Abstract
The identification of cancer stem cells in leukemia, breast, brain, colon, and other cancers suggests that many tumors are maintained by stem cells in much the same way as normal tissues are maintained. Because cancer stem cells share remarkable phenotypic and functional similarities with normal stem cells, it may be difficult to identify therapeutic approaches to kill cancer stem cells without killing the normal stem cells in the same tissue. Yet in certain tissues, like the hematopoietic system and gut epithelium, this will be critical as regenerative capacity in these tissues is acutely required for life. Components of the PI-3kinase pathway, including Akt, mTor and FoxO are critical regulators of both normal stem cell function and tumorigenesis. Intriguingly, inactivation of some pathway components, like Pten, has opposite effects on normal hematopoietic stem cells (HSCs) and leukemia-initiating cells. This raises the possibility that drugs targeting this pathway could be more effective at eliminating cancer stem cells while being less toxic against normal stem cells.
Introduction
Stem cells possess two defining characteristics: the ability to self-renew and the capacity to differentiate. In recent years, a number of major cancers including acute myeloid leukemia [1; 2], breast cancer [3], brain cancers [4; 5; 6], and colon cancer [7; 8] have been shown to follow a cancer stem cell model in which cancer cells are hierarchically organized. In each of these cancers, a small population of cancer stem cells appears uniquely capable of forming new tumors and transferring disease. These cancer stem cells both self-renew to form more cancer stem cells as well as differentiate to form phenotypically diverse cancer cells with limited proliferative potential.
Leukemic stem cells are defined as cells that can initiate leukemia after transplantation into healthy recipient mice. Unfractionated human acute myelogenous leukemia (AML) cells are inefficient at transferring disease to immunocompromised mice. However, by using a flow-cytometer to isolate phenotypically distinct subsets of leukemia cells, leukemia-initiating cells can be enriched among CD34+CD38-; AML cells that represent 0.2–4% of leukemia cells [1]. Other leukemia cells are depleted for the ability to transfer disease. This proved that leukemias follow a cancer stem cell model where some leukemia cells have a greater capacity to transfer disease than others [1; 2; 9; 10]. A recent study has found that leukemia-initiating cells represent a surprisingly high proportion of leukemia cells in various mouse models of leukemia [11]. This raises the possibility that the frequency of leukemia-initiating cells among human AMLs has been underestimated, presumably as a result of the xenogeneic immune response they encounter after transplantation into immunocompromised mice. Nonetheless, AMLs that arise after Pten deletion in mice also follow a cancer stem cell model, with leukemia-initiating cells being 400-fold enriched among cells that express hematopoietic stem cell (HSC) markers [12]. These data indicate that at least some mouse leukemias follow a cancer stem cell model in which leukemia-initiating cells are highly enriched among cells that express markers similar to normal HSCs, just as observed among human AMLs [1; 2]. Many, but perhaps not all, mouse and human AMLs thus follow a cancer stem cell model in which cells capable of extensive proliferation and leukemia initiation represent a minority of leukemia cells.
Phenotypic and functional similarities between normal and cancer stem cells
A remarkable finding in cancer biology is that normal stem cells and cancer stem cells often share phenotypic and functional similarities. This has been best characterized in the hematopoietic system where HSCs and leukemia-initiating cells, or leukemic stem cells (LSCs) express many of the same cell surface markers. In addition to these phenotypic similarities, there are also remarkable similarities in the pathways that regulate self-renewal. The best example is the polycomb family transcriptional repressor, Bmi-1. Bmi-1 promotes the self-renewal of both normal HSCs and LSCs, in part by repressing the Ink4a and Arf tumor suppressors [13; 14; 15; 16; 17]. Bmi-1 is not required for the formation of HSCs or LSCs, but is required for their maintenance [13; 18]. Thus, in the absence of Bmi-1, HSCs and LSCs can form, but they fail to transfer hematopoiesis or leukemia upon transplantation. Beyond Bmi-1, there are also a number of other regulators of self-renewal in HSCs that have been implicated in leukemogenesis [19; 20; 21; 22]. The phenotypic and functional similarities among HSCs and LSCs suggest that it may be difficult to target LSCs without also killing HSCs.
