Abstract
Deletions in chromosome (chr.) 13q occur frequently in head and neck squamous cell carcinoma (HNSCC). Previous studies failed to identify common deleted regions in chr.13q, though several candidate tumour suppressor genes (TSGs) loci, e.g. BRCA2, RB1 and BRCAX have been localized in this chromosome, as well as no prognostic significance of the deletion has been reported. Thus, in the present study, deletion mapping of chr. 13q has been done in 55 primary HNSCC samples of Indian patients using 11 highly polymorphic microsatellite markers of which three were intragenic to BRCA2 gene, one intragenic to RB1 gene and another from BRCAX locus. The deletion in chr.13q was significantly associated with progression of HNSCC. High frequencies (27–39%) of loss of heterozygosity were found in 13q13.1 (BRCA2), 13q14.2 (RB1), 13q21.2–22.1 (BRCAX) and 13q31.1 regions. Deletions in the BRCA2 and RB1 regions were significantly correlated. The four highly deleted regions were associated with clinical stage and histological grades of the tumour as well as poor patient outcome. Deletion in the 13q31.1 region was only found to be associated with HPV infection. High frequencies (11–23%) of microsatellite size alteration (MA) were seen to overlap with the highly deleted regions. Forty per cent of the samples showed rare biallelic alteration whereas loss of normal copy of chromosome 13q was seen in five tumours. Thus, it seems that the putative TSGs located in the BRCAX and 13q31.1 regions as well as the BRCA2 and RB1 genes may have some cumulative effect in progression and poor prognosis of HNSCC. Significant association between deletion in BRCA2 and RB1 gene loci may indicate functional relationship between the genes in this tumour progression.
Keywords: chromosome 13q, head and neck squamous cell carcinoma, loss of heterozygosity, microsatellite marker, survival
Head and Neck Squamous cell carcinoma (HNSCC) is an epithelial malignant disease arising from the mucosa of the upper aero-digestive tract (oral cavity, larynx, oropharynx, and hypopharynx). HNSCC is the fifth most common cancer worldwide (Parkin et al. 1999; Parkin et al. 2001). In South–Central Asia, 80% of HNSCC are found in the oral cavity and oropharynx (Sankaranarayanan et al. 1998; Parkin et al. 1999) and in most regions of India, cancer of the oral cavity is the leading malignancy accounting for 20% cancers in men and the third most common cancer in female (Sen et al. 2002). Epidemiological studies have linked tobacco, betel nut leaf quid, alcohol, some environmental factors and infection with oncogenic types of HPV to the development of HNSCC (D'Costa et al. 1998). Among HPVs, infection with high risk HPV-16 and 18 types are the most prevalent in the HNSCC tumours (D'Costa et al. 1998; Nagai 1999; Dahlgren et al. 2004).
The karyotypes of the HNSCC tumours are complex often near triploid and are composed of multiple numerical and structural abnormalities of the chromosomes (Sreekantaiah et al. 1994). Cytogenetic studies of HNSCC tumours have identified gradual increase of chromosomal aberrations from premalignant to malignant lesions and deletions in several chromosomal arms, e.g. 1p, 2q, 4q, 5q, 8p, 9p21–24, 10p, 10q, 11p, 13q, 17p, etc. (Jin et al. 1990; Hittleman et al. 1993; Sreekantaiah et al. 1994; Van Duke et al. 1994; Sauders et al. 2000).
Allelotyping studies (using microsatellite markers) of HNSCC have identified >25% loss of heterozygosity (LOH) in several chromosomal regions, e.g. 3p, 4q, 6p, 8p, 9p, 11q, 13q, etc. and among these regions, deletions in 11q, 13q and 14q were proposed to be associated with the development of carcinoma in situ (Nawroz et al. 1994; Califano et al. 1996) of this tumour. Deletion mapping of HNSCC tumours by different investigators have identified deletions in different regions of chr.13q, for example, Yoo et al. (1994) have seen highest (50%) deletion in the chr.13q14.1–14.3 region, whereas Maestro et al. (1996) have seen highest (30–65%) deletion in the chr.13q14.2–14.3 and 13q32 – ter regions. On the other hand, Gupta et al. (1999) has identified three highly deleted regions at the chr.13q12.1 (25–30%), 13q14.3 (47–85%) and 13q34 (50%) regions. Also, high frequency of LOH in the chr.13q13, and 13q14.3 regions has been reported by Li et al. (1994) and Kirkpatrick et al. (1997), respectively. Thus, there is an ambiguity in the localization of minimal deleted regions among the different investigators. The chr.13q is known to harbour two candidate tumour suppressor genes (TSGs), i.e. BRCA2 and RB1 at the 13q13.1 and 13q14.3 regions, respectively. Also, an unknown candidate TSGs' locus, named as BRCAX, associated with the development of breast cancer is localized at 13q21 region (Kainu et al. 2000).
Besides LOH, microsatellite instability (MA) was detected in different cancers and it may represent a form of genomic instability (Arzimanoglou et al. 1998). In HNSCC, the occurrence of MA in the Indian patient population has been shown to be high compared to the Western patient population (Chakraborti et al. 2001). Recently, we have detected high frequencies of MA in and around the LOH regions of chr.3, 8 and 9 in primary HNSCC, which indicates the importance of MA in the development of this tumour (Dasgupta et al. 2002; Bhattacharya et al. 2003; Chakraborty et al. 2003; Tripathi (Bhar) et al. 2003).
Thus, in the present study detailed deletion mapping has been done to localize the candidate TSGs loci in chr.13q associated with the development of HNSCC using the (i) intragenic microsatellite markers of BRCA2 and RB1 genes; (ii) microsatellite markers from the BRCAX locus and other regions of chromosome 13q. The clinicopathological correlation as well as the prognostic significance of the deleted regions has also been analysed in this tumour.
Material and methods
Sample collection and clinical data
Fifty-five freshly operated tumour samples as well as matched benign tissues or in some cases peripheral blood leucocytes (PBL) were collected from patients admitted to Chittaranjan National Cancer Institute and Cancer Center & Welfare Home, Kolkata, India. All the samples were collected during the period of 1995–99 and informed consent was obtained–from all the patients prior to the collection of the samples. No preoperative radiotherapy or chemotherapy was given to the patients. After collection the samples were frozen immediately and stored at −80 °C until further use. Tumor stage and grade were defined according to the tumour node metastasis (TNM) classification of Union Internationale Contre le cancer (UICC) (Harmer 1978). Detailed clinicopathological features of the patient are presented in Table 1. Overall patients' median age was 49.5 years (range 20–73 years).
