Abstract
Purpose
Metastasis remains an incurable common complication in patients with gastric cancer. A variety of theories have been proposed to explain the inefficiency of the metastatic process. To compare protein expression of metastasis-related genes (nm23, KISS1, KAI1 and p53) between primary tumours and metastatic tumours may be useful in illustrating these theories.
Methods
Metastasis-related tissue microarrays (including normal tissues, primary tumours, nodal metastases and liver metastases) were constructed. The protein expression of nm23, KISS1, KAI1 and p53 in lymph node and liver metastases from advanced gastric cancer specimens was mainly examined by immunohistochemical staining in relation to primary tumours.
Results
Immunohistochemical staining showed reduced protein expression of nm23, KISS1 and KAI1 in lymph node and liver metastases compared with primary tumours. Results for p53 were to the contrary.
Conclusions
Our investigations revealed a tendency of reduced protein expression of metastasis suppressor genes nm23, KISS1 and KAI1 in gastric cancer with the progress of metastasis. This means that the progression theory is an important determinant of metastatic efficiency.
Keywords: KAI1, KISS1, nm23, p53
Lymph node or/and liver metastases are diagnosed in >50% of gastric cancer patients. Despite multidisciplinary treatment including microsurgical tumour removal, radio-, and chemotherapy, survival is usually limited to several years, even months. Given improved treatment of primary tumours of gastric cancer and increasing senescence of the population, the incidence of gastric cancer lymph node or/and liver metastases will moreover rise in the future.
A variety of theories have been proposed to explain the inefficiency of the metastatic process. Three theories were mostly accepted: the progression theory, the transient compartment theory, and the allelic diversity theory (Hunter 2003). But which theory might explain the whole metastasis process remains unclear. Comparing differences of gene expression between the primary tumour and the metastatic tumour from the same patient may be helpful in finding some clues. Over the past decade, several genes have been identified, which are involved in the progression toward metastasis in vitro and in vivo (Debies and Welch 2002). We selected four of these genes for evaluation of their roles in gastric metastases using tissue microarray (TMA) and immunohistochemistry.
nm23
The first novel metastasis suppressor gene (MSG) discovered and cloned was nm23, which maps to chromosome 17q21 and encodes the nucleoside diphosphate kinase A, a member of the NDP kinase family. Nm23 expression is reduced in metastatic melanoma and breast cancer cell lines (Debies and Welch 2002). Transfection into cell lines affects invasion, motility, colonization, differentiation, and liver metastasis (Howlett et al. 1994; Arnaud-Dabernat et al. 2003; Suzuki et al. 2004). Decreased expression of nm23-H1, the human homologue, is found in advanced stage of human cancer (Tannapfel et al. 1997; Berney et al. 1999; Dursun et al. 2002).
Moreover, reduced expression of nm23-H1 within primary colorectal carcinomas could serve as a predictor of liver metastasis and lymph node metastases Berney et al. (1999). Most studies showed that there seemed to be a prognostic implication of nm23-H1 expression in gastric and colorectal carcinoma Dursun et al. (2002). Namely, well-differentiated adenocarcinomas showed significantly strong staining for nm23-H1 compared with the moderately and poorly differentiated adenocarcinomas (Yeung et al. 1998; Dursun et al. 2002).
However, results of many other researchers showed discordant evidence: nm23-H1 expression could not predict liver or lymph node metastases and clinical survival in gastric and colorectal cancer patients (Wang et al. 1998; Sarris & Lee 2001a; Dusonchet et al. 2003). So, the exact biological function of nm23 remains unclear.
KISS1
Transfection of the metastasis suppressor KISS1/Metastin (KISS1; 1q32) suppressed the metastatic capability of malignant melanoma and breast cancer cell lines Harms et al. (2003). Vice versa, the KISS1 expression is lost as melanoma cells convert from the benign to the malignant phenotype. Clinical correlations have not been extensively established for KISS1, mostly because antibodies are unavailable. However, reduced or loss of KISS1 mRNA expression is associated with cancer progression and clinical outcome in gastric, bladder and oesophageal squamous cell carcinoma (Sanchez-Carbayo et al. 2003; Dhar et al. 2004; Ikeguchi et al. 2004). But in hepatocellular carcinoma, the results were to the contrary (Ikeguchi et al. 2003).
The molecular mechanism of KISS1 action remains unclear. Recent identification of a 54 amino acid peptide of KISS1, termed metastin or kisspeptin-54, and its cognate G-protein coupled receptor (AXOR12, GPR54 or hOT7T17S) has provided additional clues and avenues of research (Kotani et al. 2001; Ohtaki et al. 2001).
