Abstract
Bacterial secondary metabolites often contain attached carbohydrates that play a significant role in conferring biological activity. A small proportion of these bioactive sugars are derived from aminosugar oxidation to ultimately provide hydroxyaminosugars, nitrososugars, and nitrosugars. Recent advances in the elucidation of hydroxyaminosugar-, nitrososugar-, and nitrosugar-containing natural product gene clusters have enabled the proposal of biosynthetic pathways, the in vitro characterization of aminosugar oxidases, and the structure determination of key enzymes. This article focuses upon the key enzymatic transformations in aminosugar, hydroxyaminosugar, nitrososugar and nitrosugar biosynthesis, as well as the potential unique chemical reactivity of alkoxyaminosugars, with a particular focus upon developments within the last two years.
Introduction
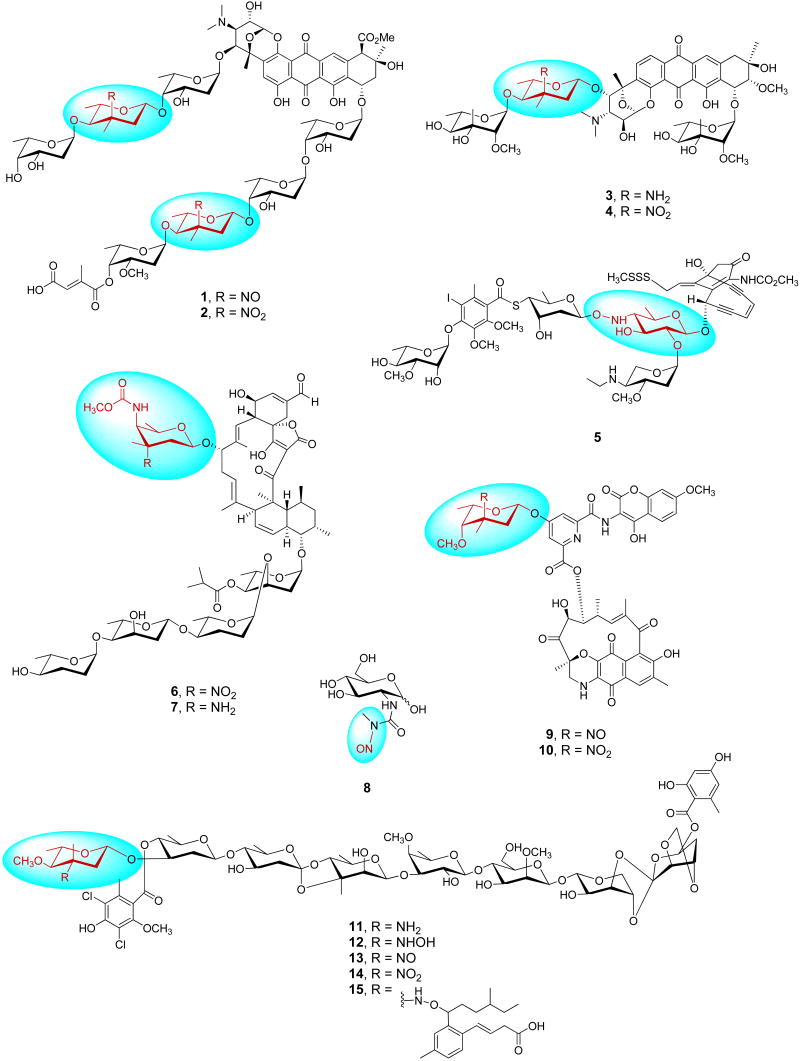
Glycosylated secondary metabolites continue to serve as an important source for drug discovery. Key to these natural pharmacophores, the attached sugars are often critical for biological activity and subtle alterations in natural product glycosylation can transform a secondary metabolite's pharmacological properties, molecular and cellular specificity, and even mechanism of action [1,2]. In terms of the carbohydrate structural diversity within glycosylated secondary metabolites, variations upon deoxy- and aminosugars are the most prevalent, the biosyntheses of which have been recently reviewed [3-6]. Although less common, aminosugar oxidation – specifically, hydroxyaminosugars, nitrososugars and nitrosugars – uniquely extends nature's glycochemical diversity. These exotic sugars are distributed among various natural product scaffolds including anthracyclines - arugomycin, viriplanins (Fig. 1, 1 and 2), cororubicin, and respinomycins (Fig. 1, 3 and 4); enediynes – calicheamicin (Fig. 1, 5) and esperamicin; spirotetronate antibiotics - tetracarcin A, kijanimicin, lobophorins, and arisostatins (Fig. 1, 6 and 7); ansamycins – rubradirins (Fig. 1, 9 and 10); and orthosomycins – evernimicins (Fig. 1, 11-14). Although strictly not a nitrososugar, the natural monosaccharide streptozocin (Fig. 1, 8) has also been included within this chemically diverse set of natural products. The attached carbohydrates of this unique natural product set are important to a broad range of biological effects exhibited by these compounds, including antibacterial, antitumor, antimalaria, anticholesterolemic, antiviral and antidiabetic activities. This article focuses upon the key enzymatic transformations in aminosugar, hydroxyaminosugar, nitrososugar and nitrosugar biosynthesis, as well as the potential unique chemical reactivity of alkoxyaminosugars, with a particular focus upon developments within the last two years.
Fig. 1.
Representative amino- hydroxyamino-, nitroso-, and nitrosugar-containing bacterial secondary metabolites - viriplanins A (1) and D (2) from Ampullariella regularis, respinomycins A (3) and D (4) from Streptomyces xanthocidicus, calicheamicin γI1 (5) from Micromonospora echinospora, arisostatins A (6) and B (7) from Micromonospora sp. TP-AO316, streptozotocin (8) from Streptomyces achromogenes, and evernimicin variants - aminosugar analog (11), hydroxylaminosugar analog (12), nitrososugar analog (13), everninomicin D (14) and Sch 49088 (15) from Micromonospora carbonacea.
