Abstract
Learned vocalization, the substrate for human language, is a rare trait. It is found in three distantly related groups of birds—parrots, hummingbirds, and songbirds. These three groups contain cerebral vocal nuclei for learned vocalization not found in their more closely related vocal nonlearning relatives. Here, we cloned 21 receptor subunits/subtypes of all four glutamate receptor families (AMPA, kainate, NMDA, and metabotropic) and examined their expression in vocal nuclei of songbirds. We also examined expression of a subset of these receptors in vocal nuclei of hummingbirds and parrots, as well as in the brains of dove species as examples of close vocal nonlearning relatives. Among the 21 subunits/subtypes, 19 showed higher and/or lower prominent differential expression in songbird vocal nuclei relative to the surrounding brain subdivisions in which the vocal nuclei are located. This included relatively lower levels of all four AMPA subunits in lMAN, strikingly higher levels of the kainite subunit GluR5 in the robust nucleus of the arcopallium (RA), higher and lower levels respectively of the NMDA subunits NR2A and NR2B in most vocal nuclei and lower levels of the metabotropic group I subtypes (mGluR1 and -5) in most vocal nuclei and the group II subtype (mGluR2), showing a unique expression pattern of very low levels in RA and very high levels in HVC. The splice variants of AMPA subunits showed further differential expression in vocal nuclei. Some of the receptor subunits/subtypes also showed differential expression in hummingbird and parrot vocal nuclei. The magnitude of differential expression in vocal nuclei of all three vocal learners was unique compared with the smaller magnitude of differences found for nonvocal areas of vocal learners and vocal nonlearners. Our results suggest that evolution of vocal learning was accompanied by differential expression of a conserved gene family for synaptic transmission and plasticity in vocal nuclei. They also suggest that neural activity and signal transduction in vocal nuclei of vocal learners will be different relative to the surrounding brain areas.
Indexing terms: song system, song nuclei, neurotransmitter
Vocal learning is the ability to imitate sounds, usually species-specific vocalizations. It has been experimentally found in only six animal groups, all of which are distantly related. These are three groups of mammals (humans, bats, and cetaceans) and three groups of birds (parrots, hummingbirds, and songbirds; see Fig. 1A; Nottebohm, 1972; Jarvis et al., 2000). In other groups tested, including nonhuman primates, vocalizations are found to be only innate (Marler and Mitani, 1988). Learned vocalizations are usually composed of a set of complex syllables forming syntax that is modifiable, compared with innate vocalizations. The three vocal learning bird groups have seven similar but not identical cerebral vocal nuclei for learned vocalization (Fig. 1B; Jarvis et al., 2000). These nuclei have not been found in closely or distantly related vocal nonlearners. Most information about cerebral vocal nuclei is known in songbirds, the zebra finch in particular, where they form two major networks: 1) a posterior vocal pathway unique to vocal learners that is involved in production of learned vocalizations (see Fig. 1B, yellow-labeled nuclei) and 2) an anterior vocal pathway also unique to vocal learners that is involved in vocal learning and vocal syntax and is differentially regulated in different social contexts (see Fig. 1B, red-labeled nuclei). A third system, an auditory pathway, is involved in audition and auditory learning and has been found in both vocal learners and vocal nonlearners (see Fig. 1B, blue-labeled nuclei; Nottebohm et al., 1976; Scharff and Nottebohm, 1991; Yu and Margoliash, 1996; Jarvis et al., 1998, 2000; Hessler and Doupe, 1999b; Ribeiro and Mello, 2000; Kobayashi et al., 2001).
Fig. 1.
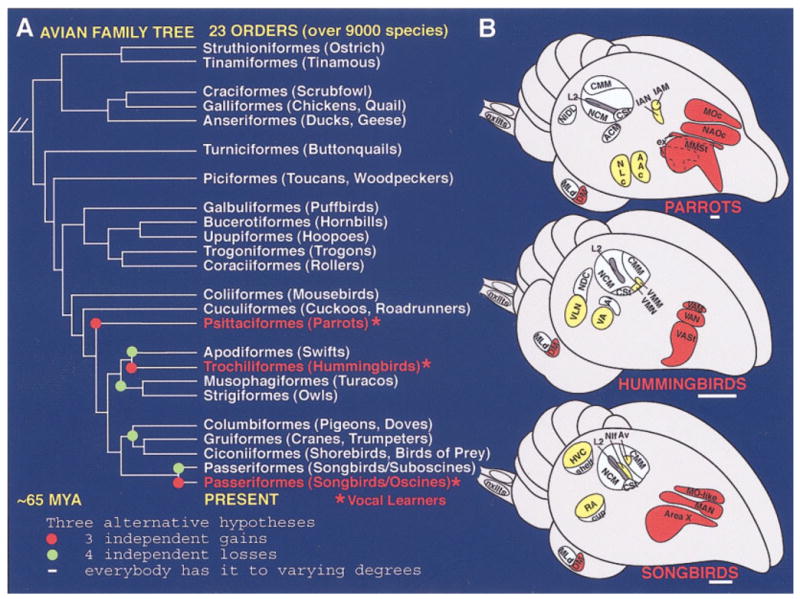
A: Phylogenetic relationships of living birds (Sibley and Ahlquist, 1990) with vocal learners highlighted in red, possible independent gains of vocal learning highlighted with red dots, or possible independent losses highlighted with green dots. B: Semi-3D view of seven cerebral vocal regions (yellow and red) found in each of the vocal learning bird groups and of auditory regions (blue) found in all birds. The three red-labeled vocal nuclei in the cerebrums are in nearly identical locations in all three vocal learning orders, and in songbirds constitute what is called the anterior vocal pathway. The four yellow-labeled cerebral vocal nuclei are in different locations across vocal learning orders, but in the same brain subdivisions relative to each other, and in songbirds constitute what is called the posterior vocal pathway. The posterior RA-like nucleus (hummingbird VA and parrot AAC) projects to the brainstem motor neurons (nXIIts) that control the syringeal muscles for production of sounds (Nottebohm et al., 1976; Paton et al., 1981; Striedter, 1994; Durand et al., 1997; Gahr, 2000). See Figure 4A,B for neural connectivity in songbirds. (Modified from Jarvis et al., 2000.) Scale bars = 1 mm.
Each vocal nucleus has its own unique temporal and spatial firing pattern to control either the muscles of the syrinx and respiratory apparatus or other nuclei during singing or the practice of singing (Yu and Margoliash, 1996; Hessler and Doupe, 1999a,b; Hahnloser et al., 2002). Furthermore, singing induces expression of action-potential-regulated genes in these nuclei with different patterns for different genes, and this has been found for all three vocal learning groups (Jarvis and Nottebohm, 1997; Kimpo and Doupe, 1997; Jarvis and Mello, 2000; Jarvis et al., 2000; Li and Jarvis, 2001; Wada and Jarvis, unpublished results). Neurotransmission of activity and the regulation of these activity-dependent genes are thought to occur by neurotransmitter release onto neurotransmitter receptors (Worley et al., 1990; Lerea, 1997; Clayton, 2000; Jarvis, 2004a). For these reasons, we considered whether the neurotransmitter receptors are different for each vocal nucleus in vocal learners.
In vertebrates, one of the most important neurotransmitter receptors for excitatory transmission with a role in learning and generation of complex behavior is the glutamate receptor family (Abel and Lattal, 2001). In mammals, in total 26 receptor subunits/subtypes have been cloned to date, and they have been classified into four subfamilies based on their functions or pharmacological agents to which their receptor subunit combinations preferentially bind: three ionotropic receptor subfamilies, 1) α-amino-3-hydroxy-5-methyl-4-isoxazole proprionic acid (AMPA), 2) kainate (KA), and 3) N-methyl-D-aspartate (NMDA); and 4) the metabotropic subfamily (Pin and Duvoisin, 1995; Dengledine et al., 1999; Hollmann, 1999; Gustinci et al., 2003). Each ionotropic receptor is typically formed from hetero- or homotetrads of different receptor subunits encoded by different genes. Each metabotropic receptor is typically a monomer encoded by a single gene and, thus, is classified as a receptor subtype. Upon binding glutamate, the receptors directly gate ion channels (ionotropic receptors) and/or change conformation (metabotropic receptors) to transmit electrical signals between neurons and regulate expression of targeted genes via second messengers. Each receptor subunit/subtype has a unique brain expression pattern indicative of different brain subdivisions or cell types having different glutamate receptor functions (Bahn et al., 1994; Conti et al., 1994; Standaert et al., 1994; Testa et al., 1994). Furthermore, some of these subunits/subtypes have different mRNA splice variants contributing further to the diversity of receptor function.
In songbirds, pharmacological studies indicate the presence of glutamate receptors in vocal nuclei; specifically, NMDA receptors have been shown to be required for vocal learning (Basham et al., 1996) and have kinetics that change during vocal learning critical periods (Stark and Perkel, 1999; White et al., 1999; Dutar et al., 2000; Livingston et al., 2000). However, to date only three glutamate receptor subunits have been cloned from the songbird brain (NR1, NR2A, and NR2B, belonging to the NMDA subfamily; Singh et al., 2000; Heinrich et al., 2002). Here we report the identification and presence of nearly all glutamate receptor subunits/subtypes discovered in mammals within the songbird brain. Furthermore, we compared expression of four genes from each of the subfamilies that had most differential expression in zebra finches (GluR1, GluR5, NR2A, and mGluR2) in hummingbirds, parrots, and doves (the latter of which are vocal nonlearners). We found that several of these subunits/subtype show unique differential expression in cerebral vocal nuclei of all three distantly related vocal learning bird groups, suggesting that differential expression of neurotransmitter receptors in vocal systems contribute to learned vocal communication.
MATERIALS AND METHODS
Animals
The following representative species were used for this study: zebra finches (Taeniopygia guttata), a songbird (n = 16; six for in situ hybridizations, six for immunohistochemistry, and four for Western blotting); sombre hummingbirds (Aphantochroa cirrochloris; n = 3); budgerigars (Melospititicus undulatus), a parrot (n = 3); and doves (vocal nonlearners)—ring doves (Stretophilia risoria; n = 3) and rock doves (pigeon; Columbia livia; n = 3). We chose these species because they are among the best studied in terms of the neuroethology of learned and innate vocal communication (Nottebohm, 1993; Striedter, 1994; Durand et al., 1997; Slabbekoorn et al., 1999; Jarvis and Mello, 2000; Jarvis et al., 2000). We examined only males, because this is the sex that most often demonstrates vocal learning. All birds were adults. The zebra finches, 150 days to ~1 year old, were obtained from our breeding colonies at the Duke University Medical Center and Tokyo Medical and Dental University. Because we wanted to examine expression of glutamate receptors in the brain in the absence of vocal behavior, birds were kept under relatively quiet conditions, not singing, for several hours to overnight before sacrifice. Brain sections of comparable quiet control sombre hummingbirds and budgerigars were obtained from our collection (Jarvis and Mello, 2000; Jarvis et al., 2000). Dove brains were collected from a parallel study conducted by Michael McElroy in the Jarvis laboratory. All animals were treated under the humane guidelines of Duke University and Tokyo Medical and Dental University, and experiments were approved by animal care committees.
RT-PCR and cloning of glutamate receptors
Detailed cloning will be described separately (Wada et al., in preparation). Briefly, we attempted to clone cDNAs of 21 of 26 known mammalian glutamate receptor subunits/subtypes (Gustinci et al., 2003) from zebra finch brain mRNA [four AMPA (GluR1–4), five kainate (GluR5–7, KA1–2), six NMDA (NR1, NR2A–D, NR3A), and six metabotropic (mGluR1–5, mGluR8)]. Degenerate primers were made to conserved, unique regions of each subfamily among mammals (mouse, rat, and human) and when available with other vertebrates [Xenopus, chicken, and the previously described zebra finch NR2B sequence (Basham et al., 1999)]. We chose primers that would amplify regions with sequences specific to that subunit/subtype, to prevent cross-hybridizations to the related subunits/subtypes in in situ hybridization. Splice-variant-specific primers were also made to the alternative Flip and Flop domains of the AMPA subunits. RT-PCR was then performed on total RNA from adult male zebra finch brains with the appropriate degenerate primer pairs. PCR products were examined on 1.5% agarose gels and, if present, cut from the gel and cloned into the pGEMTeasy plasmid (Promega, Madison, WI). These plasmids were transformed into XL-1 blue supercompetent cells (Stratagene, La Jolla, CA), and Escherichia coli colonies were selected under ampicillin (100 μg/ml) agarose plates. Colonies were then picked and grown in LB broth with ampicillin (100 μg/ml), and DNA minipreps were prepared. Inserts were 5′- and 3′-end sequenced and sequences passed through BLASTN (DNA) and BLASTX (protein) GenBank searches. If a clone revealed homology to the expected glutamate receptor subunit/subtype of other species, then the entire clone was sequenced. All sequences have been deposited into GenBank (accession Nos. in Table 1).
TABLE 1.