In addition, Yan et al. (2001) showed that KISS1 specifically reduced expression of MMP9 (not MMP2) in HT1080 cells, suggesting an alternative mechanism of action. Most data strongly suggest that KISS1 is acting at a late step in the metastatic cascade; however, definitive proof is still required.
KAI1
KAI1 (11p11.2-p13, Kangai 1/CD82, C33) is a member of the TM4SF superfamily of adhesion molecules and influences lymphocyte differentiation and function. Although KAI1 was discovered as a prostate cancer metastasis suppressor, it has been associated with the metastatic potential of breast, lung, liver, bladder and melanoma cells (Lombardi et al. 1999; Yang et al. 2000, 2001). KAI1 expression is inversely correlated with aggressive behaviour in a panel of human cancer cells lines (Yang et al. 2001). Moreover, the KAI1-transfected highly malignant colorectal carcinoma cell line LoVo showed increased homotypic cell–cell adhesion and cell aggregation in comparison with control cells (Liu et al. 2001, 2003).
Ito et al. (2005) reported that KAI-1 expression was significantly reduced in cases of clinically apparent metastasis of papillary microcarcinomas of the thyroid. Goncharuk et al. (2004) showed that co-downregulation of PTEN, KAI-1,and nm23-H1 significantly correlates with distant metastasis and predicts shortened survival in non-small cell lung cancer. In vitro experiments show that p53 binds and activates KAI1 promoter. However, in prostate cell lines, induction of p53 with DNA damaging agents did not increase KAI1 expression. So, it remains questionable whether KAI1 is the primary target of the tumour suppressor protein p53 (Adachi et al. 1996; Mashimoto et al. 1998; Maurer et al. 1999; Jackson & Puisieux 2000; Shinohara et al. 2001).
p53
Tumour suppressor p53 protein is a transcriptional factor, which regulates several cellular processes, creating limitations for tumorigenic transformation. In response to proliferation-affecting events (DNA damage, oncogenes, oxidative stress, etc.), cells utilizing p53 pathways may switch to growth arrest, programmed cell death, or cell differentiation. Mutation of p53 and aberration of p53 expression were related to human cancer development and progression. However, it remains questionable whether it can predict metastases. Kakeji et al. (1993) showed that p53 overexpression has higher potential for metastasis to lymph nodes. Graziano et al. found that p53 overexpression may be involved in early stages of gastric cancer progression (Kataoka et al. 2000; Graziano et al. 2001).
Materials and methods
Tumour samples
Formalin-fixed and paraffin-embedded gastric tumours and control specimens, which had been resected during the period 1995–2000, were from the archives of the department of pathology, Changhai hospital. Seventy-one tumours, 71 surrounding non-tumours, 64 regional lymph node and 41 liver metastatic tissues were studied. The population characteristics (age, sex, types, differentiation and T stage) are summarized in Table 1.
Table 1.
Population characteristics of the 71 patients with gastric carcinoma
| Characteristics | ||
|---|---|---|
| Age (years) | Mean | 55.7 (27–80) |
| Sex (n) | Male | 43 |
| Female | 28 | |
| Differentiation (n) | Well | 1 |
| Moderate | 31 | |
| Poor | 39 | |
| Type (n) | Adenocarcinoma | 52 |
| Mucinous | 11 | |
| Undifferentiated | 4 | |
| Ring-cell | 4 | |
| T stage (n) | T1 | 2 |
| T2 | 14 | |
| T3 | 37 | |
| T4 | 18 |
Construction of tissue microarrays
Briefly, a tissue arraying instrument (Beecher Instruments, Silver Spring, MD, USA) was used to create holes in a recipient paraffin block and to acquire tissue cores from the donor block by a thin-walled needle with an inner diameter of 2 mm, held in an x–y precision guide. The cylindrical sample was retrieved from the selected region in the donor and extruded directly into the recipient block with defined array coordinates. After the construction of the array block, multiple 5-μm sections were cut and H&E-stained sections were used for histological verification of tumour tissues on the arrayed samples Yu et al. (2004). The TMA consisted of one normal, two primary tumours and two to three nodal metastases cores from each invasive gastric cancer case placed at one line. For those with liver metastases, two cores were involved.
