Abstract
Dehydroamino acids are valuable building blocks that are a challenge to incorporate synthetically into unprotected peptides. Lantibiotic synthetases possess dehydration activity that converts Ser and Thr residues in their peptide substrates into dehydroalanine and dehydrobutyrine residues, respectively. We show here that lacticin 481 synthetase can convert the Thr analogs (R)-3-EtSer, (R)-3-vinylSer, (R)-3-ethynylSer, and (R)-3-[(E)-propenyl]Ser into the corresponding dehydro amino acids when incorporated into its peptide substrate. This relaxed substrate specificity holds promise for using the enzyme for synthetic purposes and for lantibiotic engineering. On the other hand, (R)-3-PrSer, (R)-3-iPrSer, and allo-Thr are not substrates for the enzyme.
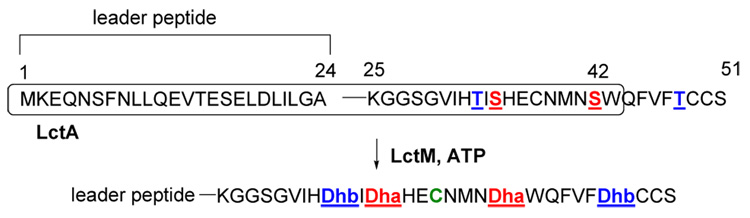
Dehydroamino acids are found in many cyclic peptide natural products,1 and they enhance proteolytic stability and biological activity of linear peptides.2 Furthermore, dehydroamino acids can be used as precursors to peptide conjugates3 and unnatural amino acids.4–6 Incorporation of dehydroamino acids into unprotected peptides using synthetic methods is currently challenging.7 One promising route to these structures is through the biosynthetic machinery for lantibiotics. These compounds are ribosomally synthesized and post-translationally modified antimicrobial peptides.5 The first step in the modification process involves the dehydration of Ser and Thr residues to dehydroalanines and Z-dehydrobutyrines, respectively. This reaction was recently reconstituted in vitro for lacticin 481 synthetase LctM (Figure 1) and the haloduracin synthetases HalM.8,9 In this study we report on the substrate selectivity of the enzymatic dehydration reaction by LctM using various Thr analogs, showing that several dehydroamino acids are readily accessible using this approach.
Figure 1.
Dehydration of LctA catalyzed by lacticin 481 synthetase (LctM). The truncated substrate used in this work is boxed.
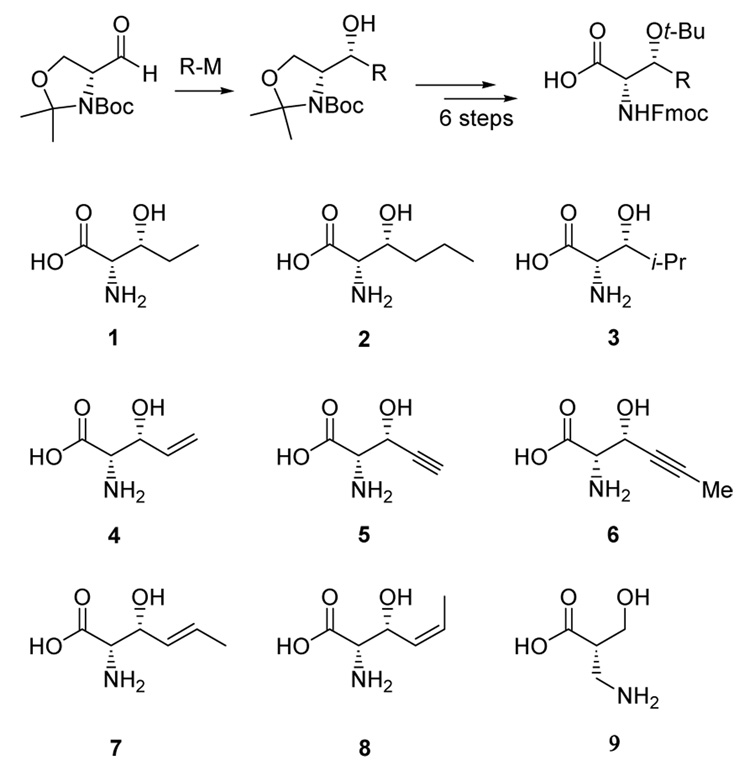
Several studies have recently shown that the lantibiotic dehydratases can process non-lantibiotic substrates as long as they are attached at the C-terminus of a leader peptide (Figure 1).10–12 During lantibiotic biosynthesis, this leader peptide is removed by a protease after completion of the post-translational modifications. The specificity of the dehydratases with respect to the structure of the hydroxyl-bearing amino acid has not been investigated to date. To evaluate this question, we prepared a series of potential substrate analogs (Scheme 1). These structures were all prepared suitably protected for Fmoc-based solid phase peptide synthesis (SPPS).13 The key step in the synthesis of the Thr analogs 1–8 involved the addition of organozinc or organocopper reagents to D-Garner aldehyde,14 which took place with excellent diastereoselectivities and without affecting the existing stereocenter as shown by chiral SFC analysis.15
Scheme 1.