Early discoveries and original biosynthetic hypotheses
The isolation of evernimicin (also referred to as everninomicin or Ziracin™) variants from M. carbonacea presented an early basis for studying the biosynthesis of hydroxyamino-, nitroso- and nitrosugars. Specifically, fermentation of M. carbonacea led to the isolation of the entire evernimicin series (amino-, hydroxyamino-, nitroso- and nitrosugar-containing) in addition to a novel alkoxyaminosugar metabolite Sch 49088 (Figure 1, 11-14) [7,8]. Consistent with this, a series of respinomycins (most notably the amino- and nitrosugar variants, Fig. 1, 3 and 4) were isolated from S. xanthocidicus [9], while a series of viriplanins (most notably the nitroso- and nitrosugar variants, Fig. 1, 1 and 2) were isolated from A. regularis [10]. Interestingly, modification of the rubradirin isolation strategy from S. achromogenes also led to the discovery of the corresponding nitrososugar variant, which readily converted photo-oxidatively to rubradirin and thereby implicated nitrosugar variants to be artifacts derived from spontaneous oxidation of their nitrososugar precursors (Fig. 1, 9 and 10) [11]. On the basis of these early studies, the biosynthesis of hydroxyamino-, nitroso-, and nitrosugars were anticipated to derive from successive oxidation of aminosugars, wherein some of the oxidative steps may be non-enzymatic, either before or after glycosyltransferase-catalyzed attachment to the natural product core structure (often referred to as the aglycon in such reactions).
Aminosugar biosynthesis – fundamentals and recent advances
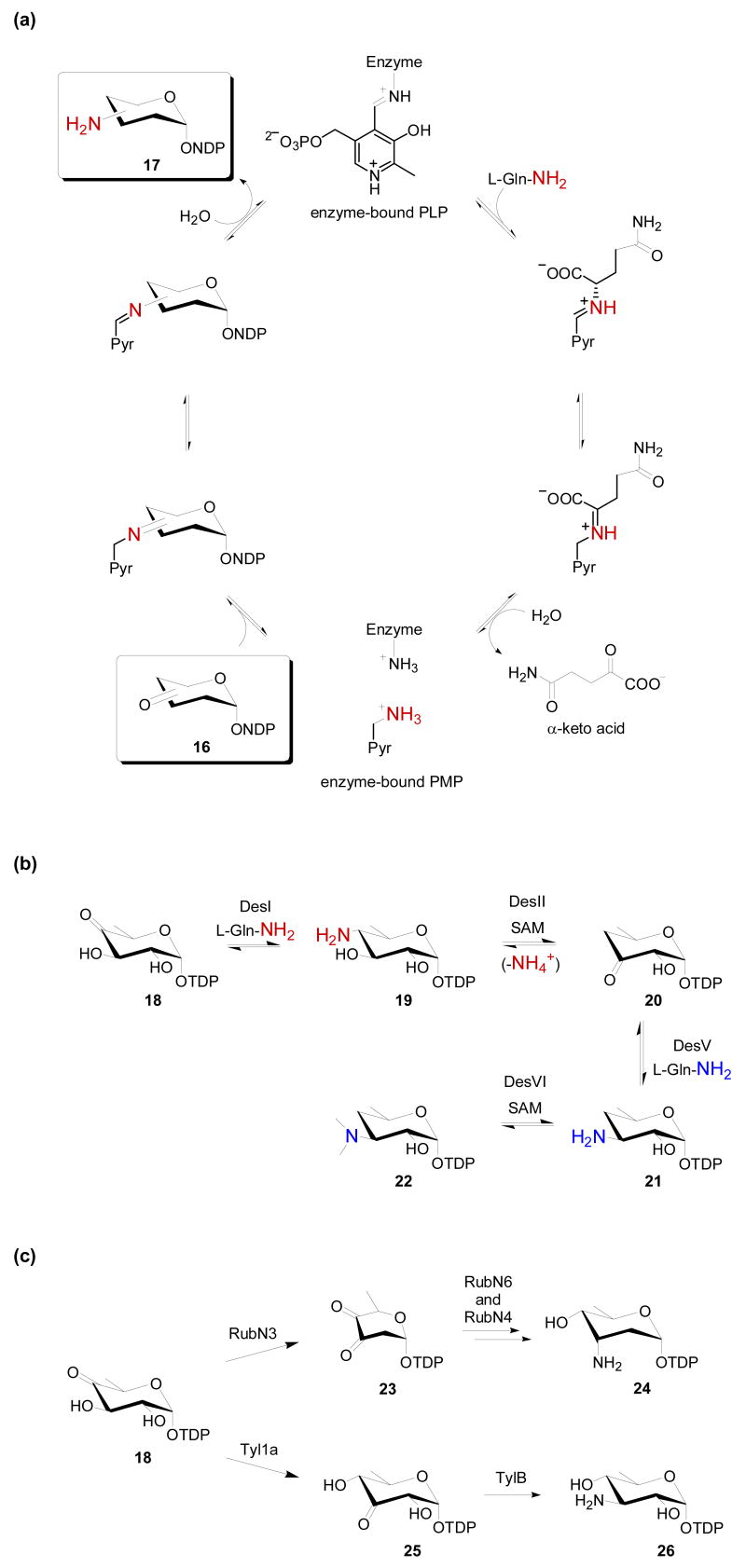
As discussed above, aminosugar nucleotides, or possibly the corresponding aminosugar glycosides, are the key starting materials for aminosugar N-oxidation. C2-, C3-, C4-, and/or C6-aminosugars are well-represented among glycosylated secondary metabolites wherein the C2- and/or C6-aminosugars are more common to aminoglycosides [12], while C3- and/or C4-aminosugars are more common to a wide range of metabolites including aromatic and macrolide polyketides, non-ribosomal peptides, polyenes, enediynes, ansamycins, indolocarbazoles, nucleosides, orthosomycins and oligosaccharides [3,6]. In all cases, amine installation is catalyzed by pyridoxal phosphate (PLP)-dependent enzymes known as aminotransferases (E.C. 2.6.1.16) using an amino acid (typically L-Gln) as the amino donor and a C2-, C3-, C4-, or C6-ketosugar nucleotide as the amino acceptor (Fig. 2a, 16). Unlike the biosynthesis of 2-aminosugars in primary metabolism, which derive from d-glucosamine-6-phosphate synthase-catalyzed amination of d-fructose-6-phosphate [13], the biosynthesis of aminosugars in secondary metabolism (including C2) derive from sugar nucleotide processes [3,6,12].
Fig. 2.