Pten deletion has opposite effects on HSCs and LSCs
Pten (Phosphatase and tensin homologue) is a phosphatase that negatively regulates signaling through the phosphatidylinositol-3-OH kinase (PI-3kinase) pathway, attenuating proliferation and survival signals. Pten is the second most frequently mutated gene in human cancers (after p53), and is inactivated by a variety of mechanisms in some leukemias [23; 24; 25]. Furthermore, the PI-3kinase pathway is usually over-activated in a variety of malignancies including leukemia.
To test its effect on HSC function, we conditionally deleted Pten from adult HSCs [12]. Loss of Pten in HSCs led to myeloproliferative disease within days and transplantable AMLs and acute lymphoblastic leukemias (ALLs) within weeks. Pten-deficiency had no discernable effect on HSC differentiation or survival but caused HSCs to go into cycle. This caused a transient increase in HSC numbers; however, by three weeks after Pten deletion HSCs became depleted. Consistent with this, Pten-deficient whole bone marrow cells or purified HSCs were able to engraft in irradiated mice and give rise to all types of blood cell lineages, but only for the first several weeks after injection. The levels of reconstitution declined over time, and recipient mice were rarely long-term multilineage reconstituted by Pten-deficient cells. Longitudinal studies of mice that were chimeric for Pten-deficient and wild-type HSCs showed that the loss of HSCs over time reflected a cell-autonomous requirement for Pten in the maintenance of HSCs [12]. Similar results were independently obtained by Li and colleagues [26]. HSCs thus require Pten to maintain quiescence and to self-renew over time.
In contrast to this requirement for Pten in the maintenance of HSCs, LSCs arose and expanded in number after Pten deletion. The LSCs were transplantable and could be enriched among cells that expressed HSC markers [12]. Most mice died with AML and ALL within 6 weeks of Pten deletion.
The observation that Pten deletion had opposite effects on normal HSCs and LSCs raised the possibility that by targeting this pathway it would be possible to eliminate LSCs without affecting normal HSCs. Pten deletion leads to increased activation of Akt and mTor, the mammalian Target of rapamycin. These signaling kinases have multiple roles within the cell that include promoting proliferation, survival, protein translation, ribosome biogenesis, and glycolysis [27]. To test whether the effects of Pten deletion were mediated by mTor activation, we administered rapamycin, a potent and specific inhibitor of mTor, to Pten-deleted mice. Rapamycin not only eliminated LSCs and maintained the health of mice, but it also rescued the depletion of Pten-deficient HSCs. Pten-deficient HSCs could even give long-term multilineage reconstitution of irradiated mice as long as the mice were maintained on rapamycin. This demonstrated that both the expansion of LSCs and the depletion of normal HSCs were mediated by increased mTor activation. Thus, many of the effects of Pten-deletion were mediated by increased mTor activation. By targeting mTor, LSCs could be eliminated while normal HSC function was rescued.
Although Pten is frequently deleted in many kinds of cancer and the PI-3kinase pathway is usually over-activated in leukemia, Pten is rarely deleted in leukemia [28; 29; 30]. Moreover, inherited germline mutations in Pten [31] are associated with hamartomas and a high risk for breast, thyroid, and endometrial cancers but not an increased risk of leukemia [32]. These observations raise the question of why Pten is rarely deleted in leukemia and why inherited mutations in Pten predispose to many other cancers but not leukemia. Our results offer a potential explanation: HSCs are efficiently depleted after Pten deletion. This means that spontaneous mutations that lead to a loss of Pten from HSCs will lead to the depletion of these HSCs before they have an opportunity to progress to leukemia. As a result, Pten deletion would not represent an efficient path to leukemic transformation. What remains unknown is whether downstream hematopoietic progenitors are also eliminated after Pten deletion, or whether this response is stem cell specific. Nonetheless, it may not be as necessary to deplete downstream progenitors that lose Pten, as the half-lives of these cells are generally much shorter than HSCs, limiting their opportunity to accumulate the additional mutations required for leukemogenesis.