Sequence Comparison of Cloned Partial Zebra Finch Glutamate Receptor Subunits with Humans1
| Subunits | zf/Human residue | zf Accession No. | Human accession No. | cDNA identity (%) | aa identity (%) |
|---|---|---|---|---|---|
| AMPA | |||||
| GluR1 | 325–749aa/907aa | AB042749 | M64752 | 85 | 93 |
| GluR2 | 1–883aa/883aa | AB042750 | I58181 | 92 | 92 |
| GluR3 | 342–767aa/894aa | AB042751 | S50128 | 86 | 96 |
| GluR4 | 333–635aa/902aa | AB042752 | P48058 | 89 | 95 |
| Kalnate | |||||
| GluR5 | 651–757aa/905aa | AB107127 | U16125 | 85 | 98 |
| GluR6 | 651–757aa/869aa | AB107128 | AJ301610 | 86 | 100 |
| GluR7 | 653–758aa/919aa | AB107129 | U16127 | 85 | 96 |
| KA1 | 535–591aa/956aa | AB107130 | S67803 | 90 | 98 |
| KA2 | 444–631aa/980aa | AB107131 | S40369 | 86 | 89 |
| NMDA | |||||
| NR1 | 705–843aa/938aa | AB042756 | Q05586 | 85 | 99 |
| NR2A | 647–819aa/1,464aa | AB042757 | Q12879 | 83 | 98 |
| NR2B | 801–937aa/1,482aa | AB107125 | U9027 | 87 | 97 |
| NR2C | 645–817aa/1,236aa | AB042758 | U77782) | 87 | 95 |
| NR2D | 675–847aa/1,336aa | AB042759 | U77783 | 86 | 90 |
| NR3A | 489–621aa/1,155aa | AB107126 | AF416558 | 78 | 86 |
| Metabotropic | |||||
| mGluR1 | 97–810aa/906aa | AB042753 | L76631 | 82 | 92 |
| mGluR2 | 167–761aa/872aa | AB042754 | AB045011 | 72 | 78 |
| mGluR3 | 165–757aa/877aa | AB107132 | Q14832 | 83 | 83 |
| mGluR4 | 97–777aa/912aa | AB042755 | Q14833 | 85 | 86 |
| mGluR5 | 445–781aa/1180aa | AB107133 | D28538 | 84 | 96 |
| mGluR8 | 172–791aa/908aa | AB107134 | U92459 | 84 | 96 |
| AMPA splice-specific | |||||
| GluR1 Flip | 763–812aa/907aa | AB120327 | M64752 | 96 | 100 |
| GluR1 Flop | 530–580aa/644aa | AB120328 | AF167332 | 95 | 100 |
| GluR2 Flip | 643–692aa/756aa | AB120329 | BC028736 | 96 | 100 |
| GluR2 Flop | 769–819aa/883aa | AB120330 | BC10574 | 98 | 100 |
| GluR3 Flip | 781–830aa/894aa | AB120331 | U10301 | 95 | 100 |
| GluR3 Flop | 530–580aa/644aa | AB120332 | AF167332 | 98 | 100 |
| GluR4 Flip | 771–820aa/902aa | AB120333 | U16129 | 96 | 100 |
| GluR4 Flop | 770–820aa/884aa | AB120334 | S943712 | 98 | 100 |
All zf clones contain coding regions. In the second column to the left of the slashes (/) are the zf aa residues matched and numbered according to human the sequence. To the right of the slashes are the aa sizes of the full-length human proteins. zf Clones ranged in size from 172 bp (KA1) to 2,146 bp (mGluR1).
Rat sequence accession No., no human cDNA sequence is yet available.
In situ hybridization
Serial sagittal (right hemisphere) 10-μm sections were cut throughout the entire brains of adult quiet control zebra finch males. For other species, only sections known to contain vocal nuclei were cut and/or selected from our collection. 35S-labeled riboprobe in situ hybridizations were conducted. Riboprobes were made from T7 and SP6 promoter sites of pGEMTeasy with Promega RNA polymerases. For pGEMTeasy, high-concentration SP6 RNA polymerase (Progema) is necessary to generate high-quality riboprobes. Frozen sections were fixed in 3% paraformaldehyde in phosphate-buffered saline (PBS; pH 7.0), acetylated, dehydrated in an ascending ethanol series, and air dried. We then used one of two different hybridization solutions: 1) 50% formamide, 2× SSPE, 2 μg/μl yeast tRNA, 1 μg/μl poly-A, 0.4 μg/μl bovine serum albumin (BSA), 100 mM dithiothreitol (DTT; Mello et al., 1997); 2) 50% formamide, 10% dextran, 1× Denhardt’s, 12 mM EDTA (pH 8.0), 10 mM Tris-HCl (pH8.0), 30 mM NaCl, 0.5 μg/μl yeast tRNA, and 10 mM DTT (Yoshida et al., 1998). Both solutions gave similar results, except that the signals were stronger for the second. Either 32 μl (hybridization solution 1) or 100 μl (hybridization solution 2) with 35S-labeled probes (1 × 106 cpm/slide) were applied to each slide, coverslipped, and hybridized at 65°C for 4 hours in solution 1 or 16 hours in solution 2 under mineral oil. The mineral oil was removed by chloroform washes and excess probe removed by washing in 2× SSPE at room temperature for 1 hour; 2× SSPE, 50% formamide, 0.1% β-mercaptoethanol at 65°C for 1 hour; and 0.1× SSPE twice at 65°C for 30 minutes each. Slides were dehydrated in an ascending ethanol series and exposed to β-max hyperfilm (Kodak, Rochester, NY) for 2–3 days. They were then dipped in NTB-2 (Kodak) emulsion for ~2 weeks and developed, sections were counterstained with cresyl violet and then coverslipped. These exposure times allowed signal detection for imaging without signal saturation for quantification.
Quantification and statistical analysis
We measured gene expression levels by one of two methods: 1) from silver grains of emulsion-dipped slides (for Fig. 4A,B) and 2) from X-ray films (for Figs. 5C, 9). For emulsion-dipped slides, we calculated the average number of silver grains per cell, following a previously described semiautomated procedure (Jarvis and Nottebohm, 1997). First, images of brain regions from the dipped slides were captured with a Spot III CCD camera coupled to a Leica DMRXA2 microscope under a ×40 objective by using Spot software v3.2.4 (Diagnostic Instruments, Inc.). NIH Image software was then used to count silver grains from one or two cell fields (~0.2 mm2 per field, containing neurons, glia, and neuropil) in the central region of each nucleus (for lMAN 70–80% of the nucleus field was captured in one field). The average background number of grains per same-size field on the glass slide without brain tissue was then subtracted. The two counts from each field were averaged and then divided by the average number of cells (neurons and glia) per field for that nucleus (~60–100 cells depending on nucleus) to obtain grains/cell; the average number of cells was calculated separately from adjacent well Nissl-stained sections from the same animals as used for the in situ quantitations. Comparison using grains/cell controls for potential regional differences resulting from differences in cell density. For brain images on X-ray films, the exposed film was placed under a high-power dissecting scope (Wild M420) and captured to the computer with the Spot III camera. Images were transferred to Photoshop (Adobe) and converted to gray scale. Vocal nuclei and adjacent nonvocal areas were outlined with a highlighting tool, and the average pixel density was calculated. Compared with counting silver grains from emulsion-dipped slides, this method allowed us to perform high-throughput scanning and quantification of gene expression levels, and the results were comparable to silver grain counts (data not shown).
To graph ratios of differential expression in vocal and auditory regions relative to their surrounding brain subdivision (for Figs. 4A,B, 5C, 9), we divided the values of silver grain or pixel density of vocal and auditory regions with the respective adjacent nonvocal or nonauditory regions. To perform statistical analysis, we first performed an ANOVA to determine whether there was an overall significant difference in expression between vocal nuclei and the surrounding brain subdivisions. We followed the ANOVA with a Fisher’s protected least significant difference (Fisher’s PLSD) post hoc test for each gene. The Fisher’s PLSD test performs pairwise comparisons (measurements from vocal nuclei and surrounding brain subdivisions come from the same animals), is less prone than other methods to making errors when considering the magnitude of differences between the means of two measures with a small n (three animals in our case), and accounts for repeated measures (more than one measurement from the same animal). After performing the Fisher’s PLSD test, we then focused on those significant differences between the brain regions of interests, such as expression levels in a vocal nucleus compared with the respective adjacent nonvocal region surround. We chose a significance cutoff of P =.01, because we had more than 20 comparisons, but less than 100, for each gene; with a cutoff of .05, there is a probability that 1 of 20 comparisons could be significantly different by chance. For statistical analysis of expression ratio relationships between vocal nuclei (see Fig. 4C,D), we used a simple regression analysis with a 95% confidence interval.
Western blotting and immunohistochemistry
Total brains of adult male zebra finches were homogenated in cold PBS, and the lysates were separated by SDS-PAGE (12.5%), transferred to a nitrocellulose membrane, and blocked with 5% skim milk in PBS for 1 hour. Membranes were then incubated with either rabbit anti-rat GluR1 or mGluR1 polyclonal antibodies made against the carboxyl terminal intracellular portions (1:500 dilution; Upstate Biotechnology, Lake Placid, NY; catalog Nos. 03-306 and 06-301, respectively). Membranes were then reacted with a horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody (1:500; Zymed Laboratories, South San Francisco, CA), and binding was detected on X-ray films by using an ECL detection system (Amersham Bioscience, Piscataway, NJ). For immunohistochemistry, anesthetized birds were perfused with PBS and then 4% paraformaldehyde in PBS. Sagittal 20-μm sections were cut on a freezing microtome, freely floated in PBS, and reacted with 0.3% hydrogen peroxide for 20 minutes at room temperature. Sections were then washed three times with PBS and incubated for 2–3 hours in PBS containing 0.4% Triton X-100, 4% normal horse serum, and 1% BSA and then in fresh solution of the same composition overnight with the primary antibody. Colorimetric staining was performed with the avidin-biotin peroxidase complex (ABC) and nickel-intensified diaminobenzidine (DAB) detection kit from Vector Laboratories (Burlingame, CA).
RESULTS
Songbird glutamate receptors
By using RT-PCR with degenerate primers to conserved regions among mammals and/or other vertebrates, we attempted to and successfully cloned 21 of 26 known glutamate receptor subunits/subtypes described in mammals (Gustinci et al., 2003), covering all four glutamate receptor subfamilies, AMPA (GluR1–4), kainate (GluR5–7, KA1–2), NMDA (NR1, NR2A–D, NR3A), and metabotropic (mGluR1–5, mGluR8), from the zebra finch brain. We did not attempt to clone mGluR6 and mGluR7, because mammalian mGluR6 is known to be expressed in only the retina (Nakajima et al., 1993), and mGluR6 and -7 were too close in sequence to be distinguished reliably by initial PCR or by in situ hybridization. We did not attempt to clone the NMDA receptor subunit NR3B, because it was just being characterized at the time when we began our study (Nishi et al., 2001), or delta 1 and delta 2 subunits, because these are known to be involved mainly in cerebellar function (Morando et al., 2001). Without fail, we found all 21 glutamate receptor subunits/subtypes in the zebra finch brain. BLASTN and BLASTX searches revealed high cDNA (82–90%) and putative amino acid (83–100%) sequence identities between zebra finch and human (Table 1). The only exception was mGluR2 with 72% cDNA and 78% amino acid identity; this was due to an apparent splice difference in zebra finch, which has been not reported in mammals (data not shown). The spliced Flip and Flop regions of the AMPA subunits were nearly 100% identical between zebra finch and human (Table 1), showing conserved splicing recipient and donor sites. These results suggest 1) that the family and presence of glutamate receptors in the brain are highly conserved between birds and mammals and, thus, that the common stemamniote ancestor of birds and mammals presumably possessed these same glutamate receptor genes over ~300 million years ago, and 2) that the physiological function of most if not all glutamate receptors in songbirds is presumably similar to that in mammals, in support of electrophysiological and pharmacological findings (Basham et al., 1996; Stark and Perkel, 1999; White et al., 1999; Dutar et al., 2000; Livingston et al., 2000).
General songbird brain expression pattern
In situ hybridization revealed that each glutamate receptor subunit/subtype has a unique expression pattern in the songbird brain (Fig. 2). Within the cerebrum, the most apparent expression differences were coincident with Nissl-defined axonal–lamina boundaries [cell-sparse regions separating the hyerpallium (H), mesopallium (M), nidopallium (N), arcopallium (A), striatum (St), and pallidum (P)]. An exception to this rule was that expression in primary sensory areas [L2-auditory, entopallium (E)-visual, and basorostralis (B)-somatosensory] was similar among these areas and often different from the surrounding nidopallium (Fig. 2; entopallium not shown). There was no recognizable subfamily-specific brain expression pattern. Rather, several different receptor subunits/subtypes across subfamilies shared similar but not identical expression patterns. For example, GluR1 (AMPA type) and GluR7 (kainate type) were both higher in the mesopallium, caudal striatum (CSt), and arcopallium; GluR4 (AMPA type), mGluR2, and mGluR8 (metabotropic type) were higher throughout pallial regions vs. subpallial regions; mGluR4 and mGluR5 (metabotropic type) were lower in pallial relative to the subpallial striatum region (Fig. 2). Most glutamate receptor subunits/subtypes were low or absent in the subpallial pallidal region (P), an inhibitory area homologous to the mammalian globus pallidus (Reiner et al., 2004). The differential pallial vs. sub-pallial expression in the songbird brain matched that of the homologous subunits/subtypes expressed in mammalian cortical (pallial) vs. basal ganglia (subpallial) regions (Bahn et al., 1994; Conti et al., 1994; Standaert et al., 1994; Testa et al., 1994; Wada et al., in preparation).
Fig. 2.
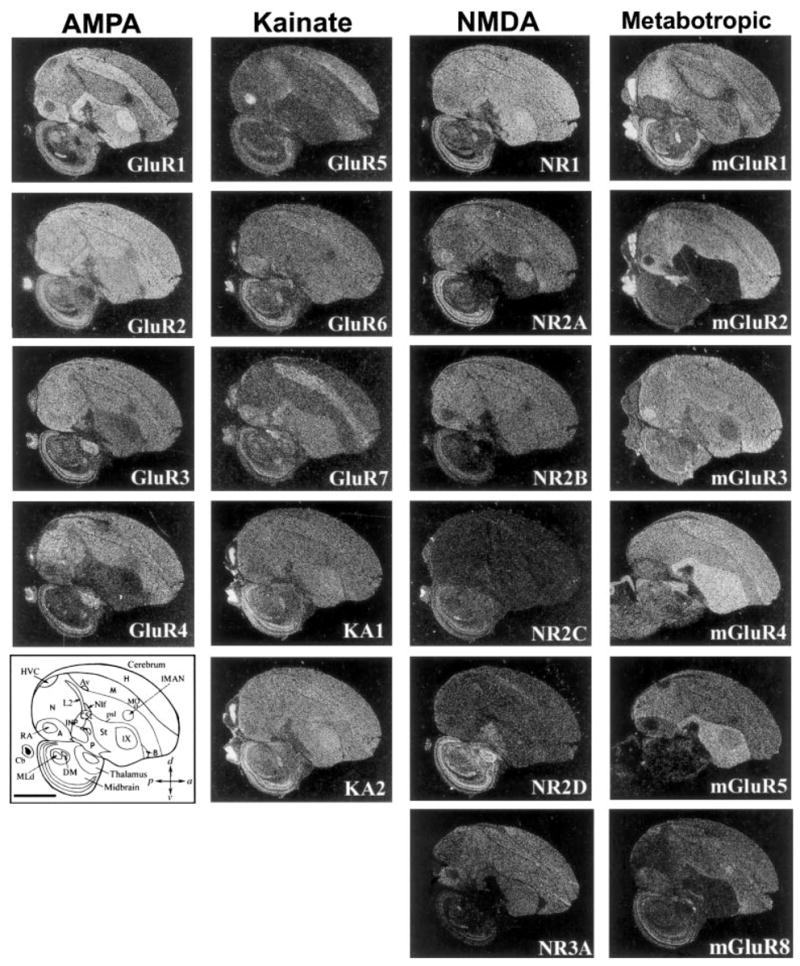
Expression profiles of 21 glutamate receptor subunits/subtypes from the four glutamate receptor subfamilies in adult zebra finch male brain. Shown are negative-image autoradiographs of in situ hybridizations, sagittal sections, ~1.5 mm from the midline, hybridized to 35S-labeled riboprobes of each glutamate receptor subunit. White label is mRNA signal. At the top is the receptor subfamily name of the column. Lower left: Camera lucida drawing of brain areas; the pallial-subpallial lamina (psl) separates pallial the from subpallial regions. Encircled regions in the cerebrum are vocal nuclei. Scale bar = 2 mm.