Table 1.
Distribution of patients' clinicopathological characteristics and molecular information
| Lymph node (numbers) | Tobacco habit (numbers) | HPV status (numbers) | |||||
|---|---|---|---|---|---|---|---|
| Clinico-pathological parameters | % of samples | Node + | Node − | Tobacco + | Tobacco − | HPV + | HPV − |
| Age | |||||||
| >49.5 | 49.1 (27/55) | 11 | 16 | 18 | 9 | 18 | 9 |
| ≤49.5 | 50.9 (28/55) | 17 | 11 | 19 | 9 | 22 | 6 |
| Sex | |||||||
| Male | 81.8 (45/55) | 21 | 24 | 31 | 14 | 33 | 12 |
| Female | 18.2 (10/55) | 7 | 3 | 6 | 4 | 7 | 3 |
| Religion | |||||||
| Hindu | 81.8 (45/55) | 23 | 22 | 30 | 15 | 31 | 14 |
| Muslim | 18.2 (10/55) | 5 | 5 | 7 | 3 | 9 | 1 |
| Site | |||||||
| OF | 20 (11/45) | 4 | 7 | 9 | 2 | 9 | 2 |
| OC | 56.4 (31/45) | 19 | 12 | 22 | 9 | 21 | 10 |
| LA | 23.6 (13/45) | 5 | 8 | 6 | 7 | 10 | 3 |
| Grade | |||||||
| WDSCC | 49.1 (27/55) | 12 | 15 | 20 | 7 | 21 | 6 |
| MDSCC | 45.5 (25/55) | 15 | 10 | 14 | 11 | 18 | 7 |
| PDSCC | 5.4 (3/55) | 1 | 2 | 3 | 0 | 1 | 2 |
| Stage | |||||||
| I | 9.1 (5/55) | 1 | 4 | 3 | 2 | 3 | 2 |
| II | 21.8 (12/55) | 5 | 7 | 11 | 1 | 10 | 2 |
| III | 40 (22/55) | 13 | 9 | 15 | 7 | 17 | 5 |
| IV | 29.1 (16/55) | 9 | 7 | 8 | 8 | 10 | 6 |
Grade: WDSCC, well differentiated squamous cell carcinoma; MDSCC, moderately differentiated squamous cell carcinoma; PDSCC, poorly differentiated squamous cell carcinoma; Site: OC, oral cavity; OF, orofacial and LA-larynx; (+) and (−): positive and negative for that aetiology and lymph node status, respectively.
Microdissection and DNA extraction
The normal cells present as contaminant in the specimens were removed by microdissection procedure and the samples containing >60% tumour cells were taken for DNA extraction as described by Dasgupta et al. (2002). DNA was extracted from the microdissected tissue sections and their corresponding normal tissue or PBL by proteinase-K (Sigma Chemicals, St. Louis, MO, USA) digestion followed by phenol:chloroform extraction (Sambrook et al. 1989). When the adjoining normal tissue of the lesion was contaminated with infiltrating tumour cells, then PBL was taken for normal DNA extraction.
Selection of microsatellite markers
To determine if there was LOH or MA on chr.13, 11 highly polymorphic microsatellite markers were taken (Figure 1). Three markers namely D13S1701, D13S1699 and D13S1695 are intragenic to BRCA2 gene (Chunder et al. 2004), the marker D13S153 is in the intron 2 of RB1 gene (Ogawara et al. 1998) and D13S1308 is from BRCA-X locus (Chunder et al. 2004) (Figure 1). The information and primer sequence of the microsatellite markers were obtained from the website http://www.ncbi.nlm.nih.gov.
Figure 1.
Allele status of the chromosome 13q markers in the primary head and neck squamous cell carcinoma samples. LOH, Loss of heterozygosity; MA1, Microsatellite size alteration of one allele; MA2, Microsatellite size alteration of both allele; LMA, Loss of one allele and size alteration of the other allele; RH, Retention of homozygosity; H: Non informative.
Analysis of microsatellite alterations
Polymerase chain reaction (PCR) was performed with all the markers in a 20 μl reaction volume containing 67 mm Tris-HCl (pH 8.7), 16.6 mm (NH4)2SO4, 1–2 mm MgCl2, 4 pmol of each primer, 0.2 mm of each dNTPs, 50–100 ng of template DNA and 0.5 units of Taq DNA polymerase. One of the paired primers in the reaction mixture was end labelled with [?32p] ATP using T4-Polynucleotide Kinase as described by Nawroz et al. (1994) and Dasgupta et al. (2002). The PCR conditions were 95 °C for 5 min, then 30 cycles at 95 °C for 1 min, annealing at appropriate temp (50–60 °C) for 1 min and extension at 72 °C for 2 min followed by final extension at 72 °C for 7 min. The labelled PCR products were electrophoresed in 7% polyacrylamide gel containing 8 m Urea and auto-radiographed. This microsatellite analysis procedure used for the allelotyping could detect LOH and MA in the presence of 50% and 10–-30% tumour DNA, respectively (Dasgupta et al. 2002).
The LOHs were interpreted for all informative cases by densitometric scanning (Shimidazu, CS-9000, Kyoto, Japan) of the autoradiographs. The allelic loss was recorded if there was a complete absence of one allele or if the relative band intensity of one allele was reduced at least 50% in the tumour in comparison to the homologous allele in the corresponding normal DNA. An LOH index of values ≥1.5 was considered as loss of small allele and values <0.67 was considered to be loss of larger allele (MacGrogan et al. 1994). The MAs were detected by a shift in the mobility of one (MA1) or both (MA2) alleles as compared to the normal DNA (Wistuba et al. 1999b). For calculation of LOH at a locus, samples showing homozygosity and MA at that locus were not considered, whereas for calculation of MAs, the samples showing LOHs only were not considered (Dillon et al. 1997). Those samples showing both LOH and MA at the same locus was considered for calculating both LOH and MA. To verify the proper pairing of the samples, additional typing was done with different RFLP markers (NAT-KpnI, Hb-HinfI, Hb-HincIII, CypIAI-MspI, ALAD-RsaI) (Dasgupta et al. 2002) and in all the cases the results were in concordance with the earlier findings (data not shown).