One prediction of this hypothesis is that tissues that give rise to cancers that commonly exhibit Pten deletion should also have stem cells that can tolerate Pten deletion without being eliminated. There is some evidence that this is the case. Pten deletion in both neural stem cells [33] and prostate stem cells [34] leads to sustained increases in self-renewal and stem cell numbers, in contrast to the effect we observed in the hematopoietic system. Furthermore, Pten deletion is frequently observed in brain tumors [35] and prostate cancer [36]. These observations suggest that Pten deletion is an efficient path to transformation only in tissues in which the stem/progenitor cells can tolerate Pten deficiency. Possible mechanisms by which Pten deletion might lead to the loss of certain stem cells are addressed in the next section.
Our observation that rapamycin selectively eliminated LSCs while not harming HSCs suggests that rapamycin and its analogues may be used to treat cancers that exhibit increased PI-3kinase pathway activation. A subset of patients with refractory/relapsed AML responded favorably to a rapamycin analogue [37; 38]. Nonetheless, trials that have tested rapamycin analogues as single agents in a variety of cancers have generally yielded disappointing results [39; 40; 41]. This raises the question of whether our studies of Pten deficient mice can provide any insight into how rapamycin analogues can be used more effectively against cancer. When rapamycin was administered immediately after Pten deletion, it was extremely effective at preventing the generation or maintenance of LSCs: as long as these mice were maintained on rapamycin, they remained healthy with no histological evidence of hematopoietic neoplasms [12]. However, when rapamycin was started weeks after Pten deletion when mice already had leukemia, rapamycin was effective at reducing LSCs and prolonging the life of mice; however, all of these mice eventually died with AML and ALL [12]. It is not clear whether this reduced response to rapamycin in advanced leukemias reflects the accumulation of additional mutations that reduce rapamycin sensitivity, or whether rapamycin simply has a better opportunity to decimate the LSC pool when the pool size is much smaller. Nonetheless, one possibility raised by these results is that rapamycin analogues will yield more encouraging results when used in patients with minimal residual disease.
Rapamycin analogues may also provide more promising results in combination with other agents. Rapamycin as a single agent showed only a modest effect when used to treat lymphomas in mice over-expressing Akt [42]. But, when these mice were treated with doxorubicin (a DNA intercalating agent) together with rapamycin, most of these mice achieved remissions lasting more than 60 days [42]. Additional data suggest that NF-kB, a transcription factor that regulates cell survival, can mediate rapamycin resistance and that concomitant inhibition of NF-kB and mTor increases the death of cancer cells [43]. These data suggest that rapamycin may sensitize tumors with activated mTor to other chemotherapeutics.
The PI-3kinase pathway, FoxO, and cellular senescence
What mechanisms are responsible for HSC depletion after Pten deletion? One obvious candidate is a senescence response. A prior study demonstrated that Pten deletion led to a p53-mediated senescence response in prostate epithelium progenitor cells [44]. The possibility that HSCs are depleted by a potent senescence response after Pten deletion is appealing because this could also potentially explain why LSCs do not exhibit this response: secondary mutations that occur during the progression of Pten-deficient cells to leukemia could inactivate this senescence response in leukemia cells. This raises the question of whether HSCs induce a senescence response after Pten deletion, and whether this is mediated by p53, Ink4a/Arf, or other gene products.