Fundamentals and recent advances relevant to aminosugar biosynthesis. (a) General catalytic mechanism of an aminotransferase. (b) The biosynthesis of TDP-d-desosamine. An unusual amination-deamination in this pathway contributes to C4 deoxygenation. (c) Aminotransferase-catalyzed transformation en route to TDP-d-rubranitrose (23→24) and TDP-d-mycaminose (18→26). PLP, pyridoxal 5′-phosphate; PMP, pyridoxamine 5′-phosphate; NDP, nucleoside diphosphate; TDP, deoxythymidine diphosphate; SAM, (S)-adenosylmethionine.
In terms of biosynthetic strategies, sugar amination is commonly also associated with C2, C3, C4 and/or C6 deoxygenation reactions. The most common of these, C6 deoxygenation, is catalyzed by a NAD+-dependent NDP-hexose-4,6-dehydratase to provide a biosynthetic intermediate common to most novel sugars - an NDP-4-keto-6-deoxyhexose [5,6]. Enzymatic deoxygenation at the remaining three positions proceeds via drastically different mechanisms [5]. Deoxygenation at C2 occurs via β-elimination followed by hydride reduction, while C3 deoxygenation occurs via an unprecedented PLP-dependent radical-based mechanism [5]. In contrast, Liu and co-workers recently revealed C4 deoxygenation to proceed via an aminosugar intermediate (Fig. 2b). Specifically, amino sugar 19, formed via DesI-mediated amination of 18, was converted by a newly characterized SAM-dependent deaminase DesII [14] to the corresponding 4,6-dideoxyhexose (20). To complete the biosynthesis of TDP-d-desosamine (22), a sugar precursor commonly employed in the biosynthesis of macrolide antibiotics such as erythromycin, a second aminotransferase-catalyzed reaction (DesV) is also required. In the absence of C4 deoxygenation, the alternative route to C3 aminosugars requires an isomerization to the requisite C3 ketosugar nucleotide precursor (Fig. 2c, 23) as elucidated in recent studies on TDP-d-rubranitrose (the nitrosugar precursor to rubradirin, Fig. 1, 10) and TDP-d-mycaminose (a precursor to the macrolide antibiotic tylosin) [15,16] (Fig. 2c). Using purified enzymes, both the d-rubranitrose and d-mycaminose studies revealed in vitro conversion of 4-keto species 23 or 18 to 3-amino species 24 or 26 in the presence of an isomerase/aminotransferase pair (RubN6/N4 or Tyl1a/TylB, respectively), while the latter study revealed Tyl1a was capable of generating the distinct 3-keto intermediate 25.
Additional advances in the last two years relevant to aminosugar biosynthesis include the reported gene clusters for the aminohexose-bearing enediynes neocarzinostatin and maduropeptin [17,18] and the aminopentose-containing indolocarbazole AT2433 [19], the in vitro characterization of the polyene (nystatin) sugar C3-aminotransferase NysDII [20] and the structure determination of sugar aminotransferases DesI and DesV [21,22].
Hydroxyaminosugar biosynthesis
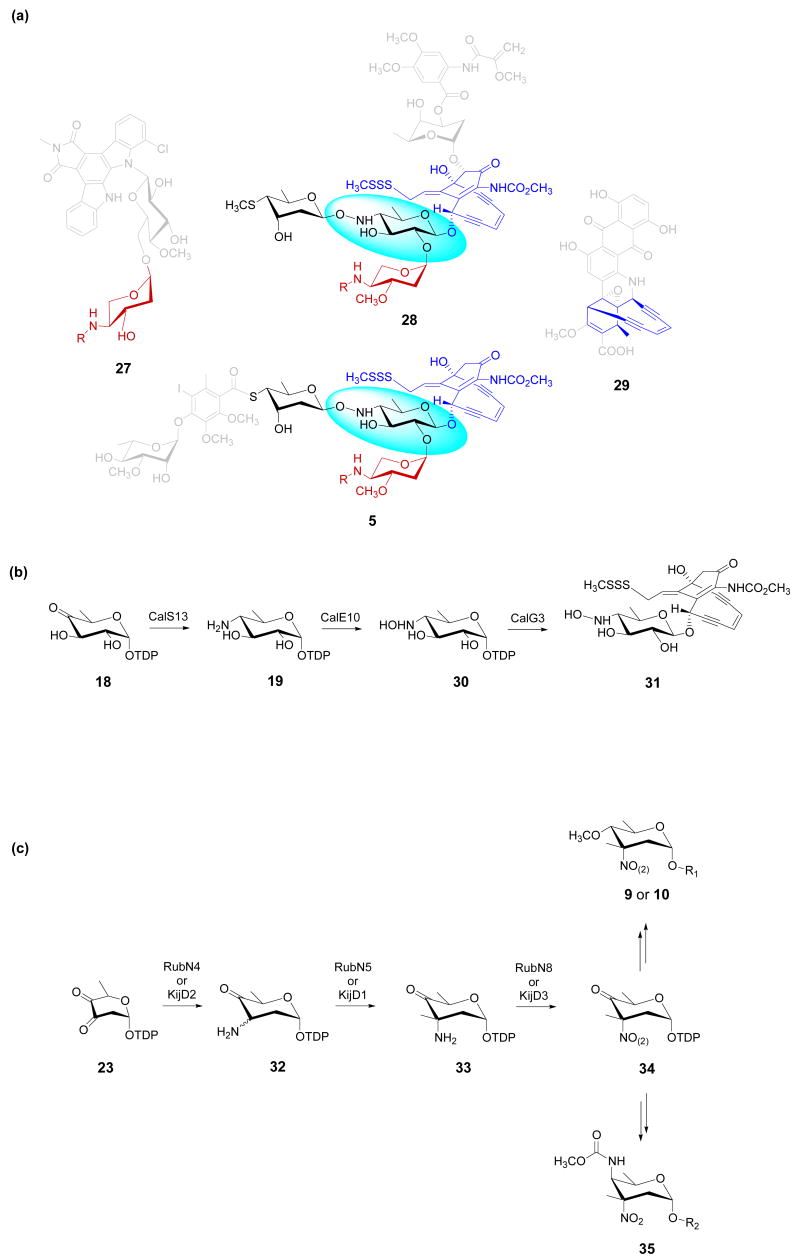
The unique conformation of the calicheamicin aryltetrasaccharide, which predominately derives from the hydroxyamino glycosidic bond (Fig. 1, 5), is essential for 5-DNA affinity and ultimately, the metabolite's remarkable ability to induce oxidative DNA strand scission [23-25]. The availability of the gene clusters encoding for the biosyntheses of the 10-membered enediynes calicheamicin [26] (Fig. 1 and 3a, 5), esperamicin (accession number AY267372) (Fig. 3a, 28), and dynemicin (Fig. 3a, 29) [27], as well as the indolocarbazole AT2433 (Fig. 3a, 27) [19] presented a genomic basis for proposing the biosynthetic pathway for the hydroxyaminosugar precursor TDP-4-hydroxyamino-6-deoxy-α-d-glucose (Fig. 3b, 30). Specifically, comparison of the three enediyne loci helped eliminate genes common to the enediyne core biosynthesis (a common element of all three enediynes), while a comparison of the calicheamicin, esperamicin, and AT2433 loci facilitated the elimination of genes involved in the biosynthesis of the aminopentose moiety common to these three metabolites. In conjunction with the known routes to aminosugar biosynthesis described in the previous paragraph, this information led to a proposed pathway wherein two P450-dependent enzymes (CalO2 and CalE10) were identified as candidates for the putative aminosugar N-oxygenase (Fig. 3b, 19→30). In vivo studies revealed calS13 (also known as calH) could complement a ΔdesV S. venezuelae disruption mutant and this complementation led to the production of a hybrid macrolide in which 4-amino-4,6-dideoxy-d-glucose was substituted for d-desosamine [28]. Based upon this in vivo study, CalS13 was assigned as the requisite C-4-ketosugar aminotransferase (Fig. 3b, 18→19), reminiscent of DesI (Fig. 2b, 18→19). Subsequent in vitro studies with CalO2 and CalE10 revealed only CalE10 could catalyze aminosugar N-oxidation and demonstrated, for the first time, the oxidation to occur at the sugar nucleotide stage to provide 30 (Fig. 3b) [29]. Furthermore, substrate specificity studies revealed CalE10-catalyzed oxidation to be both stereo- and regiospecific with only trace amounts of the corresponding nitrosugar detected. It should be noted that, while this was the first aminosugar N-oxygenase to be characterized, a related P450 enzyme (NocL, 41% identity) was recently characterized as the oxidase involved in nocardicin oxime formation [30].
