoma cells that have inducible expression of HCV core protein and cells with and without constitutive expression of CYP2E1
MATERIALS AND METHODS
Reagents and Media
High glucose Dulbecco’s Modified Eagle Medium (DMEM), F12, blasticidin S and fetal bovine serum (FBS) were purchased from Invitrogen (Carlsbad, CA). G418 and antibody to PA28α were from Calbiochem (La Jolla, CA). MeO-Suc-Phe-Leu-Phe-AFC (FLF-AFC) was from MP Biomedicals (Aurora, Ohio). Other reagents, all of analytical grade quality, were from Sigma (St. Louis, MO). PA28 was a gift from Dr. George DeMartino, University of Texas Southwestern Medical Center, Dallas, TX.
Cell Lines
Huh7 cells, which express core protein under a tetracycline-repressible promoter, either in CYP2E1-non-expressing (191-20) or in CYP2E1-expressing (L-14) cell-lines were used in this study. Both cell lines expressed core protein in the absence of tetracycline after 4 days of tetracycline withdrawal and exhibited no core expression in its presence at 2 μg/ml. (Fig. 1A). Both core protein and CYP2E1 were detected by Western blot using monoclonal anti-HCV core (Affinity BioReagents, Golden, CO), or anti-CYP2E1 (Calbiochem, La Jolla, CA), respectively. Culture conditions were as described7
Fig.1.
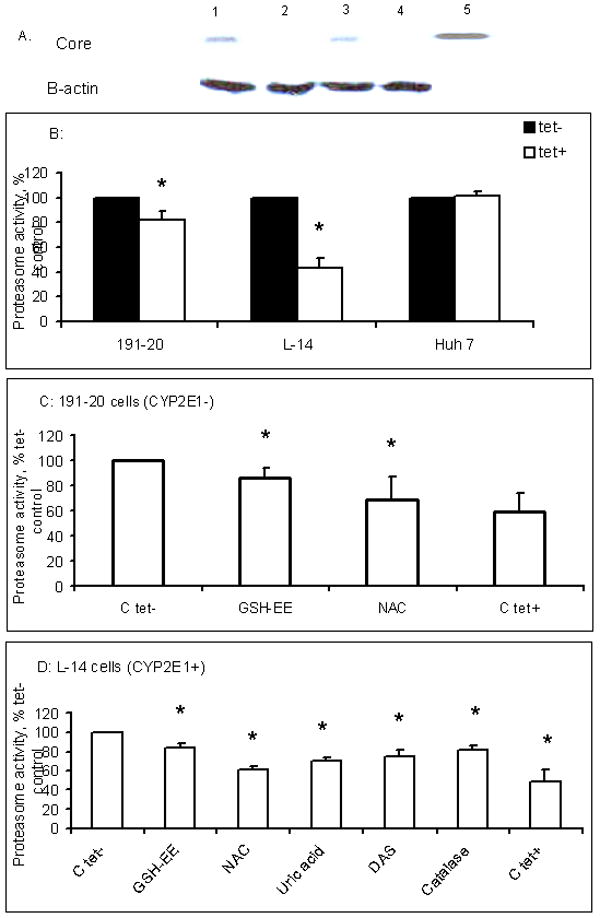
A. Expression of core protein in tet- and tet+ Huh7 cells. HCV core protein was detected in cell lysates by Western blot and B-actin was used as a loading control: 1.191-20 tet- cells; 2.191-20 tet+ cells; 3. L-14 tet- cells; 4.L-14 tet+ cells; 5.Recombinant HCV core protein. B. Proteasome activity in 191-20, L-14 and Huh7 cells before and after tetracycline treatment. All three cell lines were treated or not with 2 μg tetracycline/ml for 4 days. Core protein was detected only in cell lysates of tet- 191-20 and L-14 cells by Western blot (inset above each bar). Proteasome activity is expressed as percent of control. Percent of control is calculated as (proteasome activity in all experimental readings expressed as nmole AFC/μgDNA divided by nmole AFC/μgDNA proteasome activity in untreated tet-cells) × 100%. Data are mean values ± SD from 3 experiments. * indicates a significant difference (p<0.05) between tet- control cells and other treatments. C. Proteasome activity in 191-20 cells Tetracycline-untreated cells were treated with 5 mM GSH-EE, 20 mM NAC or 1000 U catalase for 24 hr. Proteasome activity was detected by in situ assay and compared with that in 191-20 tetracycline-treated cells. Data from 4 independent experiments are expressed as percent proteasome activity in control in tetracycline-untreated cells. * indicates a significant difference (p<0.05) between tet- control cells and other treatments. C. Proteasome activity in L-14 cells: L-14 cells were treated just as described in A with 20 μM DAS and 100 μM Uric acid for 24 hr. Data from 4 independent experiments are expressed as percent of control proteasome activity in tetracycline-untreated cells. * indicates a significant difference between tet- control cells and other treatments.
Cell Treatments
Hepatoma cells were plated onto 96-well black plates with clear bottoms at a density of 5×10 3 cells/well and were incubated in a 1:1 mixture of DMEM/F12, media supplemented with 5% FBS, penicillin-streptomycin and selective antibiotics (200 μg G418/ml for the core protein and 4 μg blasticidinS /ml for the CYP2E1 expression). After overnight attachment, cells were treated as described in the text and figure legends.
Proteasome Purification
20S proteasome was purified from rat livers according to the procedure described 15. The purification steps included a high-speed (105,000 × g) centrifugation for 60 min followed by a 16 hr centrifugation of the cytosol at 52,000 × g, DEAE-Sepharose chromatography of the resuspended proteasome pellet and hydrophobic interaction chromatography on phenyl-sepharose column. In addition to the above-described purification, the residual enzyme remaining in high-speed supernatant fraction (which was enriched with PA28) was purified, as described 16.