Our data suggest that whatever pathways lead to the depletion of HSCs after Pten deletion, that this response occurs downstream of mTor because it can be rescued by rapamycin treatment. However, the PI-3kinase pathway is a highly branched pathway, with many outputs and feedback loops, making it difficult to predict exactly which signal induces HSC depletion. mTor exists in two distinct complexes that function downstream and upstream of Akt: mTorc1 and mTorc2 [45; 46]. mTorc1 is downstream of Akt, controls the downstream effectors S6 kinase (S6K, which regulates ribosome biogenesis) and eukaryotic translation initiation factor 4E binding protein 1 (4E-BP1, which regulates protein translation), and is rapamycin sensitive [46]. mTorc2 activates Akt and is rapamycin insensitive [46]. Although Akt activation leads to mTorc1 activation, mTorc1 activation can reduce Akt activation by negative feedback mechanisms. Rapamycin treatment increases Akt signaling by disrupting the negative feedback inhibition of activated S6K on the PI-3kinase-Akt pathway ([47]; Figure 1). Rapamycin treatment can therefore activate Akt and the many effectors that lie downstream of Akt on pathways that parallel mTorc1. However, recent data suggest that in a small subset of cancers, including some AMLs, prolonged rapamycin treatment not only inhibits the mTorc1 complex but also prevents the assembly of the mTorc2 complex, thus blocking the activation of Akt [48; 49]. It is possible that prolonged rapamycin treatment inhibits Akt instead of further activating it in Pten-deficient HSCs. This raises the general question of whether HSC depletion is initiated by effectors that lie downstream of mTorc1, such as S6K or 4E-BP1, or parallel pathways that lie downstream of Akt.
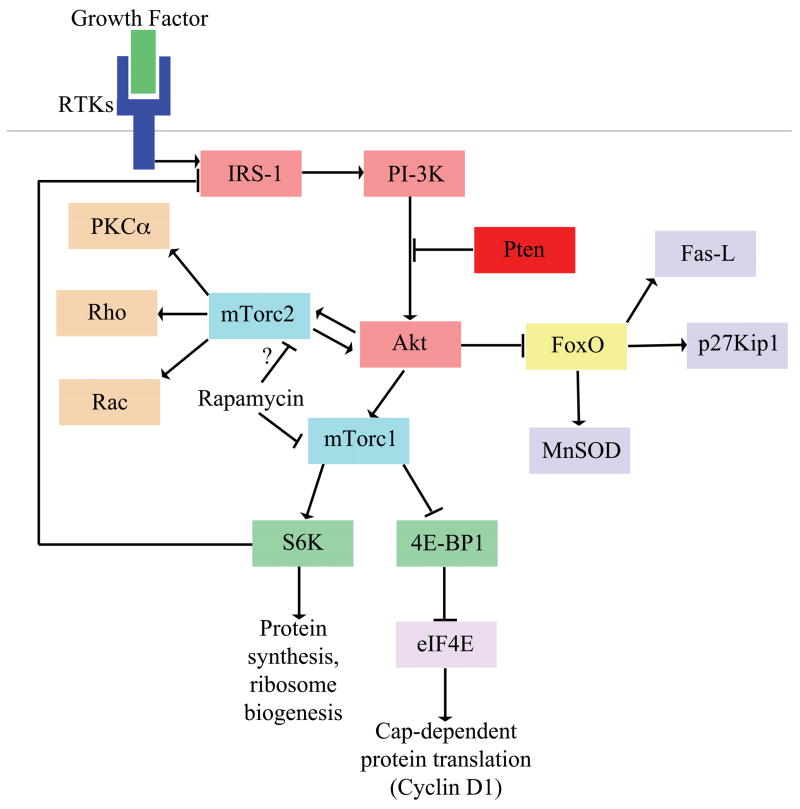
Figure 1. The PI-3kinase pathway and possible mechanisms of HSC depletion.