Unique differential expression in vocal brain areas
A striking exception to the subdivision-specific expression patterns was that nearly all glutamate receptor subunits/subtypes showed highly differential expression, higher or lower, in one or more vocal nuclei relative to expression in the surrounding cells of each of their subdivisions (Figs. 2,3). Each brain subdivision possesses at least one or more vocal nuclei (Av and MO in the mesopallium; MAN, HVC, and NIf in the nidopallium; RA in the arcopallium; Area X in the striatum; DLM in the thalamus; and DM in the midbrain). These expression differences in vocal nuclei are not the result of lamina differences, insofar as there are no known laminal boundaries separating vocal nuclei from the surrounding brain tissue. To evaluate the extent of this differential expression, we quantified in situ hybridization-labeled silver grains per cell within vocal nuclei relative to each subdivision immediately adjacent to the vocal nuclei. We found that 19 of 21 glutamate receptor subunits/subtypes had significant differential expression in one or more vocal nuclei (asterisk in Fig. 4A; the exceptions were KA1 and NR2D). In contrast, in nonvocal areas, such as auditory regions, fewer of these same subunits/subtypes had differential expression, and with lower magnitudes (Figs. 2, 4B). After further detailed examination of serial brain sections, we classified the vocal nuclei expression patterns into seven overlapping categories: 1) unique differential expression in large cerebral vocal nuclei, 2) differential expression in small cerebral vocal nuclei, 3) differential expression in noncerebral vocal nuclei, 4) correlations of expression among vocal nuclei, 5) lateral and medial differences in the anterior vocal pathway nuclei, 6) cell type uniqueness within vocal nuclei, and 7) individual animal differences.
Fig. 3.
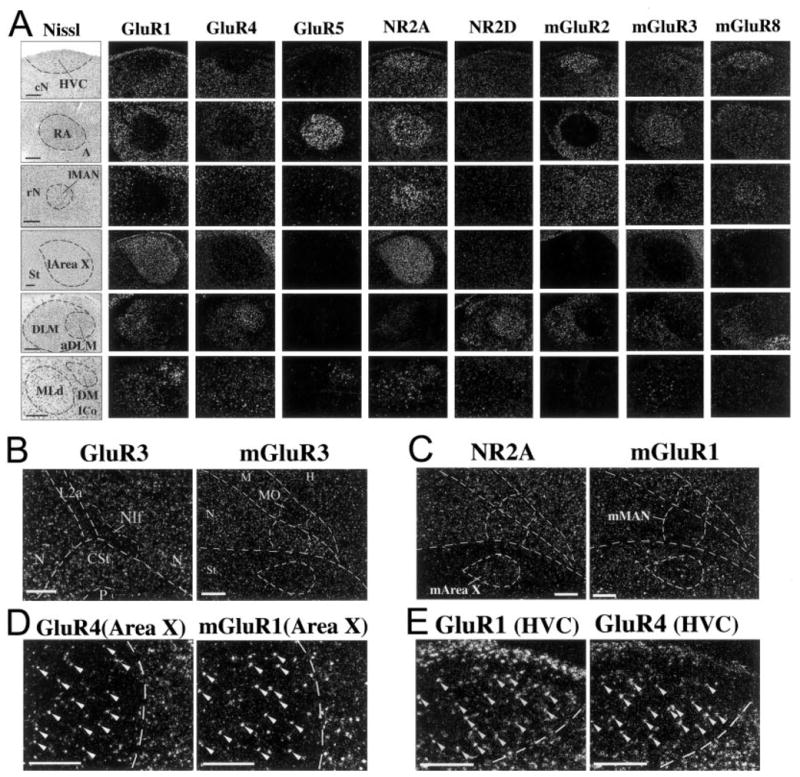
Higher magnification of differential expression in vocal nuclei. A: Nissl-stained (cresyl violet, left) and emulsion-dipped slides (darkfield microscopy, right) of higher power views of glutamate receptor subunits/subtypes in situ hybridization images that show prominent differential expression in zebra finch cerebral (upper four rows), thalamic (aDLM, fifth row), and midbrain (DM, sixth row) vocal nuclei. B: Examples of glutamate receptor subunit/subtype differential expression in small vocal nuclei NIf (low GluR3) and MO (low mGluR3). These differences are not as great as those seen in the large cerebral vocal nuclei. C: Examples of the medial part of the anterior vocal pathway nuclei with differential expression patterns that matched their lateral counterparts (higher NR2A in m-Area X and lower mGluR1 in mMAN). D: Higher power view of GluR4 and mGluR1 expression in the sparse, large cells (arrows) of l-Area X. In the striatum surrounding Area X (right of dashed lines) both large and smaller cells express high levels of these genes. E: Higher power view of differential expression of GluR1 and GluR4 in HVC. Scale bars = 300 μm in A,D,E; 200 μm in B,C.
Fig. 4.
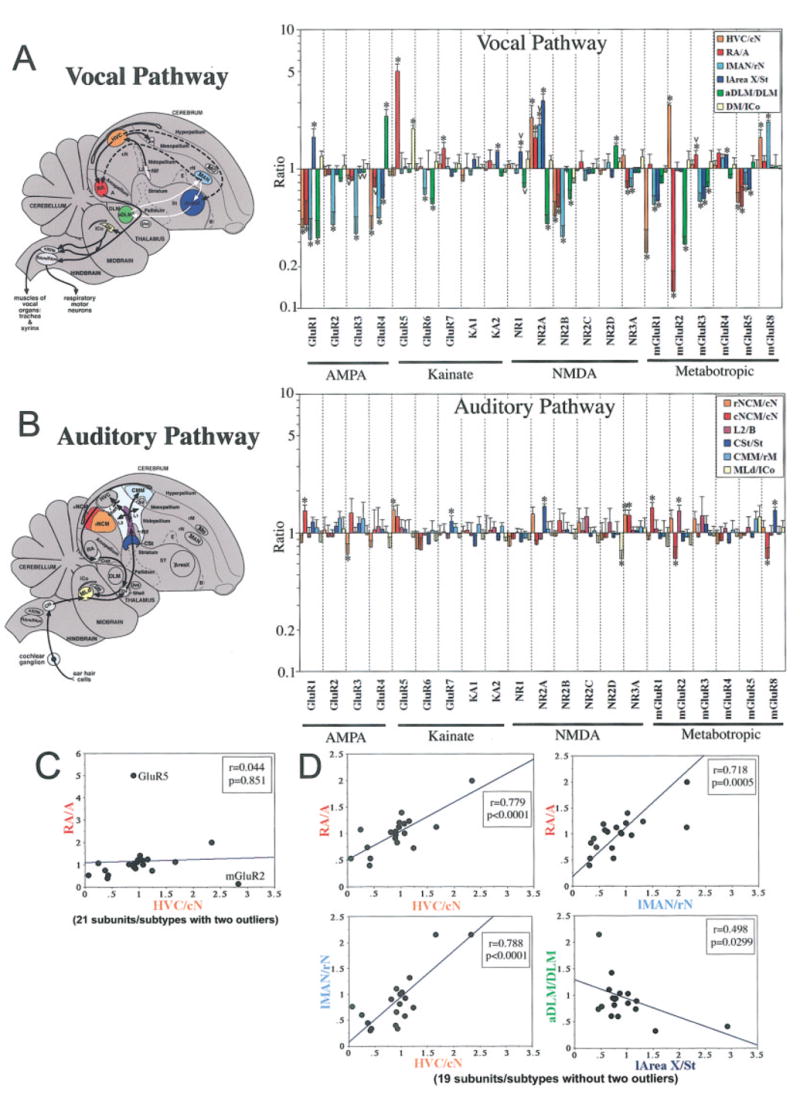
Quantitative comparisons of glutamate receptor subunits/subtypes expression in vocal and auditory areas. Shown to the left are sagittal brain diagrams of songbird vocal and auditory pathways color-coded to match color bars of graphs at right. For A, black arrows show connections of the posterior vocal pathway, white arrows shows connections of the anterior vocal pathway, and dashed lines show connections between the two. For B, caudal and rostral NCM (cNCM and rNCM) as well as CMM are more medial than in the plane shown. Graphs (A,B) show differential gene expression ratios (y-axis) of the 21 glutamate receptor subunits/subtypes (x-axis) in zebra finch vocal and auditory nuclei relative to their respective surrounding brain subdivisions (cN, caudal nidopallium immediately ventral to HVC; A, nonauditory arcopallium immediately anterior to RA; rN, nidopallium rostral to lMAN; St, striatum immediately caudal to l-Area X; DLM, thalamus dorsal and immediately caudal to aDLM; the region of ICo measured is between DM and MLd; and rM, mesopallium ~1 mm rostral to CMM; anatomical locations of these surrounding regions are labeled in the diagrams at left). The y-axis is log-scaled to view comparable ratios above and below 1 graphically. When expression is similar to the surround (not different), the ratio is ~1; when higher or lower than the surround (differential), the ratio is significantly above or below ~1 respectively. *P < .01 (Fisher’s PLSD test; n = 3 male zebra finches); v = differential expression variation in one or more individuals. Error bars show SEM. C: Comparison of gene expression ratios of 21 glutamate receptor subunits/subtypes in HVC and RA without outliers GluR5 and mGluR2 removed. D: Correlations with outliers GluR5 and mGluR2 removed. Each data point represents an expression ratio of one subunit/subtype averaged from three male zebra finches. Axes are in linear format.
Unique differential expression in large cerebral vocal nuclei
Among the seven cerebral vocal nuclei, four of them (HVC, MAN, RA, and Area X; Fig. 4A) are larger than the others. All 19 of the 21 subunits/subtypes had differential expression in one or more these larger vocal nuclei (8 subunits/subtypes in HVC, 9 subunits/subtypes in RA, 11 subunits/subtypes in MAN, and 9 subunits/subtypes in Area X; Figs. 2, 3A, 4A). Of the two subunits that did not show differential expression in cerebral vocal nuclei, one (NR2C) was barely expressed in the cerebrum, and the other (NR2D) was selectively expressed throughout the pallidum (P), a brain subdivision with no known vocal nucleus (Fig. 2). These differential expression differences ranged from 7.7-fold lower (mGluR2 in RA) to 5.1-fold higher (GluR5 in RA) than the surrounding brain subdivisions. Indeed, among the 84 possible combinations (21 subunits × 4 cerebral vocal nuclei) close to half (37/84 = 44%) showed higher or lower differential expression in vocal nuclei (Fig. 4A).
To compare the differential expression of glutamate receptors in vocal areas relative to nonvocal areas, we quantified the extent of possible differential expression within cerebral auditory areas. We chose auditory regions, because a few subunits/subtypes appeared to have some differential expression relative to the surrounding subdivision (mGluR1 in caudal N for example; Fig. 2), and these regions are involved in auditory processing of learned vocalizations (Chew et al., 1995; Mello et al., 1995). We found 10 of 21 subunits/subtypes with statistically significant differential expression in auditory areas relative to the surrounding cells of each of their subdivisions (asterisk in Fig. 4B). However, the magnitude of these differences was much smaller (1.21-fold lower to 1.75-fold higher) than that found for vocal nuclei (7.7-fold lower to 5.1-fold higher). This suggests that, although both vocal and auditory pathways contribute to learned vocal communication in songbirds, high levels of differential expression of glutamate receptor subunits/subtypes within a brain subdivision are unique to vocal nuclei.
Differential expression in small cerebral vocal nuclei
The three smaller vocal nuclei (NIf, Av, and MO; Fig. 2) appeared to have very few differences relative to the surrounding brain subdivisions. These were relatively low GluR3 and high mGluR4 in NIf, low mGluR3 in the medial part of the MO, and no recognizable differences in Av (Fig. 3B; mGluR4 in NIf not shown). The differences in MO were variable among animals. It is possible that there are more differences in the smaller nuclei that were not easily recognizable either because of: 1) the relatively smaller vocal nuclei and thus their potential for being missed in brain sections or 2) the location of NIf sandwiched between L2 and L1; the latter regions show expression differences between them, of which NIf expression could easily blend into the background with one or the other. Thus, the absence of differential expression in these small vocal nuclei should be viewed with caution.
Differential expression in noncerebral vocal nuclei
A number of glutamate receptor subunits/subtypes also showed differential expression in the noncerebral vocal nuclei, DLM and DM. In DLM, nine subunits showed differential expression that ranged from 3.7-fold lower to 2.0-fold higher in the anterior part of this dorsal thalamic region (aDLM) compared with its cells more posteriorly (Figs. 3A, 4A). This included NR2D, the subunit not differentially expressed in the cerebral vocal nuclei, which was higher in aDLM. In birds, generally, the term DLM is used to refer to the entire lateral nucleus of the dorsal thalamus. In songbirds, DLM is also often used to refer to the entire region, but as a vocal nucleus. However, the exact boundaries of a putative vocal part of DLM in songbirds, we believe, might not have been consistently delineated well in prior connectivity studies (Okuhata and Saito, 1987; Johnson et al., 1995; Vates et al., 1997; Luo and Perkel, 1999a,b; Luo et al., 2001). Apparently, the differential expression patterns of glutamate receptor subunits/subtypes may have revealed well-defined boundaries of a vocal part of DLM (Fig. 3A). Only two glutamate receptor subunits, GluR1 and GluR5, appeared to show differential expression in the midbrain vocal nucleus DM compared with the adjacent midbrain (Figs. 3A, 4A; GluR1 not significant at P =.01).
Correlations of expression among vocal nuclei
We found on regression analysis that GluR5 and mGluR2 were consistent outliers (Fig. 4C; determined by using one-by-one simple regression analysis for an outliers search). When these outliers were removed from the analysis, significant correlations were found among the differential expressions of the remaining genes (Fig. 4D). The differential expression levels in the four cerebral vocal nuclei (HVC, RA, MAN, and Area X) all showed positive relationships with each other (Fig. 4D). These relationships were strongest among the pallial vocal nuclei (HVC, RA, and MAN; Fig. 4D) and between Area X and MAN (data not shown). The differential expression levels between Area X and aDLM showed a negative relationship with each other (Fig. 4D).