HCV Core Protein Purification
Cloning, expression and purification of HCV core protein were performed as described by Kunkel et al 17. Briefly, HCV core residues 1 to 179 (HCVC 179) derived from the AG94 isolate of genotype 1a sequence was amplified by PCR and cloned into a pET30a expression vector (Novagen). Core protein was expressed by transforming E. coli BL21 (DE3) cells with the expression vector. Expression was induced by addition of 1 mM isopropyl-β-D thiogalactopyranoside (IPTG). The cells were solubilized by sonication. Lysates were then diluted with 8M Urea and incubated overnight at 4°C with 50 mM DTT. The supernatant was applied to a cation-exchange column (Poros 20 CM) pre-equilibrated with 0.25 M HEPES, pH 7.0, 8 M urea and 1.5 M NaCl. Each fraction was separated by SDS-PAGE and the gels were stained with Coomassie blue to identify HCVC 179, which was found in a single peak eluting around 650mM NaCl. These fractions were subsequently subjected to reverse-phase high-pressure liquid chromatography (YMC-ODS, reverse-phase 150×4.6mm) and eluted with a linear gradient of 20 mM sodium-phosphate-methanol, pH 3.0. Fractions were analyzed as describe above. Eluted fractions containing core protein were pooled and dialyzed overnight at 4°C against refolding buffer (20 mM Tris (pH 7.0), 100 mM NaCl). Proteinase inhibitors were added to the dialyzed sample (50 μM Leupeptin, 1 μM Pepstatin A and 0.5 mM PMSF). . The homogeneity of purified core protein was determined by SDS-PAGE and estimated to 98%. Its identity was confirmed by immunoblot using a mouse monoclonal antibody to HCV core protein (Affinity BioReagents, Golden, CO).
Mitochondrial Isolation
This was performed according to procedure of Korenaga et al 6. The mitochondrial fractions were prepared from livers of C57Bl/6 mice fed the Lieber DeCarli control or ethanol diet for 3 weeks. Briefly, liver tissue was rinsed with ice-cold PBS and lysed by 20 strokes in a Dounce homogenizer, in 40 mM Tris-HCl, 1 mM EDTA, 150 mM NaCl, (pH 7.4) Homogenates were subjected to low speed centrifugation, mitochondrial pellets were obtained by centrifugation at 10,000 × g for 10 min and then were resuspended in 200 mM mannitol, 70 mM EDTA, 10 mM HEPES (pH 7.5). Alternatively, mitochondria were prepared as described 19. Contamination of mitochondria with less than 5% of cytosol was detected. The latter was determined by the activity of lactate dehydrogenase (LDH) in the mitochondrial preparation compared with that in cytosol.
Microsome Isolation
Microsomes were isolated from livers of control and ethanol-fed mice following the fractionation procedure previously published 19.
Proteasome Activity
Proteasome chymotrypsin-like (Cht-L) activity was detected in situ (intact cells) by measuring hydrolysis of the membrane-permeable substrate, methoxy-succinyl-phe-leu-phe-7-amido-4-trifluoromethyl coumarin (MeO-Suc-FLF-AFC) 20. To compare and combine the results of several experiments, for which we use different cell passages, proteasome activity expressed as nmoleAFC/μgDNA in treated samples was divided by proteasome activity in nmoleAFC/μgDNA in control sample and this ratio was multiplied by 100%. In vitro assay of chymotrypsin-like activity purified proteasome was performed using the fluorogenic substrate Suc-LLVY-AMC. Briefly, the latter reaction was run in black 96-well plates, at 200 μl/well, containing 0.1M Tris-HCl, (pH 7.5) and 13 μM Suc-LLVY-AMC substrate. After 15-60 min incubation at 37°C, the reaction product, AMC, was detected fluorometrically on a plate reader (Victor 3, Perkin Elmer, Shelton, CT), (excitation 355nm; emission 460nm). Enzyme specific activity is expressed as nmole AMC/mg protein.
In vitro Incubations
Purified 20S proteasome (50 μg protein/ml) in 0.1 mM Tris-HCl (pH 7.5) and incubated for 10 min with increasing quantities of core/mitochondrial mixture. The ratio of core protein to mitochondrial protein in the incubation mixture was always maintained at 1:100,000. Thus, the ratio of core protein to mitochondrial proteins used for these in vitro experiments were comparable to those reported in HCV core-expressing mice 6. We tested the 20S proteasome activity using 100, 200 or 300 pg of core protein using corresponding amount of mitochondrial protein (i.e. 10, 20, and 30 μg of mitochondrial protein, respectively) in the incubation medium. For experiments, the same mass quantities of microsomal proteins were also used. After incubation, the mixtures were assayed for proteasome chymotrypsin-like activity.
Statistical Analyses
Data are expressed as mean values ± standard deviation. Multiple comparisons for significance were determined by one-way ANOVA, using a Tukey post-hoc test. Comparison between two groups used Student’s t-test. A probability value of 0.05 or less was considered significant.
RESULTS
Differential proteasome activity in core + (tet-) and core- (tet+) cells
Proteasome activity was significantly higher in both CYP2E1-negative 191-20 cells and CYP2E1-positive L-14 cells that expressed core protein in the absence of tetracycline as compared to when the core protein expression was repressed by tetracycline. Proteasome chymotrypsin-like activity was 20% higher in core-positive 191-20 cells compared with core-negative 191-20 cells. This activity was 60% higher in the constitutively expressing CYP2E1 core-positive L-14 cells compared to the same cells that do not express core protein (Fig.1B). These results clearly indicated an association between HCV core expression and proteasome activity. Tetracycline by itself had no effect on proteasome activity. This was confirmed using the parental core non-expressing Huh7 cells that were similarly treated with tetracycline (Fig.1B).
Because expression of HCV core protein has been shown to generate intracellular oxidants 7, we tested the effect of various antioxidants on in situ proteasome activity in both 191-20 and L-14 cells. The addition of GSH-EE, NAC, UA, DAS or catalase to the extracellular medium of core-positive cells caused a decrease in proteasome activity to approach the activity level in the core-negative cells (Fig. 1C, D).