Activated receptor tyrosine kinases (RTKs) signal through scaffolding adaptors (e.g. IRS-1) that activate PI-3kinase. Pten suppresses the activation of Akt. Deletion of Pten leads to the hyperactivation Akt, loss of FoxO function, and activation of the mTorc1 and mTorc2 complexes. Important target genes of FoxO include regulators of cell death (e.g. Bim-1 and Fas-L), cell cycle progression (e.g. p21Cip1 and p27Kip1), and ROS detoxification (e.g. catalase and MnSOD). There are multiple pathways downstream of activated mTorc1, including S6 kinase (which regulates protein synthesis and ribosome biogenesis) and 4E-BP1 (which regulates cap-dependent translation). Rapamycin treatment reduces the activation of S6K, reduces the inhibition of 4E-BP and rescues the function of Pten-deficient HSCs. Although Akt activation leads to mTorc1 activation, mTorc1 activation can reduce Akt activation by a negative feedback mechanism involving S6K. Rapamycin treatment can disrupt this negative feedback mechanism and therefore increase Akt signaling, which affects the many effectors that lie downstream of Akt. Downstream targets of mTorc2 include kinases such as PKCα Rho, Rac, and Akt. Prolonged rapamycin treatment has been demonstrated to also inhibit assembly of the mTorc2 complex by sequestering mTor, leading to decreased Akt activation. This finding raises the possibility that rapamycin treatment may also restore FoxO function in Pten-deficient HSCs by inhibiting mTorc2. Abbreviations (RTKs, receptor tyrosine kinases; IRS-1, insulin receptor substrate-1; PI-3K, phosphatidylinositol-3-OH kinase; Fas-L, Fas ligand; MnSOD, manganese superoxide dismutase; mTorc1, mTor complex 1; mTorc2, mTor complex 2; 4E-BP1, eukaryotic translation initiation factor 4E binding protein 1; eIF4E, eukaryotic translation initiation factor 4E; S6K, S6 kinase; PKCα, protein kinase C alpha)
One set of effectors that lie downstream of Akt are the FoxO transcription factors that are inactivated upon phosphorylation by Akt. FoxO transcription factors regulate cell proliferation, cell survival, and promote the expression of enzymes that detoxify reactive oxygen species (ROS), like manganese superoxide dismutase and catalase [50]. As a result, ROS levels go up after Akt activation and persistent activation of Akt can lead to cell death due to ROS damage. Conditional inactivation of FoxO1, FoxO3, and FoxO4 leads to increased ROS levels in HSCs and to the subsequent loss of HSCs [51]. Like Pten-deficient bone marrow cells, FoxO-deficient bone marrow cells were unable to stably engraft irradiated recipient mice. It is likely that FoxO3a is the most important regulator of HSCs as FoxO3a deletion by itself leads to HSC depletion [52]. Treatment of FoxO-deficient mice with the anti-oxidant N-acetylcystine (NAC) restored HSC quiescence and at least partially rescued the depletion of primitive hematopoietic progenitors, though it was not tested whether the long-term multilineage reconstitution capacity of bone marrow cells was rescued [51]. It is unlikely that FoxO proteins mediate all of the effects of Pten deletion because Pten deletion is much more leukemogenic than deletion of FoxO1, FoxO3, and FoxO4 [53]. Nonetheless, FoxO genes could be one important mediator of the effects of Pten deletion on HSCs. Additional studies will be required to address the relationship between the mTor and FoxO pathways.
Conclusion
The cancer stem cell model predicts that cancer therapies must destroy cancer stem cells in order to be effective [54; 55]. Since cancer stem cells and normal stem cells often share remarkable phenotypic and functional similarities, these therapies may be toxic to normal HSCs. The different responses of LSCs and normal HSCs to Pten deletion offers the possibility of eliminating LSCs without harming normal HSCs by targeting this pathway. This provides proof-of-principle that functional differences between cancer stem cells and normal stem cells can be identified and therapeutically exploited. Ultimately, by developing a sophisticated understanding of stem cell self-renewal it may be possible to identify new classes of therapies that are more effective against cancer and less toxic to normal stem cells.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute. O.H.Y. was supported by a Medical Scientist Training Grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
BIBLIOGRAPHY