Detection of HPV types 16 and 18
The presence of HPV in head and neck lesions was detected by PCR using primers (MY09 and MY011) from the consensus LI region (D'Costa et al. 1998). The typing of HPV type 16/18 in the LI positive samples was done by PCR using specific primers from HPV type 16E6 and HPV type 18E7 region (Balaram et al. 1995). The PCR products were electrophoresed in 2% agarose gel and stained with ethidium bromide for visualization under UV light and photographing. For final confirmation of the HPV types, the PCR products after gel electrophoresis were transferred to the nylon membrane for Southern Hybridization with 32P-labelled HPV type specific probes (Gillison et al. 2000). As a positive control for HPVs, the DNA from SiHa (for HPV type 16) and HeLa (for HPV type 18) cell lines (obtained from American Type Culture Collection, Manassas, VA, USA) and the HPV type specific plasmids were used.
Statistical analysis of the clinical data
Correlation of regional loss with progression of the tumours
For data analysis, we used two approaches: (i) for individual samples, we determined the frequencies of LOH at individual chromosomal regions; (ii) to determine whether the deletions in chr.13q were associated with the progression of the tumour, we determined frequencies of loss of individual markers using a fractional allele loss (FAL) index, as defined below by Wistuba et al. (1999a).
13q FAL index = Total number of chr.13q markers with LOH/Total number of informative chr.13q markers.
To correlate regional loss with progression of the tumour, we calculated the mean FAL index for each histological category. The t-statistics was used for comparison of the mean FAL index of these histological groups.
Association study between clinicopathological features and regional loss
We performed chi-square analysis to determine the association between the tumours' genetic profile (LOH/MA) and different clinicopathological features such as tobacco habits, nodes at pathology, tumour stages, grades and HPV infection.
Prognostic influence of regional loss
Survival curves were calculated according to Kaplan–Meier method. Postoperative overall survival was measured from the date of surgery to the date of last follow-up or death. The log-rank test was used to assess the differences in patient survival between cases with loss and retention of heterozygosity of the genes. Multivariate analysis of overall survival with different prognostic factors was performed using Cox's proportional hazards method (Cox 1972). A Cox regression analysis was performed to calculate the risk ratio (RR). Probability value (P-value) ≤0.05 was considered statistically significant. All the statistical analysis was performed using statistical program SPSS (SPSS Inc. Chicago, IL, USA).
Results
Molecular abnormalities (LOH/MA) on chr.13q
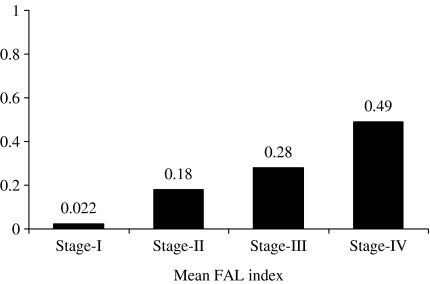
Deletion mapping of chr.13q by 11 microsatellite markers in primary HNSCC samples showed microsatellite instability (LOH/MA) in 85% (47/55) of the tumours in at least one of the markers indicating importance of chr.13q in the development of this tumour (Figures 1 and 2). The preference for deletions in larger and smaller alleles was seen in 53 and 47% of the affected alleles, respectively (data not shown), which indicates that the deletion phenomenon was independent of the size of the allele as seen in our previous studies (Dasgupta et al. 2002). The overall allelic loss of chr.13q gradually increased with progression of the clinical stage and there was a significant correlation between the mean FAL index (P < 0.01) and progression of the tumour (Figure 3).
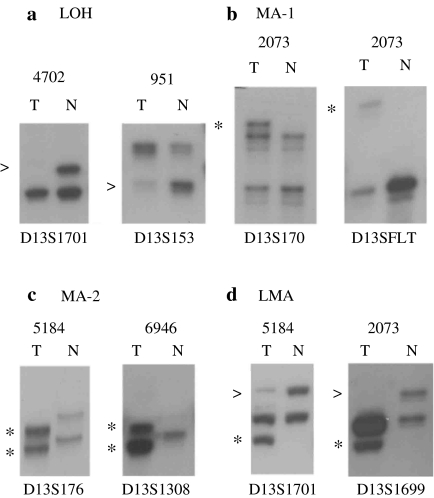
Figure 2.
Representative photographs of the autoradiograph showing (a) loss of heterozygosity, (b) microsatellite size alteration of one allele, (c) microsatellite size alteration of both allele and (c) loss of one allele and microsatellite size alteration of other allele at different marker loci on chr.13q in different head and neck squamous cell carcinoma samples. T: DNA from tumour tissue after microdissection, N: corresponding normal tissue or peripheral blood leucocyte DNA. Arrows (>) indicate loss of the corresponding allele and asterisk (*) indicates size alteration of one or both allele.
Figure 3.
Distribution of fractional allele loss index. P ≤ 0.01.
High frequencies of deletions have been detected in four chromosomal regions, i.e. 13q13.1 (D1: 33–39%), 13q14.2 (D2: 35%), 3q21.2–22.1 (D3: 27–33%) and 13q31.1 (D4: 31%) (Figure 1). High frequencies (11–23%) of MA have also been seen to overlap with the highly deleted regions (D1–D4) (Figure 1). The contraction and expansion of alleles were observed in 51 and 49% of the affected alleles, respectively (data not shown). Overall, molecular abnormalities (LOH/MA) in the D1 and D3 regions were seen in 59 and 55% samples, respectively (Figure 1). Interestingly, significant correlation (P-value: 0.042) has been found between deletions in the D1 and D2 regions.
Five samples (1542, 5184, 5219, 951 and 2785) showed LOH or MA at all the informative loci, indicating loss of normal copy of chr.13 in these tumours (Figure 1). Eight samples (558, 4200, 2073, 5090B, 615, 1640, 4702 and 1552) showed LOH and/or MA in more than two consecutive markers, particularly around the high LOH regions (Figure 1). No homozygous deletion was found in these samples. However, 40% (22/55) of the samples showed rare biallelic alterations (MA-2 or LOH + MA) in and around the high LOH regions.