Immunohistochemistry
Protein expression was analysed using immunohistochemistry. The antibodies used are summarized in Table 2. In brief, serial 4 μm slices were obtained from formalin-fixed and paraffin embedded tissue specimens. Sections were dewaxed in xylene and rehydrated in alcohol followed by wet autoclave pretreatment (4 min at 120 °C) in citrate buffer (20 mM Tris, 15 mM EDTA, 10 mM tri-Na-Citrat, pH 7.8) for antigen retrieval. After this, hydrogen peroxidase was added for 10 min (3% H2O2 in methanol) and specimens were washed three times in TBS-buffer. After washing with phosphate-buffered saline (PBS), the primary antibody was applied for overnight incubation with 1% BSA in PBS. After washing with PBS, the sections were incubated with the secondary antibody. For monoclonals, rabbit anti-mouse immunoglobulin G (IgG) (RAMHRP, Dako7P161) was applied for 30 min and then the sections were washed in PBS and finally incubated with the tertiary antibody, swine-anti rabbit IgG (RAMHRP, Dako7P217), for 30 min. For polyclonals, no tertiary antibody was used. After the final washing with PBS, staining was performed by means of 3-amino-9-ethylcarbazole (AEC) in dimethyl formamide with H2O2, followed by counterstaining with Mayer-haematoxylin for 30 s. All cases were stained in the same run for each protein. Tumour specimens that had shown positive results in former studies served as positive controls. The primary antibody was omitted for negative controls.
Table 2.
Panel of antibodies used
| Antibody | Clone | Company | Dilution | Staining type |
|---|---|---|---|---|
| nm23-H1 | sc-465 | Santa Cruz, CA, USA | 1:100 | Membrane |
| KAI1 | sc-1087 | Santa Cruz, CA, USA | 1:150 | Cytoplasmatic and membrane |
| KISS1 | sc-15400 | Santa Cruz, CA, USA | 1:150 | Cytoplasmatic |
| P53 | DO-7 | Dako, Copenhagen, Denmark | 1:200 | Nuclear and cytoplasmatic |
Two observers (YGZ and CY) evaluated the staining results. A mean percentage of positive tumour cells was determined in at least five areas at ×400 magnification (50–250 cancer cells per area) and assigned from 0 to 100. The intensity of immunostaining was scored as follows: weak, 1+; moderate, 2+; and intense, 3+. For tumours that showed heterogeneous staining, the predominant pattern was taken into account for scoring. The percentage of positive tumour cells and the staining intensity were multiplied to produce a weighted score for each case. The theoretical limits of the scores ranged from 0 (0% of cells staining) to 300 (100% of the cells staining at 3+ intensity) Lombardi et al. (1999). For convenience in reporting and before statistical analysis, a quartation (negative, weak, moderate and strong staining) was made for each antibody, as previously described. The cut-off points were based on the scores: negative, 0; weak, ≤75; moderate, 75–150; and intense, >150. Some cases were unavailable due to an insufficient amount of tumour in the specimens or to non-evaluable staining results. Expression in the primary tumour was compared with that in the paired metastasis. Concordant and discordant pairs were distinguished according to different levels of scores.
Statistical analysis
The SPSS 11.0 software package for Macintosh (SPSS Inc., Chicago, IL, USA) was used for all statistical analysis. Variable associated with metastasis-associated proteins expression as well as the correlation between them and clinical pathological factors were analysed by the chi-square test.
Results
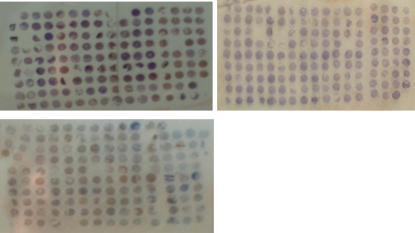
Three metastasis TMAs were constructed, which involved 153, 144 and 140 sites, respectively (Figure 1). We used conventional whole-section to compensate the lost tissue arrays.
Figure 1.
Tumour tissue microarray (TMA) slide. The TMA consisted of one normal, two primary tumour, and two to three nodal metastases cores from each invasive gastric cancer case placed at one line. For those with liver metastases, two cores were involved.
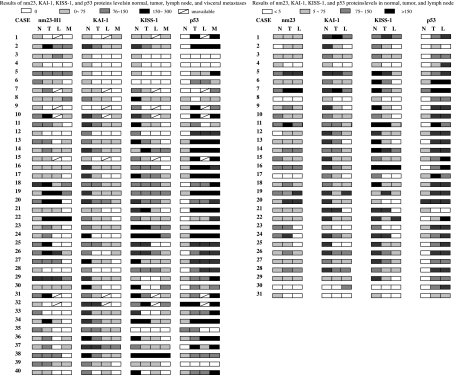
Most of the ‘normal’ adjacent gastric mucosa showed a weak or moderate expression nm23, moderate or strong expression of KAI1 and KISS1, and weak or loss expression of p53 compared with their tumours. Only a few cases showed reverse results without statistical significance (Figures 2 and 3). Most proteins showed predominantly different levels of expression in primary tumours and their metastases. nm23, KAI1 and KISS1 were downregulated with the progression of gastric carcinoma. However, p53 positive-staining seemed to be upregulated in the metastases than that in the primary tumour of gastric cancer (Figure 2, Table 3).