Lateral and medial differences in the anterior vocal pathway
Differences were observed between the lateral and the medial parts of vocal nuclei in the anterior vocal pathway. Among the 11 subunits/subtypes with differential expression in lateral MAN (lMAN; Fig. 4A), only two subtypes had the same differential expression in medial MAN (mMAN), these being lower mGluR1 (Fig. 3C) and higher mGluR8 (not shown). Among the nine subunits/subtypes with differential expression in Area X (l-Area X; Fig. 4A), only six subunits had the same differential expression in medial area X (m-Area X), these being higher NR2A (Fig. 3C), lower mGluR3, and higher mGluR4 and mGluR5 (not shown). These findings are consistent with the previous observation that singing causes different patterns of motor-driven gene expression in the lateral and medial parts of MAN and Area X (Jarvis et al., 1998). There were no differential expression patterns in mMAN or m-Area X that were not present in their lateral counterparts.
Cell type uniqueness within vocal nuclei
The vocal nuclei are known to be composed of several types of neurons that perform different physiological functions and have different projections (Alvarez-Buylla and Kirn, 1997; Mooney, 2000; Perkel and Farries, 2000; Hahnloser et al., 2002). Some glutamate receptor subunits/subtypes appeared to have differential expression restricted to a particular cell type within a vocal nucleus. For example, GluR4 (AMPA type) and mGluR1 (metabotropic type) in Area X were expressed mainly in sparse, large cells (Fig. 3D, arrowheads), suggesting expression in Area X neurons that project to DLM. In the striatum surrounding Area X (Fig. 3D, area to the right of the dashed lines), both large and smaller cells expressed high levels of these same genes. This suggests that the unique differential expression in Area X is due to lower levels of these subunit/subtype in the smaller, spiny neurons. Both GluR1 and GluR4 (AMPA types) in HVC were expressed in a sparse cellular distribution (Fig. 3E, arrowheads), suggesting that the differential expression in HVC is due to lower expression in one of its cell types (X-projecting, RA-projecting, and/or interneurons) compared with the surrounding nidopallium. Double-labeling experiments are necessary to verify these suggestions.
Individual animal differences
The expression patterns that we observed were very stable among all animals examined, except for four subunits: GluR3 (in HVC, l-Area X, and aDLM), NR1 (in l-Area X and aDLM), NR2A (in lMAN) and mGluR3 (in RA; Fig. 4A, v for variable). For GluR3, expression was higher than, lower than, or the same as that in the surrounding subdivision, depending on the individual. For NR1, NR2A, and mGluR3, expression was always higher (four of six animals examined) or similar (two of six animals) to that in the surround. The source of these individual variations has not yet been identified, but potentially they could be due to recent behavioral experience, including social factors or the types of songs the birds produce. They do not appear to be due to recent singing activity, in that all birds did not sing in the 16 hours before sacrifice.
Differential regulation of mRNA splicing in vocal nuclei
One gene can give rise to many different gene products through alternative mRNA splicing, thereby increasing diversity of the gene’s functions. Among the best studied splice variants for glutamate receptors are the AMPA subunits. In mammals, all four AMPA receptor subunits (GluR1–4) are alternatively spliced into at least two forms, called Flip and Flop (Fig. 5A). The Flop spliced forms of the AMPA receptor subunits usually causes the receptors to desensitize faster after channel opening; the Flip forms have the opposite effect (Dengledine et al., 1999; Petralia et al., 1999). We found that, in most cases, either the Flip and/or the Flop splice-specific forms had the same direction of differential expression in vocal nuclei as revealed by the general probes, which hybridize to both Flip and Flop forms (Fig. 5B,C). For example, the low differential GluR1 expression in HVC appeared to be due to the Flip form and not the Flop form (Fig. 5C). However, in some cases, the Flip and Flop forms had differential expression in opposite directions (one higher and one lower than the surrounding subdivisions) that were not revealed by the general probe alone. For example, the general probe for GluR2 showed expression levels similar to those of the surrounding brain subdivisions in HVC and l-Area X, whereas the GluR2-Flip probe showed expression that was lower in HVC and l-Area X, and the GluR2-Flop probe showed expression that was higher in HVC and l-Area X relative to the surrounding brain subdivisions (Fig. 5B,C). This suggests that the general probe canceled the detection of the Flip and Flop differential expression in vocal nuclei. Similar opposite patterns of Flip and Flop expression were found for GluR3 in HVC and l-Area X (Fig. 5C). Thus, diversity of expression in splice variants leads to more diversity of differential gene expression in vocal nuclei. This is the first finding that we are aware of demonstrating differential mRNA splicing that is specific to vocal brain areas.
Fig. 5.
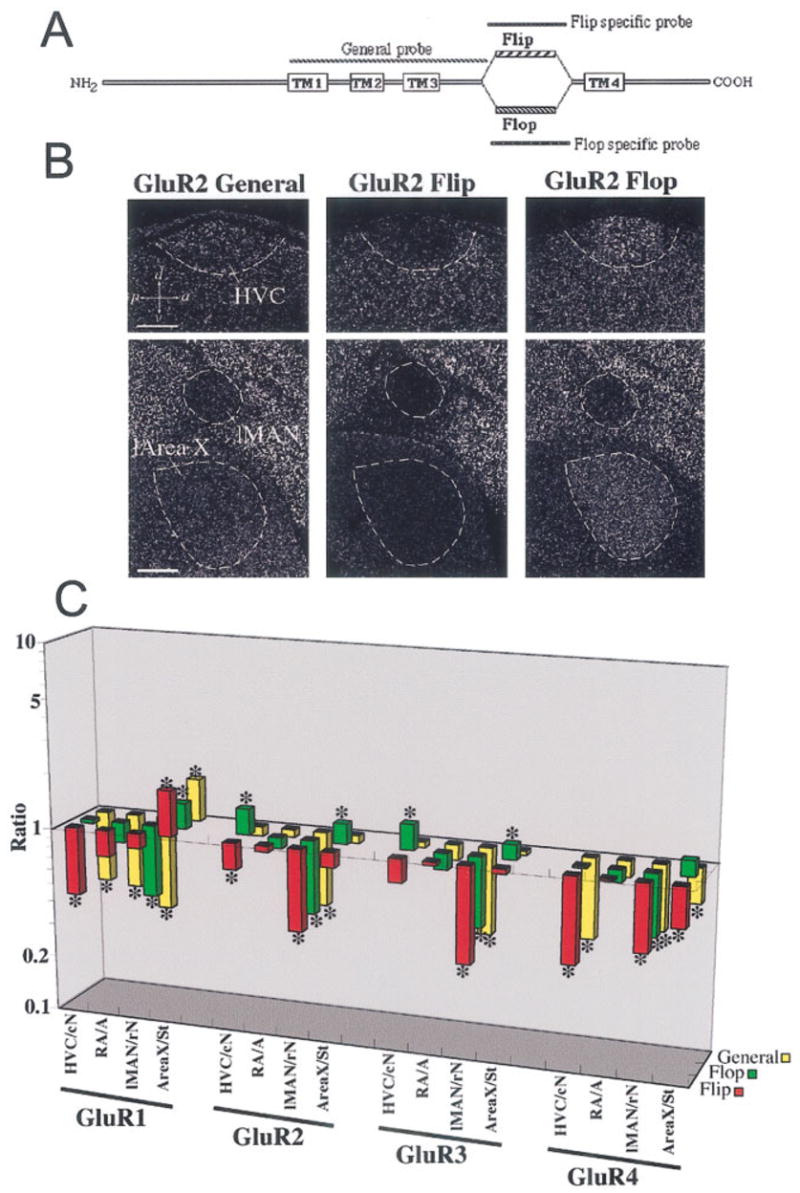
Differential regulation of mRNA splicing of AMPA subunits in vocal nuclei. A: General schematic of mammal AMPA receptor proteins with their four transmembrane domains (TM1–TM4). Between TM3 and TM4 are the alternatively spliced extracellular domains Flip or Flop. The general probe hybridizes to the TM1–TM3 region of both splice variant mRNAs. Each specific probe hybridizes to the Flip or Flop spliced form only. B: Example of differences in expression of AMPA receptor splice variants revealed by comparing hybridizations with the GluR2 general, Flip, and Flop probes in zebra finch vocal nuclei. There is lower expression of GluR2 Flip and higher expression of GluR2 Flop mRNA in HVC and l-Area X compared with the surrounding brain subdivision. C: Quantification of differential expression of splice variants of all four AMPA subunits (GluR1–4; gene expression level on the z-axis, brain region on the x-axis, and gene on the y-axis). The z-axis is log scaled to view comparable ratios above and below 1 graphically. Further explanation of ratio interpretations is given in the legend to Figure 4. *P < 0.01 (Fisher’s PLSD test; n = 3 male zebra finches). Scale bars = 300 μm.
Differential expression at the protein level
We obtained several antibodies that recognize specific subunits/subtypes of several glutamate receptors in mammals (GluR1 and mGluR1). We found that anti-rat GluR1 and anti-rat mGluR1 antibodies recognized proteins of similar molecular weight in Western blots of zebra finch and rat brains (Fig. 6A). For brain sections, we observed zebra finch protein expression patterns similar to the mRNA expression patterns, including differential expression in vocal nuclei (Fig. 6A). However, GluR1 protein levels appeared to be more differentially expressed in l-Area X (higher) and RA (lower), but less so in HVC and lMAN (Fig. 6A) relative to the mRNA (Fig. 2); mGluR1 protein levels appeared to be similar to the mRNA, with the exception that protein in RA was much lower than the surrounding arcopallium, whereas the mRNA was not. Both GluR1 and mGluR1 proteins were present in neuropil and cell bodies (Fig. 6B). However, GluR1 protein in Area X was much higher in its neuropil than in the neuropil of the surrounding striatum; the opposite pattern was seen for mGluR1 (Fig. 6B) with cell body expression restricted to sparse large neurons as seen with the mRNA (Fig. 3D). Expression in neuropil is expected of glutamate receptors, given that they are usually transferred to dendrites as parts of functional receptor proteins. In general, unique differential expression in vocal nuclei at the mRNA level appears to be reflected at the protein level.
Fig. 6.
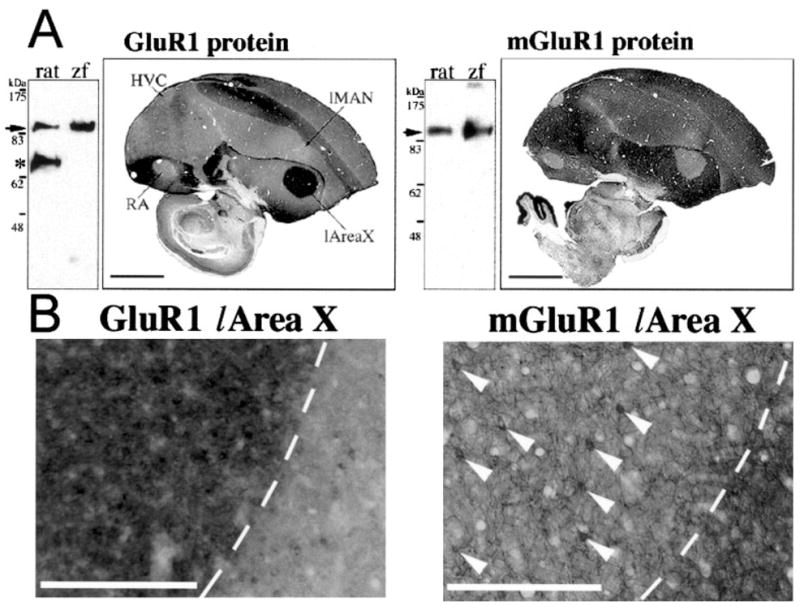
Differential expression at the protein level. A: Right: Western blots of rat and zebra finch (zf) whole-brain extracts reacted with antibodies that recognize rat GluR1 and mGluR1 carboxy termini. Cross-reactivity to similarly sized zebra finch proteins are found (arrows). *Additional protein band detected in rat but not in zebra finch brains. It is unknown whether this additional band is a specific variant of rat GluR1. Left: Immunohistochemistry for GluR1 and mGluR1 protein expression in sagittal zf male brain sections with the same antibodies. B: Higher magnification showing protein expression of GluR1 and mGluR1 in l-Area X (left of the dashed lines) and surrounding striatum. Arrowheads point to the sparse, large neurons in Area X. The clear white areas seen in the mGluR1 image of B are blood vessels that have expanded upon perfusion of the animal. Scale bars = 2 mm in A; 300 μm in B.
Other vocal learners
We examined whether differential expression of glutamate receptor subunits/subtypes in vocal nuclei relative to the surrounding brain subdivisions is unique to songbirds or occurs in vocal nuclei of other vocal learners, hummingbirds and parrots. We selected one subunit/subtype that had the most differential expression in zebra finch vocal nuclei from each of the four families (GluR1 from the AMPA family, GluR5 from the kainate family, NR2A from the NMDA family, and mGluR2 from the metabotropic family) and hybridized them to brain sections of the sombre hummingbird and budgerigar. The zebra finch subunits/subtype cross-hybridized to the hummingbird and parrot homologs under the same high-stringency in situ hybridization conditions as used for zebra finch brain sections (Figs. 7, 8). Although they are not identical, the general expression patterns of these four subunits/subtype in regions outside the vocal systems of the hummingbirds and parrots was similar to the pattern in the zebra finch, including in auditory areas (Figs. 7, 8). As with the zebra finch, these other vocal learning species had differential expression of the glutamate receptor subunits/subtype in their vocal nuclei, with some similarities and some differences among them as follows.
Fig. 7.