Detection of HPV
Using consensus L1 primers, 73% (40/55) of the HNSCC samples were found to be positive for HPV infection (Figure 2). All the HPV positive samples were seen to be positive for HPV type-16 using both HPV type-16/18 specific primers. Significant correlation was found between HPV infection and deletion in the D4 region (P-value: 0.0003) and MA in the D1 region (P-value: 0.046) (Table 2).
Table 2.
Clinicopathological correlations of LOH and MA in the highly deleted regions of chr.13q in HNSCC
| D1 | D2 | D3 | D4 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinico-pathological parameters | LOH+ | LOH− | Total | P value | LOH+ | LOH− | Total | P value | LOH+ | LOH− | Total | P value | LOH+ | LOH− | Total | P value |
| WDSCC | 9 | 18 | 27 | 0.005 | 3 | 24 | 27 | 0.021 | 9 | 18 | 27 | 0.027 | 5 | 22 | 27 | 0.034 |
| MDSCC | 16 | 9 | 25 | 7 | 18 | 25 | 13 | 12 | 25 | 6 | 19 | 25 | ||||
| PDSCC | 3 | 0 | 3 | 2 | 1 | 3 | 3 | 0 | 3 | 3 | 0 | 3 | ||||
| Stage I & II | 2 | 15 | 17 | 0.00029 | 0 | 17 | 17 | 0.00939 | 4 | 13 | 17 | 0.00339 | 2 | 15 | 17 | 0.013 |
| Stage III | 14 | 8 | 22 | 6 | 16 | 22 | 9 | 13 | 22 | 4 | 18 | 22 | ||||
| Stage IV | 12 | 4 | 16 | 6 | 10 | 16 | 6 | 10 | 16 | 8 | 8 | 16 | ||||
| Lymph node − | 16 | 11 | 27 | 0.22 | 5 | 22 | 27 | 0.56 | 12 | 15 | 27 | 0.88 | 7 | 20 | 27 | 0.93 |
| Lymph node + | 12 | 16 | 28 | 7 | 21 | 28 | 13 | 15 | 28 | 7 | 21 | 28 | ||||
| HPV + | 21 | 19 | 40 | 0.69 | 11 | 29 | 40 | 0.095 | 18 | 22 | 40 | 0.91 | 5 | 35 | 40 | 0.0003 |
| HPV − | 7 | 8 | 15 | 1 | 14 | 15 | 7 | 8 | 15 | 9 | 6 | 15 | ||||
| Tobacco + | 19 | 18 | 37 | 0.92 | 9 | 28 | 37 | 0.51 | 17 | 20 | 37 | 0.91 | 8 | 29 | 37 | 0.34 |
| Tobacco − | 9 | 9 | 18 | 3 | 15 | 18 | 8 | 10 | 18 | 6 | 12 | 18 | ||||
| D1 | D2 | D3 | D4 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinico-pathological parameters | MA+ | MA− | Total | P value | MA+ | MA− | Total | P value | MA+ | MA− | Total | P value | MA+ | MA− | Total | P value |
| WDSCC | 10 | 15 | 25 | 0.69 | 3 | 22 | 25 | 0.3 | 7 | 18 | 25 | 0.73 | 7 | 18 | 25 | 0.014 |
| MDSCC | 6 | 21 | 27 | 1 | 26 | 27 | 2 | 25 | 27 | 1 | 26 | 27 | ||||
| PDSCC | 2 | 1 | 3 | 2 | 1 | 3 | 2 | 1 | 3 | 0 | 3 | 3 | ||||
| Stage I & II | 2 | 15 | 17 | 0.019 | 1 | 16 | 17 | 0.085 | 2 | 15 | 17 | 0.07 | 1 | 16 | 17 | 0.011 |
| Stage III | 8 | 14 | 22 | 1 | 21 | 22 | 3 | 19 | 22 | 1 | 21 | 22 | ||||
| Stage IV | 8 | 8 | 16 | 4 | 12 | 16 | 6 | 10 | 16 | 6 | 10 | 16 | ||||
| Lymph node − | 9 | 18 | 27 | 0.92 | 4 | 23 | 27 | 0.36 | 5 | 22 | 27 | 0.78 | 4 | 23 | 27 | 0.95 |
| Lymph node + | 9 | 19 | 28 | 2 | 26 | 28 | 6 | 22 | 28 | 4 | 24 | 28 | ||||
| HPV + | 10 | 30 | 40 | 0.046 | 3 | 37 | 40 | 0.18 | 9 | 31 | 40 | 0.44 | 7 | 33 | 40 | 0.31 |
| HPV − | 8 | 7 | 15 | 3 | 12 | 15 | 2 | 13 | 15 | 1 | 14 | 15 | ||||
| Tobacco + | 11 | 26 | 37 | 0.49 | 4 | 33 | 37 | 0.97 | 6 | 31 | 37 | 0.31 | 4 | 33 | 37 | 0.26 |
| Tobacco − | 7 | 11 | 18 | 2 | 16 | 18 | 5 | 13 | 18 | 4 | 14 | 18 | ||||
Association of molecular abnormalities with clinicopathological variables and patient survival
Among the various clinicopathological parameters tested, deletions in the D1–D4 regions were significantly correlated (P-value: 0.00029–0.034) with both pathological grades and TNM stage of the tumours (Table 2). MAs in the D1 and D4 regions were seen to be significantly associated (P-value: 0.011–0.019) with TNM stage of the tumour, whereas MA in the D4 region was only found to be associated (P-value: 0.014) with clinical grade of the tumour (Table 2).
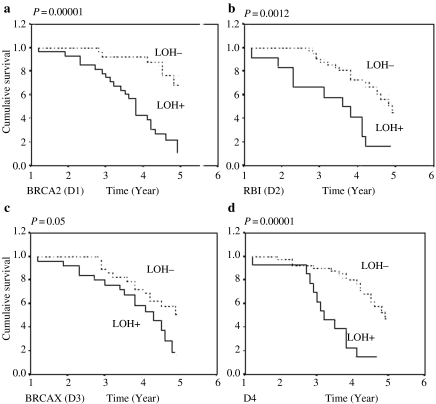
The clinical outcome of the HNSCC patients was investigated for a period up to 5 years. Log-rank test uncovered a statistically significant difference in overall patient survival between cases with LOH and cases without LOH for all the deleted regions (P-value: 0.00001–0.05) (Figure 4).
