Abstract
Aging is a pleiotropic and stochastic process influenced by both genetics and environment. As a result the fundamental underlying causes of aging are controversial and likely diverse. Genome maintenance and in particular the repair of DNA damage is critical to ensure longevity needed for reproduction and as a consequence imperfections or defects in maintaining the genome may contribute to aging. There are many forms of DNA damage with double-strand breaks (DSBs) being the most toxic. Here we discuss DNA DSBs as a potential causative factor for aging including factors that generate DNA DSBs, pathways that repair DNA DSBs, consequences of faulty or failed DSB repair and how these consequences may lead to age-dependent decline in fitness. At the end we compare mouse models of premature aging that are defective for repairing either DSBs or UV light-induced lesions. Based on these comparisons we believe the basic mechanisms responsible for their aging phenotypes are fundamentally different demonstrating the complex and pleiotropic nature of this process.
1. Introduction
For mammals, aging is the time-dependent deterioration of an individual that decreases fitness and ultimately causes death and is caused by many factors that are influenced by both genetics and environment. Aging is not greatly influenced by natural selection since evolutionary pressure subsides once the individual has lived long enough for procreation (Kirkwood, 2002). Thus, aging is subject to little regulation making the aging process appear highly variable, stochastic and pleiotropic. As a result age-related decline is subject to a large number of potential influences and identifying these influences can be difficult and controversial. One potential aging target is nuclear DNA since it is a permanent blueprint that controls cellular processes. Thus, DNA replication and genome maintenance mechanisms are highly regulated to ensure faithful reproduction and maintenance of the blueprint and these pathways assure sufficient longevity for procreation and survival of the species. There are many of these longevity assurance mechanisms that are necessary to maintain this blueprint and imperfections in any of them can cause lasting changes called mutations. Therefore, aging may be a consequence of imperfect longevity assurance mechanisms designed to protect the genome from damage and to prevent the accumulation of mutations. The most deleterious damage to DNA is a DSB; thus, presenting the intriguing possibility that imperfections or defects in DSB repair pathways contribute to the aging process.
2. Factors that generate DNA DSBs
There are two general types of DNA damage: single-strand DNA (ssDNA) and double-strand DNA (dsDNA) lesions. ssDNA lesions include base lesions, intrastrand crosslinks and single-strand breaks (SSBs) while dsDNA lesions include DSBs and interstrand crosslinks (ICLs). Therefore, a wide range of DNA lesions exist and any of them may contribute to the aging process, but DSBs and ICLs are much more toxic than ssDNA lesions because they are incompatible with DNA replication (Bessho, 2003). A replication fork will stall and may collapse when encountering a DSB, a catastrophic event that can lead to translocations, death or senescence (Michel et al., 2007). During mitosis DSBs can lead to cancer-causing instability (Lobrich and Jeggo, 2007). In addition, faulty repair of DSBs can lead to genomic rearrangements that are incompatible with mitosis. For example, a rearrangement that that joins two centromeres into a single chromosome (called a dicentric) will be torn apart during mitosis.
Agents that cause DNA breaks may come from endogenous and exogenous sources. Common endogenous agents are reactive oxygen species (ROS), by-products of oxygen metabolism that are produced in the mitochondria and peroxisomes. ROS include superoxide, hydrogen peroxide and hydroxy radicals and are important for multiple biological processes that include cell signaling (Valko et al., 2007). However, ROS are highly reactive due to unpaired electrons that can react with biomolecules including DNA. These reactions may cause a wide range of genetic damage (Friedberg et al., 1995), mostly base lesions and DNA single-strand breaks (SSBs) but also an occasional DSB. Exogenous sources also give rise to many agents that cause genetic damage including DNA DSBs. These agents are everywhere from water, air and soil and may be both natural and man made (White, 2007). Some well known exogenous agents include anti-cancer chemotherapeutics (Wyrobek et al., 2005), cigarette smoke (Wu et al., 2004) and ionizing radiation (Benkhaled et al., 2007; Lucas, 1998). In addition to agent-induced genetic damage, DSBs may occur as a consequence of closely aligned SSBs on complementary DNA strands, intermediates of DNA repair, eroded telomeres and replication errors. For example H2O2 commonly causes thymidine glycols that may lead to DSBs at a replication fork. In addition, DSBs may be generated by site-specific endonucleases important for developmental programs like DSBs induced by the Rag heterodimer (composed of Rag-1 and Rag-2) in lymphocyte precursors. Rag-1 and Rag-2 expression is largely restricted to developing T and B lymphocytes (Yamamoto et al., 1992) and Rag induces a DSB at the recombination signal sequences to initiate V(D)J [Variable(Diverse)Joining] recombination for assembling antigen receptor genes (Lieber et al., 2004). Rag is also capable of generating multiple nicks at non-B DNA structures at the major breakpoint region in the Bcl-2 gene (Raghavan et al., 2005; Raghavan et al., 2004). Thus, DNA DSBs may come from many sources and are inescapable; therefore, pathways must exist to repair them to ensure longevity.
3. Pathways that repair or suppress DNA DSBs
DNA repair pathways correct DNA lesions to prevent mutations and preserve genomic integrity (Hoeijmakers, 2001). These pathways are regarded as longevity assurance mechanisms important for suppressing tumor-causing mutations. Specific pathways are specialized for repairing specific damage, even though there is some functional overlap. For example, a variety of excision repair pathways correct ssDNA lesions including BER (Almeida and Sobol, 2007; Barnes and Lindahl, 2004), nucleotide excision repair (NER) (de Boer and Hoeijmakers, 2000) and mismatch repair (MMR) (Kolodner and Marsischky, 1999). Of these, BER is prominent for repairing ROS-induced DNA damage, NER is important for repairing UV light-induced lesions and MMR is critical for postreplication repair. The major pathways that repair DNA DSBs are homologous recombination (HR) and nonhomologous end joining (NHEJ). HR is an error free pathway that utilizes the sister chromatid as a template during S/G2 phase (West, 2003) while NHEJ is considered to be an error prone pathway that joins ends together without a template and functions during both G1 and S phases (Lieber et al., 2003). Both pathways are frequently used in mammalian cells (Jasin, 2000) and disruption of either leads to gross chromosomal rearrangements (GCRs) that may cause cancer (Hoeijmakers, 2001) and perhaps aging (Hasty et al., 2003; Lombard et al., 2005; Vijg and Dolle, 2002). In addition, telomere length maintenance ensures chromosomal ends do not become open DSBs. These pathways are critical for ensuring sufficient health span for reproduction. Some data shows they decline as mammals age suggesting a causal factor for aging (Gorbunova et al., 2007). For this review, we focus on pathways that repair or suppress DNA DSBs: HR, NHEJ and telomere maintenance.