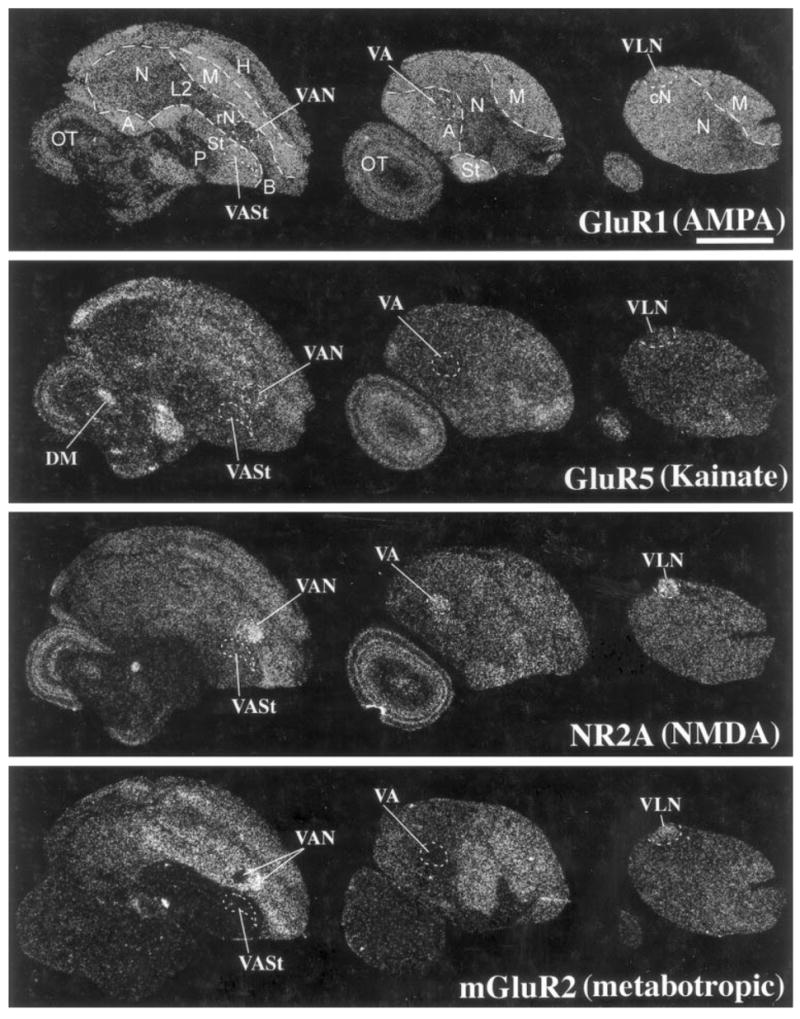
Glutamate receptor subunit/subtype expression in the sombre hummingbird brain. Brain sections are parasagittals in the same orientation as in Figure 2, ~1.9, ~3.5, ~4.0 mm, left to right, from the midline. Scale bar = 2 mm.
Fig. 8.
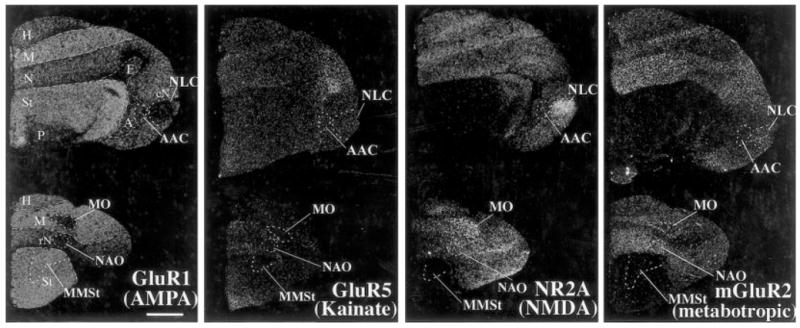
Glutamate receptor subunit/subtype expression in the budgerigar brain. Brain sections are frontals, which allow easier visualization of the parrot posterior pathway nuclei NLC and AAC than in sagittals. The sections are ~2.0 mm (top) and ~5.2 mm (bottom) from zero point. Scale bar = 2 mm.
Hummingbird
We observed that two (NR2A and mGluR2) of the four subunits had differential expression in the sombre hummingbird cerebral vocal nuclei with some similarities to zebra finches (Figs. 7, 9A,B). This included higher NR2A expression in VLN (HVC-like nucleus), VA (RA-like), and VAN (MAN-like); higher mGluR2 expression in VLN (HVC-like); and lower mGluR2 expression in VA (Figs. 7, 9B). In addition, for mGluR2, the posterior part of the hummingbird VAN nucleus (VAN-P) had lower expression, whereas the anterior part (VAN-A) had higher expression relative to the surrounding nidopallium, and this dual pattern was unique to hummingbirds (Figs. 7, 9B). The magnitude of the differential expression levels in hummingbird vocal nuclei was not as great as that seen for zebra finches (Fig. 9A,B); however, the differential expressions in vocal nuclei were still prominent (Fig. 7, NR2A and mGluR2). Unlike the case for zebra finches, there was no detectable differential expression of the four subunits/subtypes in the hummingbird VASt vocal nucleus (Area X-like structure in the striatum; Figs. 7, 9B). For noncerebral brain areas, similar to zebra finches, GluR5 expression was differentially and prominently expressed in DM (Fig. 7).
Fig. 9.
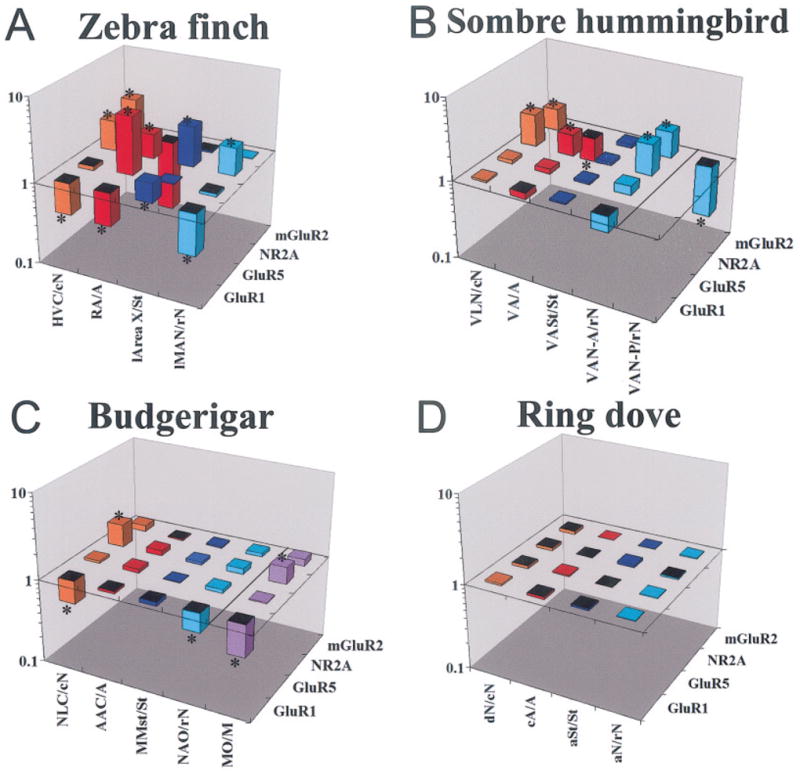
Quantitative comparisons of glutamate receptor subunit/subtype expression in vocal nuclei of all three distantly related vocal learners—zebra finch (A), sombre hummingbird (B), and budgerigar (C)—and comparable brain locations in a vocal nonlearner, the ring dove (D; gene expression level on the z-axis, brain region on the x-axis, and gene on the y-axis). The z-axis is log scaled to view comparable ratios above and below 1.0 graphically. Further explanation of ratios is given in the legend to Figure 4. The bars are color coded according to vocal nucleus as in Figure 4A. The line in B separates the quantified values in the anterior part of hummingbird VAN (VAN-A) from the posterior part (VAN-P). The line in C separates the values of parrot MO from the other cerebral vocal nuclei; MO was quantified only in parrots, being much larger and easier to identify in them. *P <.01 (Fisher’s PLSD test; n = 3 male birds per species). Abbreviations not in abbreviations list are given in the legend to Figure 4; otherwise, for hummingbirds and parrots: lN, lateral nidopallium adjacent to VLN or NLC, respectively; mN, medial nidopallium adjacent to VAN or NAO, respectively; for doves: aSt, anterior striatum at a location where Area X is found in songbirds; cA, caudal arcopallium at a location where RA is found in songbirds; dN, dorsal nidopallium at a location where HVC is found in songbirds.
Parrot
We observed that two (NR2A and GluR1) of the four subunits had differential expression in budgerigar cerebral vocal nuclei with some similarities to zebra finches (Figs. 8, 9A,C). This included higher NR2A expression in NLC (HVC-like) and lower GluR1 in NLC (HVC-like) and NAO (MAN-like) nuclei relative to the surrounding brain subdivisions (Figs. 8, 9A,C). There were, however, no other differences similar to the zebra finch. The MO vocal nucleus in parrots is much larger than in songbirds and hummingbirds, and this nucleus in parrots showed relatively prominent lower GluR1 and higher NR2A differential expression (Figs. 8, 9C). As with hummingbirds, there was no detectable differential expression of any of the four subunits/subtypes in the parrot MMSt vocal nucleus (Area X-like structure in the striatum; Figs. 8, 9C).
In considering together the results from the three species, whenever NR2A was differentially expressed in vocal nuclei, it was always higher than in the surrounding brain subdivision; mGluR2 was higher in the nidopallium vocal nuclei (HVC-like and/or MAN-like) and lower in the arcopallium vocal nuclei (RA-like); and GluR1 tended to be lower, except in the striatal vocal nucleus of zebra finches (higher in Area X). Zebra finches showed more differential expression differences than the hummingbird and parrot species examined, with GluR5 being exceptionally high in RA of zebra finches.
Vocal nonlearners
We wondered whether differential expression of glutamate receptors in vocal nuclei is unique to vocal learners. We hybridized the same set of four glutamate receptor subunits/subtype to pigeon and ring dove brain sections containing the midbrain vocal nucleus DM, the thalamic region DLM, and cerebral regions in which forebrain vocal nuclei have been found in vocal learners. Doves produce only innate vocalizations (Nottebohm and Nottebohm, 1971; Baptista, 1996). The general brain expression pattern, including auditory areas and primary sensory regions (L2, entopallium, and basorotralis) of these vocal nonlearning species, was similar to that of songbirds, hummingbirds, and parrots (Fig. 10). However, there were no cerebral differential expression patterns in areas where vocal nuclei are found in vocal learners (Figs. 9D, 10). Thus, highly differential expression within cerebral subdivisions appears to be unique to vocal nuclei of vocal learners. The doves had differential expression in thalamic and midbrain regions as found in the vocal learners. This included lower GluR1 and higher NR2A in the anterior part of DLM in a location similar to aDLM of songbirds and the conserved higher GluR5 in DM (Fig. 10, insets; DM data not shown). The lower GluR1 in the aDLM-like region was similar to that in the zebra finch, but the higher NR2A was in the opposite direction to that found in the zebra finch. Thus, differential expression in thalamic and midbrain vocal nuclei appears not to be unique to vocal learners.
Fig. 10.
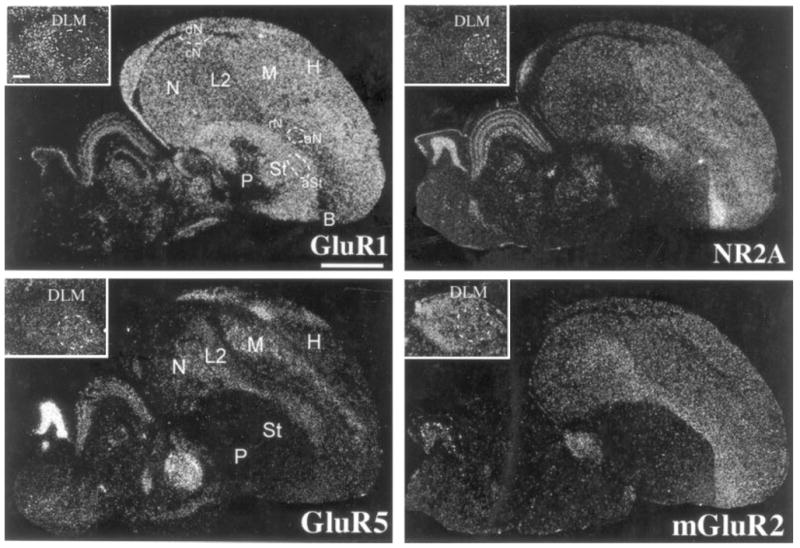
Glutamate receptor subunit/subtype expression in the ring dove brain. Brain sections are parasagittals in the same orientation as in Figure 2. The ring dove sections are ~2.0 mm from the midline. The regions aSt, aN, and dN are locations where one would expect to find vocal nuclei in songbirds. The insets show dove aDLM specializations of GluR1 and NR2A. Scale bars = 2 mm; 300 μm for insets.
DISCUSSION
We believe these findings to be a striking example of differential gene expression of nearly an entire gene family in systems that control a complex behavior, learned vocalization. Nearly all glutamate receptor subunits/subtypes showed differential expression in one or more cerebral vocal nuclei. Prior studies have shown differential expression of some genes in songbird and parrot vocal nuclei (Arnold et al., 1976; Bottjer, 1993; Ball, 1994; Casto and Ball, 1994; Aamodt et al., 1995; Holzenberger et al., 1997; Durand et al., 1998; Denisenko-Nehrbass et al., 2000), including the three previously cloned glutamate receptor subunits, NR1, NR2A, and NR2B from songbirds (Singh et al., 2000; Heinrich et al., 2002). However, none have shown systematic differential expression of nearly one entire gene family and across all vocal learning orders. We note that, in these other studies, the differential expression tends to be of genes involved in neural transmission (tyrosine hydroxylase, D1 dopamine receptor, and enkephalin). We suggest that the neural transmission machinery is generally specialized in the vocal nuclei of vocal learners. Below we present the significance of our findings for zebra finch vocal nuclei synaptic transmission and plasticity, for the possible mechanism of vocal nuclei differential gene expression, and for the evolution of vocal learning.
Zebra finch vocal nuclei synaptic transmission and plasticity
When glutamate binds to its receptors, two types of events occur: 1) chemical transmission of electrical activity for perception and production of behavior and 2) plasticity via second messengers that alters molecules and gene expression. For the ionotropic receptors (AMPA, kainate, and NMDA), this involves influx of cations (K+, Na+, and Ca2+), whereas, for the metabotropic receptors (mGluR1–8), this involves activation of intracellular signals via G-proteins coupled to their intracellular side (Pin and Duvoisin, 1995; Dengledine et al., 1999). Because glutamate receptors are highly conserved across species, as reinforced by this study, their functions across species are thought to be homologous (Dengledine et al., 1999). Thus, because we determined the brain expression patterns of nearly the entire known gene family in the zebra finch, although not in specific cell types, we believe that it is possible to make informative and testable predictions about relative synaptic transmission functions in zebra finch vocal nuclei. We discuss each receptor subfamily separately.