Figure 4.
Kaplan–Meier 5-year survival probability curves with cumulative survival of head and neck squamous cell carcinoma patient by loss of heterozygosity (LOH) status in (a) D1 (BRCA2), (b) D2 (RB1), (c) D3 (BRCA-X) and (d) D4 region. Survival time was defined as the time from surgery to the patient's death, known recurrence or the last time the patient was known to be alive. The dotted line represents survival probability without LOH and smooth line represents with LOH probability.
We used the Cox-proportional hazards model to determine which factors were jointly predictive of overall survival of the patients. Among the different clinicopathological variables used in our analysis, lymph node status (RR = 0.1388, P = 0.0159) and deletion in all the four highly deleted regions (D1: RR = 0.1705, P = 0.0053, D2: RR = 0.155, P = 0.0091, D3: RR = 0.1744, P = 0.004, D4: RR = 0.1940, P = 0.0021) were significantly predictive of overall survival time (Table 3).
Table 3.
Multivariate analysis of overall survival of 55 head and neck squamous cell carcinoma patients with different clinicopathological parameters using Cox-proportional hazards model
| Overall survival | ||
|---|---|---|
| Variables | P-value | Hazard ratio |
| D1 (BRCA2) | 0.0053 | 0.1705 |
| D2 (RB1) | 0.0091 | 0.1556 |
| D3 (BRCAX) | 0.0046 | 0.1744 |
| D4 | 0.0021 | 0.1940 |
| Histological grade | 0.1829 | 0.00001 |
| Clinical stage | 0.8203 | 0.00001 |
| Node | 0.0159 | 0.1388 |
| Tobacco | 0.3870 | 0.00001 |
| HPV | 0.5311 | 0.00001 |
Discussion
In the present study, we have analysed the allelic alterations (LOH/MA) in 11 microsatellite loci spanning whole chr.13q in 55 primary HNSCC samples at different clinical and histological stages. We found gradual increase in the mean FAL index (Figure 3) from stage I to more aggressive stage IV tumours, which is an indication of the accumulative nature of the deletions occurring in different chr.13q regions during progression of HNSCC. All the four highly deleted regions (D1–D4) were associated with higher stages and grades of the tumour as well as poor patient outcome (Table 2, Figure 4), which is supportive to the fact, that chr.13q deletion is a later event in progression of the disease and is an important prognostic determinant. In addition, according to multivariate analysis, deletions in the D1–D4 regions along with positive lymph node were significantly predictive of over all survival (Table 3). We have not analysed the microsatellite alterations of chr.13q in dysplastic lesions and CIS of head and neck due to low incidence of such alterations in stage I/II tumours. However, Califano et al. (1996) showed the association between chr.13q deletion and the development of carcinoma in situ (CIS) in head and neck cancer using the microsatellite markers D13S133 and D13S170 from the chromosomal 13q14.3 and 13q31.1 regions, respectively. In our analysis, D13S170 marker showed high deletions mainly in the stage III/IV tumours (Figure 1). This discrepancy might be due to the difference in sample preparation, methodology used and ethnicity.
It is evident from Figure 5 that the deletions in the D1 and D2 regions overlap with the deletions reported previously in some HNSCC as well as other tumours. Deletions in the D3 and D4 regions have not yet reported earlier in HNSCC, while deletions in these regions have been reported in breast carcinoma and ovarian and fallopian tube cancers (Jongsma et al. 2002; Chunder et al. 2004). The significant association between deletions in the D1 and D2 region indicates that deletion in one region may impose selective pressure on the other regions to be deleted or size altered for development of the disease. The distance between the D1 and D2 is about 17.1 Mb and there are several samples in our study where either D1 or D2 alterations have been seen. This indicates that D1 and D2 are not epiphenomena due to large deletion. The microsatellite markers analysed in the D1 and D2 regions are intragenic to the putative TSGs BRCA2 and RB1, respectively. The co-operativity between BRCA2 and RB1 loci in HNSCC has not yet been reported earlier. In parathyroid carcinoma, tumours that showed loss of RB1 by allelotyping also showed loss of BRCA2 (Shattuck et al. 2003). Similar results have also been reported in prostrate cancer (Cooney et al. 1996; Latil et al. 1996; Melamed et al. 1997). The overall 59% LOH seen in HNSCC samples in our study is comparable to that has been seen in early onset breast tumours of Indian Patients (Chunder et al. 2004). Also, high frequency of LOH has been reported in the BRCA2 loci using flanking microsatellite markers in oesophageal carcinoma, hepatocellular carcinoma and ovarian/fallopian tube carcinoma (Kuroki et al. 1995;Hu et al. 2002; Jongsma et al. 2002) (Figure 5). However, Gupta et al. (1999) suggested that neither BRCA2 nor RB1 were associated with the development of HNSCC. Kirkpatrick et al. 1997 had seen infrequent deletion in the microsatellite markers flanking to the BRCA2 gene in HNSCC. On the other hand, Maestro et al. (1996) have found overall 65% (28/43) LOH in the RB1 locus using a VNTR marker in HNSCC, but they did not found any changes in the expression pattern of the gene using both immunohistochemical and RT-PCR techniques, suggesting the involvement of any other candidate TSG in this region. Similar conclusion has also been made by Ogawara et al. (1998) in this tumour. However, no one has sequenced the RB1 cDNA as well as genomic region of the gene in HNSCC to see whether the functional RB1 protein has been produced or not. Thus, the discrepancy of the association of BRCA2 and RB1 between our study and other Western studies in HNSCC might be due to the difference in ethnicity and aetiological factors. However, we have not analysed the mutation as well as methylation of BRCA2 and RB1 genes to see if these inactivation mechanisms of TSGs have any role in the down-regulation of these genes. A candidate TSG known as DICE1 has been found to be located in chr.13q14.1–13q14.2 regions, but its association with HNSCC tumours has not yet been analysed (Li et al. 2003). Ogawara et al. (1998) also reported a significant correlation between the LOH in chr.13q14.3 and lymph node metastasis in HNSCC contrary to that has been seen in our analysis (Table 2). This discrepancy might be due to the presence of normal cell contaminants in the surgical specimens used in the allelotyping study of Ogawara et al. (1998). Till date no biochemical correlation between BRCA2 and RB1 has been established, however, both the proteins could interact in the BRCT domain (transactivation) of BRCA1 protein associated with the cell cycle regulation and DNA repair (Welcsh & King 2001). Thus, it suggests that inactivation of BRCA2 and RB1 genes might develop gross chromosomal abnormalities in later stages of HNSCC for selective growth advantage of the tumour and poorer outcome of the patients (Figure 4).