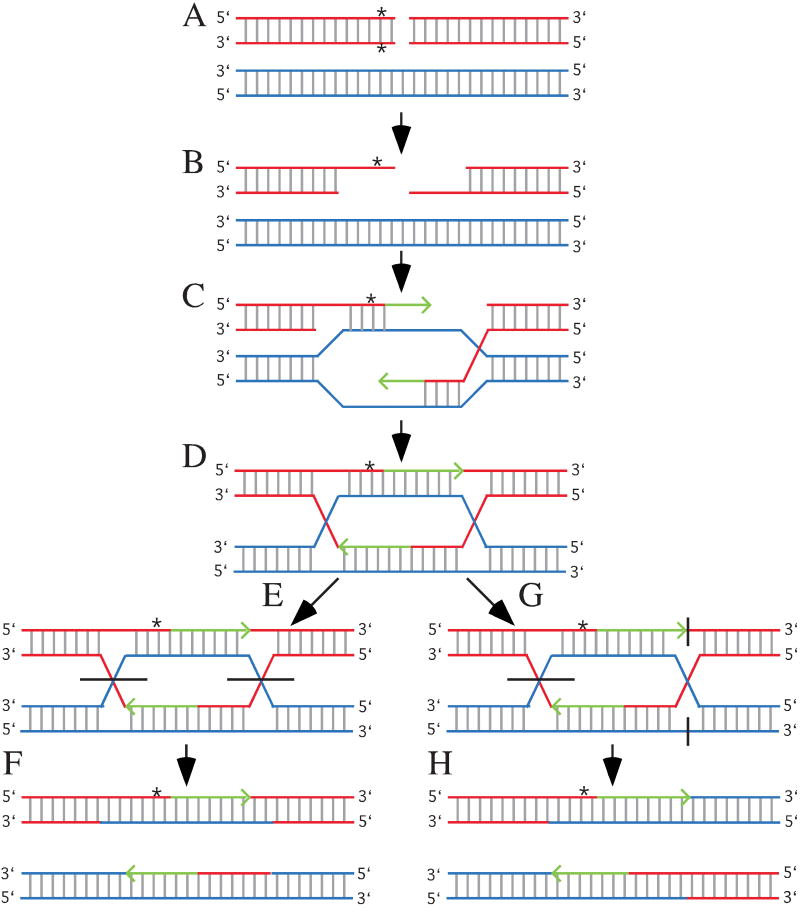
HR repairs DNA DSBs by using a homologous template to ensure fidelity; therefore, HR occurs primarily during S or G2 (West, 2003). There are many proteins required for efficient HR and many more that influence its function. Central to HR is Rad51, a homolog to the prokaryotic recombination protein, RecA. The ends of a DSB are resected to expose single strands and Rad51 forms a nucleoprotein filament on these strands to induce invasion to a homologous template usually provided by the sister chromatid but potentially provided by either the homologous chromosome or by a repeat on the same or different chromosome. Invading strands serve as primers for DNA replication and subsequent intermediates may be resolved in a noncrossover plane such that the DSB is repaired without recombination or in a crossover plane such that the DSB is repaired in addition to recombination (Fig. 1). While HR repairs DNA DSBs with fidelity, it may also be mutagenic if it occurs between repeats on the same or a different chromosome to generate inversions, deletions or translocations. In addition, HR between homologous chromosomes may induce loss of heterozygosity by gene conversion since the ends are resected and since heteroduplexes are formed (Luo et al., 2000). Thus, HR must be tightly monitored to prevent these mutations. Thus, it is possible an age-related decline in this regulation contributes to aging.
Fig. 1.
Homologous recombination and gene conversion. (a) Homologous templates are aligned. This usually occurs between sister chromatids during DNA replication, but may also occur between chromosomes as shown here. The red chromosome has a DSB and a sequence difference (*) located close to the break. (b) The ends are resected to form single-strands. Note the sequence difference from the bottom red strand is removed. (c) The red single-strands invade the blue homologous template to form a joint molecule and primers for DNA replication (green arrows). Note the blue strands are used as templates for DNA replication; therefore, the green strands contain the same information as the blue strands. This means sequence differences on the resected red strand are lost (gene conversion). (d) The formation of Holliday junctions (a mobile attachment between four strands of DNA) and heteroduplexes (double-strand DNA composed of a strand from each homologue). Note the sequence difference is lost for one strand and a mismatch is formed for the other strand. (e) Noncrossover resolution of the Holliday junctions. Horizontal black lines represent locations where strands are cut. (f) The final products. For this diagram the top chromosome contains a mismatched that may be resolved by MMR. (g) Crossover resolution. The left Holliday junction is resolved as shown for the noncrossover event; however, the outer strands are cut to resolve the right Holliday junction. (g) The final products showing the chromosomes recombine. The heteroduplex and mismatch are the same as described for the noncrossover.
There is some evidence that HR regulation changes with age. For example, the Drosophila male germline shows an age-dependent increase in HR. However, there is little evidence for increased or decreased HR in mammals probably because the majority of cells in an adult mammal are postmitotic and HR does not function in G0/G1 cells. Yet, the levels of nonallelic homologous recombination appear to increase with age since rearrangements in human blood cells are elevated from newborns to adults (Flores et al., 2007). Thus, it is possible these rearrangements accumulate with age due to diminished HR regulation.
NHEJ repairs DNA DSBs by joining open ends together without the aid of a homologous template and can therefore occur during G1 in addition to S phase. In mammals, at least seven proteins are required: Ku70, Ku80, DNA-PKCS, Artemis, Xrcc4, DNA Ligase IV (Lig4) and Xrcc4-like factor (Lieber et al., 2004). Ku70 and Ku80 form a heterodimer called Ku that binds to DNA ends (Walker et al., 2001) and together with a PI-3 kinase catalytic subunit, DNA-PKCS, forms a holoenzyme referred to as DNA-PK (DNA dependent – protein kinase). Artemis opens hairpins and processes overhangs in a complex with DNA-PKCS and these ends are ligated by the Xrcc4-Lig4 heterodimer in a complex with Xrcc4-like factor (Ahnesorg et al., 2006; Buck et al., 2006). Mice deleted for Ku or Xrcc4/Lig4 exhibit phenotypes that result from defective repair of DNA DSBs such as hypersensitivity to clastogenic agents and GCRs (Ferguson et al., 2000; Gu et al., 1997; Lim et al., 2000; Nussenzweig et al., 1996). NHEJ also repairs the DSBs formed during the assembly of V(D)J [Variable(Diverse)Joining] segments of antigen receptor genes; thus, NHEJ-deletion causes failed lymphocyte development resulting in severe combined immunodeficiency (scid).
There is evidence that NHEJ functionally declines with age suggesting a process that limits life span and contributes to aging (Gorbunova et al., 2007). As rats age Ku levels diminish in the testis and Ku70 or Ku80 levels are differentially expressed in various tissues (Um et al., 2003). Similarly as humans age, Ku nuclear localization and DNA binding is impaired in blood mononuclear cells (Doria et al., 2004; Frasca et al., 1999) and Ku70, but not Ku80, levels decline in lymphocytes (Ju et al., 2006). By correlation, Ku levels decline and their cellular distribution is altered as human fibroblasts approach senescence (Seluanov et al., 2007b). In keeping with these results, NHEJ function declines in the brains of aging rats (Ren and de Ortiz, 2002; Vyjayanti and Rao, 2006) and in Alzheimer's patients (Shackelford, 2006) and becomes less efficient and more error-prone in senescent cells (Seluanov et al., 2004). Thus, NHEJ declines with age supporting the possibility that defective NHEJ will lead to early aging.
Telomeres are important structures that cap and maintain chromosome ends by forming higher order structures called telomeric loops (de Lange, 2002). This cap is composed of many TTAGGG repeats that end in a single-stranded overhang used by a telomere-specific enzyme, telomerase, as a template to extend and maintain telomere length. This means the natural ends of chromosomes are not the same as DNA DSBs. However, when telomeres erode the chromosomal end is much like a DSB and available for end joining by NHEJ such that chromosomes can become fused together (Artandi et al., 2000; Chin et al., 1999). Similar to HR, telomerase is important for proliferating cells and telomerase activity is restricted to only a few cells in an adult, the germ cells and stem cells (Flores et al., 2006); thus, telomerase can only impact these cell types with age. Importantly, telomerase activity is insufficient to protect telomeres in stem cells suggesting that tissue renewal can become compromised with age due to eroded telomeres that may appear as DSBs.
Some species, but not all, express telomerase at very low levels in quiescent somatic cells in the adult permitting speculation that low telomerase levels may be important for determining the health of quiescent cells with age. However, telomerase expression may also promote cancer by enabling cellular proliferation since eroded telomeres induce anti-tumor responses that either kill the cell or stop proliferation. Recent data show telomerase levels in quiescent somatic cells are inversely proportional to species body mass with no correlation to life span (Seluanov et al., 2007a). Thus, the larger the body mass the less telomerase is expressed in adult quiescent cells indicating that extra measures are needed to prevent cancer for species of increased body mass. This is likely the reason somatic cells express low levels of telomerase in mice but not humans. Therefore, human somatic cells that express low telomerase levels are cancer prone.
Besides telomerase, there are many proteins needed to maintain telomeres including the NHEJ proteins Ku70, Ku80 and DNA-PKCS. These NHEJ proteins are proposed to maintain telomeres since they associate with telomeres (d'Adda di Fagagna et al., 2001; Hsu et al., 1999), suppress telomere fusions (Bailey et al., 1999; Hsu et al., 2000; Li et al., 2007; Samper et al., 2000) and impact telomere length maintenance (d'Adda di Fagagna et al., 2001; Espejel et al., 2002). Thus, Ku70, Ku80 and DNA-PKCS are important for both DSB repair and telomere maintenance. Since these protein levels decline with age it is possible that telomere maintenance, as well as NHEJ, undergoes an age-dependent decline.