Using SPPS, the protected amino acids were then incorporated into the synthetic heptapeptides CysAsnMetAsnXxxTrpAla corresponding to residues 38–44 of the LctA substrate for LctM. The amino acids 1–9 replaced a Ser in these peptides that is usually present at position 42 of LctA (Figure 1). The synthetic peptides were then ligated to a truncated LctA peptide corresponding to residues 1–37 containing a thioester at its C-terminus. This peptide was obtained by expression in Escherichia coli fused at the C-terminus to an intein-chitin binding domain, subsequent purification using affinity chromatography, and elution with the sodium salt of mercaptoethanesulfonic acid (MES).16–18 The resulting MES thioester was then ligated with the synthetic heptapeptides using native chemical ligation.19,20
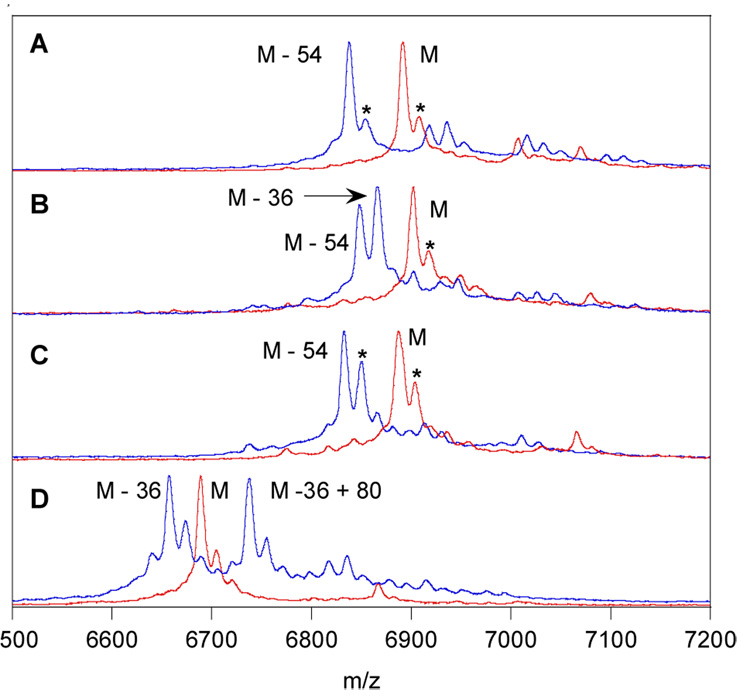
After HPLC purification, the LctA analogs that correspond to LctA1-43 were incubated with LctM in the presence of ATP and Mg2+. Previous studies have shown that the enzyme uses ATP for phosphorylation of the hydroxyl groups of Ser and Thr that undergo elimination.18 The assays with the LctA analogs were then analyzed by MALDI-TOF mass spectrometry. As a control, Thr was first incorporated at position 42 of the truncated LctA peptide (LctA1-43-S42T). Clean conversion to a product with a mass decreased by 54 Da (3 H2O) with respect to the substrate was observed, resulting from dehydration of Thr33, Ser35, and Thr42 (see Figure S1 in the Supporting Information). Substitution of Ser42 with (R)-3-ethylserine also led to clean threefold dehydration (Figure 2A), demonstrating that LctM can tolerate an ethyl group and thereby install dehydronorvalines into peptides. However, both propyl and isopropyl groups proved too large for the enzyme as the LctA analogs with (R)-3-propylserine (2) and (R)-3-isopropylserine (3) at position 42 resulted in just two dehydrations after incubation with LctM (Figures S2 and S3). Treatment of the two-fold dehydrated product with cyanogens bromide, which cleaves the peptide after Met1 and Met40, and subsequent analysis by MALDI-MS conclusively showed that analogs 2 and 3 at position 42 were not dehydrated (Figures S6 and S7). On the other hand, a three-carbon substituent that is less sterically demanding such as the propynyl group in 6 was accepted by the enzyme (Figure 2B) as were the vinyl and ethynyl analogs 4 and 5 (eg Figure 2C for ethynyl). Upon dehydration, these substrates result in interesting α,β-γ,δ-unsaturated amino acids with extended conjugation.
Figure 2.
MALDI-MS spectra of assays in which analogs of a truncated LctA peptide (LctA1-43) were incubated with LctM. Substrates are shown in red and assay products in blue. Substrates contained the following Ser/Thr analogs at position 42: (A) 1, (B) 6, (C) 5, and (D) 9. The asterisks denote oxidation products (+16) involving Met.