Figure 3.
(a) Schematic representation of comparative genomics analysis for delineation of the genes involved in hydroxyaminosugar biosynthesis. Specifically, genes common to the esperamicin (28), calicheamicin (5) and dynemicin (29) loci are anticipated to be involved in the biosynthesis of the enediyne core (highlighted in blue), while genes common to the esperamicin (28), calicheamicin (5) and AT2433 (27) loci are anticipated to be involved in the biosynthesis of the common aminopentose moiety (highlighted in red). The remaining genes common to the esperamicin and calicheamicin loci are anticipated to be involved in thiosugar and hydroxyaminosugar biosynthesis. (b) The biosynthesis and attachment of 4,6-dideoxy-4-hydroxyamino-d-glucose in the calicheamicin-producer M. echinospora. All steps have been confirmed via in vitro biochemical studies. (c) Proposed biosynthetic pathways for the rubradirin nitroso/nitrosugar (rubranitrose) (9 or 10) and the kijanimicin nitrosugar (kijanose) (35). TDP, deoxythymidine diphosphate.
Nitrososugar and nitrosugar biosynthesis
The elucidation of the biosynthetic gene clusters encoding the nitrosugar-containing bacterial secondary metabolites kijanimicin (a spirotetronate structurally related to the arisostatins, Fig. 1, 7) [31] and rubradirins (Fig. 1, 10) [32] presented a basis from which to propose the corresponding nitrosugar biosynthetic pathways. While the sequence of proposed events differ between the two proposed pathways, both conceptually build upon well-established precedent for ketosugar nucleotide amination (Fig. 3c, 23→32) and C-methylation (Fig. 3c, 32→33) to provide a reasonable, potentially common, N-oxidase substrate (Fig. 3c, 33). The key N-oxidation step in these respective pathways (Fig. 3c, 33→34) was proposed to be catalyzed by FAD-dependent oxidoreductases RubN8 and KijD3. Consistent with this postulation, KijD3 shares high sequence identity with both RubN8 (57% identity, 68% similarity) and the evernimicin EvdC (61% identity, 72% similarity) [31]. At some point, the pathways diverge with subsequent elaboration, ultimately differentiating the core sugar architecture in rubradirins (Fig. 3c, 9 or 10) and kijanimicin (Fig. 3c, 35). It is important to note that, with the exception of the RubN6/N4 in vitro study discussed in the previous aminosugar section of this review, no additional biochemical support exists for either proposed route. With this in mind, a few key distinguishing features of the current postulations should be noted. First, in contrast to the established precedent of methyltransferase-catalyzed ketosugar-α-C-methylation [33,34], the formally proposed rubranitrose pathway lacks a critical C4-carbonyl for activation of the putative C3 carbon nucleophile. Second, N-oxidation in the rubranitrose pathway was formally proposed to occur after glycosyltransfer while the corresponding kijanimicin N-oxidation was suggested to occur at the sugar nucleotide stage. Third, on the basis of the previous isolation of a nitrososugar variant (Fig. 1, 9) [11], enzyme-catalyzed N-oxidation in the rubranitrose pathway was postulated to provide the nitrososugar, followed by spontaneous auto-oxidation to the nitrosugar. In contrast, kijanimicin N-oxidation was proposed to lead directly to the nitrosugar. Finally, although the absolute stereochemistry of d-rubranitrose has been a subject of some controversy, the chemical synthesis of both enantiomers and the absence of a 5-epimerase in the rubradirin biosynthetic gene cluster are consistent with the original d-sugar assignment [32,35].
Amino-, hydroxyamino-, nitroso- and nitrosugar glycosyltransferases
Although most glycosyltransferases are single polypeptides, it was recently discovered that the macrolide desosaminyltransferase (an aminosugar glycosyltransferase) DesVII required an ‘auxiliary’ or ‘accessory’ protein, Des VIII, for in vitro and in vivo activity [36,37]. Since this seminal discovery, other aminosugar glycosyltransferase/activating protein pairs have been discovered, such as AknS/AknT (aclacinomycin) [38], TylM2/TylM3 (tylosin) [39], MycB/MydC (mycinamycin) [39], EryCIII/EryCII (erythromycin) [40], and additional putative homologs [41]. While the ‘auxiliary/accessory’ protein was initially proposed to serve as an aminosugar nucleotide chaperone [36], in vitro characterization of a variety of auxiliary/accessory protein-independent aminosugar glycosyltransferases, including the vancomycin and teichoplanin GtfA and GtfE [42,43], vicenistatin VinC [44], amphotericin and nystatin AmphDI and NysDI [45], and the calicheamicin aminopentosyltransferase CalG4 [46], is inconsistent with this early hypothesis. More recent studies suggest a glycosyltransferase-activating role for these auxiliary/accessory proteins [40].