4. Accumulation of damage/mutations
Increasing evidence demonstrates that DNA damage and mutations accumulate with age in mice and humans including the type of damage/mutations that result from imperfect DSB repair. For example unrepaired DNA DSBs (Sedelnikova et al., 2004) and GCRs (Dolle et al., 1997) accumulate in a variety of tissues as mice age. In addition, GCRs (both spontaneous and H2O2 induced) appear in replicating and quiescent cells while point mutations (both spontaneous and UV-induced) are highly replication-dependent (Busuttil et al., 2007a; Busuttil et al., 2007b). These data suggest that GCRs, unlike small mutations, are not influenced by DNA replication suggesting that DSBs and GCRs could be more important for age-related decline since most cells are postmitotic in adult mammals. Naturally, accumulation of both point mutations and GCRs could be important for aging stem cells.
Interestingly, in human and mouse cells (lymphocytes, kidneys, liver, skin) the frequency of chromosomal aberrations (Crowley and Curtis, 1963; Li et al., 2007; Martin et al., 1985; Ramsey et al., 1995; Tucker et al., 1999), as well as mutations at the HPRT locus (Dempsey et al., 1993; Jones et al., 1995a), increases with age as a function of their life span rather than chronological time. This suggests that age-dependent mutation accumulation is related to the rate of aging and could be a function of repair capacity (Hart and Setlow, 1974). Indeed, mutation accumulation at HPRT has been found to accelerate in a mouse model of premature aging (Odagiri et al., 1998) and decelerate in mice whose life span is extended by caloric restriction (Dempsey et al., 1993). Deletion of either Ku70 or Ku80 causes early aging and cells derived from these mice show an early onset of chromosomal aberrations that also increase in control mice with age (Li et al., 2007). Thus, DNA damage and mutations increase as a function of biological age (not chronological time) suggesting that genetic damage and mutations contribute to aging.
5. DNA damage defective mammalian models of aging
A number of premature aging syndromes have been described in humans called segmental progeroid syndromes. These syndromes are called segmental since they display only a subset of age-related pathologies and were described based on clinical observations. Therefore, it is striking that most of them result from defective chromosomal metabolism. A complete description of these syndromes and their phenotypes has been provided (Bohr, 2002).
The best-known segmental progeroid syndrome is Werner's syndrome (WS) (Goto et al., 1997). An inactivating mutation in WRN, a homolog of the E. coli RECQ gene, causes WS (Yu et al., 1996). WRN is both a 3′→5′ DNA helicase and a 3′→5′ DNA exonuclease (Huang et al., 1998) and is likely important for several DNA metabolic pathways including replication and repair including homologous recombination (Otterlei et al., 2006). Cells deficient in WRN exhibit genetic instability that includes large chromosomal deletions suggesting a defect in DSB repair. Individuals with WS develop, two to three decades prematurely, atrophic skin, thin gray hair, osteoporosis, type II diabetes, cataracts, arteriosclerosis, and cancer. Interestingly, about half the cancers are mesenchymal in origin, in contrast to cancers that develop in normal individuals, of which 90% are epithelial in origin. WS individuals typically die in the fifth decade of life, primarily of cardiovascular disease or cancer. There are five RECQ-like genes in mammals. All encode 3′→5′ DNA helicases, and at least three, WRN, RTS (Rothmund Thomson syndrome) and BLM (Bloom syndrome gene) are associated with premature aging and/or cancer prone syndromes in humans (Mohaghegh and Hickson, 2001).
A variety of genetically altered mice have also been described with early aging phenotypes (Hasty et al., 2003; Lombard et al., 2005). Many of these mice were originally generated in an attempt to better understand cancer predisposition and like in humans are defective for DNA repair. These DNA repair pathways include those that repair double-stranded lesions: HR, NHEJ and cross-link repair (CLR). Defects in specific genes that result in premature aging include Brca1 for HR (Cao et al., 2003); Ku80 (Holcomb et al., 2007; Vogel et al., 1999), Ku70 (Li et al., 2007), Xrcc4 (Chao et al., 2006) and DNA-PKCS (Espejel et al., 2004b) for NHEJ, and Ercc1 and Xpf for CLR (Tian et al., 2004; Weeda et al., 1997). Furthermore, telomere attrition is similar to a DSB and telomere-defective mice exhibit an aging phenotype, especially when crossed with Atm-mutant (Wong et al., 2003), Wrn-mutant (Chang et al., 2004; Du et al., 2004) and NHEJ-mutant mice (Espejel et al., 2004a). Epimutations may also play a role in aging since a defect in PASG, a SNF-2-like protein that facilitates methylation, induces an early aging phenotype similar to mice defective for repairing DNA DSBs (Sun et al., 2004). In addition, histone acetylation may influence the repair of DNA DSBs since Ku80- and DNA ligase IV-mutant cells are hypersensitive to a histone deacetylase inhibitor (Yaneva et al., 2005). Phenotypes for these aging models have been reviewed and will not be discussed in detail here (Hasty et al., 2003; Hofer et al., 2005; Lombard et al., 2005; Warner and Sierra, 2003). Briefly, there are common phenotypes that include reduced stress resistance, kyphosis, skin atrophy, skin ulcers, poor wound healing and premature cellular senescence for fibroblasts. In addition, a variety of these aging models exhibit reduced haematopoietic stem cell function (Nijnik et al., 2007; Rossi et al., 2007).
6. DNA damage responses and cytotoxicity
DNA damage checkpoints respond to many forms of DNA damage, including DSBs, to facilitate repair or removal of these lesions (Campisi and d'Adda di Fagagna, 2007). These checkpoint machineries monitor the genome for problems that adversely affect DNA replication or mitosis and halt cell cycle progression when such problems are encountered to allow time for the damage to be repaired. If damage is severe or irreparable, these machineries engage either cell death (apoptosis) or cellular senescence pathways. The latter is a condition that refers to the limited proliferation potential (replicative life span) and eventual permanent arrest exhibited by primary cells in tissue culture (Hayflick, 1965). These responses are anti-cancer pathways utilizing classical tumor suppressors. Both apoptosis and cellular senescence occur in response to oxidative stress, DNA damage and telomere erosion (Collado et al., 2007; Parrinello et al., 2003; Reed, 1999; Sharpless and DePinho, 2007) and are important for reducing mutations (Busuttil et al., 2003; Collado and Serrano, 2006; Sharpless and DePinho, 2005). DNA damage responses to replication associated DSBs may also induce cellular senescence to inhibit tumorigenesis (Bartkova et al., 2005; Bartkova et al., 2006; Di Micco et al., 2006). These same or similar responses may contribute to aging by reducing the population of healthy cells (Bree et al., 2002; Campisi, 1997; Campisi, 2000; Pelicci, 2004). Therefore, apoptosis and cellular senescence are important anti-cancer mechanisms needed to ensure longevity and may contribute to aging by depleting or altering cells with age (Janzen et al., 2006; von Zglinicki et al., 2005).
It is possible these responses contribute to aging more than the accumulation of DNA damage and mutations. Evidence for this possibility comes from human and mouse aging models that display similar phenotypes even though they are defective for different DNA repair pathways. For example, premature cellular senescence is observed for fibroblasts derived from aging models defective for NHEJ (Lim et al., 2000), ICL repair (Grillari et al., 2007) or telomere maintenance (Chin et al., 1999). Furthermore, NHEJ suppresses premature cellular senescence and apoptosis caused by exposure to a histone deacetylase inhibitor (Yaneva et al., 2005). This likely occurs since the status of histone acetylation influences the repair of DNA DSBs in Saccharomyces cerevisiae (Choy and Kron, 2002) (Jazayeri et al., 2004) and possibly mammals (Ikura et al., 2000). Elevated responses are also observed in aging models that are defective for aspects of DNA metabolism not directly involved in repairing dsDNA lesions. For example, apoptosis is elevated for early aging mice expressing a defective mitochondrial DNA polymerase (Kujoth et al., 2006). In addition, premature cellular senescence along with elevated levels of the cell cycle regulator p16INK4a occur in early aging mice defective for a SNF2-like factor that facilitates DNA methylation called PASG (Sun et al., 2004). Thus, defects in any one of a number of genome integrity pathways may cause a similar aging phenotype suggesting that responses common to a diverse range of DNA conditions can cause premature ageing.