Differential effects of ethanol treatment in 191-20 and L-14 cells
In core-negative and core-positive 191-20 cells, treatment with 50 mM ethanol for 72 hr did not affect proteasome activity (data not shown). However, exposure of CYP2E1-positive L-14 cells to ethanol suppressed proteasome activity by 33 % in the presence of core protein (Fig. 2A). Suppression of proteasome activity by ethanol in core positive cells was blocked by simultaneous exposure to DAS, catalase or GSH-EE (Fig. 2B).
Fig.2.
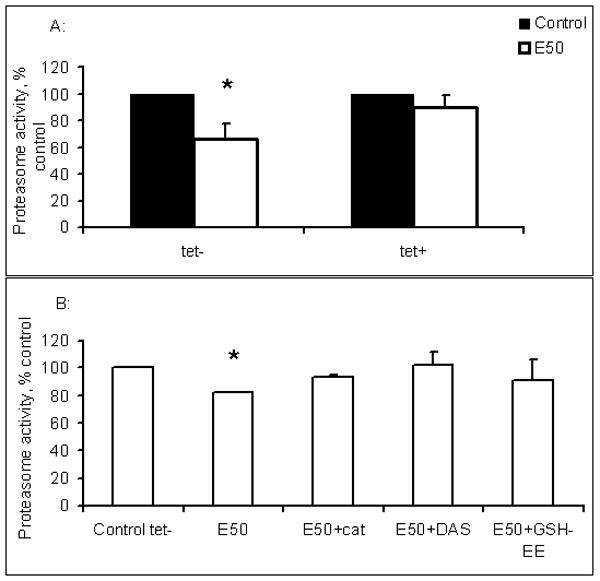
A. Effects of ethanol on L-14 cells. Cells were treated or not with 50 mM ethanol for 72 hr. Proteasome activity was measured by in situ assay. Data from 4 independent experiments are presented as percent of control proteasome activity in tetracycline-untreated and treated cells. * indicates a significant difference (p<0.05) between proteasome activity in control (untreated cells) and ethanol-treated cells. B. Effects of ethanol on L-14 cells in the presence of glutathione precursors, GSH-EE and NAC, and ethanol metabolism inhibitors. Tetracycline-untreated L-14 cells were exposed to 50 mM ethanol in the presence or absence of catalase, DAS, or GSH-EE at the indicated concentrations for 72 hr. Proteasome activity was measured by in situ assay. Data from 3 independent experiments are presented as percent of control proteasome activity in ethanol-untreated L-14 cells. * indicates a significant difference between control and treated cells.
Biphasic response of 20S proteasome to t-BOOH
To provide an explanation for the observed differential regulation of proteasome activity by core protein, we tested whether the proteasome activity is affected by various doses of the oxidant, t-butyl hydroperoxide (t-BOOH). Core-positive, CYP2E1 negative 191-20 cells were treated with increasing concentrations of t-BOOH (10 μM to 100 μM) for 5 hr and in situ proteasome activity was measured. Ten μM t-BOOH increased proteasome activity in these cells by 75%, 30 μM t-BOOH had no significant effect and 50 and 100 μM t-BOOH completely abolished enzyme activity (Fig.3). We had previously reported a similar biphasic regulation of proteasome in HepG2 cells exposed to the peroxynitrite donor, SIN-1, which elevated proteasome activity at low doses and decreased its in situ activity at high doses 21.
Fig.3. Effects of various concentrations of t-BOOH on proteasome activity in core+ 191-20 cells.
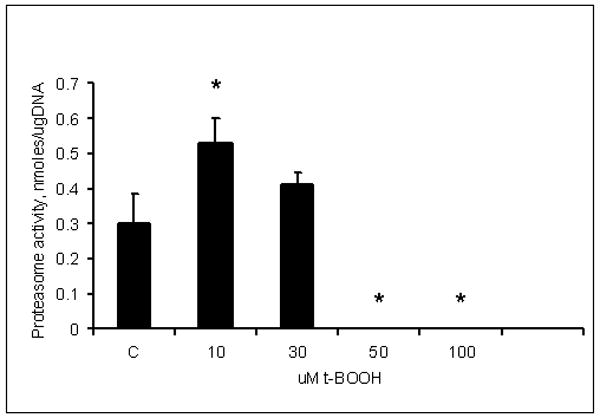
Cells were exposed to indicated concentrations of t-BOOH for 5 hr and then proteasome activity was measured by in situ assay. Data from 2 experiments are presented as specific proteasome activity, nmoles/μg DNA. * indicates a significant difference (p<0.05) between control and treated cells.
In vitro activation of proteasome with core protein
To determine whether core protein directly affects proteasome activity in vitro (in the absence of oxidants), we incubated purified 20S proteasome with core protein, as detailed in Materials and Methods. The purity of core protein was confirmed (shown in Fig.4A). However, we utilized two different proteasome preparations. One preparation (Pr.1) contained both 20S proteasome as well as the proteasome activator PA28, as revealed by Western blot analysis (Figure 4B). The other preparation (Pr. 2) had undetectable levels of PA28 (Fig. 4 B). We observed that the core protein caused a dose-dependent activation of the proteasome only with Pr. 1 preparation and not with Pr. 2 (Fig. 4C). Such activation was rather specific for core protein, as the addition of same mass of BSA or recombinant NS3 protein (Virogen, Inc) failed to activate Pr.1 preparation (Fig. 4C). The effect of HCV core protein on proteasome preparation was similar to the effect of commercially available HCV core protein (Virogen Inc.) on commercially available 20S proteasome obtained from Boston Biochem (Fig. 4C). Thus, Pr. 1 preparation was used in all subsequent in vitro incubations.
Fig.4.