With respect to glycosyltransferases associated with N-oxidized sugars, the in vitro characterization and crystal structure of the calicheamicin hydroxyaminosugartransferase CalG3 (which transfers 4,6-dideoxy-4-hydroxyamino-α-D-glucose to the enediyne warhead) was recently reported [47]. In this same study, CalG2 was demonstrated to form the corresponding hydroxyamino-glycosidic bond to the adjacent thiosugar in the aryltetrasaccharide. Although biochemical studies pertaining to nitroso- or nitrosugar glycosyltransferases are lacking, disruption of rubG2 (the putative rudradirin nitrososugar glycosyltransferase) led to the production of the rubradirin aglycon in vivo, consistent with RubG2 as the requisite glycosyltransferase in this pathway [32].
The utility of alkoxyamine-appended natural products
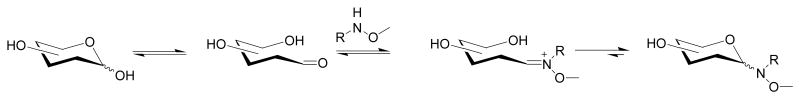
As highlighted in the previous paragraphs, nature has devised ingenious methods for expanding the chemical diversity of sugars to provide natural products appended with nitroso-, nitro- and hydroxy/alkoxyaminosugars, the latter of which offer unique opportunities for additional natural product diversification. Specifically, Peri and co-workers first revealed that simple methoxyamine-appended model compounds could readily react, under slightly acidic conditions, with free sugars to form ‘neoglycosides’ [48]. This reaction, which is particularly advantageous over classical glycosylation strategies as it does not require protecting groups or anomeric activation, has subsequently been exploited for the diversification (referred to ‘neoglycorandomization’ [49,50]) of a variety of natural product scaffolds including the cardenolide digitoxin [51], the alkaloid colchicine [52], and the nonribosomal peptide vancomycin [53]. In each of these cases, differentially-glycosylated natural product analogs with improved activity and/or selectivity were discovered. More recently, Langenhan and co-workers revealed that the neoglycosylation reaction is not simply limited to methoxyamine-appended molecules but, in fact, tolerates a variety of alkoxy substituents [54]. In this context, it is intriguing to consider the potential reactivity of naturally-occurring alkoxyamine-appended metabolites such as calicheamicin (Fig. 1, 5), esperamicin (Fig. 3a, 28), or the evernimicin analog Sch 49088 (Fig. 1, 15) in such chemoselective glycosylation reactions.
Concluding remarks
Given the importance of sugar attachments in mediating natural product bioactivity, advances in the study of novel sugar biosynthetic pathways will continue to provide new opportunities for in vivo pathway engineering [6]. In particular, the elucidation of the genes responsible for the key oxidation of aminosugar nucleotides to ultimately provide hydroxyamino-, nitroso-, and nitrosugars, clearly present a new set of genes to increase the sugar structural diversity of a vast array of natural products via in vivo pathway engineering. Although aminotransferase-catalyzed amine installation proceeds via a highly conserved mechanism, it is interesting to note that, like the variant routes to sugar deoxygenation, aminosugar nucleotide oxidation can be mediated via distinct oxidative enzymes (flavin-containing oxidases or P450s). However, the scope of naturally occurring sugar N-oxidation elucidated to date is limited to the C3 or C4 of the sugar (Fig. 1) and consistent with this, the first N-oxygenase to be characterized in vitro (CalE10) displayed notable regio- and stereospecificity. Thus, expanding sugar N-oxidation toward C2- or C6-aminosugar may ultimately depend upon structure-based engineering or enzyme evolution strategies, which will clearly be enabled via structural elucidation of aminosugar nucleotide oxidases. Finally, these unique aminosugar oxidases also potentially present enzymatic routes to the installation of chemoselective handles for differential glycosylation. Continued study of the structure and function of hydroxyamino-, nitroso-, and nitrosugar biosynthetic enzymes, as well as the many potential utilities of the corresponding novel sugars, are anticipated to offer many exciting opportunities.
Figure 4.
General schematic of a neoglycosylation reaction. In ‘neoglycorandomization’, R can be any complex natural product scaffold.
Acknowledgments
This contribution was supported in part by National Institutes of Health Grants CA84374 and U19 CA113297 (JST). JST is an H. I. Romnes fellow. The authors thank Dr. James A. Watson, Jr. for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
• of special interest
•• of outstanding interest
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 2.Weymouth-Wilson AC. The role of carbohydrates in biologically active natural products. Nat Prod Rep. 1997;14:99–110. doi: 10.1039/np9971400099. [DOI] [PubMed] [Google Scholar]
- •3.Nedal A, Zotchev SB. Biosynthesis of deoxyaminosugars in antibiotic-producing bacteria. Appl Microbiol Biotechnol. 2004;64:7–15. doi: 10.1007/s00253-003-1535-9. [DOI] [PubMed] [Google Scholar]; In this mini-review, the proposed biosynthetic pathways for the deoxyaminosugar components of both macrolide and non-macrolide antibiotics are highlighted.
- •4.Rupprath C, Schumacher T, Elling L. Nucleotide deoxysugars: essential tools for the glycosylation engineering of novel bioactive compounds. Curr Med Chem. 2005;12:1637–1675. doi: 10.2174/0929867054367167. [DOI] [PubMed] [Google Scholar]; This review provides a survey of the synthesis of TDP-activated sugars by chemical and chemoenzymatic approaches and discusses the promiscuity of glycosyltransferases. It summarizes the most important enzymes in the field of synthesis using enzymes from biosynthetic pathways.
- ••5.Thibodeaux CJ, Melançon CE, Liu Hw. Unusual sugar biosynthesis and natural product glycodiversification. Nature. 2007;446:1008–1016. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]; This review provides a contemporary review of nature's common biosynthetic strategies toward generating carbohydrate diversity and also highlights key advances for altering the glycosylation of natural product scaffolds.