Figure 2.
Results of nm23, KAI-1, KISS-1 and p53 protein levels in normal, tumour, lymph node and liver metastases.
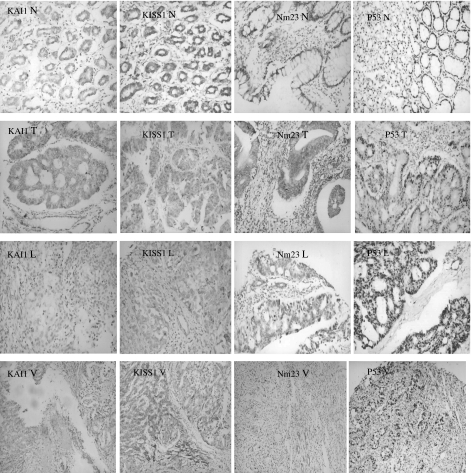
Figure 3.
Representative examples of IHC staining for KAI1, KISS1, nm23 and p53 (IHC ×200). N, normal tissue. The percentage and intensity of positive cells of KAI1, KISS1 and nm23 were gradually lower in metastases (especially in liver lesions) than that in primary tumour and normal tissues. T, primary tumour; L, nodal metastases; V, Lver metastases.
Table 3.
Staining results of metastases compared with primary tumours and concordance scores and discordance scores in 71 normal mucosa, 71 primary tumour (P), 64 nodal metastases (L), and 40 liver metastases (V)
| Positive expression | Concordance (scores) positive/negative | Discordance (scores) (higher:lower/lower:higher) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Proteins | Normal +(%) | Primary +(%) | Nodal +(%) | Liver +(%)* | P/L | P/V | L/V | P/L | P/V | L/V |
| nm23 | 66 (93.0) | 59 (83.1) | 45 (70.3) | 18 (45.0)†† | 32/9 | 5/7 | 7/8 | 21/2 | 27/1 | 14/4 |
| KAI-1 | 68 (95.8) | 53 (74.6)§ | 42 (65.6) | 20 (50)¶ | 22/13 | 6/10 | 16/10 | 26/3 | 22/2 | 6/1 |
| KISS-1 | 68 (95.8) | 53 (74.6)§ | 39 (60.9) | 23 (57.5) | 22/16 | 7/8 | 10/7 | 24/2 | 22/3 | 13/3 |
| P53 | 36 (50.7) | 65 (91.6)** | 58 (90.6) | 36 (90) | 40/4 | 20/3 | 18/4 | 6/14 | 4/13 | 1/10 |
+(%): +numbers of positive cases; %: numbers of positive cases/numbers of positive cases and negative cases ×100
P = 0.000 (χ2 = 17.478) compared with primary tumours
P = 0.010 (χ2 = 6.604) compared with nodal tumours
P = 0.000 (χ2 = 12.574) compared with normal mucosas
P = 0.009 (χ2 = 6.904) compared with primary tumours
P = 0.000 (χ2 = 28.839) compared with normal mucosas.
Expression of nm23 was seen in 59 of 71 (83.1%) of the primary tumours compared with 45 of 64 (70.3%) in lymph node metastases and 18 of 40 (45.0%) in liver metastases (Table 3). Among the discordance, the combination of high scores in the primary tumour with lower scores in lymph node and liver metastases (n = 21 and 27, respectively) occurred significantly more often than the combination of lower scores in the primary tumour with high in the metastasis (n = 2 and 1, respectively). High scores were also seen in lymph node metastases (n = 14) compared with lower scores in liver metastases (n = 4).
For KAI1, lower expressions were seen in 26 cases in lymph node metastases and 22 cases in liver metastases than the primary tumour, in contrast to three and two cases with higher expression in the lymph node and liver metastases, respectively. The number of cases with lower expression in the metastases was significantly greater than that with high expression. Most of the staining scores in the metastases were at the same level. Only seven cases showed different expression. Six of them had higher levels in the lymph node metastases than that in the liver metastases. Positive staining results for KAI1 were found in 53 of 71 (74.6%), 42 of 64 (65.6%) and 20 of 40 (50.0%) of the primary tumour, lymph node metastases and liver metastases (Table 3).