- 1.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukemia after transplantation into SCID mice. Nature. 1994;17:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 2.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nature Medicine. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 3.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100:15178–83. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–8. [PubMed] [Google Scholar]
- 6.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 7.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 8.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–5. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 9.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 10.Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 11.Kelly PN, Dakic A, Adams JM, Nutt SL, Strasser A. Tumor growth need not be driven by rare ‘cancer stem cells’. Science. 2007 doi: 10.1126/science.1142596. In Press. [DOI] [PubMed] [Google Scholar]
- 12.Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–82. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 13.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 14.Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–5. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 15.van der Lugt NMT, Alkema MJ, Berns A, Deschamps J. The Polycomb-group homolog Bmi-1 is a regulator of murine Hox gene expression. Mechanisms of Development. 1996;58:153–164. doi: 10.1016/s0925-4773(96)00570-9. [DOI] [PubMed] [Google Scholar]
- 16.Molofsky AV, He S, Kruger GM, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–7. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park IK, Qian D, Kiel M, Becker M, Pihalja M, Weissman IL, Morrison SJ, Clarke M. Bmi-1 is required for the maintenance of adult self-renewing hematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 19.Hock H, Hamblen MJ, Rooke HM, Schindler JW, Saleque S, Fujiwara Y, Orkin SH. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431:1002–7. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 20.Hock H, Meade E, Medeiros S, Schindler JW, Valk PJ, Fujiwara Y, Orkin SH. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 2004;18:2336–41. doi: 10.1101/gad.1239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato Y, Iwama A, Tadokoro Y, Shimoda K, Minoguchi M, Akira S, Tanaka M, Miyajima A, Kitamura T, Nakauchi H. Selecive activation of STAT5 unveils its role in stem cell self-renewal in normal and leukemic hematopoiesis. J Exp Med. 2005;202:169–179. doi: 10.1084/jem.20042541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Passegue E, Wagner EF, Weissman IL. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell. 2004;119:431–43. doi: 10.1016/j.cell.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 23.Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100:387–390. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- 24.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6:184–92. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 25.Levine AJ, Feng Z, Mak TW, You H, Jin S. Coorination and communication between the p53 and IGF-1-AKT-TOR signal transduction pathways. Genes & Development. 2006;20:267–275. doi: 10.1101/gad.1363206. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, Haug JS, Rupp D, Porter-Westpfahl KS, Wiedemann LM, Wu H, Li L. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–22. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- 27.Inoki K, Corradetti MN, Guan KL. Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet. 2005;37:19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- 28.Liu TC, Lin PM, Chang JG, Lee JP, Chen TP, Lin SF. Mutation analysis of PTEN/MMAC1in acute myeloid leukemia. Am J Hematol. 2000;63:170–175. doi: 10.1002/(sici)1096-8652(200004)63:4<170::aid-ajh2>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 29.Aggerholm A, Gronbaek K, Guldberg P, Hokland P. Mutational analysis of the tumour suppressor gene MMAC1/PTEN in malignant myeloid disorders. Eur J Haematol. 2000;65:109–13. doi: 10.1034/j.1600-0609.2000.90181.x. [DOI] [PubMed] [Google Scholar]
- 30.Sakai A, Thieblemont C, Wellmann A, Jaffe ES, Raffeld M. PTEN gene alterations in lymphoid neoplasms. Blood. 1998;92:3410–3415. [PubMed] [Google Scholar]
- 31.Simpson P. Notch and the choice of cell fate in Drosophila neuroepithelium. Trends Genet. 1990;6:343–345. doi: 10.1016/0168-9525(90)90260-d. [DOI] [PubMed] [Google Scholar]