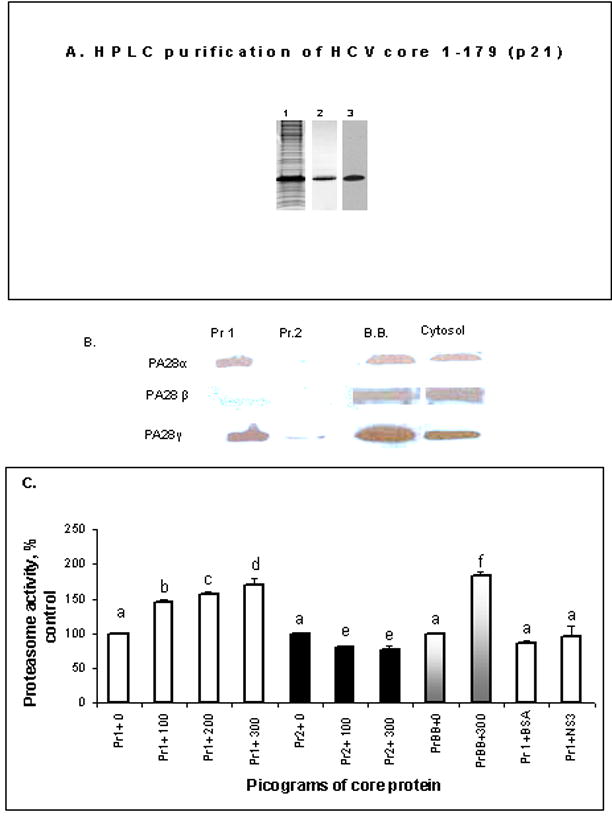
A. HPLC purification of core protein. Samples were subjected to SDS-PAGE and visualized by coomassie blue stain (lanes 1 and 2) or immunoblot for core protein (lane 3). Lane 1: lysed bacterial cell pellet; Lane 2: purified HCVC 179; Lane 3: purified HCVC 179. B.Expression of PA28 subunits in various proteasome preparations (Pr.1-proteasome 1; Pr.2- proteasome 2; BB-proteasome from Boston Biochem). A representative blot is presented for two proteasome preparations that were obtained as described, then subjected to Western blot analysis. 20S proteasome preparations (Pr 1 and Pr 2) were probed for PA28 α, β and γ using specific antibodies (Biomol International) BB proteasome preparations and crude cytosolic proteasome preparation were used as controls. C. Effects of core protein on proteasome activity in two proteasome preparations. 20S proteasome (Pr1 and Pr2) preparations were incubated with 100, 200 and 300 pg core protein for 5 min at 25°C and with 300 pg BSA or NS3 protein. Proteasome activity was then detected by Suc-LLVY-AMC hydrolysis. Data from 3 experiments are presented as percent of 20S proteasome activity. As the control, exposure of commercial HCV core protein (Virogen Inc) to commercial 20S proteasome (Boston Biochem) was used (Pr1-white bars, Pr2- black bars, BB- gray bars). Bar columns with different letters are significantly different from each other, while columns with the same letters are not.
Effects of mitochondria and microsomes on proteasome activation by core protein
It has been reported that in vivo, HCVcore protein associates with the mitochondrial membrane 4, 5. Therefore, we examined the effects of isolated mitochondria on the 20S proteasome activity modulation by HCV core on the premise that the core protein might function in association with other mitochondrial components. Pr.1 was incubated with core protein as well as mitochondrial preparations obtained from livers of control or ethanol-fed C57Bl/6 mice. Inclusion of 30 μg mitochondrial protein in the incubation mixture (containing 300 pg core) enhanced the activation of proteasome by core protein up to 6.3 fold (Fig. 5A). In contrast, a 3.2-fold increase in proteasome activation was observed when mitochondria fractions from ethanol-fed mice were used. This indicates a blunted response with mitochondria from ethanol-fed mice (Fig 5A). No differences were observed between freshly isolated and frozen mitochondria on core protein-induced proteasome activation. Furthermore, mitochondria used at a 10 μg level significantly enhanced the activating effect of recombinant PA28 α, which, by itself, increased proteasome activity by 1.8-fold (Fig.5B).
Fig.5.
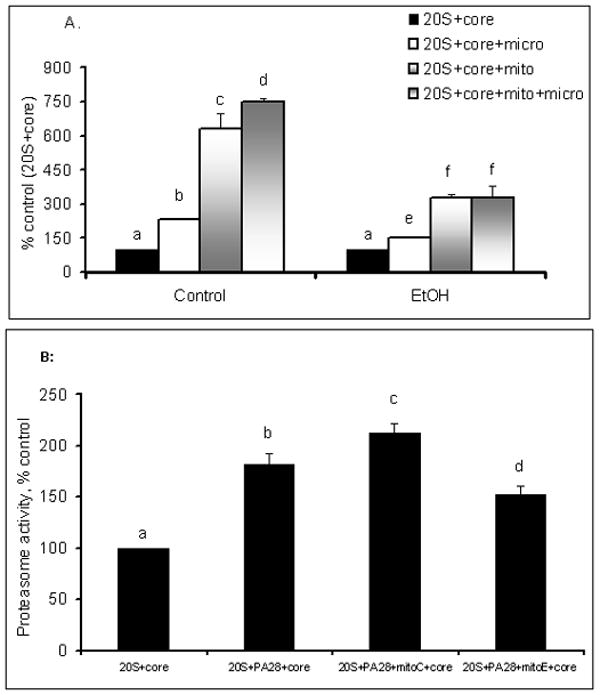
A.The effects of mitochondria and microsomes on core-activated 20S proteasome activity. Mitochondrial and microsome fractions were obtained from livers of control and ethanol-fed mice. 20S proteasome was incubated with 300 pg core protein and an equal mixture of mitochondrial and microsomal proteins (Material and Methods). Proteasome activity was then assayed. Data from 3 experiments are expressed as percent of 20S proteasome activity with core protein alone. Bar columns with different letters are significantly different from each other, while columns with the same letters are not. By themselves, mitochondria and microsomes possess little-to-no proteasome activity (not shown).
B. Effects of mitochondria+core on 20S proteasome (1) after addition of recombinant PA28α. 20S proteasome (Pr1) was exposed to PA28 in the presence or absence of core protein and of mitochondria isolated from livers of control (MitoC) or ethanol-fed (MitoE) mice (100pg core/10 μg mitochondria). Data from 3 experiments are expressed as percent of 20S proteasome activity (without additions). Bar columns with different letters are significantly different from each other, while columns with the same letters are not.