Reduced expression was also seen in KISS1. Positive expressions of KISS1 in the primary tumour, lymph node metastases and liver metastases were 53 of 71 (74.6%), 39 of 64 (60.9%) and 23 of 40 (57.5%), respectively. Similar to KAI1 and nm23, lower expressions were seen in 24 cases in nodal metastases and 22 cases in liver metastases than the primary tumour, in contrast to two and three cases with higher expression in the lymph node and liver metastases, respectively. The number of cases with high scores in lymph node metastases (n = 13) was also more than that in the liver metastases (n = 3).
Of all the markers, the results of p53 were remarkably similar in the primary tumour and metastases. However, discordant levels of the results were also found. In these cases, 14 in the lymph node metastases and 13 in the liver metastases had higher expression of p53 than that in the primary tumours. Six and four in the distinct metastases had a higher expression. Remarkably 10 cases had high expression in the liver metastases and low expression in the lymph node metastases.
Discussion
According to the progression theory, inactivation of MSG and activation of metastasis promoting gene (MPG) are meant to play a crucial role in human cancer metastasis (Debies & Welch 2002; Hunter 2003). We present protein expression of the MSGs nm23, KISS1 and KAI1 and candidate MPG p53 in gastric cancer lymph node and liver metastases by immunohistochemistry. These proteins expression in all metastases was compared with that in primary tumours. So, tendencies in expression differences could be noticed, which might reflect actual changes in metastasis-related gene expression in the metastases lesions.
As a result, the expression of nm23, KISS1 and KAI1 was found to be reduced in the metastases, especially in the liver metastases (statistically significant) compared with the primary tumours, suggesting a relevant role in the process of lymph node and liver metastasis. This is not unexpected, as they are supposed to be MSGs. p53 was highly expressed in the primary tumours and the metastases with no significant difference in the expression. But in several cases, the level of p53 expression in the metastases was higher than that in the primary tumour, suggesting that p53 plays roles of both tumour growth suppression and metastasis suppression and the former is the major one. Tumours or tumour subclones with reduced expression of MSG and mutation of MPG will therefore tend to metastasize.
Reduced expression of nm23 was found to be related to the presence of metastasis in several tumours and this relationship has also been described in gastric carcinoma. Most studies focused on nm23 expression in the primary tumour and the relationship between the loss expression and the clinical malignant behaviours. A few researchers systemically studied nm23 in the primary tumour and matched lymph node and liver metastases and results remain controversial. Kumar et al. (1999) reported that over-expression of nm23 was noted in 71% (42/59) of the primary tumours and 18% (10/55) of the metastatic tumours. Similar results were also seen in breast and head and neck squamous cell cancer (Lee et al. 1996; Stark et al. 2005). These results are consistent with our finding: loss of expression in 16.9% (12/71) of the primary tumours and 29.9% (19/64) of the nodal metastases and 65% (22/40) of the liver metastases. In addition, a trend of lower expression rate and level of nm23 were found in metastases compared with primary tumours. As shown in Figure 3, the percentage of nm23-positive cells was lower in metastases, especially in liver metastases than in primary tumour, suggesting its involvement in the process of metastasis and supporting the putative role of nm23 as a metastasis suppressor molecule.
KISS1 and KAI1 have not been studied extensively in gastric carcinoma. Dhar et al. (2004) studied KISS expression in 40 gastric cancer tissues in situ hybridization and found gastric cancers with low KiSS-1 had frequent venous invasion, distant metastasis and tumour recurrence. Donald et al. studied a subset of 12 colorectal patients with stage IV metastatic disease and observed a progressive downregulation of KAI1, from the normal adjacent colonic mucosa (KMS 193) to the primary tumour (KMS 72; P = 0.0001) to the liver metastasis (KMS 25; tumour compared with metastasis, P = 0.0135) (Lombardi et al. 1999). Similar study of comparing MSG expression in the primary tumour and metastases was seen in breast cancer brain metastases. Stark et al. (2005) found that nm23, KISS1, KAI1, BRMS1 and Mkk4 expression was overall reduced in breast cancer brain metastases. The above studies mainly focused on RNA level, not on protein level. Our studies were first based on the level of protein and showed similar results: KISS1 and KAI1 protein expression have a tendency to be reduced from normal mucosa to primary tumour then to metastases. Loss of KAI1 was found in 25.4% (18/71) of the primary tumours, 34.4% (22/64) of the nodal metastases, and 50% (20/40) of the liver metastases. The rates of loss of KISS1 were 25.4% (18/71), 39.1% (22/64) and 42.5% (17/40). The implication was that KISS1 and KAI1 gradually lost their functions with the progress of metastasis of gastric cancer. Even both primary tumours and matched metastases have positive expressions of KISS1 and KAI1, the levels between them have remarkable difference in some cases (Figure 3). This phenomenon in gastric carcinomas suggested that higher expression of KISS1 and KAI1 would reduce the metastatic potential.