Even though deficits in diverse DNA repair pathways increase various types of DNA damage, cellular responses to these different types of damage have a similar outcome; that is, they induce either apoptosis or cellular senescence. The question is do these responses normally contribute to aging? Some evidence analyzing the tumor suppressor p53, suggests they do not. p53 is well known for inducing checkpoints in response DNA damage that can lead to either apoptosis or cellular senescence (Meek, 2004). Complete deletion of p53 function enhances tumor susceptibility and reduces longevity in mice (Donehower et al., 1992) while overexpression of full-length p53 genomic sequences from a BAC lowers cancer incidence but does not influence aging (Garcia-Cao et al., 2002) and overexpression of Arf/p53 extends life span by improving cancer resistance (Matheu et al., 2007). In addition, mice with constitutively high p53 caused by reduced MDM2 are resistant to tumor formation but do not exhibit early aging (Mendrysa et al., 2006). Thus, overexpression of p53 reduces cancer without negatively impacting life span or accelerating aging.
However, other data suggests that overexpression of tumor suppressors is toxic supporting the hypothesis that cellular responses designed to prevent cancer-causing mutations contribute to aging (Bree et al., 2002; Campisi, 2000; Pelicci, 2004). Strangely enough, support for this hypothesis also comes from p53. Studies show that p53 levels influence cellular replication capacity since p53-deletion increases replicative potential (Harvey et al., 1993) while p53 overexpression decreases replicative potential and promotes cellular senescence (Sugrue et al., 1997). In fact diminished negative regulation of p53 is extremely toxic since deletion of a p53 negative regulator, MDM2 or MDM4, is embryonic lethal and this lethality is rescued by deletion of p53 (Jones et al., 1995b; Montes de Oca Luna et al., 1995; Parant et al., 2001). The role p53 dosage plays in aging is most clearly shown for Brca1-mutant mice because their early aging phenotype is only seen in a p53-heterozygous mutant background (Cao et al., 2003). In a p53 wild type background Brca1-defective mice die early during embryogenesis while in a p53-homozygous mutant background they exhibit a high cancer incidence (Xu et al., 2001). Furthermore, p53 is required for premature replicative senescence and low tumor incidence associated with Ku80-deletion (Holcomb et al., 2006; Lim et al., 2000) and for the adverse cellular and organismal problems and low cancer incidence associated with telomere erosion (Chin et al., 1999). These observations suggest conditions that initiate a p53-dependent checkpoint, like DNA damage, induce cell cycle arrest that may ultimately result in apoptosis or cellular senescence, especially if the DNA damage persists. Therefore, p53 activity may be mildly increased in DNA repair deficient humans and mice that exhibit precocious aging.
In human, the p53 gene encodes a variety of isoforms that are expressed in a tissue-specific manner (Bourdon et al., 2005) and the ratio of these isoforms may contribute to cancer prevention and aging. To support a role for isoforms in cancer prevention, variable expression of these isoforms is observed in breast tumor compared to normal breast. These isoforms may be important for DNA damage responses since at least one human-specific isoform is essential for the ATR-intra-S phase checkpoint in response to DNA damage (Rohaly et al., 2005). Some of these isoforms may be important for aging as suggested by two mouse models that express N-terminally truncated p53 (Maier et al., 2004; Tyner et al., 2002). Both models exhibit increased cancer resistance but decreased life span accompanied by an early onset of aging phenotypes similar to the DNA repair deficient mice. One of these models expresses an artificial C-terminal p53 fragment called the M protein (Tyner et al., 2002) while the other expresses a naturally produced isoform called p44 (mouse) or Δ40p53 (human) (Maier et al., 2004)(Scrable et al., 2005). For both mouse models full-length p53, in combination with the N-terminal deleted p53, is required to observe early aging. Since p53 naturally forms a tetramer, it is likely these truncated isoforms associate with full-length p53 to influence its function. In support, the M protein interacts with wild-type p53, increases its stability, and facilitates its nuclear localization in the absence of stress (Moore et al., 2007). Another isoform stabilizes p53 in the presence of Mdm2 and alters the expression levels of p53-induced gene products (Yin et al., 2002). Thus, overexpression of these p53 isoforms likely increase some aspects of p53 function that reduces tissue function or regeneration which is consistent with an accelerated loss of stem cell functional capacity (Dumble et al., 2004). Therefore, these mouse models suggest that activation of p53 by a dominant-N-terminally truncated isoform accelerates the aging process perhaps in response to DNA damage and highlights the importance of p53 isoform ratios.
7. Comparison of GCRs to small mutations
Are dsDNA lesions and GCRs more likely to impact aging than ssDNA lesions and point mutations? Both GCRs and point mutations increase with age in a tissue specific manner, so either could be important for at least some tissues. However, current data suggests that GCRs are more important. For example, aging does not always correlate to point mutation levels since mice defective for antioxidant defense clearly show elevated point mutations but do not show signs of early aging with the exception of cancer (Busuttil et al., 2005). Even though cancer is an age-related disease, it is the outcome of a single altered cell while general aging is wide spread cellular decline. Thus, point mutations by themselves do not mandate general widespread cellular decline. However, GCRs may. At this time the data are uncertain, but GCRs appear to correlate with aging since they increase with age (Dolle et al., 1997; Vijg and Dolle, 2002) and defects that increase GCRs frequently accelerate aging in both mouse and human (Hasty et al., 2003; Lombard et al., 2005). Large deletions would obviously cause haploinsufficiency of many genes and this reduced dosage may impair cell function, especially in areas of imprinted genes. To support this possibility age-associated deletions have been observed in mammalian ribosomal RNA genes (Strehler, 1995). In addition, one could imagine GCRs impacting more than just the genes within a deletion or at the junctions of the translocation or inversion. The GCRs may influence mitosis and induce mitotic checkpoints. BubR1, Bub3 and Rae1 are mitotic checkpoint proteins that impact aging (Baker et al., 2004; Baker et al., 2006). Mice deleted for BubR1 die early during development (Wang et al., 2004); however, expression of a hypomorphic allele results in reduced life span and an early onset of aging features similar to some DNA repair deficient mice. Combined haploinsufficiency for Bub3/Rae1 show a similar but more sever phenotype. These mice exhibit both increased aneuploidy and cellular senescence with elevated levels of p53 and other stress response proteins, but the severity of their aging phenotype correlates with cellular senescence not aneuploidy. Thus, alterations in chromosome structure could induce senescence and aging by enhancing mitotic checkpoints (Fernandez-Capetillo and Nussenzweig, 2004).
Comparing NHEJ-defective mice that suppress GCRs to NER-defective mice that suppress point mutations suggest that the mechanisms leading to their aging phenotypes are fundamentally different. Early aging is observed in NER-defective mice with a subtle defect in Xpd, a helicase for transcription factor IIH (de Boer et al., 2002). Deletion of Xpd is lethal, however, subtle changes that hinder but do not ablate function cause an aging phenotype in humans (trichothiodystrophy, xeroderma pigmentosum combined with Cockayne syndrome) as well as in mice (Andressoo et al., 2006; de Boer et al., 1998). On the surface, the Xpd- and NHEJ-defective mouse phenotypes appear similar including kyphosis and early loss of reproductive function. However, there are also important differences. First, fibroblasts derived from NER-defective mice do not undergo premature cellular senescence; thus, there is no evidence for a p53-mediated DNA damage response (Kipling and Faragher, 1997; van de Ven et al., 2006). By comparison NHEJ-defective fibroblasts undergo rapid senescence that is dependent on p53 (Lim et al., 2000). Second, NER-mutant fibroblasts are not hypersensitive to acute oxidative stress unlike NHEJ-mutant fibroblasts (Parrinello et al., 2003). Third, there is no correlation to genomic instability and aging for NER-defective aging models (Dolle et al., 2006) while Ku70- and Ku80-mutant mice exhibit an early onset of chromosomal aberrations that are similar to aged control mice (Li et al., 2007). Fourth, NER aging models exhibit modest reductions in life span (Wijnhoven et al., 2005) compared to mice deleted for either Ku70 or Ku80 (Li et al., 2007). This is surprising considering their early onset of aging pathologies occurs at about the same time. Fifth, NER-mutant mice, but not Ku80-mutant mice, exhibit a temporary adaptive stress response at 2 weeks that is similar to the constitutive response shown by long-lived endocrine defective mice and calorie-restricted mice including reduced post natal growth, hypoglycemia, and perturbation of the growth hormone/insulin-like growth factor 1 neuroendocrine axis (van de Ven et al., 2006). This adaptive stress response is designed to aid survival during a period of stress and may also share underlying mechanisms of extended longevity with calorie restriction. Thus, the fundamental cause of these aging phenotypes is likely different for the NER- and NHEJ-defective aging models highlighting the possibility that multiple mechanisms may cause age-related decline. For the NHEJ-mutant mice the cause of early aging appears to be a combination of GCRs and induced anti-cancer responses (Figure 2). However, the cause of early aging in the NER-defective mice is less clear since the aging phenotype is not associated with mutations and since these genetic defects do not induce cellular senescence. It is possible that impaired transcription contributes to aging due to the intimate relationship between NER and transcription. With age there is increased transcriptional variation between cells (Bahar et al., 2006); yet this could occur for many reasons including deficient NER, GCRs or cellular senescence. Therefore, the future goal will be to determine the relationship of these underlying causes found in NER- and NHEJ-deficient mice to that of control mice and humans keeping in mind that aging is likely influenced greatly by both genetics and environment.