- 32.Eng C. PTEN: one gene, many syndromes. Hum Mutat. 2003;22:183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- 33.Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X, Wu H. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- 34.Wang S, Garcia AJ, Wu M, Lawson DA, Witte ON, Wu H. Pten deletion leads to the expansion of a prostate stem/progenitor cell subpopulation and tumor initiation. Proc Natl Acad Sci U S A. 2006;103:1480–5. doi: 10.1073/pnas.0510652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knobbe CB, Merlo A, Reifenberger G. Pten signaling in gliomas. Neuro Oncol. 2002;4:196–211. [PMC free article] [PubMed] [Google Scholar]
- 36.Sulis ML, Parsons R. PTEN: from pathology to biology. Trends Cell Biol. 2003;13:478–83. doi: 10.1016/s0962-8924(03)00175-2. [DOI] [PubMed] [Google Scholar]
- 37.Recher C, Beyne-Rauzy O, Demur C, Chicanne G, Dos Santos C, Mas VM, Benzaquen D, Laurent G, Huguet F, Payrastre B. Antileukemic activity of rapamycin in acute myeloid leukemia. Blood. 2005;105:2527–34. doi: 10.1182/blood-2004-06-2494. [DOI] [PubMed] [Google Scholar]
- 38.Recher C, Dos Santos C, Demur C, Payrastre B. mTOR, a new therapeutic target in acute myeloid leukemia. Cell Cycle. 2005;4:1540–9. doi: 10.4161/cc.4.11.2159. [DOI] [PubMed] [Google Scholar]
- 39.Abraham RT, Gibbons JJ. The Mammalian Target of Rapamycin Signaling Pathway: Twists and Turns in the Road to Cancer Therapy. Clin Cancer Res. 2007;13:3109–3114. doi: 10.1158/1078-0432.CCR-06-2798. [DOI] [PubMed] [Google Scholar]
- 40.Margolin K, Longmate J, Baratta T, Synold T, Christensen S, Weber J, Gajewski T, Quirt I, Doroshow JH. CCI-779 in metastatic melanoma: a phase II trial of the California Cancer Consortium. Cancer. 2005;104:1045–8. doi: 10.1002/cncr.21265. [DOI] [PubMed] [Google Scholar]
- 41.Chang SM, Wen P, Cloughesy T, Greenberg H, Schiff D, Conrad C, Fink K, Robins HI, De Angelis A, Raizer J, Hess K, Aldape K, Lamborn KR, Kuhn J, Dancey J, Prados MD. Phase II study of CCI-779 in patients with recurrent glioblastoma multiforme. Invest New Drugs. 2005;23:357–61. doi: 10.1007/s10637-005-1444-0. [DOI] [PubMed] [Google Scholar]
- 42.Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW. Survival signalling by Akt and elF4E in oncogenesis and cancer therapy. Nature. 2004;428q:332–7. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 43.Ghosh S, Tergaonkar V, Rothlin CV, Correa RG, Bottero V, Bist P, Verma IM, Hunter T. Essential role of tuberous sclerosis genes TSC1 and TSC2 in NF-kappaB activation and cell survival. Cancer Cell. 2006;10:215–26. doi: 10.1016/j.ccr.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 44.Chen BP, Fraser C, Reading C, Murray L, Uchida N, Galy A, Sasaki D, Tricot G, Jagannath S, Barlogie B, et al. Cytokine-mobilized peripheral blood CD34+Thy-1+Lin- human hematopoietic stem cells as target cells for transplantation-based gene therapy. Leukemia. 1995;9(Suppl 1):S17–25. [PubMed] [Google Scholar]
- 45.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 46.Guertin DA, Sabatini DM. An expanding role for mTOR in cancer. Trends Mol Med. 2005;11:353–61. doi: 10.1016/j.molmed.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 47.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 48.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–68. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 49.Zeng Z, Sarbassov dos D, Samudio IJ, Yee KW, Munsell MF, Ellen Jackson C, Giles FJ, Sabatini DM, Andreeff M, Konopleva M. Rapamycin derivatives reduce mTORC2 signaling and inhibit AKT activation in AML. Blood. 2007;109:3509–12. doi: 10.1182/blood-2006-06-030833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 51.Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, Armstrong SA, Passegue E, DePinho R, Gilliland DG. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–39. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 52.Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, Matsuoka S, Miyamoto T, Ito K, Ohmura M, Chen C, Hosokawa K, Nakauchi H, Nakayama K, Nakayama KI, Harada M, Motoyama N, Suda T, Hirao A. Foxo3a is Essential for Maintenance of the Hematopoietic Stem Cell Pool. Cell Stem Cell. 2007;1:101–12. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 53.Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, Jiang S, Gilliland DG, Chin L, Wong WH, Castrillon DH, DePinho R. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–23. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 55.Rossi DJ, Weissman IL. Pten, tumorigenesis, and stem cell self-renewal. Cell. 2006;125:229–231. doi: 10.1016/j.cell.2006.04.006. [DOI] [PubMed] [Google Scholar]