In cells, core protein is associated not only with mitochondria, but also with ER membrane. To determine the effects of ER preparations (i.e. microsomes) on core-mediated proteasome activation, we conducted similar experiments as with mitochondrial fractions. Only a 2.3-fold increase in proteasome activity was observed when microsomes from control mice were incubated in the presence of core protein and Pr.1 proteosome preparation (Fig. 5A). Microsomes combined with mitochondria caused a 7.4-fold elevation of proteasome activity. Proteasome activation was again attenuated when microsomes from ethanol-fed mice were used with core protein (1.5-fold stimulation). Moreover, there was no further elevation of proteasome activity when microsomes and mitochondria from ethanol-fed mice were combined with core protein (Fig 5A). Neither mitochondria nor microsomes by themselves possessed significant proteasome (not shown).
DISCUSSION
HCV core protein is degraded by the ubiquitin-proteasome pathway and E6AP has been identified as the specific ubiquitin ligase that marks the protein for degradation 22, 23. Here, we demonstrated that core protein also regulates proteasome activity, which potentially would influence protein catabolism in HCV-infected hepatocytes. Further, this study identifies a dual mechanism of core-mediated proteasome activation via core protein-induced oxidant generation as was revealed from our cell culture experiments and evidence of cross-talk between core protein and PA28-20S complex supported by the cell-free studies.
As has been already demonstrated by others 6, 7, 191-20 and L-14 cells and core-expressing or non-expressing mice had differential levels of ROS production/glutathione content in mitochondria. Furthermore, Otani et al 7 showed that the ratio between pro-and anti-oxidative factors (ROS and GSH, respectively) is lowest in core-positive 191-20 cells, with moderate increase in core-positive L-14 cells, followed by an increase in ethanol and/or t-butyl hydroperoxide (tBOOH)-treated core-positive L-14. Thus, in core-positive/CYP2E1-negative cells, core protein caused predominant generation of oxidants in mitochondria, while in CYP2E1-expressing cells, oxidant generation was initiated by core protein both at the mitochondrial and the ER levels and was potentiated by ethanol treatment.
Here, we utilized the same cell lines, which were exposed or not to ethanol, to create varying gradations of oxidant stress. We presumed that the differential levels of oxidant stress differentially regulated the proteasome activity. To mimic this situation, we exposed core-positive 191-20 cells to various doses of the oxidant, t-BOOH, which at low doses increased proteasome function, while at high doses it suppressed proteasome activity. This findings was not limited to t-BOOH and similar results were reported using various concentrations of hydrogen peroxide as well as the peroxynitrite donor, SIN1, in HepG2 cells 24, 21.
Here, we observed enhanced proteasome activity in both core-positive 191-20 and L-14 cells, with a much higher magnitude of proteasome activation in CYP2E1-expressing L-14 cells. These results, taken together with previous reports of low to moderate ROS generation /GSH depletion in core-positive 191-20 and L-14 cells 7, indicated that these levels of oxidative stress activate proteasome. This conjecture was confirmed by data, which showed that treatment with GSH analogs/precursors reversed core-induced proteasome activation in both cell lines. In addition, inclusion of antioxidants (uric acid or catalase) or the CYP2E1 inhibitor, DAS, lowered proteasome activity in CYP2E1-positive/core-positive L-14 cells to approach the same levels in core-negative cells. However, when L-14 cells were exposed to ethanol (portending a much higher level of oxidant generation), proteasome activity was suppressed. Indeed, a reciprocal relationship between CYP2E1 expression and proteasome activity has been previously reported 10, 25, 26 and we have also shown a decline in proteasome function by ethanol exposure12, 27. The mechanism of this decline in proteasome activity can be attributed, in part, to the inability of PA28 to activate the 20S enzyme, when proteasome is heavily oxidatively modified, though low amounts of oxidants facilitate the 20S-PA28 interactions 21.
However, even in the absence of oxidative stress, core protein was able to activate 20S proteasome, which formed the complex with PA28 α and γ. This means that, in addition to indirect core-mediated regulation of proteasome activity via oxidant stress, the possibility of direct core-proteasome interactions cannot be excluded. To clarify whether core protein also affects proteasome function via direct protein-proteasome interactions, we used a cell-free system, where proteasome is exposed to core protein in the absence of oxidative stress. We observed a link between activating effects of core protein only on 20S proteasome co-purified with PA28 subunits, indicating that core protein affected 20S proteasome activity by facilitating its interaction with assembled PA28 (α, β and γ subunits.). In fact, PA28α and to lesser extent, β are known as classical activators of 20S proteasome Cht-like activity in the cytosol and that γ subunit regulates proteasome Trypsin-like activity, mainly, in the nucleus 28,29. In our study, PA28-20S proteasome complex responded to core protein by increased Cht-like activity, while, as reported earlier, core protein forms a complex with PA28γ, but not with PA28α and β 30. However, even if core protein initially attaches to PA28γ, we cannot exclude that γ subunit subsequently activates α subunit in PA28-20S proteasome preparation, thereby increasing Cht-like proteasome activity. This core-induced proteasome activation was further enhanced by addition of exogenous recombinant PA28α.
In HCV-expressing cells, core protein is located between the outer mitochondrial membrane and the endoplasmic reticulum 6. To mimic this cellular interaction in cell free system, we measured whether proteasome activation by core protein was further modulated by mitochondria and/or microsomes. Each subcellular fraction, either alone or in combination, potentiated proteasome activation by core protein. The activating effect of mitochondria was not dependent on their structural integrity, as both frozen and freshly isolated mitochondrial preparations equally enhanced core protein-mediated 20S proteasome activation. Therefore, proteasomal activation was likely the result of interactions between 20S-PA28 complex and specific mitochondrial protein(s), possibly, in/on the outer membrane. This is consistent with findings of others that core protein has a strong tendency to associate with various proteins, such as STAT1 and Jaks, affecting their functions 31, 32. Interestingly, the cross talk between core-activated proteasome and mitochondria or/and microsomes was blunted in the presence of mitochondria or microsomes from ethanol-fed mice, indicating that ethanol-induced dysregulation of protein expression in these organelles may be attributed to their adduction by oxidants.