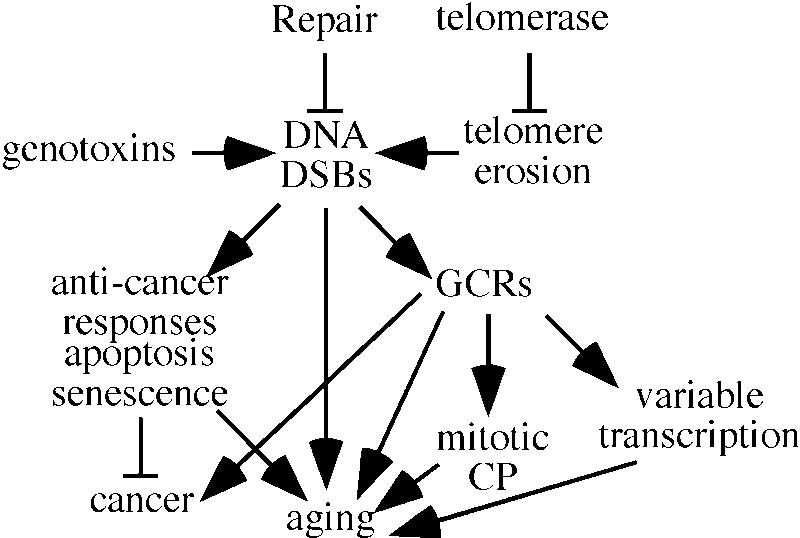
Fig. 2.
DSBs as a causative factor in aging. DSBs are generated by a variety of sources including exposure to genotoxins and telomere erosion. Telomerase suppresses telomere erosion while HR and NHEJ repair DNA DSBs. Unrepaired DSBs induce anti-cancer responses that may induce apoptosis or cellular senescence. As an indirect consequence these responses may contribute to aging. DSBs may be incorrectly repaired to generate gross chromosomal rearrangements (GCRs). These rearrangements may lead to cancer but may also contribute to aging by inducing mitotic checkpoints or increasing transcriptional variation.
8. Summary
Aging is a pleiotropic and stochastic process heavily influenced by genetic and environmental factors making aging-related processes difficult to understand and controversial. Here we argue that DNA repair pathways are essential for longevity and imperfections or defects in these pathways have the potential to cause early aging. As a result DNA damage and mutations may heavily influence the aging process either directly or perhaps indirectly by inducing either apoptosis or cellular senescence. DNA DSBs may be particularly harmful for longevity because they are more cytotoxic than other forms of damage and can lead to GCRs that negatively impact cell viability or promote cancer. Mice defective for NHEJ and other DNA repair pathways support the notion that DNA repair is important for longevity and delaying aging. However, a close comparison of mice defective for NHEJ and NER suggest the fundamental basis for aging is different in each model highlighting the complexity of aging and the need for deeper analysis.
Acknowledgments
This work was supported by P01 AG17242, R01 CA76317-05A1, UO1 ES11044 to PH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This PDF receipt will only be used as the basis for generating PubMed Central (PMC) documents. PMC documents will be made available for review after conversion (approx. 2-3 weeks time). Any corrections that need to be made will be done at that time. No materials will be released to PMC without the approval of an author. Only the PMC documents will appear on PubMed Central -- this PDF Receipt will not appear on PubMed Central.
References
- Ahnesorg P, Smith P, Jackson SP. XLF Interacts with the XRCC4-DNA Ligase IV Complex to Promote DNA Nonhomologous End-Joining. Cell. 2006;124:301–13. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst) 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andressoo JO, Mitchell JR, de Wit J, Hoogstraten D, Volker M, Toussaint W, Speksnijder E, Beems RB, van Steeg H, Jans J, de Zeeuw CI, Jaspers NG, Raams A, Lehmann AR, Vermeulen W, Hoeijmakers JH, van der Horst GT. An Xpd mouse model for the combined xeroderma pigmentosum/Cockayne syndrome exhibiting both cancer predisposition and segmental progeria. Cancer Cell. 2006;10:121–32. doi: 10.1016/j.ccr.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–5. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dolle ME, Calder RB, Chisholm GB, Pollock BH, Klein CA, Vijg J. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–4. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- Bailey SM, Meyne J, Chen DJ, Kurimasa A, Li GC, Lehnert BE, Goodwin EH. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc Natl Acad Sci U S A. 1999;96:14899–904. doi: 10.1073/pnas.96.26.14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, van Deursen JM. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–9. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Jeganathan KB, Malureanu L, Perez-Terzic C, Terzic A, van Deursen JM. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J Cell Biol. 2006;172:529–40. doi: 10.1083/jcb.200507081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–76. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, Takaoka M, Nakagawa H, Tort F, Fugger K, Johansson F, Sehested M, Andersen CL, Dyrskjot L, Orntoft T, Lukas J, Kittas C, Helleday T, Halazonetis TD, Bartek J, Gorgoulis VG. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–7. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Benkhaled L, Xuncla M, Caballin MR, Barrios L, Barquinero JF. Induction of complete and incomplete chromosome aberrations by bleomycin in human lymphocytes. Mutat Res. 2007 doi: 10.1016/j.mrfmmm.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Bessho T. Induction of DNA replication-mediated double strand breaks by psoralen DNA interstrand cross-links. J Biol Chem. 2003;278:5250–4. doi: 10.1074/jbc.M212323200. [DOI] [PubMed] [Google Scholar]
- Bohr VA. Human premature aging syndromes and genomic instability. Mech Ageing Dev. 2002;123:987–93. doi: 10.1016/s0047-6374(02)00039-8. [DOI] [PubMed] [Google Scholar]
- Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, Saville MK, Lane DP. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122–37. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bree RT, Stenson-Cox C, Grealy M, Byrnes L, Gorman AM, Samali A. Cellular longevity: role of apoptosis and replicative senescence. Biogerontology. 2002;3:195–206. doi: 10.1023/a:1016299812327. [DOI] [PubMed] [Google Scholar]
- Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, Fischer A, Durandy A, de Villartay JP, Revy P. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–99. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- Busuttil R, Bahar R, Vijg J. Genome dynamics and transcriptional deregulation in aging. Neuroscience. 2007a;145:1341–7. doi: 10.1016/j.neuroscience.2006.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busuttil RA, Garcia AM, Cabrera C, Rodriguez A, Suh Y, Kim WH, Huang TT, Vijg J. Organ-specific increase in mutation accumulation and apoptosis rate in CuZn-superoxide dismutase-deficient mice. Cancer Res. 2005;65:11271–5. doi: 10.1158/0008-5472.CAN-05-2980. [DOI] [PubMed] [Google Scholar]
- Busuttil RA, Garcia AM, Reddick RL, Dolle ME, Calder RB, Nelson JF, Vijg J. Intra-organ variation in age-related mutation accumulation in the mouse. PLoS ONE. 2007b;2:e876. doi: 10.1371/journal.pone.0000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busuttil RA, Rubio M, Dolle ME, Campisi J, Vijg J. Oxygen accelerates the accumulation of mutations during the senescence and immortalization of murine cells in culture. Aging Cell. 2003;2:287–94. doi: 10.1046/j.1474-9728.2003.00066.x. [DOI] [PubMed] [Google Scholar]
- Campisi J. Aging and cancer: the double-edged sword of replicative senescence. J Am Geriatr Soc. 1997;45:482–8. doi: 10.1111/j.1532-5415.1997.tb05175.x. [DOI] [PubMed] [Google Scholar]
- Campisi J. Cancer, aging and cellular senescence. In Vivo. 2000;14:183–8. [PubMed] [Google Scholar]
- Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–40. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- Cao L, Li W, Kim S, Brodie SG, Deng CX. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev. 2003;17:201–13. doi: 10.1101/gad.1050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Multani AS, Cabrera NG, Naylor ML, Laud P, Lombard D, Pathak S, Guarente L, DePinho RA. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat Genet. 2004;36:877–82. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- Chao C, Herr D, Chun J, Xu Y. Ser18 and 23 phosphorylation is required for p53-dependent apoptosis and tumor suppression. Embo J. 2006;25:2615–22. doi: 10.1038/sj.emboj.7601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ, Greider CW, DePinho RA. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–38. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- Choy JS, Kron SJ. NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol Cell Biol. 2002;22:8215–25. doi: 10.1128/MCB.22.23.8215-8225.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–33. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Collado M, Serrano M. The power and the promise of oncogene-induced senescence markers. Nat Rev Cancer. 2006;6:472–6. doi: 10.1038/nrc1884. [DOI] [PubMed] [Google Scholar]