AMPA
AMPA receptors are responsible for fast synaptic transmission (milliseconds). They are thought to form homo- and heterotetramers (Dengledine et al., 1999), where GluR2 combines with GluR1, GluR3, or GluR4. In the mammalian cerebrum, GluR2 is the most uniform and ubiquitous, whereas GluR1, -3, and -4 are more selectively expressed (Petralia et al., 1999). This is similar to the zebra finch pattern found here (Fig. 2, AMPA panel). GluR4-containing receptors generally have the fastest desensitization time and GluR1 the slowest (Dengledine et al., 1999), and GluR2 combined with GluR3 has a faster desensitization time than GluR2 combined with GluR1 or GluR4 (Dengledine et al., 1999). Thus, given the zebra finch expression patterns (Fig. 2, AMPA panel), the motor vocal nuclei HVC and RA potentially could be dominated by GluR2/GluR3 heteromers resulting in faster desensitization times than the surrounding brain areas, with higher GluR2/GluR1 and GluR2/GluR4 heteromers. The zebra finch vocal nucleus lMAN has less expression of all four AMPA subunits relative to the surround, which would suggest that fast transmission is not as prominent a feature of zebra finch lMAN. AMPA currents have been found in zebra finch lMAN, HVC, and RA (Stark and Perkel, 1999), but relative desensitization and synaptic transmission times have not been compared. Area X would be expected to have a lower number of GluR2/GluR4 receptors relative to the surrounding striatum that is compensated for by a higher number of GluR2/GluR1 receptors (Fig. 2, AMPA panel), potentially giving Area X cells (probably its small spiny neurons) more slowly desensitized AMPA receptors than those in the surrounding striatum. The large cells of Area X, presumably the projection neurons to DLM (Perkel et al., 2002), appear to have high levels of GluR4, as does the surround (Fig. 3D) and, thus, should have as fast a desensitization time as the surrounding striatum. The anterior part of DLM, with its selectively high expression of GluR4 (Fig. 3A), would be expected to have faster desensitization times. The Flop spliced form of the AMPA receptor subunits usually causes the receptors to desensitize faster after channel opening than the Flip form (Dengledine et al., 1999; Petralia et al., 1999). Zebra finch vocal nuclei tend to have higher levels of the Flop forms and lower levels of the Flip forms relative to the surrounding brain subdivisions in HVC and Area X (except GluR1 in Area X; Fig. 5C), and this may lead to even faster desensitization than the surrounding subdivisions. Both forms are consistently low in lMAN.
Kainate
Kainate receptors also form homo- and heterotetramers, where GluR5–7 usually combine with either KA1 or KA2 (Hollmann, 1999). In the mammalian cerebrum, only KA2 is abundant and uniformly distributed (Wisden and Seeburg, 1993). This is the case for KA2 in the zebra finch cerebrum found here (Fig. 2, kainate panel). A difference from mammals is that zebra finch KA1 is also relatively abundant and uniform. As with AMPA receptors, kainate receptors are rapidly activated (milliseconds); GluR5 desensitizes the most quickly but takes the longest to recover (seconds; Dengledine et al., 1999). Thus, it is likely that cells in zebra finch RA, with a high level of GluR5, have rapid activation and slow recovery of kainate receptors relative to the surrounding arcopallium. In the mammalian hippocampus, kainate receptors down-regulate GABAergic interneuron inhibition (Rodriquez-Moreno and Lerma, 1998). During singing, zebra finch RA has phasic firing governed by brief bursts (Yu and Margoliash, 1996). RA’s GABAergic interneurons were proposed to control this phasic firing, transforming afferent nonphasic HVC activity into phasic RA activity (Spiro et al., 1999). Perhaps GluR5 is regulating RA’s GABAergic inhibition in RA as it does in the mammalian hippocampus.
NMDA
NMDA receptors are in part responsible for slow synaptic transmission (milliseconds to seconds) and also form homo- and heterotetramers, where NR1 combines with NR2A, -B, -C, or -D (Cull-Candy et al., 2001). As in mammals (Standaert et al., 1994), zebra finch NR1 expression is the most uniform and ubiquitous, whereas NR2A and NR2B are more restricted and are present mainly in the cerebrum, and NR2C and NR2D are in mainly cerebral pallidal cells, as well as in the thalamus, brainstem, and cerebellum (Fig. 2, NMDA panel; cerebellum not shown). The different NR1 receptor combinations from fastest to slowest in deactivation times are NR1/NR2A > NR1/NR2B > NR1/NR2C > NR1/NR2D (Dengledine et al., 1999). NR3A has been proposed to modulate the activity of NR1/NR2A and NR1/NR2B receptors (Das et al., 1998). Thus, given that adult zebra finch pallial vocal nuclei (HVC, RA, and lMAN) have higher NR2A and lower NR2B relative to their respective brain subdivisions, they may have faster NMDA deactivation times relative to the rest of the cerebrum. This suggestion is supported by differences we noted from another study (White et al., 1999) in which adult NMDA excitatory postsynaptic potential (EPSP) decay times are on average three to four times faster in vocal nuclei than in the hyperpallium (or Wulst; 58 msec in RA, 48 msec in lMAN, and 185 msec in the hyperpallium). Pharmacological studies do indicate that zebra finch lMAN axons synapse onto NMDA receptors of RA neurons and HVC axons synapse onto AMPA and NMDA receptors of RA neurons (Mooney and Konishi, 1991; Stark and Perkel, 1999). Given this knowledge, our expression findings suggest that the lMAN input will be found mainly on NR1/NR2A NMDA receptors in RA, whereas the HVC input mainly on both NR1/NR2A NMDA and GluR2/GluR3 AMPA receptors in RA. The zebra finch striatal vocal nucleus (l-Area X) has NR2A and NR1 levels usually higher than those in the surround. This would theoretically result in l-Area X having slower NMDA deactivation times than the pallial vocal nuclei.
Our findings also provide insight into behaviorally induced immediate early gene synthesis. NMDA receptors in mammals regulate activity-dependent induction of ZENK (an acronym for Zif-268, Egr-1, NGFI-A, and Krox-24) and c-fos synthesis in neurons (Lerea, 1997). As singing induces expression of these genes in cerebral vocal nuclei (Jarvis and Nottebohm, 1997; Kimpo and Doupe, 1997), our results suggest that the induction is likely mediated through NR1/NR2A receptors for all four large cerebral vocal nuclei, and additionally through NR1/NR2B receptors for Area X (Fig. 2, NMDA panel). Interestingly, all brain areas that do not express high ZENK regardless of activity (i.e. DLM, Ov, L2, E, B, and P; Jarvis et al., 1998) have selective high NR2D expression (Fig. 2, NMDA panel, Ov and E not shown).
Metabotropic receptors
These receptors are classified into three groups. Group I (mGluR1 and 5) couples phosphatidylinositol hydrolysis to increased intracellular Ca2+ release, cAMP activation, and K+-channel inhibition (Pin and Duvoisin, 1995). Groups II (mGluR2 and 3) and III (mGluR4, -6, -7, and -8) tend to have the opposite effect. Because of these indirect actions, metabotropic glutamate receptors take longer to affect membrane currents than ionotropic receptors. Furthermore, they are usually farther away from sites of glutamate release and as such need high-frequency presynaptic stimulation to be activated postsynaptically. In this manner, metabotropic receptors have a strong modulatory role in synaptic transmission via signal transduction (Geiger et al., 1999). We found that group I receptors are generally lower in zebra finch cerebral vocal nuclei relative to the surround, whereas groups II and III are higher, mainly in HVC and/or RA. This suggests that the overall effect of metabotropic receptor activity in zebra finch vocal nuclei will be stronger inhibition of cAMP and intracellular Ca2+ release and higher activation of K+-channel outflux. Such modulation would result in stronger hyperpolarization relative to the surrounding brain areas. This prediction is supported by strong and lasting inhibition seen in HVC and RA after singing (McCasland and Konishi, 1981; McCasland, 1987; Yu and Margoliash, 1996) and by relatively unique pharmacological findings in HVC, where group II and III agonists generate long-lasting hyperpolarizing currents, whereas group I agonists have no effect (Dutar et al., 2000).
Possible mechanisms and consequences of differential expression in vocal nuclei
Many groups have reported that large numbers of genes are expressed with region specific patterns in the vertebrate brain (Eberwine et al., 1992; Watson and Margulies, 1993; Livesey and Hunt, 1996; Berke et al., 1998; Zirlinger et al., 2001). Some of this variability is thought to be due to differences in promoter regions among genes. Some studies have found unique species-specific differences in promotor regions in the same gene, such as the vasopressin neurotransmitter receptor, which control species-specific differential brain expression patterns (Young et al., 1999). In the case of glutamate neurotransmitter receptors, many receptor subunits/subtypes are thought to possess multipromotor sequences that are regulated by various combinations of transcription factors and CpG methyl islands, the latter of which are regulated by methylation and acetylation (Myers et al., 1998; Huang et al., 1999, 2002). Our findings suggest that cells located in the vocal nuclei may possess a unique set of transcription factors and/or genetic modifications to glutamate receptor promoters allowing for unique differential expression in those cells. In support of this idea, a vocal-nucleus-specific protein has been found in Estrilidine finches (Akutagawa and Konishi, 2001). Neurotransmitter receptors also regulate expression of different genes via cell signal transduction cascades (Sheng and Greenberg, 1990; Bading, 1999), so the differential expression of glutamate receptors in vocal nuclei is expected to lead to downstream consequences in the expression of the genes they regulate. In support of this idea, the BDNF gene, which requires activation through NMDA receptors for its expression (West et al., 2001), is synthesized at lower basal levels in songbird nuclei HVC, RA, and lMAN than in the surrounding brain areas (Li et al., 2000; Li and Jarvis, 2001).
Evolution of vocal learning
Several hypotheses have been proposed for the evolution of cerebral brain nuclei for vocal learning. One hypothesis is that, because cerebral vocal nuclei have not been found in vocal nonlearners, then songbirds, hummingbirds, and parrots each evolved their similar cerebral vocal nuclei independently from a common ancestor (Fig. 1A, red dots; Brenowitz, 1997; Jarvis et al., 2000). If true, our results then further suggest that differential expression of glutamate receptors in vocal nuclei coevolved in manners similar but not identical to the vocal neural systems of each group independently. This hypothesis is supported by the absence of such cerebral differential expression in vocal nonlearners. The second hypothesis is that, because the vocal nuclei are similar among vocal learners, then either vocal nonlearners lost their cerebral vocal systems multiple independent times or vocal non-learners have cerebral forebrain nuclei that have not been detected in prior studies (Fig. 1A, green dots; Brenowitz, 1997; Jarvis et al., 2000). If true, then our results would further suggest that differential expression of glutamate receptor subunits/subtypes in cerebral vocal nuclei were lost in vocal nonlearners and that the differences found in the vocal learners were the result of divergence.
The differential expression levels found in vocal nuclei of hummingbirds and parrots are not as prominent as they are in zebra finches. These differences could be related to the type of learned vocal behavior they display, and we are currently testing this hypothesis. Some differences could also be related to differences in cell types in comparable vocal nuclei among vocal learning orders. Given these possibilities, though, the commonality among vocal learners is still that the vocal nuclei show prominent differential gene expression relative to the brain subdivision in which they are located, a property not common in other brain regions of each subdivision.
Of the above two hypotheses, independent evolution of cerebral vocal nuclei and vocal learning is the most commonly accepted. If this is correct, then the mechanisms of how such differential expression in vocal nuclei evolved require explanation. We propose an explanation that we call the vocal connectional evolution hypothesis: To date, in vocal nonlearners, no cerebral motor connections have been found to project to brainstem vocal nuclei (nXIIts and DM; Wild, 1997; Wild et al., 1997). However, vocal non-learners have differential glutamate receptor expression in noncerebral vocal nuclei. We suggest that differential expression in cerebral vocal nuclei of vocal learners could have been stimulated after a mutational event caused a connection from the arcopallium into the brainstem vocal nucleus DM. Because connectivity of brain pathways outside the vocal system is similar to that within the vocal system (Karten, 1967, 1968; Margoliash et al., 1994; Mello et al., 1998; Iyengar et al., 1999; Farries, 2001; Iyengar and Bottjer, 2002), once the arcopallium is connected with the brainstem vocal nuclei, this presumably can induce a feedback cascade of specializations in a preexisting cerebral network. This hypothesis requires only a few mutational changes that can be selected upon (selection for specific neural connections), followed by normal interactions of neural activity and gene expression in pre- and postsynaptic neurons. This hypothesis is consistent with our correlation findings. When one vocal nucleus has higher or lower expression of a particular subunit, one or more of the connecting vocal nuclei tend to have similar specializations.
Parallels have been noted between avian vocal learning brain pathways with human language brain regions (Doupe and Kuhl, 1999; Jarvis, 2004a,b), including the presence of a connection from the human face motor cortex to the brainstem vocal motor neurons (nucleus ambiguous); this connection is weak to absent in nonhuman primates (Kuypers, 1958a,b; Deacon, 1997). Given our findings of vocal nuclei specializations in vocal learning birds, it is not going too far to suggest that vocal learning mammals, including humans, also evolved unique differential expression of glutamate receptors in vocal areas of their cerebrums and that these will not be found in vocal non-learning mammals.
Acknowledgments
Grant sponsor: the Whitehall Foundation (E.D.J.); Grant sponsor: the National Science Foundation (E.D.J.); Grant sponsor: the Japan Ministry of Education, Science, and Culture (M.H.); Grant sponsor: Japan Scholarship Association Award (K.W.).
We thank Michael McElroy for sectioning ring doves brains and Jarvis laboratory members for critical reading of the manuscript. We also thank Drs. Richard Mooney, Michael Ehlers, and V. Anne Smith for critical reading and useful comments and Dr. Minmin Luo for consultation on aDLM location in songbirds.