- •6.Salas JA, Méndez C. Engineering the glycosylation of natural products in actinomycetes. Trends in Microbiol. 2007;15:219–232. doi: 10.1016/j.tim.2007.03.004. [DOI] [PubMed] [Google Scholar]; This review provides a contemporary survey of some of the common biosynthetic routes toward generating carbohydrate diversity and also highlights key in vivo strategies for engineering the natural product glycosylation.
- 7.Ganguly AK, Girijavallabhan VM, Miller GH, Sarre OZ. Chemical modification of everninomicins. J Antibiot. 1982;35:561–570. doi: 10.7164/antibiotics.35.561. [DOI] [PubMed] [Google Scholar]
- 8.Saksena AK, Jao E, Murphy B, Schumacher D, Chan TM, Puar MS, Jenkins JK, Maloney D, Cordero M, Pramanik BN, Bartner P, Das PR, McPhail AT, Girijavallabhan VM, Ganguly AK. Structure elucidation of SCH 49088, a novel everninomicin antibiotic containing an unusual hydroxylamino-ether sugar, everhydroxylaminose. Tetrahedron Lett. 1998;39:8441–8444. [Google Scholar]
- 9.Ubukata M, Uzawa J, Osada H, Isono K. Respinomycins A1, A2, B, C and D, a novel group of anthracycline antibiotics. II. Physico-chemical properties and structure elucidation. J Antibiot. 1993;46:942–951. doi: 10.7164/antibiotics.46.942. [DOI] [PubMed] [Google Scholar]
- 10.Hütter K, Baader E, Frobel K, Zeeck A. Viriplanin A, a new anthracycline antibiotic of the nogalamycin group. I. Isolation, characterization, degradation reactions and biological properties. J Antibiot. 1986;39:1193–1204. doi: 10.7164/antibiotics.39.1193. [DOI] [PubMed] [Google Scholar]
- 11.Bannister B, Zapotocky BA. Protorubradirin, an antibiotic containing a C-nitrososugar fragment, is the true secondary metabolite produced by Streptomyces achromogenes var. rubradiris. Rubradirin, described earlier, is its photo-oxidation product. J Antibiot. 1992;45:1313–1324. doi: 10.7164/antibiotics.45.1313. [DOI] [PubMed] [Google Scholar]
- •12.Flatt PM, Mahmud T. Biosynthesis of aminocyclitol-aminoglycoside antibiotics and related compounds. Nat Prod Rep. 2007;24:358–392. doi: 10.1039/b603816f. [DOI] [PubMed] [Google Scholar]; This topical review covers recent efforts toward understanding aminoglycoside biosynthesis.
- 13.Milewski S, Gabriel I, Olchowy J. Enzymes of UDP-GlcNAc biosynthesis in yeast. Yeast. 2006;23:1–14. doi: 10.1002/yea.1337. [DOI] [PubMed] [Google Scholar]
- •14.Szu Ph, He X, Zhao L, Liu Hw. Biosynthesis of TDP-d-desosamine: identification of a strategy for C4 deoxygenation. Angew Chem Int Ed. 2005;44:6742–6746. doi: 10.1002/anie.200501998. [DOI] [PubMed] [Google Scholar]; The biochemical characterization of purified DesII and the mechanistic implications for the overall C4 deoxygenation are reported. In this study, DesII was demonstrated to be a member of the S-adenosylmethionine (SAM) family of radical enzymes and a new strategy for sugar deoxygenation was identified.
- 15.Lamichhane J, Liou K, Lee HC, Kim CG, Sohng JK. Functional characterization of ketoreductase (rubN6) and aminotransferase (rubN4) genes in the gene cluster of Streptomyces achromogenes var. rubradiris. Biotechnol Lett. 2006;28:545–553. doi: 10.1007/s10529-006-0013-8. [DOI] [PubMed] [Google Scholar]
- 16.Melançon CE, Hong L, White J, Liu Yn, Liu Hw. Characterization of TDP-4-keto-6-deoxy-d-glucose-3,4-ketoisomerase from the d-mycaminose biosynthetic pathway of Streptomyces fradiae: in vitro activity and substrate specificity studies. Biochem. 2007;46:577–590. doi: 10.1021/bi061907y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W, Nonaka K, Nie L, Zhang J, Christenson SD, Bae J, Van Lanen SG, Zazopoulos E, Farnet CM, Yang CF, Shen B. The neocarzinostatin biosynthetic gene cluster from Streptomyces carzinostaticus ATCC 15944 involving two iterative type I polyketide synthases. Chem Biol. 2005;12:293–302. doi: 10.1016/j.chembiol.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 18.Van Lanen SG, Oh Tj, Liu W, Wendt-Pienkowski E, Shen B. Characterization of the maduropeptin biosynthetic gene cluster and Actinomadura madurae ATCC 39144 supporting a unifying paradigm for enediyne biosynthesis. J Am Chem Soc. 2007;129:13082–13094. doi: 10.1021/ja073275o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Q, Zhang C, Blanchard S, Thorson JS. Deciphering indolocarbazole and enediyne aminodideoxypentose biosynthesis through comparative genomics: insights from the AT2433 biosynthetic locus. Chem Biol. 2006;13:733–743. doi: 10.1016/j.chembiol.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Nedal A, Sletta H, Brautaset T, Borgos SEF, Sekurova ON, Ellingsen TE, Zotchev SB. Analysis of the mycosamine biosynthesis and attachment genes in the nystatin biosynthetic gene cluster of Streptomyces noursei ATCC 11455. Appl Environ Microbiol. 2007;73:7400–7407. doi: 10.1128/AEM.01122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •21.Burgie ES, Holden HM. Molecular architecture of DesI: a key enzyme in the biosynthesis of desosamine. Biochem. 2007;46:8999–9006. doi: 10.1021/bi700751d. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report describes the X-ray crystallographic structure of the PLP-dependent aminotransferase DesI, which is involved in the biosynthesis of TDP-desosamine. Upon comparison with PseC, the only other structurally-characterized sugar-modifying aminotransferase, an approximately 180° difference in sugar orientation was consistent with the stereochemical difference in amino transfer exhibited by these two biocatalysts.