- Crowley C, Curtis HJ. The Development Of Somatic Mutations In Mice With Age. Proc Natl Acad Sci U S A. 1963;49:626–8. doi: 10.1073/pnas.49.5.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Adda di Fagagna F, Hande MP, Tong WM, Roth D, Lansdorp PM, Wang ZQ, Jackson SP. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr Biol. 2001;11:1192–6. doi: 10.1016/s0960-9822(01)00328-1. [DOI] [PubMed] [Google Scholar]
- de Boer J, Andressoo JO, de Wit J, Huijmans J, Beems RB, van Steeg H, Weeda G, van der Horst GT, van Leeuwen W, Themmen AP, Meradji M, Hoeijmakers JH. Premature aging in mice deficient in DNA repair and transcription. Science. 2002;296:1276–9. doi: 10.1126/science.1070174. [DOI] [PubMed] [Google Scholar]
- de Boer J, de Wit J, van Steeg H, Berg RJ, Morreau H, Visser P, Lehmann AR, Duran M, Hoeijmakers JH, Weeda G. A mouse model for the basal transcription/DNA repair syndrome trichothiodystrophy. Mol Cell. 1998;1:981–90. doi: 10.1016/s1097-2765(00)80098-2. [DOI] [PubMed] [Google Scholar]
- de Boer J, Hoeijmakers JH. Nucleotide excision repair and human syndromes. Carcinogenesis. 2000;21:453–60. doi: 10.1093/carcin/21.3.453. [DOI] [PubMed] [Google Scholar]
- de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–40. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- Dempsey JL, Pfeiffer M, Morley AA. Effect of dietary restriction on in vivo somatic mutation in mice. Mutat Res. 1993;291:141–5. doi: 10.1016/0165-1161(93)90153-q. [DOI] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d'Adda di Fagagna F. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–42. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Dolle ME, Busuttil RA, Garcia AM, Wijnhoven S, van Drunen E, Niedernhofer LJ, van der Horst G, Hoeijmakers JH, van Steeg H, Vijg J. Increased genomic instability is not a prerequisite for shortened lifespan in DNA repair deficient mice. Mutat Res. 2006;596:22–35. doi: 10.1016/j.mrfmmm.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Dolle ME, Giese H, Hopkins CL, Martus HJ, Hausdorff JM, Vijg J. Rapid accumulation of genome rearrangements in liver but not in brain of old mice [see comments] Nat Genet. 1997;17:431–4. [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–21. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Doria G, Barattini P, Scarpaci S, Puel A, Guidi L, Frasca D. Role of immune responsiveness and DNA repair capacity genes in ageing. Ageing Res Rev. 2004;3:143–51. doi: 10.1016/j.arr.2003.04.001. [DOI] [PubMed] [Google Scholar]
- Du X, Shen J, Kugan N, Furth EE, Lombard DB, Cheung C, Pak S, Luo G, Pignolo RJ, DePinho RA, Guarente L, Johnson FB. Telomere shortening exposes functions for the mouse werner and bloom syndrome genes. Mol Cell Biol. 2004;24:8437–46. doi: 10.1128/MCB.24.19.8437-8446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumble M, Gatza C, Tyner S, Venkatachalam S, Donehower LA. Insights into aging obtained from p53 mutant mouse models. Ann N Y Acad Sci. 2004;1019:171–7. doi: 10.1196/annals.1297.027. [DOI] [PubMed] [Google Scholar]
- Espejel S, Franco S, Rodriguez-Perales S, Bouffler SD, Cigudosa JC, Blasco MA. Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres. Embo J. 2002;21:2207–19. doi: 10.1093/emboj/21.9.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejel S, Klatt P, Menissier-de Murcia J, Martin-Caballero J, Flores JM, Taccioli G, de Murcia G, Blasco MA. Impact of telomerase ablation on organismal viability, aging, and tumorigenesis in mice lacking the DNA repair proteins PARP-1, Ku86, or DNA-PKcs. J Cell Biol. 2004a;167:627–38. doi: 10.1083/jcb.200407178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejel S, Martin M, Klatt P, Martin-Caballero J, Flores JM, Blasco MA. Shorter telomeres, accelerated ageing and increased lymphoma in DNA-PKcs-deficient mice. EMBO Rep. 2004b;5:503–9. doi: 10.1038/sj.embor.7400127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson DO, Sekiguchi JM, Chang S, Frank KM, Gao Y, DePinho RA, Alt FW. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc Natl Acad Sci U S A. 2000;97:6630–3. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Nussenzweig A. Aging counts on chromosomes. Nat Genet. 2004;36:672–4. doi: 10.1038/ng0704-672. [DOI] [PubMed] [Google Scholar]
- Flores I, Benetti R, Blasco MA. Telomerase regulation and stem cell behaviour. Curr Opin Cell Biol. 2006;18:254–60. doi: 10.1016/j.ceb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Flores M, Morales L, Gonzaga-Jauregui C, Dominguez-Vidana R, Zepeda C, Yanez O, Gutierrez M, Lemus T, Valle D, Avila MC, Blanco D, Medina-Ruiz S, Meza K, Ayala E, Garcia D, Bustos P, Gonzalez V, Girard L, Tusie-Luna T, Davila G, Palacios R. Recurrent DNA inversion rearrangements in the human genome. Proc Natl Acad Sci U S A. 2007;104:6099–106. doi: 10.1073/pnas.0701631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasca D, Barattini P, Tirindelli D, Guidi L, Bartoloni C, Errani A, Costanzo M, Tricerri A, Pierelli L, Doria G. Effect of age on DNA binding of the ku protein in irradiated human peripheral blood mononuclear cells (PBMC) Exp Gerontol. 1999;34:645–58. doi: 10.1016/s0531-5565(99)00026-1. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA repair and mutagenesis. American Society of Microbiology; Washington, D. C.: 1995. [Google Scholar]
- Garcia-Cao I, Garcia-Cao M, Martin-Caballero J, Criado LM, Klatt P, Flores JM, Weill JC, Blasco MA, Serrano M. “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. Embo J. 2002;21:6225–35. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Seluanov A, Mao Z, Hine C. Changes in DNA repair during aging. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M, Imamura O, Kuromitsu J, Matsumoto T, Yamabe Y, Tokutake Y, Suzuki N, Mason B, Drayna D, Sugawara M, Sugimoto M, Furuichi Y. Analysis of helicase gene mutations in Japanese Werner's syndrome patients. Hum Genet. 1997;99:191–3. doi: 10.1007/s004390050336. [DOI] [PubMed] [Google Scholar]
- Grillari J, Katinger H, Voglauer R. Contributions of DNA interstrand crosslinks to aging of cells and organisms. Nucleic Acids Res. 2007;35:7566–76. doi: 10.1093/nar/gkm1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Seidl KJ, Rathbun GA, Zhu C, Manis JP, van der Stoep N, Davidson L, Cheng HL, Sekiguchi JM, Frank K, Stanhope-Baker P, Schlissel MS, Roth DB, Alt FW. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7:653–65. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- Hart RW, Setlow RB. Correlation between deoxyribonucleic acid excision-repair and life-span in a number of mammalian species. Proc Natl Acad Sci U S A. 1974;71:2169–73. doi: 10.1073/pnas.71.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey M, Sands AT, Weiss RS, Hegi ME, Wiseman RW, Pantazis P, Giovanella BC, Tainsky MA, Bradley A, Donehower LA. In vitro growth characteristics of embryo fibroblasts isolated from p53- deficient mice. Oncogene. 1993;8:2457–67. [PubMed] [Google Scholar]
- Hasty P, Campisi J, Hoeijmakers J, van Steeg H, Vijg J. Aging and genome maintenance: lessons from the mouse? Science. 2003;299:1355–9. doi: 10.1126/science.1079161. [DOI] [PubMed] [Google Scholar]
- Hayflick L. The limited in vitro lifetime of human diploid cell strains. Experimental Cell Research. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–74. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- Hofer AC, Tran RT, Aziz OZ, Wright W, Novelli G, Shay J, Lewis M. Shared phenotypes among segmental progeroid syndromes suggest underlying pathways of aging. J Gerontol A Biol Sci Med Sci. 2005;60:10–20. doi: 10.1093/gerona/60.1.10. [DOI] [PubMed] [Google Scholar]
- Holcomb VB, Vogel H, Hasty P. Deletion of Ku80 causes early aging independent of chronic inflammation and Rag-1-induced DSBs. Mech Ageing Dev. 2007 doi: 10.1016/j.mad.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcomb VB, Vogel H, Marple T, Kornegay RW, Hasty P. Ku80 and p53 suppress medulloblastoma that arise independent of Rag-1-induced DSBs. Oncogene. 2006;25:7159–65. doi: 10.1038/sj.onc.1209704. [DOI] [PubMed] [Google Scholar]