The nature of the putative proteasome-interacting protein(s) in mitochondria and ER is not clear. Heat shock proteins (HSP) are potential candidates for providing communication between mitochondria, microsomes and the proteasome, as the role of HSP in interactions between the microsomal enzyme, CYP2E1, and the proteasome has been established 33. Mitochondria may also affect the activity of 20S-PA28 complex via interactions between mitochondrial HSP70 and PA28, because mitochondria express HSP70 34 and the association between HSP70 and PA28 during substrate refolding has been reported 35
The physiological and pathogenic relevance of proteasome activation by core protein is in an enhanced ability of proteasome to cleave the substrate proteins. Since some HCV proteins (such as NS5B as well as nuclear core protein) are targeted by proteasome 36, their rapid degradation may be favorable for HCV elimination. However, these proteins, in turn, form stable complexes with other important signaling proteins, thereby facilitating the removal of HCV-signaling factor complexes by proteasome. For example, in HCV protein-expressing cells, the depletion of STAT1, a signal transduction factor for interferons, may be potentially attributed to enhanced proteasome function as demonstrated by suppression of STAT1 disappearance in the presence of the proteasome inhibitor, MG132 37. Similarly, activation of proteasome by viral proteins, leads to degradation of protective factors, as observed in other viral infections 38. However, even if core protein enhances proteasome activity, this may not result in tremendously increased generation of peptides for MHC class I-restricted antigen presentation, because the most antigen processing enzymes (including proteasome) are activated by IFN and to be effective, peptide processing requires unaltered IFN signaling, while STAT1 is degraded by proteasome. If core-positive/CYP2E1-expressing liver cells are exposed to ethanol, ethanol metabolism suppresses proteasome activity. This may prevent the degradation of the “protective” proteins, as STAT1, but will ultimately block STAT1 phosphorylation. 27. The processing of viral peptides for antigen presentation by liver cells will be further suppressed, because in this case, ethanol metabolism not only suppresses proteasome-dependent cleavage of antigenic peptides, but also prevents IFN signaling by altering STAT1 phosphorylation. The outcomes of core-induced proteasome activation and the suppressing effects of ethanol exposure for innate and adaptive immunity, as well as the identification of proteins involved in HCV core-proteasome-mitochondrion-ER interactions will be the subject of future investigations. A proposed mechanism of core-proteasome interactions in liver cells is depicted in Fig. 6.
Fig.6. Proposed mechanism of regulation of proteasome function by core protein in liver cells.
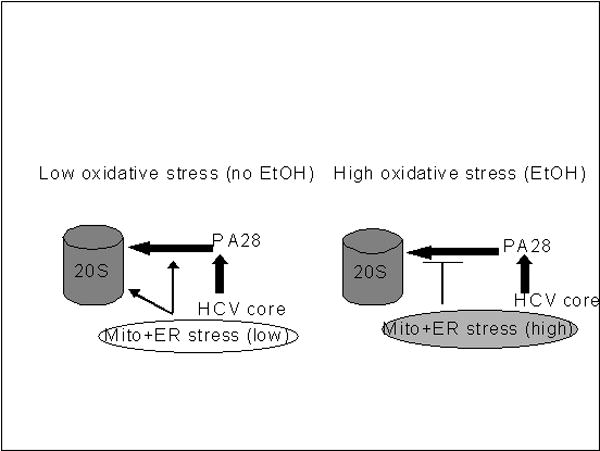
Core protein stimulates proteasome, directly facilitating PA28-20S interaction. These effects are potentiated by mitochondrial and microsomal protein (s), which further enhance proteasome activation by core protein. In addition, HCV core protein induces a low level of oxidative stress and also increases proteasome activity. Treatment of core-expressing CYP2E1+ cells by ethanol disrupts interactions between 20S proteasome-PA28 complex and mitochondria and creates a high level of oxidative stress, which suppresses proteasome activity.
In summary, HCV core protein enhanced 20S proteasome activity directly, by facilitating interactions between 20S proteasome, PA28 and mitochondria/microsomes and indirectly, via generation of low levels of oxidants. Elevated oxidant generation by ethanol metabolism and disruption of core-20S-PA28-mitochondrial interactions ultimately results in reduced liver proteasome activity that may influence the propagation of HCV clinical course in alcoholic patients.
Acknowledgments
Supported by grant 5R21 AA015379-02 and AA012863 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA), and, in part, by Medical Research Funds from the Department of Veterans Affairs. We thank Sandra Todero and John Evans for excellent technical assistance, Tiana Curry-McCoy for providing proteasome2 preparation, Dr. George DeMartino, University of Texas Southwestern Medical Center, Dallas, TX, for providing recombinant PA28α and Dr. Kusum Kharbanda, VA Medical Center, Omaha, for fruitful discussion of the manuscript.