Our studiers also compared the different rates of the four proteins expression between the nodal metastases and the liver metastases. Results revealed a higher level of nm23, KAI1 and KISS1 expression and lower level of p53 in the liver metastases than that in the nodal metastases, suggesting different mechanisms involved in the development of lymph node metastasis and liver metastasis.
There may be some arguments on the role of nm23 and p53 in the process of metastasis. Sarris and Lee (2001) reported strong nm23 immunoreactivity in 17% (12/71) of non-neoplastic colorectal epithelia, 60% (43/71) of the primary colorectal, 56% (40/71) of the lymph-node metastases and 67% (24/36) of the liver metastases. Several other studies also did not find that loss of nm23 expression was associated with metastatic behaviour and clinical survival in gastric and colorectal cancer patients. And for p53, its role in tumorigenesis is clear. But its role in metastasis is controversial. Many studies regarded p53 overexpression as an early event in tumour progression. But Kim et al. (1995) reported that p53 overexpression was correlated with depth of invasion, lymphatic invasion, lymph node metastasis and distant metastasis in colorectal cancer patients.
Some of the discrepancies in the results between the different studies discussed above may be due to a number of factors related to immunohistochemistry. Differences in antibodies and staining techniques may affect the results, and differences in scoring categories may also influence in particular. The notion that metastases arise from rare cells within the primary tumour means that a small part of the primary tumour will not reflect the assessment and scoring of the marker in that primary tumour as a whole, and yet these cells may still be responsible for the metastasis. This makes the choice of cut-off points rather arbitrary and may affect the results. If it is not the reason of immnohistochemistry, nm23 and p53 may have different functions under different conditions.
In conclusion, the expression of nm23, KAI1 and KISS1 is more often reduced than increased in the process of tumour progressing to metastases, suggesting a role for these markers in the process of metastasis, while the condition of p53 is to the contrary. This observation is in agreement with the hypothesis that downregulation of MSG is a late event in tumorigenesis, which is consistent with the progression theory. According to the assumed function of nm23, KAI1 and KISS1 as suppressors of metastases, KISS1 and KAI1 can be especially regarded as a promising target in the future treatment of gastric cancer metastasis.
References
- Adachi M, Taki T, Leki Y, Huang CL, Higashiyama M, Miyake M. Correlation of Kai1/CD82 gene expression with good prognosis in patients with non-small cell lung cancer. Cancer Res. 1996;56:1751–1755. [PubMed] [Google Scholar]
- Arnaud-Dabernat S, Bourbon PM, Dierich A, Le Meur M, Daniel JY. Knockout mice as model system for studying nm23/NDP kinase gene functions. Application to the nm23-M1 gene. J. Bioenerg. Biomembr. 2003;35:19–30. doi: 10.1023/a:1023561821551. [DOI] [PubMed] [Google Scholar]
- Berney CR, Fisher RJ, Yang J, Russell PJ, Crowe PJ. Protein markers in colorectal cancer: predictors of liver metastasis. Ann. Surg. 1999;230:179–184. doi: 10.1097/00000658-199908000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debies MT, Welch DR. Genetic basis of human breast cancer metastasis. J. Mamm. Gliand. Biol. Neopl. 2002;6:441–451. doi: 10.1023/a:1014739131690. [DOI] [PubMed] [Google Scholar]
- Dhar DK, Naora H, Kubota H, et al. Downregulation of KiSS-1 expression is responsible for tumor invasion and worse prognosis in gastric carcinoma. Int. J. Cancer. 2004;111:868–872. doi: 10.1002/ijc.20357. [DOI] [PubMed] [Google Scholar]
- Dursun A, Akyurek N, Gunel N, Yamac D. Prognostic implication of Nm23-H1 expression in colorectal carcinoma. Pathology. 2002;34:427–432. doi: 10.1080/0031302021000009342. [DOI] [PubMed] [Google Scholar]