The restrictions of the active side pocket with respect to the substituent at the β-carbon of Thr analogs is also shown with amino acids 7 and 8. Whereas the substrate peptide incorporating the E-alkene 7 was dehydrated, the Z-alkene 8 was not (Figure S4).
In addition to evaluating the tolerance to variation of the methyl group of Thr, the importance of its stereochemistry was investigated. Dehydration of Thr results in Z-dehydrobutyrine in all lantibiotics investigated to date indicating an anti elimination mechanism.5 Substitution of Ser42 with allo-Thr, if tolerated by the enzyme, would result in formation of an E-dehydrobutyrine. In the event, a substrate analog peptide with allo-Thr at position 42 resulted in two dehydrations demonstrating that allo-Thr is not a substrate and hence that (R)-stereochemistry at the β-carbon is essential. The result with allo-Thr also strongly suggests that the products with amino acids 1, 4, 5, 6, and 7 have the Z-configuration as the proton and the alkyl substituent on the β-carbon apparently cannot switch binding pockets.
In a final experiment, the possibility to dehydrate (S)-β2-homoserine (9) was investigated. Incorporation of 9 into position 42 of the LctA analog and subsequent incubation with LctM led to two major products. One corresponds to two dehydrations and the second to a peptide that has undergone two dehydrations and one phosphorylation (Figure 2D). Thus the incorporation of a β-homoserine at the position of dehydration still results in phosphorylation by LctM, but the enzyme-catalyzed elimination is prohibited.
In summary, this study describes the stereoselective synthesis of 8 Thr analogs appropriately protected for SPPS. Use of these compounds demonstrates that the dehydratase domain of LctM has relaxed substrate specificity with respect to the structure of the residue to be dehydrated. Coupled with the overall substrate promiscuity of lantibiotic dehydratases, which have been shown to dehydrate Ser/Thr incorporated into non-lantibiotic peptides10,11 or into lantibiotic peptides at non-native positions,12 this class of enzymes provides a powerful tool for engineering dehydroamino acids into peptides and reengineering of the structures of lantibiotics.
Supplementary Material
Supporting Information Available: Synthetic routes and experimental procedures for all transformations that produced previously unknown compounds as well as their full spectral characterization.
ACKNOWLEDGMENT
This work was supported by the National Institutes of Health (GM58822). We thank S.E. Denmark for use of the SFC equipment.
REFERENCES
- 1.For selected examples, seeBodanszky M, Scozzie JA, Muramatsu I. J. Antibiot. 1970;23:9–12. doi: 10.7164/antibiotics.23.9.Berdy J. Adv. Appl. Microbiol. 1974;18:309–406.Tori K, Tokura K, Okabe K, Ebata M, Otsuka H, Lukacs G. Tetrahedron Lett. 1976;3:185–188.Pascard C, Ducruix A, Lunel J, Prangé T. J. Am. Chem. Soc. 1977;99:6418–6423. doi: 10.1021/ja00461a039.Pearce CJ, Rinehart KL., Jr. J. Am. Chem. Soc. 1979;101:5069–5070.Pedras MSC, Taylor JT, Nakashima TT. J. Org. Chem. 1993;58:4778–4780.Hamann MT, Scheuer PJ. J. Am. Chem. Soc. 1993;115:5825–5826.Lau RC, Rinehart KL., Jr. J. Antibiot. 1994;47:1466–1472. doi: 10.7164/antibiotics.47.1466.For selected synthetic studies, seeSnider BB, Zeng H. J. Org. Chem. 2003;68:545–563. doi: 10.1021/jo0264980.Nicolaou KC, Zak M, Safina BS, Lee SH, Estrada AA. Angew Chem Int Ed Engl. 2004;43:5092–5097. doi: 10.1002/anie.200461341.Ley SV, Priour A, Heusser C. Org. Lett. 2002;4:711–714. doi: 10.1021/ol017184m.