Financial support: Supported from the National Institute on Alcohol Abuse and Alcoholism (NIAAA), grants 5R21 AA015379-02 and AA012863 and in part, by AA09384 and by Medical Research Funds from the Department of Veteran Affairs
List of Abbreviations
- HCV
hepatitis C virus
- CYP2E1
cytochrome P450 2E1
- DAS
diallyl sulfide
- GSH-EE
glutathione ethyl ester
- NAC
N-acetyl cysteine
- UA
uric acid
- MHC
major histocompatibility complex
- t-BOOH
t-butyl hydroperoxide
- 4MP
4-methylpyrazole
- ER
endoplasmic reticulum
- HSP70
heat shock protein 70
- Jak
Janus kinase
- STAT1
signal transducer and activator of transcription 1
- IFNα
interferon alpha
- IFNγ
interferon gamma
- PN
peroxynitrite
Footnotes
No conflict of interests exist
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ulrich HD, Vogel S, Davies AA. SUMO keeps a check on recombination during DNA replication. Cell Cycle. 2005;4:1699–702. doi: 10.4161/cc.4.12.2194. [DOI] [PubMed] [Google Scholar]
- 2.Kim TK, Maniatis T. Regulation of interferon-gamma-activated STAT1 by the ubiquitin-proteasome pathway. Science. 1996;273:1717–9. doi: 10.1126/science.273.5282.1717. [DOI] [PubMed] [Google Scholar]
- 3.Rock KL, York IA, Saric T, Goldberg AL. Protein degradation and the generation of MHC class I-presented peptides. Adv Immunol. 2002;80:1–70. doi: 10.1016/s0065-2776(02)80012-8. [DOI] [PubMed] [Google Scholar]
- 4.Wen F, Abdalla MY, Aloman C, Xiang J, Ahmad IM, Walewski J, McCormick ML, Brown KE, Branch AD, Spitz DR, Britigan BE, Schmidt WN. Increased prooxidant production and enhanced susceptibility to glutathione depletion in HepG2 cells co-expressing HCV core protein and CYP2E1. J Med Virol. 2004;72:230–40. doi: 10.1002/jmv.10567. [DOI] [PubMed] [Google Scholar]
- 5.Wang T, Weinman SA. Causes and consequences of mitochondrial reactive oxygen species generation in hepatitis C. J Gastroenterol Hepatol. 2006;21(Suppl 3):S34–7. doi: 10.1111/j.1440-1746.2006.04591.x. [DOI] [PubMed] [Google Scholar]
- 6.Korenaga M, Okuda M, Otani K, Wang T, Li Y, Weinman SA. Mitochondrial dysfunction in hepatitis C. J Clin Gastroenterol. 2005;39:S162–6. doi: 10.1097/01.mcg.0000155517.02468.46. [DOI] [PubMed] [Google Scholar]
- 7.Otani K, Korenaga M, Beard MR, Li K, Qian T, Showalter LA, Singh AK, Wang T, Weinman SA. Hepatitis C virus core protein, cytochrome P450 2E1, and alcohol produce combined mitochondrial injury and cytotoxicity in hepatoma cells. Gastroenterology. 2005;128:96–107. doi: 10.1053/j.gastro.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 8.Osna NA, Clemens DL, Donohue TM., Jr Interferon gamma enhances proteasome activity in recombinant Hep G2 cells that express cytochrome P4502E1: modulation by ethanol. Biochem Pharmacol. 2003;66:697–710. doi: 10.1016/s0006-2952(03)00252-1. [DOI] [PubMed] [Google Scholar]
- 9.Bardag-Gorce F, Li J, French BA, French SW. The effect of ethanol-induced CYP2E1 on proteasome activity: the role of 4-hydroxynonenal. Exp Mol Pathol. 2005;78:109–15. doi: 10.1016/j.yexmp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Kessova IG, Cederbaum AI. The effect of CYP2E1-dependent oxidant stress on activity of proteasomes in HepG2 cells. J Pharmacol Exp Ther. 2005;315:304–12. doi: 10.1124/jpet.105.088047. [DOI] [PubMed] [Google Scholar]
- 11.Donohue TM, Osna NA, Clemens DL. Recombinant Hep G2 cells that express alcohol dehydrogenase and cytochrome P450 2E1 as a model of ethanol-elicited cytotoxicity. Int J Biochem Cell Biol. 2006;38:92–101. doi: 10.1016/j.biocel.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Osna NA, White RL, Todero S, McVicker BL, Thiele GM, Clemens DL, Tuma DJ, Donohue TM., Jr Ethanol-induced oxidative stress suppresses generation of peptides for antigen presentation by hepatoma cells. Hepatology. 2007;45:53–61. doi: 10.1002/hep.21442. [DOI] [PubMed] [Google Scholar]
- 13.Nevins CL, Malaty H, Velez ME, Anand BS. Interaction of alcohol and hepatitis C virus infection on severity of liver disease. Dig Dis Sci. 1999;44:1236–42. doi: 10.1023/a:1026605130185. [DOI] [PubMed] [Google Scholar]
- 14.Khan KN, Yatsuhashi H. Effect of alcohol consumption on the progression of hepatitis C virus infection and risk of hepatocellular carcinoma in Japanese patients. Alcohol Alcohol. 2000;35:286–95. doi: 10.1093/alcalc/35.3.286. [DOI] [PubMed] [Google Scholar]
- 15.Beyette JR, Hubbell T, Monaco JJ. Purification of 20S proteasomes. Methods Mol Biol. 2001;156:1–16. doi: 10.1385/1-59259-062-4:1. [DOI] [PubMed] [Google Scholar]
- 16.Chu-Ping M, Slaughter CA, DeMartino GN. Purification and characterization of a protein inhibitor of the 20S proteasome (macropain) Biochim Biophys Acta. 1992;1119:303–11. doi: 10.1016/0167-4838(92)90218-3. [DOI] [PubMed] [Google Scholar]
- 17.Kunkel M, Lorinczi M, Rijnbrand R, Lemon SM, Watowich SJ. Self-assembly of nucleocapsid-like particles from recombinant hepatitis C virus core protein. J Virol. 2001;75:2119–29. doi: 10.1128/JVI.75.5.2119-2129.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown EA, Zhang H, Ping LH, Lemon SM. Secondary structure of the 5’ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donohue TM, Jr, McVicker DL, Kharbanda KK, Chaisson ML, Zetterman RK. Ethanol administration alters the proteolytic activity of hepatic lysosomes. Alcohol Clin Exp Res. 1994;18:536–41. doi: 10.1111/j.1530-0277.1994.tb00906.x. [DOI] [PubMed] [Google Scholar]