- Dusonchet L, Corsale S, Migliavacca M, et al. Nm23-H1 expression does not predict clinical survival in colorectal cancer patients. Oncol. Rep. 2003;10:1257–1263. [PubMed] [Google Scholar]
- Goncharuk VN, del-Rosario A, Kren L, et al. Co-downregulation of PTEN, KAI-1, and nm23-H1 tumor/metastasis suppressor proteins in non-small cell lung cancer. Ann. Diagn. Pathol. 2004;8:6–16. doi: 10.1016/j.anndiagpath.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Graziano F, Cascinu S, Staccioli MP, et al. Potential role and chronology of abnormal expression of the Deleted in Colon Cancer (DCC) and the p53 proteins in the development of gastric cancer. BMC Cancer. 2001;1:9. doi: 10.1186/1471-2407-1-9. e-pub 1 August 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms JF, Welch DR, Miele ME. KISS1 metastasis suppression and emergent pathways. Clin. Exp. Metastasis. 2003;20:11–18. doi: 10.1023/a:1022530100931. [DOI] [PubMed] [Google Scholar]
- Howlett AR, Petersen OW, Steeg PS, Bissell MJ. A novel function for Nm23: overexpression in human brast carcinoma cells leads to the formation of basement membrane and growth arrest. J. Natl Cancer Inst. 1994;86:1838–2844. doi: 10.1093/jnci/86.24.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter KW. Allelic diversity in the host genetic background may be an important determinant in tumor metastatic dissemination. Cancer Lett. 2003;200:97–105. doi: 10.1016/s0304-3835(03)00420-8. [DOI] [PubMed] [Google Scholar]
- Ikeguchi M, Hirooka Y, Kaibara N. Quantitative reverse transcriptase polymerase chain reaction analysis for KiSS-1 and orphan G-protein-coupled receptor (hOT7T175) gene expression in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2003;129:531–535. doi: 10.1007/s00432-003-0469-z. E-pub 25 July 2003. [DOI] [PubMed] [Google Scholar]
- Ikeguchi M, Yamaguchi K, Kaibara N. Clinical significance of the loss of KiSS-1 and orphan G-protein-coupled receptor (hOT7T175) gene expression in esophageal squamous cell carcinoma. Clin. Cancer Res. 2004;10:1379–1383. doi: 10.1158/1078-0432.ccr-1519-02. [DOI] [PubMed] [Google Scholar]
- Ito Y, Uruno T, Takamura Y, et al. Papillary microcarcinomas of the thyroid with preoperatively detectable lymph node metastasis show significantly higher aggressive characteristics on immunohistochemical examination. Oncology. 2005;68:87–96. doi: 10.1159/000085701. E-pub 9 May 2005. [DOI] [PubMed] [Google Scholar]
- Jackson P, Puisieux A. Is the KAI1 metastasis suppressor gene a cellular target of p53? A review of current evidence. Biochem. Biophys. Res. Commun. 2000;278:499–502. doi: 10.1006/bbrc.2000.3799. [DOI] [PubMed] [Google Scholar]
- Kakeji Y, Korenaga D, Tsujitani S, et al. Gastric cancer with p53 overexpression has high potential for metastasis to lymph nodes. Br. J. Cancer. 1993;64:589–592. doi: 10.1038/bjc.1993.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka M, Okabayashi T, Johira H, et al. Aberration of p53 and DCC in gastric and colorectal cancer. Oncol. Rep. 2000;7:99–103. [PubMed] [Google Scholar]
- Kim KM, Lee A, Chae HS, Shim SI. Expression of p53 and NDP-K/nm23 in gastric carcinomas – association with metastasis and clinicopathologic parameters. J. Korean Med. Sci. 1995;10:406–413. doi: 10.3346/jkms.1995.10.6.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani M, Detheux M, Vandenbogaerde A, et al. The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J. Biol. Chem. 2001;276:34631–34636. doi: 10.1074/jbc.M104847200. E-pub 16 July 2001. [DOI] [PubMed] [Google Scholar]
- Kumar DD, Kubota H, Tabara H, et al. nm23 in the primary and metastatic sites of gastric carcinoma. Relation to AFP-producing carcinoma. Oncology. 1999;56:122–128. doi: 10.1159/000011952. [DOI] [PubMed] [Google Scholar]
- Lee CS, Redshaw A, Boag G. nm23-H1 protein immunoreactivity in laryngeal carcinoma. Cancer. 1996;77:2246–2250. doi: 10.1002/(SICI)1097-0142(19960601)77:11<2246::AID-CNCR10>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Liu H, Mao H, Fu X. Expression of Nm23 in breast cancer–correlation with distant metastasis and prognosis. Zhonghua Zhong Liu Za Zhi. 2001;23:224–227. [PubMed] [Google Scholar]