Figure 5.
Illustration showing the approximate location of highly deleted regions of chr.13q in head and neck squamous cell carcinoma and other human cancers in different studies.
Deletion in the D3 (13q21.2–22.1) region is about 14.1 Mb wide and encompasses the Common Fragile Sites FRA13B and FRA13C at 13q21.2 and 13q22.2 regions, respectively, and also harbours a candidate breast cancer susceptibility locus BRCAX at 13q21.33 region (Gilles et al. 1996; Kainu et al. 2000; Thorland et al. 2003). In cervical carcinoma, HPV-16 integration site has been detected in the FRA13B and FRA13C regions (Wentzensen et al. 2004), though we have not seen any association between the deletion in this region and HPV infection in HNSCC. It seems that in HNSCC, HPV may not integrate in this region. Similar to our data, Ogawara et al. (1998) have seen 31% LOH using the D13S176 marker (used in our study) at 13q21.2 region in HNSCC. Using the D13S1308 marker (BRCAX: 13q21.33) comparable frequency of deletion has been reported in early and late onset breast tumours of Indian patients (Chunder et al. 2004). Moreover, high frequency (67%) of LOH has been reported in 13q21 region in ovarian and fallopian tube cancer indicating the location of a candidate TSG in this region (Jongsma et al. 2002) (Figure 5). No candidate TSG has yet been identified in this region. However, a significant association has been found between deletion in the BRCAX locus with ATM and TP53 loci in early and late onset breast tumours, respectively, indicating the possible function of the candidate TSG in the BRCAX locus in cell cycle regulation and DNA repair pathways (Chunder et al. 2004). The deletion in the D4 region (13q31.1) is comparatively high (31%) in our study than other studies using the same microsatellite marker D13S170 (Yoo et al. 1994). However, high LOH (69%) has been reported in 13q22–31 regions in ovarian and fallopian tube cancer (Jongsma et al. 2002) (Figure 5), indicating the location of candidate TSG in this region. No candidate TSG has yet been identified in this region. The significant association of deletions in D3 and D4 region with different clinicopathological parameters including poor patient outcome indicates the presence of candidate TSGs in these regions (Table 2, Figure 4).
The occurrence of high MA in and around the high LOH regions of chr.13q seen in our study (Figure 1) is comparable to that has been seen in our previous allelotyping studies of other chromosomes in HNSCC (Chakraborty et al. 2003). Though, the actual significance of MA in the development of the tumour is not clear, still MA has been suggested to be important parameters for molecular detection of cancers (Mao et al. 1994). The presence of comparatively high frequency (40%) of rare biallelic alterations in the highly deleted region indicate that the LOH/MA in one allele of this region may impose selective pressure on the other allele of the same locus for deletion or size alteration. In the later stages of the tumours (stage III and IV), loss of normal copy of the chr.13 and interstitial alterations suggest that these chromosomal alterations are needed for progression of the disease. Similar phenomenon has been reported in HNSCC in the allelotyping studies of other chromosomes and chr.13 also (Maestro et al. 1996; Dasgupta et al. 2002; Bhattarcharya et al. 2003; Chakraborty et al. 2003; Tripathi (Bhar) et al. 2003). The high prevalence of HPV infection seen in our study is comparable (74%) to that have been seen by Balaram et al. (1995). The significance of the association among the HPV infection and deletion in the D4 region and MA in the D1 region is not clear, but it seems that HPV might have some role in the occurrence of genomic instability and tumour progression (Gillison et al. 1999).
Thus, it can be concluded that the deletions in the D1–D4 regions are needed for the development of later stages of HNSCC and can be used as prognostic indicators. The loss of function of known and putative TSGs in these regions may have cumulative effect in this tumour development. Along with these deletions, other molecular changes in the chromosome such as MA, interstitial alterations and loss of normal copy of chr.13 may impose selective pressure for growth advantage of the tumour.
Acknowledgments
Authors are grateful to Director, Chittaranjan National Cancer Institute (CNCI), Kolkata for encouragement and support, Dr S. Gupta, Director, Cancer Center and Welfare Home, Kolkata for providing tumour samples and Dr N Chunder for technical help. We are also grateful to Prof H. Z. Hausen and Dr E. M. de Villiers for the gift of HPV type 16/18 plasmids. Financial support was provided by research grants from the Department of Biotechnology, Govt. of India (Grant Number: BT/MB/05/002/94).