- 22.Burgie ES, Tholden JB, Holden HM. Molecular architecture of DesV from Streptomyces venezuelae: a PLP-dependent transaminase involved in the biosynthesis of the unusual sugar desosamine. Prot Sci. 2007;16:887–896. doi: 10.1110/ps.062711007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar RA, Ikemoto N, Patel DJ. Solution structure of the esperamicin A(1)-DNA complex. J Mol Biol. 1997;265:173–186. doi: 10.1006/jmbi.1996.0719. [DOI] [PubMed] [Google Scholar]
- 24.Kumar RA, Ikemoto N, Patel DJ. Solution structure of the calicheamicin gamma(I)(1)-DNA complex. J Mol Biol. 1997;265:187–201. doi: 10.1006/jmbi.1996.0718. [DOI] [PubMed] [Google Scholar]
- 25.Galm U, Hager MH, Van Lanen SG, Ju JH, Thorson JS, Shen B. Antitumor antibiotics: Bleomycin, enediynes, and mitomycin. Chem Rev. 2005;105:739–758. doi: 10.1021/cr030117g. [DOI] [PubMed] [Google Scholar]
- 26.Ahlert J, Shepard EM, Lomovskaya N, Zazopoulos E, Staffa A, Bachmann BO, Huang K, Yang X, Fonstein L, Czisny A, Whitwam RE, Farnet CM, Thorson JS. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science. 2002;297:1173–1176. doi: 10.1126/science.1072105. [DOI] [PubMed] [Google Scholar]
- 27.Gao Q, Thorson JS. The biosynthetic genes encoding for the production of the dynemicin enediyne core in Micromonospora chersina ATCC 53710. FEMS Microbiol Lett. 2008 doi: 10.1111/j.1574-6968.2008.01112.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao L, Ahlert J, Xue Y, Thorson JS, Sherman DH, Liu Hw. Engineering a methymycin/pikromycin-calicheamicin hybrid: construction of two new macrolides carrying a designed sugar moiety. J Am Chem Soc. 1999;121:9881–9882. [Google Scholar]
- 29.Johnson HD, Thorson JS. Characterization of CalE10, the N-oxygenase involved in calicheamicin hydroxyaminosugar formation. doi: 10.1021/ja807557a. manuscript submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly WL, Townsend CA. Role of the cytochrome P450 NocL in nocardicin biosynthesis. J Am Chem Soc. 2002;124:8186–8187. doi: 10.1021/ja025926g. [DOI] [PubMed] [Google Scholar]
- ••31.Zhang H, White-Phillip JA, Melançon CE, III, Kwon Hj, Yu WI, Liu Hw. Elucidation of the kijanimicin gene cluster: insights into the biosynthesis of spirotetronate antibiotics and nitrosugars. J Am Chem Soc. 2007;129:14670–14683. doi: 10.1021/ja0744854. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report describes the cloning and in vitro characterization of genes involved in kijanimicin biosynthesis. Of particular interest is the identification of genes involved in the biosynthesis of the rare nitrosugar component, d-kijanose, and the proposal of a putative pathway for nitrogen incorporation and oxidation.
- ••32.Kim CG, Lamichhane J, Song KI, Nguyen VD, Kim DH, Jeong TS, Kang SH, Kim KW, Maharjan J, Hong YS, et al. Biosynthesis of rubradirin as an ansamycin antibiotic from Streptomyces achromogenes var. rubradiris NRRL3061. Arch Microbiol. 2007 doi: 10.1007/s00203-007-0337-3. [DOI] [PubMed] [Google Scholar]; A biosynthetic pathway for rubradirin was proposed based upon an analysis of the putative functions of gene products, the in vitro characterization of enzymes involved in sugar biosynthesis, and the evaluation of mutant strains. Of significant interest is the proposal of a sugar biosynthetic pathway for d-rubranitrose, a nitrosugar component of rubradirin.
- 33.Chen H, Zhao Z, Hallis TM, Guo Z, Liu Hw. Insights into the branched-chain formation of mycarose: methylation catalyzed by an (S)-adenosylmethionine-dependent methyltransferase. Angew Chem Int Ed. 2001;40:607–610. doi: 10.1002/1521-3773(20010202)40:3<607::AID-ANIE607>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 34.Chen H, Thomas MG, Hubbard BK, Losey HC, Walsh CT, Burkart MD. Deoxysugars in glycopeptide antibiotics: enzymatic synthesis of TDP-l-epivancosamine in chloroeremomycin biosynthesis. Proc Natl Acad Sci USA. 2000;97:11942–11947. doi: 10.1073/pnas.210395097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brimacombe JS, Rahman KMM. Syntheses of l-rubranitrose (2,3,6-trideoxy-3-C-methyl-4-O-methyl-3-nitro-l-xylo-hexopyranose) and the naturally occurring d enantiomer. Carbohydr Res. 1983;114:C1–C2. [Google Scholar]
- ••36.Borisova SA, Zhao L, Melancon CE, III, Kao CL, Liu Hw. Characterization of the glycosyltransferase activity of DesVII: analysis of and implications for the biosynthesis of macrolide antibiotics. J Am Chem Soc. 2004;126:6534–6535. doi: 10.1021/ja049967j. [DOI] [PubMed] [Google Scholar]; This is the first report to describe the in vitro activity of DesVII from S. venezuelae, the glycosyltransferase responsible for the attachment of TDP-desosamine to two macrolactones. DesVII was shown to require the additional protein component DesVIII for its activity and stands as the first disclosure of a two-component glycosyltransferase system.
- 37.Hong JSJ, Kim WS, Lee SK, Koh HS, Park HS, Park SJ, Kim YS, Yoon YJ. The role of a second protein (DesVIII) in glycosylation for the biosynthesis of hybrid macrolide antibiotics in Streptomyces venezuelae. J Microbiol Biotechnol. 2005;15:640–645. [Google Scholar]
- 38.Lu W, Leimkuhler C, Gatto GJ, Kruger RG, Oberthur M, Kahne D, Walsh CT. AknT is an activating protein for the glycosyltransferase AknS in l-aminodeoxysugar transfer to the aglycone of aclacinomycin A. Chem Biol. 2005;12:527–534. doi: 10.1016/j.chembiol.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 39.Melancon CE, III, Takahashi H, Liu Hw. Characterization of TylM3/TylM2 and MydC/MycB pairs required for efficient glycosyltransfer in macrolide antibiotic biosynthesis. J Am Chem Soc. 2004;126:16726–16727. doi: 10.1021/ja043900e. [DOI] [PubMed] [Google Scholar]
- •40.Yuan Y, Chung HS, Leimkuhler C, Walsh CT, Kahne D, Walker S. In vitro reconstitution of EryCIII activity for the preparation of unnatural macrolides. J Am Chem Soc. 2005;127:14128–14129. doi: 10.1021/ja053704n. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report describes the in vitro use of EryCIII, an aminosugar transferase, to prepare glycodiversified macrolides. The first biochemical support for the role of the accessory protein EryCII as an EryCIII-activating protein was also provided.