- Hsu HL, Gilley D, Blackburn EH, Chen DJ. Ku is associated with the telomere in mammals. Proc Natl Acad Sci U S A. 1999;96:12454–8. doi: 10.1073/pnas.96.22.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HL, Gilley D, Galande SA, Hande MP, Allen B, Kim SH, Li GC, Campisi J, Kohwi-Shigematsu T, Chen DJ. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev. 2000;14:2807–12. doi: 10.1101/gad.844000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3′-->5′ exonuclease [letter] Nat Genet. 1998;20:114–6. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–73. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, Cheng T, DePinho RA, Sharpless NE, Scadden DT. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–6. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- Jasin M. Chromosome breaks and genomic instability. Cancer Invest. 2000;18:78–86. doi: 10.3109/07357900009023065. [DOI] [PubMed] [Google Scholar]
- Jazayeri A, McAinsh AD, Jackson SP. Saccharomyces cerevisiae Sin3p facilitates DNA double-strand break repair. Proc Natl Acad Sci U S A. 2004;101:1644–9. doi: 10.1073/pnas.0304797101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones IM, Thomas CB, Tucker B, Thompson CL, Pleshanov P, Vorobtsova I, Moore DH., 2nd Impact of age and environment on somatic mutation at the hprt gene of T lymphocytes in humans. Mutat Res. 1995a;338:129–39. doi: 10.1016/0921-8734(95)00018-2. [DOI] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995b;378:206–8. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- Ju YJ, Lee KH, Park JE, Yi YS, Yun MY, Ham YH, Kim TJ, Choi HM, Han GJ, Lee JH, Lee J, Han JS, Lee KM, Park GH. Decreased expression of DNA repair proteins Ku70 and Mre11 is associated with aging and may contribute to the cellular senescence. Exp Mol Med. 2006;38:686–93. doi: 10.1038/emm.2006.81. [DOI] [PubMed] [Google Scholar]
- Kipling D, Faragher RG. Progeroid syndromes: probing the molecular basis of aging? Mol Pathol. 1997;50:234–41. doi: 10.1136/mp.50.5.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood TB. Evolution of ageing. Mech Ageing Dev. 2002;123:737–45. doi: 10.1016/s0047-6374(01)00419-5. [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- Kujoth GC, Leeuwenburgh C, Prolla TA. Mitochondrial DNA mutations and apoptosis in mammalian aging. Cancer Res. 2006;66:7386–9. doi: 10.1158/0008-5472.CAN-05-4670. [DOI] [PubMed] [Google Scholar]
- Li H, Vogel H, Holcomb VB, Gu Y, Hasty P. Deletion of Ku70, Ku80, or Both Causes Early Aging without Substantially Increased Cancer. Mol Cell Biol. 2007;27:8205–8214. doi: 10.1128/MCB.00785-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–20. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst) 2004;3:817–26. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Lim DS, Vogel H, Willerford DM, Sands AT, Platt KA, Hasty P. Analysis of ku80-mutant mice and cells with deficient levels of p53. Mol Cell Biol. 2000;20:3772–80. doi: 10.1128/mcb.20.11.3772-3780.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobrich M, Jeggo PA. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat Rev Cancer. 2007;7:861–9. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Lucas JN. Cytogenetic signature for ionizing radiation. Int J Radiat Biol. 1998;73:15–20. doi: 10.1080/095530098142662. [DOI] [PubMed] [Google Scholar]
- Luo G, Santoro IM, McDaniel LD, Nishijima I, Mills M, Youssoufian H, Vogel H, Schultz RA, Bradley A. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat Genet. 2000;26:424–9. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T, Sutherland A, Thorner M, Scrable H. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18:306–19. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GM, Smith AC, Ketterer DJ, Ogburn CE, Disteche CM. Increased chromosomal aberrations in first metaphases of cells isolated from the kidneys of aged mice. Isr J Med Sci. 1985;21:296–301. [PubMed] [Google Scholar]
- Matheu A, Maraver A, Klatt P, Flores I, Garcia-Cao I, Borras C, Flores JM, Vina J, Blasco MA, Serrano M. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–9. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- Meek DW. The p53 response to DNA damage. DNA Repair (Amst) 2004;3:1049–56. doi: 10.1016/j.dnarep.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Mendrysa SM, O'Leary KA, McElwee MK, Michalowski J, Eisenman RN, Powell DA, Perry ME. Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev. 2006;20:16–21. doi: 10.1101/gad.1378506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B, Boubakri H, Baharoglu Z, LeMasson M, Lestini R. Recombination proteins and rescue of arrested replication forks. DNA Repair (Amst) 2007;6:967–80. doi: 10.1016/j.dnarep.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Mohaghegh P, Hickson ID. DNA helicase deficiencies associated with cancer predisposition and premature ageing disorders. Hum Mol Genet. 2001;10:741–6. doi: 10.1093/hmg/10.7.741. [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–6. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- Moore L, Lu X, Ghebranious N, Tyner S, Donehower LA. Aging-associated truncated form of p53 interacts with wild-type p53 and alters p53 stability, localization, and activity. Mech Ageing Dev. 2007;128:717–30. doi: 10.1016/j.mad.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, Rodrigues NP, Crockford TL, Cabuy E, Vindigni A, Enver T, Bell JI, Slijepcevic P, Goodnow CC, Jeggo PA, Cornall RJ. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–90. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig MC, Li GC. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996;382:551–5. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- Odagiri Y, Uchida H, Hosokawa M, Takemoto K, Morley AA, Takeda T. Accelerated accumulation of somatic mutations in the senescence-accelerated mouse. Nat Genet. 1998;19:116–7. doi: 10.1038/468. [DOI] [PubMed] [Google Scholar]
- Otterlei M, Bruheim P, Ahn B, Bussen W, Karmakar P, Baynton K, Bohr VA. Werner syndrome protein participates in a complex with RAD51, RAD54, RAD54B and ATR in response to ICL-induced replication arrest. J Cell Sci. 2006;119:5137–46. doi: 10.1242/jcs.03291. [DOI] [PubMed] [Google Scholar]
- Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29:92–5. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol. 2003;5:741–7. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicci PG. Do tumor-suppressive mechanisms contribute to organism aging by inducing stem cell senescence? J Clin Invest. 2004;113:4–7. doi: 10.1172/JCI200420750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan SC, Swanson PC, Ma Y, Lieber MR. Double-strand break formation by the RAG complex at the bcl-2 major breakpoint region and at other non-B DNA structures in vitro. Mol Cell Biol. 2005;25:5904–19. doi: 10.1128/MCB.25.14.5904-5919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan SC, Swanson PC, Wu X, Hsieh CL, Lieber MR. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- Ramsey MJ, Moore DH, 2nd, Briner JF, Lee DA, Olsen L, Senft JR, Tucker JD. The effects of age and lifestyle factors on the accumulation of cytogenetic damage as measured by chromosome painting. Mutat Res. 1995;338:95–106. doi: 10.1016/0921-8734(95)00015-x. [DOI] [PubMed] [Google Scholar]
- Reed JC. Mechanisms of apoptosis avoidance in cancer. Curr Opin Oncol. 1999;11:68–75. doi: 10.1097/00001622-199901000-00014. [DOI] [PubMed] [Google Scholar]
- Ren K, de Ortiz SP. Non-homologous DNA end joining in the mature rat brain. J Neurochem. 2002;80:949–59. doi: 10.1046/j.0022-3042.2002.00776.x. [DOI] [PubMed] [Google Scholar]
- Rohaly G, Chemnitz J, Dehde S, Nunez AM, Heukeshoven J, Deppert W, Dornreiter I. A novel human p53 isoform is an essential element of the ATR-intra-S phase checkpoint. Cell. 2005;122:21–32. doi: 10.1016/j.cell.2005.04.032. [DOI] [PubMed] [Google Scholar]
- Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–9. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- Samper E, Goytisolo FA, Slijepcevic P, van Buul PP, Blasco MA. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 2000;1:244–52. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrable H, Sasaki T, Maier B. DeltaNp53 or p44: priming the p53 pump. Int J Biochem Cell Biol. 2005;37:913–9. doi: 10.1016/j.biocel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Sedelnikova OA, Horikawa I, Zimonjic DB, Popescu NC, Bonner WM, Barrett JC. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat Cell Biol. 2004;6:168–70. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- Seluanov A, Chen Z, Hine C, Sasahara TH, Ribeiro AA, Catania KC, Presgraves DC, Gorbunova V. Telomerase activity coevolves with body mass not lifespan. Aging Cell. 2007a;6:45–52. doi: 10.1111/j.1474-9726.2006.00262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluanov A, Danek J, Hause N, Gorbunova V. Changes in the level and distribution of Ku proteins during cellular senescence. DNA Repair (Amst) 2007b doi: 10.1016/j.dnarep.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluanov A, Mittelman D, Pereira-Smith OM, Wilson JH, Gorbunova V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc Natl Acad Sci U S A. 2004;101:7624–9. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DA. DNA end joining activity is reduced in Alzheimer's disease. Neurobiol Aging. 2006;27:596–605. doi: 10.1016/j.neurobiolaging.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. Cancer: crime and punishment. Nature. 2005;436:636–7. doi: 10.1038/436636a. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol. 2007;8:703–13. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- Strehler BL. Deletional mutations are the basic cause of aging: historical perspectives. Mutat Res. 1995;338:3–17. doi: 10.1016/0921-8734(95)00006-r. [DOI] [PubMed] [Google Scholar]
- Sugrue MM, Shin DY, Lee SW, Aaronson SA. Wild-type p53 triggers a rapid senescence program in human tumor cells lacking functional p53. Proc Natl Acad Sci U S A. 1997;94:9648–53. doi: 10.1073/pnas.94.18.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LQ, Lee DW, Zhang Q, Xiao W, Raabe EH, Meeker A, Miao D, Huso DL, Arceci RJ. Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes Dev. 2004;18:1035–46. doi: 10.1101/gad.1176104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Shinkura R, Shinkura N, Alt FW. Growth retardation, early death, and DNA repair defects in mice deficient for the nucleotide excision repair enzyme XPF. Mol Cell Biol. 2004;24:1200–5. doi: 10.1128/MCB.24.3.1200-1205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker JD, Spruill MD, Ramsey MJ, Director AD, Nath J. Frequency of spontaneous chromosome aberrations in mice: effects of age. Mutat Res. 1999;425:135–41. doi: 10.1016/s0027-5107(99)00036-6. [DOI] [PubMed] [Google Scholar]
- Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, Hee Park S, Thompson T, Karsenty G, Bradley A, Donehower LA. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- Um JH, Kim SJ, Kim DW, Ha MY, Jang JH, Kim DW, Chung BS, Kang CD, Kim SH. Tissue-specific changes of DNA repair protein Ku and mtHSP70 in aging rats and their retardation by caloric restriction. Mech Ageing Dev. 2003;124:967–75. doi: 10.1016/s0047-6374(03)00169-6. [DOI] [PubMed] [Google Scholar]
- Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- van de Ven M, Andressoo JO, Holcomb VB, von Lindern M, Jong WM, De Zeeuw CI, Suh Y, Hasty P, Hoeijmakers JH, van der Horst GT, Mitchell JR. Adaptive stress response in segmental progeria resembles long-lived dwarfism and calorie restriction in mice. PLoS Genet. 2006;2:e192. doi: 10.1371/journal.pgen.0020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijg J, Dolle ME. Large genome rearrangements as a primary cause of aging. Mech Ageing Dev. 2002;123:907–15. doi: 10.1016/s0047-6374(02)00028-3. [DOI] [PubMed] [Google Scholar]
- Vogel H, Lim DS, Karsenty G, Finegold M, Hasty P. Deletion of Ku86 causes early onset of senescence in mice. Proc Natl Acad Sci U S A. 1999;96:10770–5. doi: 10.1073/pnas.96.19.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zglinicki T, Saretzki G, Ladhoff J, d'Adda di Fagagna F, Jackson SP. Human cell senescence as a DNA damage response. Mech Ageing Dev. 2005;126:111–7. doi: 10.1016/j.mad.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Vyjayanti VN, Rao KS. DNA double strand break repair in brain: reduced NHEJ activity in aging rat neurons. Neurosci Lett. 2006;393:18–22. doi: 10.1016/j.neulet.2005.09.053. [DOI] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–14. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- Wang Q, Liu T, Fang Y, Xie S, Huang X, Mahmood R, Ramaswamy G, Sakamoto KM, Darzynkiewicz Z, Xu M, Dai W. BUBR1 deficiency results in abnormal megakaryopoiesis. Blood. 2004;103:1278–85. doi: 10.1182/blood-2003-06-2158. [DOI] [PubMed] [Google Scholar]
- Warner HR, Sierra F. Models of accelerated ageing can be informative about the molecular mechanisms of ageing and/or age-related pathology. Mech Ageing Dev. 2003;124:581–7. doi: 10.1016/s0047-6374(03)00008-3. [DOI] [PubMed] [Google Scholar]
- Weeda G, Donker I, de Wit J, Morreau H, Janssens R, Vissers CJ, Nigg A, van Steeg H, Bootsma D, Hoeijmakers JHJ. Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr Biol. 1997;7:427–39. doi: 10.1016/s0960-9822(06)00190-4. [DOI] [PubMed] [Google Scholar]
- West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–45. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- White PA. The sources and potential hazards of mutagens in complex environmental matrices-Part II. Mutat Res. 2007 doi: 10.1016/j.mrrev.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Wijnhoven SW, Beems RB, Roodbergen M, van den Berg J, Lohman PH, Diderich K, van der Horst GT, Vijg J, Hoeijmakers JH, van Steeg H. Accelerated aging pathology in ad libitum fed Xpd(TTD) mice is accompanied by features suggestive of caloric restriction. DNA Repair (Amst) 2005;4:1314–24. doi: 10.1016/j.dnarep.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Wong KK, Maser RS, Bachoo RM, Menon J, Carrasco DR, Gu Y, Alt FW, DePinho RA. Telomere dysfunction and Atm deficiency compromises organ homeostasis and accelerates ageing. Nature. 2003;421:643–8. doi: 10.1038/nature01385. [DOI] [PubMed] [Google Scholar]
- Wu PA, Loh CH, Hsieh LL, Liu TY, Chen CJ, Liou SH. Clastogenic effect for cigarette smoking but not areca quid chewing as measured by micronuclei in exfoliated buccal mucosal cells. Mutat Res. 2004;562:27–38. doi: 10.1016/j.mrgentox.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Wyrobek AJ, Schmid TE, Marchetti F. Relative susceptibilities of male germ cells to genetic defects induced by cancer chemotherapies. J Natl Cancer Inst Monogr. 2005:31–5. doi: 10.1093/jncimonographs/lgi001. [DOI] [PubMed] [Google Scholar]
- Xu X, Qiao W, Linke SP, Cao L, Li WM, Furth PA, Harris CC, Deng CX. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat Genet. 2001;28:266–71. doi: 10.1038/90108. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Atsuta M, Hamatani K. Restricted expression of recombination activating gene (RAG-1) in mouse lymphoid tissues. Cell Biochem Funct. 1992;10:71–7. doi: 10.1002/cbf.290100202. [DOI] [PubMed] [Google Scholar]
- Yaneva M, Li H, Marple T, Hasty P. Non-homologous end joining, but not homologous recombination, enables survival for cells exposed to a histone deacetylase inhibitor. Nucleic Acids Res. 2005;33:5320–30. doi: 10.1093/nar/gki821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Stephen CW, Luciani MG, Fahraeus R. p53 Stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat Cell Biol. 2002;4:462–7. doi: 10.1038/ncb801. [DOI] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner's syndrome gene [see comments] Science. 1996;272:258–62. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]