Footnotes
This list incorporates new terminology recommended by the Avian Brain Nomenclature Forum (Reiner et al., 2004). Online translation from the old terminology can be found at www.avianbrain.org
Abbreviations: A, arcopallium; AAC, central nucleus of the anterior arcopallium; Area X, Area X of the striatum; Av, avalanch; B, nucleus basorostralis; Cb, cerebellum; CM, caudal mesopallium; CMM, caudomedial mesopallium; CMS, caudal medial striatum; CN, cochlear nuclei; CSt, caudal striatum; DLM, medial nucleus of dorsolateral thalamus; DLMv, medial nucleus of dorsolateral thalamus, vocal part; DM, dorsal medial nucleus of the midbrain; DMm, magnocellular nucleus of the dorsomedial thalamus; E, nucleus entopallium; H, hyperpallium; HVC, (a letter based name); MOc, oval nucleus of the mesopallium complex; ICo, intercollicular nucleus; INP, intrapeduncular nucleus; L2, field L2; MMSt, magnocellular nucleus of the anterior striatum; M, mesopallium; MAN, magnocellular nucleus of anterior nidopallium; MLd, mesencephalic lateral dorsal nucleus; N, nidopallium; NAOc, oval nucleus of the anterior nidopallium complex; NCM, caudal medial nidopallium; NIf, interfacial nucleus of the nidopallium; NLC, central nucleus of the lateral nidopallium; nXIIts, tracheosyringeal subdivision of the hypoglossal nucleus; OT, optic tectum; Ov, nucleus oviodalis; P, pallidum; PAm, nucleus parambiguus; PSL, pallial subpallial lamina; RA, robust nucleus of the arcopallium; RAm, nucleus retroambiguus; St, Striatum; Uva, nucleus uvaeformis; VA, vocal nucleus of the arcopallium; VAN, vocal nucleus of the anterior nidopallium; VASt, vocal nucleus of the anterior striatum; VLN, vocal nucleus of the lateral nidopallium
LITERATURE CITED
- Aamodt SM, Nordeen EJ, Nordeen KW. Early isolation from conspecific song does not affect the normal developmental decline of N-methyl-D-aspartate receptor binding in an avian song nucleus. J Neurobiol. 1995;27:76–84. doi: 10.1002/neu.480270108. [DOI] [PubMed] [Google Scholar]
- Abel T, Lattal KM. Molecular mechanisms of memory acquisition, consolidation and retrieval. Curr Opin Neurobiol. 2001;11:180–187. doi: 10.1016/s0959-4388(00)00194-x. [DOI] [PubMed] [Google Scholar]
- Akutagawa E, Konishi M. A monoclonal antibody specific to a song system nuclear antigen in estrilidine finches. Neuron. 2001;31:545–556. doi: 10.1016/s0896-6273(01)00388-9. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla A, Kirn JR. Birth, migration, incorporation, and death of vocal control neurons in adult songbirds. J Neurobiol. 1997;33:585–601. [PubMed] [Google Scholar]
- Arnold AP, Nottebohm F, Pfaff DW. Hormone concentrating cells in vocal control and other areas of the brain of the zebra finch (Poephila guttata) J Comp Neurol. 1976;165:487–511. doi: 10.1002/cne.901650406. [DOI] [PubMed] [Google Scholar]
- Bading H. Nuclear calcium-activated gene expression: possible roles in neuronal plasticity and epileptogenesis. Epilepsy Res. 1999;36:225–231. doi: 10.1016/s0920-1211(99)00053-4. [DOI] [PubMed] [Google Scholar]
- Bahn S, Volk B, Wisden W. Kainate receptor gene expression in the developing rat brain. J Neurosci. 1994;14:5525–5547. doi: 10.1523/JNEUROSCI.14-09-05525.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball GF. Neurochemical specializations associated with vocal learning and production in songbirds and budgerigars. Brain Behav Evol. 1994;44:234–246. doi: 10.1159/000113579. [DOI] [PubMed] [Google Scholar]
- Baptista L. Nature and its nurturing in avian vocal development. In: Kroodsma D, Miller E, editors. Ecology and evolution of acoustic communication in birds. Ithaca, NY: Cornell University Press; 1996. pp. 39–60. [Google Scholar]
- Basham M, Nordeen E, Nordeen K. Blockade of NMDA receptors in the anterior forebrain impairs sensory acquisition in the zebra finch (Poephila guttata) Neurobiol Learn Mem. 1996;66:295–304. doi: 10.1006/nlme.1996.0071. [DOI] [PubMed] [Google Scholar]
- Basham ME, Sohrabji F, Singh TD, Nordeen EJ, Nordeen KW. Developmental regulation of NMDA recptor 2B subunit mRNA and Ifenprodil binding in the zebra finch anterior forebrain. J Neurobiol. 1999;39:155–167. [PubMed] [Google Scholar]
- Berke JD, Paletzki RF, Aronson GJ, Hyman SE, Gerfen CR. A complex program of striatal gene expression induced by dopaminergic stimulation. J Neurosci. 1998;18:5301–5310. doi: 10.1523/JNEUROSCI.18-14-05301.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottjer SW. The distribution of tyrosine hydroxylase immunoreactivity in the brains of male and female zebra finches. J Neurobiol. 1993;24:51–69. doi: 10.1002/neu.480240105. [DOI] [PubMed] [Google Scholar]
- Brenowitz EA. Comparative approaches to the avian song system. J Neurobiol. 1997;33:517–531. doi: 10.1002/(sici)1097-4695(19971105)33:5<517::aid-neu3>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Casto JM, Ball GF. Characterization and localization of D1 dopamine receptors in the sexually dimorphic vocal control nucleus, area X, and the basal ganglia of European starlings. J Neurobiol. 1994;25:767–780. doi: 10.1002/neu.480250703. [DOI] [PubMed] [Google Scholar]
- Chew SJ, Mello C, Nottebohm F, Jarvis E, Vicario DS. Decrements in auditory responses to a repeated conspecific song are long-lasting and require two periods of protein synthesis in the songbird forebrain. Proc Natl Acad Sci U S A. 1995;92:3406–3410. doi: 10.1073/pnas.92.8.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton DF. The genomic action potential. Neurobiol Learn Mem. 2000;74:185–216. doi: 10.1006/nlme.2000.3967. [DOI] [PubMed] [Google Scholar]
- Conti F, Minelli A, Brecha NC. Cellular localization and laminar distribution of AMPA glutamate receptor subunits mRNAs and proteins in the rat cerebral cortex. J Comp Neurol. 1994;350:241–259. doi: 10.1002/cne.903500208. [DOI] [PubMed] [Google Scholar]
- Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: diversity, development and disease. Curr Opin Neurobiol. 2001;11:327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- Das S, Sasaki YF, Rothe T, Premkumar LS, Takasu M, Crandall JE, Dikkes P, Conner DA, Rayudu PV, Cheung W, Chen HS, Lipton SA, Nakanishi N. Increased NMDA current and spine density in mice lacking the NMDA receptor subunit NR3A. Nature. 1998;393:377–381. doi: 10.1038/30748. [DOI] [PubMed] [Google Scholar]
- Deacon TW. The symbolic species. New York: W.W. Norton & Company, Inc; 1997. pp. 225–253. [Google Scholar]
- Dengledine R, Borges K, Bowie D, Traynelis S. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- Denisenko-Nehrbass NI, Jarvis E, Scharff C, Nottebohm F, Mello CV. Site-specific retinoic acid production in the brain of adult songbirds. Neuron. 2000;27:359–370. doi: 10.1016/s0896-6273(00)00043-x. [DOI] [PubMed] [Google Scholar]
- Doupe AJ, Kuhl PK. Birdsong and human speech: common themes and mechanisms. Annu Rev Neurosci. 1999;22:567–631. doi: 10.1146/annurev.neuro.22.1.567. [DOI] [PubMed] [Google Scholar]
- Durand SE, Heaton JT, Amateau SK, Brauth SE. Vocal control pathways through the anterior forebrain of a parrot (Melopsittacus undulatus) J Comp Neurol. 1997;377:179–206. doi: 10.1002/(sici)1096-9861(19970113)377:2<179::aid-cne3>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Durand SE, Liang W, Brauth SE. Methionine enkephalin immunoreactivity in the brain of the budgerigar (Melopsittacus undulatus): similarities and differences with respect to oscine songbirds. J Comp Neurol. 1998;393:145–168. doi: 10.1002/(sici)1096-9861(19980406)393:2<145::aid-cne2>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Dutar P, Petrozzino J, Vu H, Schmidt M, Perkel D. Slow synaptic inhibition mediated by metabotropic glutamate receptor activation of GIRK channels. J Neurophysiol. 2000;84:2284–2290. doi: 10.1152/jn.2000.84.5.2284. [DOI] [PubMed] [Google Scholar]
- Eberwine J, Yeh H, Miyashiro K, Cao Y, Nair S, Finnell R, Zettel M, Coleman P. Analysis of gene expression in single live neurons. Proc Natl Acad Sci U S A. 1992;89:3010–3014. doi: 10.1073/pnas.89.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farries MA. The oscine song system considered in the context of the avian brain: lessons learned from comparative neurobiology. Brain Behav Evol. 2001;58:80–100. doi: 10.1159/000047263. [DOI] [PubMed] [Google Scholar]
- Gahr M. Neural song control system of hummingbirds: comparison to swifts, vocal learning (songbirds) and nonlearning (suboscines) passerines, and vocal learning (budgerigars) and nonlearning (dove, owl, gull, quail, chicken) nonpasserines. J Comp Neurol. 2000;426:182–196. [PubMed] [Google Scholar]
- Geiger J, Roth A, Taskin B, Jonas P. Glutamate-mediated synaptic excitation of cortical interneurons. In: Jonas P, Monyer H, editors. Ionotropic glutamate receptors in the CNS. Berlin: Springer; 1999. pp. 363–398. [Google Scholar]
- Gustinci S, Batalov S, Beisel KW, Bono H, Carninci P, Fletcher CF, Grimmond S, Hirokawa N, Jarvis ED, Jegla T, Kawasawa Y, LeMieux J, Miki H, Raviola E, Teasdale RD, Tominaga N, Yagi K, Zimmer A, Hayashizaki Y, Okazaki Y. Analysis of the mouse transcriptome for genes involved in the function of the nervous system. Genome Res. 2003;13:1395–1401. doi: 10.1101/gr.1135303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahnloser RHR, Kozhevnikov AA, Fee MS. An ultra-sparse code underlies the generation of neural sequences in a songbird. Nature. 2002;495:65–70. doi: 10.1038/nature00974. [DOI] [PubMed] [Google Scholar]
- Heinrich J, Singh T, Sohrabji F, Nordeen K, Nordeen E. Developmental and hormonal regulation of NR2A mRNA in forebrain regions controlling avian vocal learning. J Neurobiol. 2002;51:149–159. doi: 10.1002/neu.10046. [DOI] [PubMed] [Google Scholar]
- Hessler NA, Doupe AJ. Singing-related neural activity in a dorsal forebrain-basal ganglia circuit of adult zebra finches. J Neurosci. 1999a;19:10461–10481. doi: 10.1523/JNEUROSCI.19-23-10461.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessler NA, Doupe AJ. Social context modulates singing-related neural activity in the songbird forebrain. Nat Neurosci. 1999b;2:209–211. doi: 10.1038/6306. [DOI] [PubMed] [Google Scholar]
- Hollmann M. Structure of ionotropic receptors. In: Jonas P, Monyer H, editors. Ionotropic glutamate receptors in the CNS. Berlin: Springer; 1999. pp. 1–98. [Google Scholar]
- Holzenberger M, Jarvis ED, Chong C, Grossman M, Nottebohm F, Scharff C. Selective expression of insulin-like growth factor II in the songbird brain. J Neurosci. 1997;17:6974–6987. doi: 10.1523/JNEUROSCI.17-18-06974.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Myers SJ, Dingledine R. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat Neurosci. 1999;2:867–872. doi: 10.1038/13165. [DOI] [PubMed] [Google Scholar]
- Huang Y, Doherty JJ, Dingledine R. Altered histone acetylation at glutamate receptor 2 and brain-derived neurotrophic factor genes is an early event triggered by status epilepticus. J Neurosci. 2002;22:8422–8428. doi: 10.1523/JNEUROSCI.22-19-08422.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Bottjer SW. The role of auditory experience in the formation of neural circuits underlying vocal learning in zebra finches. J Neurosci. 2002;22:946–958. doi: 10.1523/JNEUROSCI.22-03-00946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Viswanathan SS, Bottjer SW. Development of topography within song control circuitry of zebra finches during the sensitive period for song learning. J Neurosci. 1999;19:6037–6057. doi: 10.1523/JNEUROSCI.19-14-06037.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED. Brains and birdsong. In: Marler P, Slaberkorn H, editors. Nature’s music: the science of birdsong. New York: Elsevier-Academic Press; 2004a. pp. 239–275. [Google Scholar]
- Jarvis ED. Birdsong and the neurobiology of human language. Ann N Y Acad Sci. 2004b;1016:746–777. doi: 10.1196/annals.1298.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Mello CV. Molecular mapping of brain areas involved in parrot vocal communication. J Comp Neurol. 2000;419:1–31. doi: 10.1002/(sici)1096-9861(20000327)419:1<1::aid-cne1>3.0.co;2-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Nottebohm F. Motor-driven gene expression. Proc Natl Acad Sci U S A. 1997;94:4097–4102. doi: 10.1073/pnas.94.8.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Scharff C, Grossman MR, Ramos JA, Nottebohm F. For whom the bird sings: context-dependent gene expression. Neuron. 1998;21:775–788. doi: 10.1016/s0896-6273(00)80594-2. [DOI] [PubMed] [Google Scholar]
- Jarvis ED, Ribeiro S, Vielliard J, Da Silva ML, Ventura D, Mello CV. Behaviorally driven gene expression reveals song nuclei in hummingbird brain. Nature. 2000;406:628–632. doi: 10.1038/35020570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson F, Sablan MM, Bottjer SW. Topographic organization of a forebrain pathway involved with vocal learning in zebra finches. J Comp Neurol. 1995;358:260–278. doi: 10.1002/cne.903580208. [DOI] [PubMed] [Google Scholar]
- Karten JH. The organization of the ascending auditory pathway in the pigeon (Columba livia) I. Diencephalic projection of the inferior colliculus (nucleus mesencephali lateralis, pars dorsalis) Brain Res. 1967;6:409–427. doi: 10.1016/0006-8993(67)90055-8. [DOI] [PubMed] [Google Scholar]
- Karten JH. The ascending auditory pathway in the pigeon (Columba livia) II. Telencephalic projections of the nucleus ovoidalis thalami. Brain Res. 1968;11:134–153. doi: 10.1016/0006-8993(68)90078-4. [DOI] [PubMed] [Google Scholar]