References
- Arzimanoglou IT, Gilbert F, Barber HR. Microsatellite instability in human solid tumors. Cancer. 1998;82:1808–1820. doi: 10.1002/(sici)1097-0142(19980515)82:10<1808::aid-cncr2>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Balaram P, Nalinakumari KR, Abraham E, et al. Human papillomaviruses in 91 oral cancers from Indian betel quid chewers – high prevalence and multiplicity of infections. Int. J. Cancer. 1995;61:450–454. doi: 10.1002/ijc.2910610403. [DOI] [PubMed] [Google Scholar]
- Bhattarcharya N, Tripathi S, Dasgupta S, et al. Association of deletion in the chromosomal 8p21.3–23 region with the development of invasive head and neck squamous cell carcinoma in Indian patients. Indian J. Med. Res. 2003;118:77–85. [PubMed] [Google Scholar]
- Califano J, Van der Riet P, Westra W, et al. Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res. 1996;56:2488–2492. [PubMed] [Google Scholar]
- Chakraborti S, Dasgupta S, Roy S, et al. Microsatellite instability in squamous cell carcinoma of head and neck from the Indian patient population. Int. J. Cancer. 2001;92:555–561. doi: 10.1002/ijc.1233. [DOI] [PubMed] [Google Scholar]
- Chakraborty SB, Dasgupta S, Roy A, et al. Differential deletions in chromosome 3p are asociated with the development of head and neck squamous cell carcinoma from Indian patients. Cancer Genet. Cytogenet. 2003;145:1–9. doi: 10.1016/s0165-4608(03)00127-4. [DOI] [PubMed] [Google Scholar]
- Chunder N, Mandal S, Roy A, Roychoudhury S, Panda CK. Differential association of BRCA1 and BRCA2 genes with some breast cancer-associated genes in early and late onset breast tumors. Ann. Surg. Oncol. 2004;11:1045–1055. doi: 10.1245/ASO.2004.02.022. [DOI] [PubMed] [Google Scholar]
- Cooney K, Wetzel J, Merajver S, Macoska J, Singleton T, Wojno K. Distinct regions of allelic loss on 13q in prostrate cancer. Cancer Res. 1996;56:1142–1145. [PubMed] [Google Scholar]
- Cox DR. Regression models and life tables. J. R. Stat. Soc. B. 1972;34:187–220. [Google Scholar]
- Dahlgren L, Dahlstrand H, Lindquist D, et al. Human papilloma virus is more common in base of tongue than in mobile tongue cancer and is a favorable prognostic factor in base of tongue patients. Int. J. Cancer. 2004;112:1015–1019. doi: 10.1002/ijc.20490. [DOI] [PubMed] [Google Scholar]
- Dasgupta S, Mukherjee N, Roy S, et al. Mapping of the candidate tumor suppressor genes' loci on human chromosome 3 in head and neck squamous cell carcinoma of an Indian patient population. Oral Oncol. 2002;38:6–15. doi: 10.1016/s1368-8375(00)00131-7. [DOI] [PubMed] [Google Scholar]
- D'Costa J, Saranath D, Dedhia P, Sanghvi V, Mehta AR. Detection of HPV-16 gnome in human oral cancers and potentially malignant lesions from India. Oral Oncol. 1998;34:413–420. doi: 10.1016/s1368-8375(98)00028-1. [DOI] [PubMed] [Google Scholar]
- Dillon EK, de Boer WB, Papadimitiou JM, Turbett GR. Microsatellite instability and loss of heterozygosity in mammary carcinoma and its probable precursors. Br. J. Cancer. 1997;76:156–162. doi: 10.1038/bjc.1997.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles C, Piette J, Ploton D, Doco-Fenzy M, Foidart JM. Viral integration sites in human papilloma virus –33-immortalized cervical keratinocyte cell lines. Cancer Genet. Cytogenet. 1996;90:63–69. doi: 10.1016/0165-4608(96)00060-x. [DOI] [PubMed] [Google Scholar]
- Gillison ML, Koch WM, Copone RB, et al. Evidence for a causal association between human papilloma virus and a subset of head and neck cancers. J. Natl. Cancer Inst. 2000;92:709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- Gillison ML, Koch WM, Shah KV. Human papilloma virus in head and neck squamous cell carcinoma: are some head and neck cancers a sexually transmitted disease? Curr. Opin. Oncol. 1999;11:191–199. doi: 10.1097/00001622-199905000-00010. [DOI] [PubMed] [Google Scholar]
- Gupta VK, Schmdt AP, Pashia ME, Sunwoo JB, Scholnick SB. Multiple regions of deletion on chromosome arm 13q in head and neck squamous cell carcinoma. Int. J. Cancer. 1999;84:453–457. doi: 10.1002/(sici)1097-0215(19991022)84:5<453::aid-ijc1>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Harmer MH. UICC TNM Classification of Malignant Tumors. 3. Geneva: Union Internationale Contre le Cancer (UICC); 1978. [Google Scholar]
- Hittleman WN, Voravud N, Shin DM, Lee JS, Ro JY, Hong WK. Early changes during upper aerodigestive tract tumorigenesis. J. Cell Biochem. 1993;17F:233–236. doi: 10.1002/jcb.240531034. [DOI] [PubMed] [Google Scholar]
- Hu N, Li G, Li WJ, et al. Infrequent mutation in the BRCA2 gene in esophageal squamous cell carcinoma. Clin. Cancer Res. 2002;8:1121–1126. [PubMed] [Google Scholar]
- Jin YS, Heim S, Mandahl N, Biorkund A, Wennerberg J, Mittleman F. Nonrandom chromosome abnormalities in short term cultured primary squamous cell carcinomas of the head and neck. Genes Chromosomes Cancer. 1990;1:209–215. [Google Scholar]
- Jongsma APM, Piek JMJ, Zweemer RP, et al. Molecular evidence for putative tumor suppressor genes on chromosome 13q specific to BRCA1 related ovarian and fallopian tube cancer. J. Clin. Pathol. Mol. Pathol. 2002;55:303–309. doi: 10.1136/mp.55.5.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainu T, Juo SH, Desper R, et al. Somatic deletions in hereditary breast cancers implicate a putative novel breast cancer susceptibility locus. Proc. Natl. Acad. Sci. USA. 2000;97:9603–9608. doi: 10.1073/pnas.97.17.9603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick H, Waber P, Hoa-Thai T, et al. Infrequency of BRCA2 alteration in head and neck squamous cell carcinoma. Oncogene. 1997;14:2189–2193. doi: 10.1038/sj.onc.1201060. [DOI] [PubMed] [Google Scholar]