- Liu L, Wu DH, Li ZG, Yang GZ, Ding YQ. Effects of Kai1/CD82 on biological behaviour of human colorectal carcinoma cell line. World J. Gastroenterol. 2003;9:1231–1236. doi: 10.3748/wjg.v9.i6.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi DP, Geradts J, Foley JF, Chiao C, Lamb PW, Barrett JC. Loss of KAI1 expression in progression of colorectal cancer. Cancer Res. 1999;59:5724–5731. [PubMed] [Google Scholar]
- Mashimoto T, Wanabe M, Hirota S, et al. The expression of the Kai1 gene, a tumor metastasis suppressor, is directly activated by P53. Proc. Natl Acad. Sci. U.S.A. 1998;95:11307–11311. doi: 10.1073/pnas.95.19.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer CA, Graber HU, Firess H, et al. Reduced expression of the metastasis suppressor gene Kai1 in advanced colon cancer and its metastases. Surgery. 1999;126:869–880. [PubMed] [Google Scholar]
- Ohtaki T, Shintani Y, Honda S, et al. Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature. 2001;411:613–617. doi: 10.1038/35079135. [DOI] [PubMed] [Google Scholar]
- Sanchez-Carbayo M, Capodieci P, Cordon-Cardo C. Tumor suppressor role of KiSS-1 in bladder cancer: loss of KiSS-1 expression is associated with bladder cancer progression and clinical outcome. Am. J. Pathol. 2003;62:609–617. doi: 10.1016/S0002-9440(10)63854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarris M, Lee CS. nm23 protein expression in colorectal carcinoma metastasis in regional lymph nodes and the liver. Eur. J. Surg. Oncol. 2001;27:170–174. doi: 10.1053/ejso.2000.1070. [DOI] [PubMed] [Google Scholar]
- Shinohara T, Nishimura N, Hanibuchi M, et al. Transduction of KAI1/CD82 cDNA promotes hematogenous spread of human lung-cancer cells in natural killer cell-depleted SCID mice. Int. J. Cancer. 2001;94:16–23. doi: 10.1002/ijc.1445. [DOI] [PubMed] [Google Scholar]
- Stark AM, Tongers K, Maass N, Mehdorn HM, Held-Feindt J. Reduced metastasis-suppressor gene in breast cancer brain metastases. J. Cancer Res. Clin. Oncol. 2005;131:191–198. doi: 10.1007/s00432-004-0629-9. [DOI] [PubMed] [Google Scholar]
- Suzuki E, Ota T, Tsukuda K, et al. nm23-H1 reduces in vitro cell migration and the liver metastatic potential of colon cancer cells by regulating myosin light chain phosphorylation. Int. J. Cancer. 2004;108:207–211. doi: 10.1002/ijc.11546. [DOI] [PubMed] [Google Scholar]
- Tannapfel A, Katalinic A, Kockerling F, Wittekind C. The prediction of lymph node metastases in colorectal cancer by expression of the nucleoside diphosphate kinase/nm23-H1 and histopathological variables. Am. J. Gastroenterol. 1997;92:1182–1186. [PubMed] [Google Scholar]
- Wang CS, Lin KH, Hsu YC, Hsueh S. Distant metastasis of gastric cancer is associated with elevated expression of the antimetastatic nm23 gene. Cancer Lett. 1998;128:23–29. doi: 10.1016/s0304-3835(98)00043-3. [DOI] [PubMed] [Google Scholar]
- Yan C, Wang H, Boyd DD. KiSS-1 represses 92-kDa type IV collagenase expression by down-regulating NF-kappa B binding to the promoter as a consequence of Ikappa Balpha-induced block of p65/p50 nuclear translocation. J. Biol. Chem. 2001;276:1164–1172. doi: 10.1074/jbc.M008681200. [DOI] [PubMed] [Google Scholar]
- Yang X, Wei L, Tang C, Slack R, Montgomery E, Lippman M. Kai1 protein is down-regulated during the progression of human breast cancer. Clin. Cancer Res. 2000;6:3424–3429. [PubMed] [Google Scholar]
- Yang X, Wei L, Tang C, Slack R, Mueller S, Lippman ME. Overexpression of KAI1 suppresses in vitro invasiveness and in vivo metastasis in breast cancer cells. Cancer Res. 2001;61:284–5288. [PubMed] [Google Scholar]
- Yeung P, Lee CS, Marr P, Sarris M, Fenton-Lee D. Nm23 gene expression in gastric carcinoma: an immunohistochemical study. Aust. N. Z. J. Surg. 1998;68:180–182. doi: 10.1111/j.1445-2197.1998.tb04740.x. [DOI] [PubMed] [Google Scholar]
- Yu G, Zhu MH, Zhu Z, Ni CR, Zheng JM, Li FM. Expression of ATM protein and its relationship with p53 in pancreatic carcinoma with tissue array. Pancreas. 2004;28:421–426. doi: 10.1097/00006676-200405000-00011. [DOI] [PubMed] [Google Scholar]