- Kimpo RR, Doupe AJ. FOS is induced by singing in distinct neuronal populations in a motor network. Neuron. 1997;18:315–325. doi: 10.1016/s0896-6273(00)80271-8. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Uno H, Okanoya K. Partial lesions in the anterior forebrain pathway affect song production in adult Bengalese finches. Neuroreport. 2001;12:353–358. doi: 10.1097/00001756-200102120-00034. [DOI] [PubMed] [Google Scholar]
- Kuypers HGJM. Corticobulbar connexions to the pons and lower brain-stem in man. Brain. 1958a;81:364–388. doi: 10.1093/brain/81.3.364. [DOI] [PubMed] [Google Scholar]
- Kuypers HGJM. Some projections from the peri-central cortex to the pons and lower brainstem in monkey and chimpanzee. J Comp Neurol. 1958b;100:221–255. doi: 10.1002/cne.901100205. [DOI] [PubMed] [Google Scholar]
- Lerea L. Glutamate receptors and gene induction: signalling from receptor to nucleus. Cell Signal. 1997;9:219–226. doi: 10.1016/s0898-6568(96)00134-9. [DOI] [PubMed] [Google Scholar]
- Li X-C, Jarvis E. Soc Neurosci Abstr. 2001. Sensory- and motor-driven BDNF expression in a vocal communication system; pp. 538–8. [Google Scholar]
- Li XC, Jarvis ED, Alvarez-Borda B, Lim DA, Nottebohm F. A relationship between behavior, neurotrophin expression, and new neuron survival. Proc Natl Acad Sci U S A. 2000;97:8584–8589. doi: 10.1073/pnas.140222497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey FJ, Hunt SP. Identifying changes in gene expression in the nervous system: mRNA differential display. TINS. 1996;19:84–88. doi: 10.1016/s0166-2236(96)80032-4. [DOI] [PubMed] [Google Scholar]
- Livingston FS, White SA, Mooney R. Slow NMDA-EPSCs at synapses critical for song development are not required for song learning in zebra finches. Nat Neurosci. 2000;3:482–488. doi: 10.1038/74857. [DOI] [PubMed] [Google Scholar]
- Luo M, Perkel DJ. A GABAergic, strongly inhibitory projection to a thalamic nucleus in the zebra finch song system. J Neurosci. 1999a;19:6700–6711. doi: 10.1523/JNEUROSCI.19-15-06700.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Perkel DJ. Long-range gabaergic projection in a circuit essential for vocal learning. J Comp Neurol. 1999b;403:68–84. [PubMed] [Google Scholar]
- Luo M, Ding L, Perkel DJ. An avian basal ganglia pathway essential for vocal learning forms a closed topographic loop. J Neurosci. 2001;21:6836–6845. doi: 10.1523/JNEUROSCI.21-17-06836.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margoliash D, Fortune ES, Sutter ML, Yu AC, Wren-Hardin BD, Dave A. Distributed representation in the song system of oscines: evolutionary implications and functional consequences. Brain Behav Evol. 1994;44:247–264. doi: 10.1159/000113580. [DOI] [PubMed] [Google Scholar]
- Marler P, Mitani J. Vocal communication in primates and birds: parallels and contrasts. In: Todt D, Goedeking P, Symmes D, editors. Primate vocal communication. Berlin: Springer; 1988. pp. 3–14. [Google Scholar]
- McCasland JS. Neuronal control of bird song production. J Neurosci. 1987;7:23–39. doi: 10.1523/JNEUROSCI.07-01-00023.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCasland JS, Konishi M. Interaction between auditory and motor activities in an avian song control nucleus. Proc Natl Acad Sci U S A. 1981;78:7815–7819. doi: 10.1073/pnas.78.12.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CV, Nottebohm F, Clayton D. Repeated exposure to one song leads to a rapid and persistent decline in an immediate early gene’s response to that song in zebra finch telencephalon. J Neurosci. 1995;15:6919–6925. doi: 10.1523/JNEUROSCI.15-10-06919.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CV, Jarvis ED, Denisenko N, Rivas M. Isolation of song-regulated genes in the brain of songbirds. In: Liang P, Pardee AB, editors. Methods in molecular biology: differential display methods and protocols. Totowa, NJ: Humana Press; 1997. pp. 205–217. [DOI] [PubMed] [Google Scholar]
- Mello CV, Vates GE, Okuhata S, Nottebohm F. Descending auditory pathways in the adult male zebra finch (Taeniopygia guttata) J Comp Neurol. 1998;395:137–160. [PubMed] [Google Scholar]
- Mooney R. Different subthreshold mechanisms underlie song selectivity in identified HVc neurons of the zebra finch. J Neurosci. 2000;20:5420–5436. doi: 10.1523/JNEUROSCI.20-14-05420.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney R, Konishi M. Two distinct inputs to an avian song nucleus activate different glutamate receptor subtypes on individual neurons. Proc Natl Acad Sci U S A. 1991;88:4075–4079. doi: 10.1073/pnas.88.10.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morando L, Cesa R, Rasetti R, Harvey R, Strata P. Role of glutamate delta-2 receptors in activity-dependent competition between heterologous afferent fibers. Proc Natl Acad Sci USA. 2001;98:9954–9959. doi: 10.1073/pnas.171098398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SJ, Peters J, Huang Y, Comer MB, Barthel F, Dingledine R. Transcriptional regulation of the GluR2 gene: neural-specific expression, multiple promoters, and regulatory elements. J Neurosci. 1998;18:6723–6739. doi: 10.1523/JNEUROSCI.18-17-06723.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima Y, Iwakabe H, Akazawa C, Nawa H, Shigemoto R, Mizuno N, Nakanishi S. Molecular characterization of a novel retinal metabotropic glutamate receptor mGluR6 with a high agonist selectivity for L-2-amino-4-phosphonobutyrate. J Biol Chem. 1993;268:11868–11873. [PubMed] [Google Scholar]
- Nishi M, Hinds H, Lu HP, Kawata M, Hayashi Y. Motoneuron-specific expression of NR3B, a novel NMDA-type glutamate receptor subunit that works in a dominant-negative manner. J Neurosci. 2001;21:RC185. doi: 10.1523/JNEUROSCI.21-23-j0003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottebohm F. The origins of vocal learning. Am Naturalist. 1972;106:116–140. [Google Scholar]
- Nottebohm F. The search for neural mechanisms that define the sensitive period for song learning in birds. Neth J Zool. 1993;43:193–234. [Google Scholar]
- Nottebohm F, Nottebohm ME. Vocalizations and breeding behaviour of surgically deafened ring doves (Streptopelia risoria) Anim Behav. 1971;19:313–327. doi: 10.1016/s0003-3472(71)80012-x. [DOI] [PubMed] [Google Scholar]
- Nottebohm F, Stokes TM, Leonard CM. Central control of song in the canary, Serinus canarius. J Comp Neurol. 1976;165:457–486. doi: 10.1002/cne.901650405. [DOI] [PubMed] [Google Scholar]
- Okuhata S, Saito N. Synaptic connection of thalamocerebral vocal nuclei of the canary. Brain Res Bull. 1987;18:35–44. doi: 10.1016/0361-9230(87)90031-1. [DOI] [PubMed] [Google Scholar]
- Paton JA, Manogue KR, Nottebohm F. Bilateral organization of the vocal control pathway in the budgerigar, Melopsittacus undulatus. J Neurosci. 1981;1:1279–1288. doi: 10.1523/JNEUROSCI.01-11-01279.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkel D, Farries M. Complementary “bottom-up” and “top-down” approaches to basal ganglia function. Curr Opin Neurobiol. 2000;10:725–731. doi: 10.1016/s0959-4388(00)00156-2. [DOI] [PubMed] [Google Scholar]
- Perkel DJ, Farries MA, Luo M, Ding L. Electrophysiological analysis of a songbird basal ganglia circuit essential for vocal plasticity. Brain Res Bull. 2002;57:529–532. doi: 10.1016/s0361-9230(01)00690-6. [DOI] [PubMed] [Google Scholar]
- Petralia R, Rubio M, Wenthold R. Cellular and subcellular distribution of glutamate receptors. In: Jonas P, Monyer H, editors. Ionotropic glutamate receptors in the CNS. Berlin: Springer; 1999. pp. 143–174. [Google Scholar]
- Pin J-P, Duvoisin R. Neurotransmitter receptors. I. The metabotropic receptors: structure and functions. Neuropharmacology. 1995;34:1–26. doi: 10.1016/0028-3908(94)00129-g. [DOI] [PubMed] [Google Scholar]
- Reiner A, Bruce L, Butler A, Csillag A, Kuenzel W, Medina L, Paxinos G, Perkel D, Shimizu T, Striedter G, Wild M, Ball G, Durand S, Gunturkun O, Lee D, Mello C, Powers A, White S, Hough G, Kubikova L, Smulders TV, Wada K, Dugas-Ford J, Husband S, Yamamoto K, Yu J, Siang C, Jarvis ED. Revised nomenclature for avian telencephalon and some related brainstem nuclei. J Comp Neurol. 2004;473:377–414. doi: 10.1002/cne.20118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro S, Mello CV. Gene expression and synaptic plasticity in the auditory forebrain of songbirds. Learn Mem. 2000;7:235–243. doi: 10.1101/lm.34400. [DOI] [PubMed] [Google Scholar]
- Rodriquez-Moreno A, Lerma J. Kainate receptor modulation of GABA release involves a metabotropic function. Neuron. 1998;20:1211–1218. doi: 10.1016/s0896-6273(00)80501-2. [DOI] [PubMed] [Google Scholar]
- Scharff C, Nottebohm F. A comparative study of the behavioral deficits following lesions of various parts of the zebra finch song system: implications for vocal learning. J Neurosci. 1991;11:2896–2913. doi: 10.1523/JNEUROSCI.11-09-02896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Greenberg ME. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron. 1990;4:477–485. doi: 10.1016/0896-6273(90)90106-p. [DOI] [PubMed] [Google Scholar]
- Sibley CG, Ahlquist JE. Phylogeny and classification of birds: a study in molecular evolution. New Haven, CT: Yale University Press; 1990. [Google Scholar]
- Singh T, Basham M, Nordeen E, Nordeen K. Early sensory and harmonal experience modulate age-related changes in NR2B mRNA within a forebrain region controlling avian vocal learning. J Neurobiol. 2000;44:82–94. doi: 10.1002/1097-4695(200007)44:1<82::aid-neu8>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Slabbekoorn H, De Kort S, Ten Cate C. Comparative analysis of perch-coo vocalizations in Stretopelia doves. Auk. 1999;116:737–748. [Google Scholar]
- Spiro JE, Dalva MB, Mooney R. Long-range inhibition within the zebra finch song nucleus RA can coordinate the firing of multiple projection neurons. J Neurophysiol. 1999;81:3007–3020. doi: 10.1152/jn.1999.81.6.3007. [DOI] [PubMed] [Google Scholar]
- Standaert D, Testa C, Young A, Penney J. Organization of N-methyl-D-aspartate glutamate receptor gene expression in the basal ganglia of the rat. J Comp Neurol. 1994;343:1–16. doi: 10.1002/cne.903430102. [DOI] [PubMed] [Google Scholar]
- Stark L, Perkel D. Two-stage, input-specific synaptic maturation in a nucleus essential for vocal production in the zebra finch. J Neurosci. 1999;19:9107–9116. doi: 10.1523/JNEUROSCI.19-20-09107.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striedter GF. The vocal control pathways in budgerigars differ from those in songbirds. J Comp Neurol. 1994;343:35–56. doi: 10.1002/cne.903430104. [DOI] [PubMed] [Google Scholar]
- Testa C, Standaert D, Young A, Penney J. Metabotropic glutamate receptor mRNA expression in the basal ganglia of the rat. J Neurosci. 1994;14:3005–3018. doi: 10.1523/JNEUROSCI.14-05-03005.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vates GE, Vicario DS, Nottebohm F. Reafferent thalamo-“cortical” loops in the song system of oscine songbirds. J Comp Neurol. 1997;380:275–290. [PubMed] [Google Scholar]
- Watson JB, Margulies JE. Differential cDNA screening strategies to identify novel stage-specific proteins in the developing mammalian brain. Dev Neurosci. 1993;15:77–86. doi: 10.1159/000111319. [DOI] [PubMed] [Google Scholar]
- West AE, Chen WG, Dalva MB, Dolmetsch RE, Kornhauser JM, Shaywitz AJ, Takasu MA, Tao X, Greenberg ME. Calcium regulation of neuronal gene expression. Proc Natl Acad Sci U S A. 2001;98:11024–11031. doi: 10.1073/pnas.191352298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SA, Livingston FS, Mooney R. Androgens modulate NMDA receptor-mediated EPSCs in the zebra finch song system. J Neurophysiol. 1999;82:2221–2234. doi: 10.1152/jn.1999.82.5.2221. [DOI] [PubMed] [Google Scholar]
- Wild JM. Neural pathways for the control of birdsong production. J Neurobiol. 1997;33:653–670. doi: 10.1002/(sici)1097-4695(19971105)33:5<653::aid-neu11>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Wild JM, Li D, Eagleton C. Projections of the dorsomedial nucleus of the intercollicular complex (DM) in relation to respiratory-vocal nuclei in the brainstem of pigeon (Columba livia) and zebra finch (Taeniopygia guttata) J Comp Neurol. 1997;377:392–413. doi: 10.1002/(sici)1096-9861(19970120)377:3<392::aid-cne7>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Wisden W, Seeburg P. A complex mosaic of high-affinity kainate receptors in the rat brain. J Neurosci. 1993;13:3582–2598. doi: 10.1523/JNEUROSCI.13-08-03582.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley P, Cole A, Saffen D, Baraban J. Regulation of immediate early genes in the brain: role of NMDA receptor activation. Prog Brain Res. 1990;86:277–285. doi: 10.1016/s0079-6123(08)63184-2. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Imaki J, Okamoto Y, Iwakabe H, Fujisawa H, Matsuda A, Nakanisi S, Matsuda H, Hagiwara M. CREB-induced transcriptional activation depends on mGluR6 in rod bipolar cells. Brain Res Mol Brain Res. 1998;57:241–247. doi: 10.1016/s0169-328x(98)00089-8. [DOI] [PubMed] [Google Scholar]
- Young L, Nilsen R, Waymire K, MacGregor G, Insel T. Increased affiliative response to vasopressin in mice expressing the V1a receptor from a monogomous vole. Nature. 1999;400:766–768. doi: 10.1038/23475. [DOI] [PubMed] [Google Scholar]
- Yu AC, Margoliash D. Temporal hierarchical control of singing in birds. Science. 1996;273:1871–1875. doi: 10.1126/science.273.5283.1871. [DOI] [PubMed] [Google Scholar]
- Zirlinger M, Kreiman G, Anderson D. Amygdala-enriched genes identified by microarray technology are restricted to specific amygdaloid subnuclei. Proc Natl Acad Sci U S A. 2001;98:5270–5275. doi: 10.1073/pnas.091094698. [DOI] [PMC free article] [PubMed] [Google Scholar]