- 20.Hamel FG, Bennett RG, Harmon KS, Duckworth WC. Insulin inhibition of proteasome activity in intact cells. Biochem Biophys Res Commun. 1997;234:671–4. doi: 10.1006/bbrc.1997.6693. [DOI] [PubMed] [Google Scholar]
- 21.Osna NA, Haorah J, Krutik VM, Donohue TM., Jr Peroxynitrite alters the catalytic activity of rodent liver proteasome in vitro and in vivo. Hepatology. 2004;40:574–82. doi: 10.1002/hep.20352. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki R, Tamura K, Li J, Ishii K, Matsuura Y, Miyamura T, Suzuki T. Ubiquitin-mediated degradation of hepatitis C virus core protein is regulated by processing at its carboxyl terminus. Virology. 2001;280:301–9. doi: 10.1006/viro.2000.0785. [DOI] [PubMed] [Google Scholar]
- 23.Shirakura M, Murakami K, Ichimura T, Suzuki R, Shimoji T, Fukuda K, Abe K, Sato S, Fukasawa M, Yamakawa Y, Nishijima M, Moriishi K, Matsuura Y, Wakita T, Suzuki T, Howley PM, Miyamura T, Shoji I. E6AP Ubiquitin Ligase Mediates Ubiquitylation and Degradation of Hepatitis C Virus Core Protein. J Virol. 2007;81:1174–85. doi: 10.1128/JVI.01684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grune T, Klotz LO, Gieche J, Rudeck M, Sies H. Protein oxidation and proteolysis by the nonradical oxidants singlet oxygen or peroxynitrite. Free Radic Biol Med. 2001;30:1243–53. doi: 10.1016/s0891-5849(01)00515-9. [DOI] [PubMed] [Google Scholar]
- 25.Perez MJ, Cederbaum AI. Proteasome inhibition potentiates CYP2E1-mediated toxicity in HepG2 cells. Hepatology. 2003;37:1395–404. doi: 10.1053/jhep.2003.50228. [DOI] [PubMed] [Google Scholar]
- 26.Bardag-Gorce F, Yuan QX, Li J, French BA, Fang C, Ingelman-Sundberg M, French SW. The effect of ethanol-induced cytochrome p4502E1 on the inhibition of proteasome activity by alcohol. Biochem Biophys Res Commun. 2000;279:23–9. doi: 10.1006/bbrc.2000.3889. [DOI] [PubMed] [Google Scholar]
- 27.Osna NA, Clemens DL, Donohue TM., Jr Ethanol metabolism alters interferon gamma signaling in recombinant HepG2 cells. Hepatology. 2005;42:1109–17. doi: 10.1002/hep.20909. [DOI] [PubMed] [Google Scholar]
- 28.Brychcy M, Kuckelkorn U, Hausdorf G, Egerer K, Kloetzel PM, Burmester GR, Feist E. Anti-20S proteasome autoantibodies inhibit proteasome stimulation by proteasome activator PA28. Arthritis Rheum. 2006;54:2175–83. doi: 10.1002/art.21970. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Gao X, Ortega J, Nazif T, Joss L, Bogyo M, Steven AC, Rechsteiner M. Lysine 188 substitutions convert the pattern of proteasome activation by REGgamma to that of REGs alpha and beta. Embo J. 2001;20:3359–69. doi: 10.1093/emboj/20.13.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moriishi K, Okabayashi T, Nakai K, Moriya K, Koike K, Murata S, Chiba T, Tanaka K, Suzuki R, Suzuki T, Miyamura T, Matsuura Y. Proteasome activator PA28gamma-dependent nuclear retention and degradation of hepatitis C virus core protein. J Virol. 2003;77:10237–49. doi: 10.1128/JVI.77.19.10237-10249.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin W, Kim SS, Yeung E, Kamegaya Y, Blackard JT, Kim KA, Holtzman MJ, Chung RT. Hepatitis C virus core protein blocks interferon signaling by interaction with the STAT1 SH2 domain. J Virol. 2006;80:9226–35. doi: 10.1128/JVI.00459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hosui A, Ohkawa K, Ishida H, Sato A, Nakanishi F, Ueda K, Takehara T, Kasahara A, Sasaki Y, Hori M, Hayashi N. Hepatitis C virus core protein differently regulates the JAK-STAT signaling pathway under interleukin-6 and interferon-gamma stimuli. J Biol Chem. 2003;278:28562–71. doi: 10.1074/jbc.M210485200. [DOI] [PubMed] [Google Scholar]
- 33.Goasduff T, Cederbaum AI. CYP2E1 degradation by in vitro reconstituted systems: role of the molecular chaperone hsp90. Arch Biochem Biophys. 2000;379:321–30. doi: 10.1006/abbi.2000.1870. [DOI] [PubMed] [Google Scholar]
- 34.Lu X, Michaud C, Orlowski M. Heat shock protein-90 and the catalytic activities of the 20 S proteasome (multicatalytic proteinase complex) Arch Biochem Biophys. 2001;387:163–71. doi: 10.1006/abbi.2001.2270. [DOI] [PubMed] [Google Scholar]
- 35.Minami Y, Kawasaki H, Minami M, Tanahashi N, Tanaka K, Yahara I. A critical role for the proteasome activator PA28 in the Hsp90-dependent protein refolding. J Biol Chem. 2000;275:9055–61. doi: 10.1074/jbc.275.12.9055. [DOI] [PubMed] [Google Scholar]
- 36.Munakata T, Liang Y, Kim S, McGivern DR, Huibregtse J, Nomoto A, Lemon SM. Hepatitis C virus induces E6AP-dependent degradation of the retinoblastoma protein. PLoS Pathog. 2007;3:1335–47. doi: 10.1371/journal.ppat.0030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin W, Choe WH, Hiasa Y, Kamegaya Y, Blackard JT, Schmidt EV, Chung RT. Hepatitis C virus expression suppresses interferon signaling by degrading STAT1. Gastroenterology. 2005;128:1034–41. doi: 10.1053/j.gastro.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 38.Hampson L, Kitchener HC, Hampson IN. Specific HIV protease inhibitors inhibit the ability of HPV16 E6 to degrade p53 and selectively kill E6-dependent cervical carcinoma cells in vitro. Antivir Ther. 2006;11:813–25. [PubMed] [Google Scholar]