- 2.(a) Palmer DE, Pattaroni C, Nunami K, Chadha RK, Goodman M, Wakamiya T, Fukase K, Horimoto S, Kitazawa M, Fujita H, Kubo A, Shiba T. J. Am. Chem. Soc. 1992;114:5634–5642. [Google Scholar]; (b) Lombardi A, D'Agostino B, Nastri F, D'Andrea LD, Filippelli A, Falciani M, Rossi F, Pavone V. Bioorg. Med. Chem. Lett. 1998;8:1153–1156. doi: 10.1016/s0960-894x(98)00178-4. [DOI] [PubMed] [Google Scholar]; (c) Murkin AS, Tanner ME. J. Org. Chem. 2002;67:8389–8394. doi: 10.1021/jo0204653. [DOI] [PubMed] [Google Scholar]; (d) Li B, Yu J-PJ, Brunzelle JS, Moll GN, van der Donk WA, Nair SK. Science. 2006;311:1464–1467. doi: 10.1126/science.1121422. [DOI] [PubMed] [Google Scholar]
- 3.(a) Zhu Y, van der Donk WA. Org. Lett. 2001;3:1189–1192. doi: 10.1021/ol015648a. [DOI] [PubMed] [Google Scholar]; (b) Galonic D, van der Donk WA, Gin DY. Chem.- Eur. J. 2003;24:5997–6006. doi: 10.1002/chem.200305290. [DOI] [PubMed] [Google Scholar]
- 4.(a) Schmidt U, Lieberknecht A, Wild J. Synthesis. 1988:159–172. [Google Scholar]; (b) Bonauer C, Walenczyk T, König B. Synthesis. 2006:1–20. [Google Scholar]
- 5.Chatterjee C, Paul M, Xie L, van der Donk WA. Chem. Rev. 2005;105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- 6.Cotter PD, O'Connor PM, Draper LA, Lawton EM, Deegan LH, Hill C, Ross RP. Proc. Natl. Acad. Sci. USA. 2005;102:18584–18589. doi: 10.1073/pnas.0509371102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul M, van der Donk WA. Minirev. Org. Chem. 2005;2:23–37. [Google Scholar]
- 8.Xie L, Miller LM, Chatterjee C, Averin O, Kelleher NL, van der Donk WA. Science. 2004;303:679–681. doi: 10.1126/science.1092600. [DOI] [PubMed] [Google Scholar]
- 9.McClerren AL, Cooper LE, Quan C, Thomas PM, Kelleher NL, van der Donk WA. Proc. Natl. Acad. Sci. U.S.A. 2006;103:17243–17248. doi: 10.1073/pnas.0606088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rink R, Kuipers A, de Boef E, Leenhouts KJ, Driessen AJ, Moll GN, Kuipers OP. Biochemistry. 2005;44:8873–8882. doi: 10.1021/bi050081h. [DOI] [PubMed] [Google Scholar]
- 11.Kluskens LD, Kuipers A, Rink R, de Boef E, Fekken S, Driessen AJ, Kuipers OP, Moll GN. Biochemistry. 2005;44:12827–12834. doi: 10.1021/bi050805p. [DOI] [PubMed] [Google Scholar]
- 12.Chatterjee C, Patton GC, Cooper L, Paul M, van der Donk WA. Chem. Biol. 2006;13:1109–1117. doi: 10.1016/j.chembiol.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 13.For detailed experimental procedures, see the Supporting Information and for synthesis of 9 seeZhang X, Ni W, van der Donk WA. J. Org Chem. 2005;70:6685–6692. doi: 10.1021/jo051182o.
- 14.Garner P, Park JM. Org. Synth. 1991;70:18. [Google Scholar]
- 15.In addition to their application for probing the dehydratase substrate specificity, these procedures provide convenient access to the natural products 2-amino-3-hydroxy pentanoic acid 1 (or 3-hydroxynorvaline, HNV), 5, and 6. 1 is produced by bacteria and incorporated into antimicrobial peptides such as cycloheptamycin as well as into tRNA, and 5 and 6 have been isolated from the fungi Sclerotium rolfsii and Tricholomopsis rutilans. SeeGodtfredsen WO, Vangedal S, Thomas DW. Tetrahedron. 1970;26:4931–4946. doi: 10.1016/s0040-4020(01)93145-x.Reddy DM, Crain PF, Edmonds CG, Gupta R, Hashizume T, Stetter KO, Widdel F, McCloskey JA. Nucleic Acids Res. 1992;20:5607–5615. doi: 10.1093/nar/20.21.5607.Niimura Y, Hatanaka S-I. Phytochemistry. 1974;13:175.Potgieter HC, Vermeulen NMJ, Potgieter DJJ, Strauss HF. Phytochemistry. 1977;16:1757.Minoru S, Tetsuji M, Shuichi I.J. Antibiotics 1986393043082841
- 16.Evans TC, Jr., Xu MQ. Biopolymers. 1999;51:333–342. doi: 10.1002/(SICI)1097-0282(1999)51:5<333::AID-BIP3>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 17.Ayers B, Blaschke UK, Camarero JA, Cotton GJ, Holford M, Muir TW. Biopolymers. 1999;51:343–354. doi: 10.1002/(SICI)1097-0282(1999)51:5<343::AID-BIP4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 18.Chatterjee C, Miller LM, Leung YL, Xie L, Yi M, Kelleher NL, van der Donk WA. J. Am. Chem. Soc. 2005;127:15332–15333. doi: 10.1021/ja0543043. [DOI] [PubMed] [Google Scholar]
- 19.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 20.Dawson PE, Kent SBH. Annu. Rev. Biochem. 2000;69:923–960. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Available: Synthetic routes and experimental procedures for all transformations that produced previously unknown compounds as well as their full spectral characterization.