- 41.Hong JSJ, Park SJ, Parajuli N, Park SR, Koh HS, Jung WS, Choi CY, Yoon YJ. Functional analysis of DesVIII homologues involved in glycosylation of macrolide antibiotics by interspecies complementation. Gene. 2007;386:123–130. doi: 10.1016/j.gene.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 42.Losey HC, Jiang J, Biggins JB, Oberthür M, Ye XY, Dong SD, Kahne D, Thorson JS, Walsh CT. Incorporation of glucose analogs by GtfE and GtfD from the vancomycin biosynthetic pathway to generate variant glycopeptides. Chem Biol. 2002;9:1305–1314. doi: 10.1016/s1074-5521(02)00270-3. [DOI] [PubMed] [Google Scholar]
- 43.Oberthür M, Leimkuhler C, Kruger RG, Lu W, Walsh CT, Kahne D. A systematic investigation of the synthetic utility of glycopeptides glycosyltransferase. J Am Chem Soc. 2005;127:10747–10752. doi: 10.1021/ja052945s. [DOI] [PubMed] [Google Scholar]
- 44.Minami A, Uchida R, Eguchi T, Kakinuma K. Enzymatic approach to unnatural glycosides with diverse aglycon scaffolds using glycosyltransferase VinC. J Am Chem Soc. 2005;127:6148–6149. doi: 10.1021/ja042848j. [DOI] [PubMed] [Google Scholar]
- 45.Zhang C, Jiang J, Moretti R, Thorson JS. The in vitro characterization of polyene glycosyltransferases and implications for polyene biosynthesis and chemoenzymatic diversification. manuscript submitted. [Google Scholar]
- ••46.Zhang C, Griffith BR, Fu Q, Albermann C, Fu X, Lee IK, Li L, Thorson JS. Exploiting the reversibility of natural product glycosyltransferase-catalyzed reactions. Science. 2006;313:1291–1294. doi: 10.1126/science.1130028. [DOI] [PubMed] [Google Scholar]; This seminal report describes the ability to harness the reversibility of four glycosyltransferases to prepare libraries of NDP-sugars and exchange sugars and aglycons with ease. More than 70 differentially glycosylated calicheamicin and vancomycin analogs were prepared, demonstrating the power of this enzymatic approach for increasing the chemical diversity of natural products.
- 47.Zhang C, Bitto E, Goff RD, Griffith BR, Albermann C, Phillips G, Jr, Thorson JS. Biochemical and structural insights into calicheamicin glycosylation: studies on the ‘internal’ glycosyltransferases CalG2 and CalG3. manuscript submitted. [Google Scholar]
- •48.Peri F, Dumy P, Mutter M. Chemo- and stereoselective glycosylation of hydroxylamino derivatives: a versatile approach to glycoconjugates. Tetrahedron. 1998;54:12269–12278. [Google Scholar]; This notable report describes a general reaction between alkoxyamines and free sugars to generate ‘neoglycosides’. This synthetic strategy is particularly advantageous because it proceeds chemoselectively without prior activation of the sugar anomeric center and requires no protecting groups.
- 49.Langenhan JM, Griffith BR, Thorson JS. Neoglycorandomization and chemoenzymatic glycorandomization: two complementary tools for natural product diversification. J Nat Prod. 2005;68:1696–1711. doi: 10.1021/np0502084. [DOI] [PubMed] [Google Scholar]
- •50.Griffith BR, Langenhan JM, Thorson JS. ‘Sweetening’ natural products via glycorandomization. Curr Opin Biotechnol. 2005;16:622–630. doi: 10.1016/j.copbio.2005.10.002. [DOI] [PubMed] [Google Scholar]; This report highlights recent advances relevant to two complementary glycorandomization strategies. Chemoenzymatic glycorandomization is a biocatalytic approach dependent upon the substrate promiscuity of enzymes to activate and attach sugars to natural products. Neoglycorandomization is an efficient one-step chemical sugar ligation reaction that does not require prior sugar protection or activation.
- ••51.Langenhan JM, Peters NR, Guzei IA, Hoffmann FM, Thorson JS. Enhancing the anticancer properties of cardiac glycosides by neoglycorandomization. Proc Natl Acad Sci USA. 2005;102:12305–12310. doi: 10.1073/pnas.0503270102. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report describes the first application of neoglycorandomization to a natural product scaffold. The installation of a methoxyamine functional handle facilitated the direct glycosylation of the digitoxin aglycon, which provided access to 78 neoglycosides. These analogs exhibited significantly enhanced potency and tumor specificity, as compared to the parent natural product.
- 52.Ahmed A, Peters NR, Fitzgerald MK, Watson JA, Jr, Hoffmann FM, Thorson JS. Colchicine glycorandomization influences cytotoxicity and mechanism of action. J Am Chem Soc. 2006;128:14224–14225. doi: 10.1021/ja064686s. [DOI] [PubMed] [Google Scholar]
- 53.Griffith BR, Krepel C, Fu X, Blanchard S, Ahmed A, Edmiston CE, Thorson JS. Model for antibiotic optimization via neoglycosylation: synthesis of liponeoglycopeptides active against VRE. J Am Chem Soc. 2007;129:8150–8155. doi: 10.1021/ja068602r. [DOI] [PubMed] [Google Scholar]
- •54.Langenhan JM, Engle JM, Slevin LK, Fay LR, Lucker RW, Smith KR, Endo MM. Modifying the glycosidic linkage in digitoxin analogs provides selective cytotoxins. Bioorg Med Chem Lett. 2008;18:670–673. doi: 10.1016/j.bmcl.2007.11.058. [DOI] [PubMed] [Google Scholar]; This report describes the application of neoglycorandomization to prepare a small library of linkage-diversified digitoxin neoglycosides. Interestingly, the chemical composition of the secondary oxyamine linkage was shown to influence both the potency and selectivity of digitoxin analogs.