- Kuroki T, Fujinara Y, Nakamori S, et al. Evidence for the presence of two tumor suppressor genes for hepatocellular carcinoma on chromosome 13q. Br. J. Cancer. 1995;72:383–385. doi: 10.1038/bjc.1995.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latil A, Cussenot O, Fournier G, Lidereau R. The BRCA2 gene is not relevant to sporadic prostrate tumors. Int. J. Cancer. 1996;60:282–283. doi: 10.1002/1097-0215(19960410)66:2<282::aid-ijc2910660202>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Li WJ, IIu N, Su I, et al. Allelic loss on chromosome 13q14 and mutation in deleted in cancer 1 gene in esophageal squamous cell carcinoma. Oncogene. 2003;22:314–318. doi: 10.1038/sj.onc.1206098. [DOI] [PubMed] [Google Scholar]
- Li X, Lee NK, Ye YW, et al. Allelic loss at chromosome 3p, 8p, 13q, and 17p associated with poor prognosis in head and neck cancer. J. Natl. Cancer Inst. 1994;86:1524–1529. doi: 10.1093/jnci/86.20.1524. [DOI] [PubMed] [Google Scholar]
- MacGrogan D, Levy A, Bostwick D, Wagner M, Wells D, Bookstein R. Loss of chromosome arm 8p loci in prostrate cancer: mapping by quantitative allelic imbalance. Genes Chromosomes Cancer. 1994;10:151–159. doi: 10.1002/gcc.2870100302. [DOI] [PubMed] [Google Scholar]
- Maestro R, Piccinin S, Doglioni C, et al. Chromosome 13q deletion mapping in head and neck squamous cell carcinoma: identification of two distinct regions of preferential loss. Cancer Res. 1996;56:1146–1150. [PubMed] [Google Scholar]
- Mao L, Lee DJ, Tockman MS, Erozan YS, Askin F, Sidransky D. Microsattelite alterations as clonal markers fro the detection of human cancer. Proc. Natl. Acad. Sci. USA. 1994;91:9871–9875. doi: 10.1073/pnas.91.21.9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed J, Einhorn JM, Ittmann MM. Allelic loss on chromosome 13q in human prostrate carcinoma. Clin. Cancer Res. 1997;3:1867–1872. [PubMed] [Google Scholar]
- Nagai MA. Genetic alterations in head and neck carcinomas. Braz. J. Med. Biol. Res. 1999;32:897–904. doi: 10.1590/s0100-879x1999000700015. [DOI] [PubMed] [Google Scholar]
- Nawroz H, Riet PVD, Hruban RH, Koch W, Ruppert JM, Sidransky D. Allelotype of head and neck squamous cell carcinoma. Cancer Res. 1994;54:1152–1155. [PubMed] [Google Scholar]
- Ogawara K, Miyakawa A, Shiba M, et al. Allelic loss of chromosome 13q14.3 in human oral cancer: correlation with lymph node metastasis. Int. J. Cancer. 1998;79:312–317. doi: 10.1002/(sici)1097-0215(19980821)79:4<312::aid-ijc2>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int. J. Cancer. 2001;94:153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- Parkin DM, Pisani P, Ferlay J. Estimates of the worldwide incidence of 25 major cancers in 1990. Int. J. Cancer. 1999;80:827–841. doi: 10.1002/(sici)1097-0215(19990315)80:6<827::aid-ijc6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. 2. New York: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Sankaranarayanan R, Masuyer E, Swaminathan R, Ferlay J, Wehlan S. Head and neck cancer: a global perspective on epidemiology and prognosis. Anticancer Res. 1998;18:4779–4786. [PubMed] [Google Scholar]
- Sauders WS, Shuster S, Huang X, et al. Chromosomal instability and cytoskeletal defects in oral cancer cells. Proc Natl. Acad. Sci. U.S.A. 2000;97:303–308. doi: 10.1073/pnas.97.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen U, Sankaranarayanan R, Mandal S, Ramanakumar AV, Parkin DM, Siddiqi M. Cancer patterns in eastern India: the first report of the Kolkata Cancer Registry. Int. J. Cancer. 2002;100:86–91. doi: 10.1002/ijc.10446. [DOI] [PubMed] [Google Scholar]
- Shattuck TM, Klm TS, Costa J, et al. Mutational analysis of RB and BRCA2 as candidate tumor supressor genes in parathyroid carcinoma. Clin. Endocrinol.(Oxf) 2003;59:180–189. doi: 10.1046/j.1365-2265.2003.01814.x. [DOI] [PubMed] [Google Scholar]
- Sreekantaiah C, Rao PH, Xu L, Sacks PG, Schantz SP, Chaganti RS. Consistent chromosomal losses in head and neck squamous cell carcinoma cell lines. Genes Chromosomes Cancer. 1994;11:29–39. doi: 10.1002/gcc.2870110106. [DOI] [PubMed] [Google Scholar]
- Thorland EC, Myers SL, Gostout BS, Smith DI. Common Fragile sites are preferential targets for HPV16 integration in cervical tumors. Oncogene. 2003;22:1225–1237. doi: 10.1038/sj.onc.1206170. [DOI] [PubMed] [Google Scholar]
- Tripathi (Bhar) A, Dasgupta S, Roy A, et al. Sequential deletions in both arms of chromosome 9 are responsible for the development of head and neck squamous cell carcinoma from Indian Patients. J. Exp. Clin. Cancer Res. 2003;22:289–297. [PubMed] [Google Scholar]
- Van Duke DL, Worsham MJ, Bennninger MS, et al. Recurrent cytogenetic abnormalities in squamous cell carcinoma of the head and neck region. Genes Chromosomes Cancer. 1994;9:192–206. doi: 10.1002/gcc.2870090308. [DOI] [PubMed] [Google Scholar]
- Welesh PL, King MC. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum. Mol. Genet. 2001;10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
- Wentzensen N, Vinokurova S, Doeberitz MVK. Systematic review of genomic integration sites of Human papillomavirus genome in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res. 2004;64:3878–3884. doi: 10.1158/0008-5472.CAN-04-0009. [DOI] [PubMed] [Google Scholar]
- Wistuba II, Behrens C, Milchgrub S, et al. Sequential molecular abnormalities are involved in the multistage development of squamous cell lung carcinoma. Oncogene. 1999a;18:643–650. doi: 10.1038/sj.onc.1202349. [DOI] [PubMed] [Google Scholar]
- Wistuba II, Behrens C, Virmani AK, et al. Allelic losses at chromosome 8p, 21-23 are early and frequent events in the pathogenesis of lung cancer. Cancer Res. 1999b;59:1973–1979. [PubMed] [Google Scholar]
- Yoo GH, Xu HJ, Brennan JA, et al. Infrequent inactivation of the retinoblastoma gene despite frequent loss of chromosome 13q in head and neck squamous cell carcinoma. Cancer Res. 1994;54:4603–4606. [PubMed] [Google Scholar]