Abstract
Using a low-copy nuclear gene region (LEAFY second intron) we show multiple instances of allopolyploid speciation in Persicaria (Polygonaceae), which includes many important weeds. Fifteen species seem to be allopolyploids, which is higher than the number found in previous comparisons of chloroplast DNA and nuclear ribosomal internal transcribed spacer (nrITS) phylogenies. This underestimation of the extent of allopolyploidy is due in at least three cases to homogenization of nrITS toward the maternal lineage. One of the diploid species, P. lapathifolia, has been involved in at least six cases of allopolyploid speciation. Of the diploids, this species is the most widespread geographically and ecologically and also bears more numerous and conspicuous flowers, illustrating ecologic factors that may influence hybridization frequency. With a few exceptions, especially the narrowly endemic hexaploid, P. puritanorum, the allopolyploid species also are widespread, plastic, ecological generalists. Hybridization events fostered by human introductions may be fueling the production of new species that have the potential to become aggressive weeds.
Keywords: hybridization, LEAFY intron, phylogeny, Polygonum, invasive species
Polyploidy after hybridization (allopolyploidy) has long been known to play an important role in plant evolution (1–3). Immediate reproductive isolation from parental lineages through polyploidization ensures the ability to maintain a new genetic make-up (4, 5). Although estimates have varied widely on the frequency of polyploid events in angiosperm evolution (2, 6, 7), allopolyploid speciation seems to be fairly common in some plant groups through chromosome doubling after the hybridization of diploid parents, or through triploid bridging to produce new tetraploids (2, 8, 9).
Incongruence between gene trees from chloroplast DNA (cpDNA; inherited maternally) vs. nuclear DNA (inherited from both parents) has increased our ability to recognize hybridization in plants (10, 11). However, the often relatively low variation in cpDNA at the intraspecific level, and the susceptibility of the commonly used nuclear ribosomal internal transcribed spacer (nrITS) region to concerted evolution (12, 13), have limited the precision with which allopolyploidy can be identified. In contrast, the use of low-copy nuclear genes can provide more information when particular gene copies in allopolyploids can be linked with genes in the maternal and paternal lineages (14–18). Despite technical difficulties in identifying appropriate markers and in comparing proper orthologs, several useful nuclear genes have been tested in studies of allopolyploidy (16).
Persicaria, a clade of Polygonaceae containing approximately 120 species, is well known for its weedy species occupying disturbed areas and crop fields. Persicaria plants are highly variable in morphology (19–21). This has been attributed by some authors to hybridization (22–24), which is consistent with variation in chromosome numbers [supporting information (SI) Table S1]. However, Persicaria plants commonly self-fertilize, and some are even cleistogamous (25, 26). Furthermore, morphological studies have not provided compelling evidence of hybridization in Persicaria (27, 28). Instead, norm of reaction studies have shown individual genotypes in these taxa to be highly phenotypically plastic (29, 30).
Our molecular phylogenetic study of Persicaria, focusing on Eupersicaria (or Persicaria sect. Persicaria), suggested many cases of allopolyploid speciation on the basis of significant incongruence between cpDNA and nrITS trees (see ref. 31 and Fig. S1). Although this revealed substantial reticulation in Eupersicaria, such incongruence can only identify hybrids when nrITS has been homogenized to the paternal lineage; homogenization to the maternal parent will not yield incongruence with the cpDNA (13). Phylogenetic analyses, using a low-copy nuclear gene are therefore necessary to reveal the full extent of allopolyploidy in Eupersicaria.
In this study, we use LEAFY second intron (PL2int) to test our previous hypotheses of allopolyploid speciation in Eupersicaria and to identify additional cases. LEAFY is regarded as a single-copy gene in angiosperms (32) and has proven to be useful for phylogeny at the intraspecific level (33–35). On the basis of our analyses, we consider variation in the frequency of reticulation and the potential consequences of allopolyploid speciation for invasiveness.
Results
PL2int Sequences and Aligned Dataset.
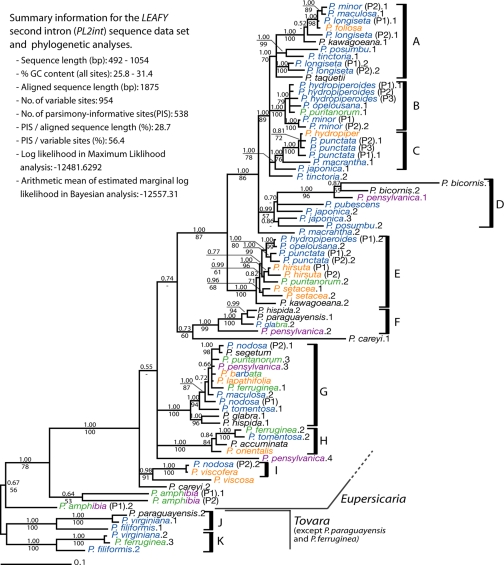
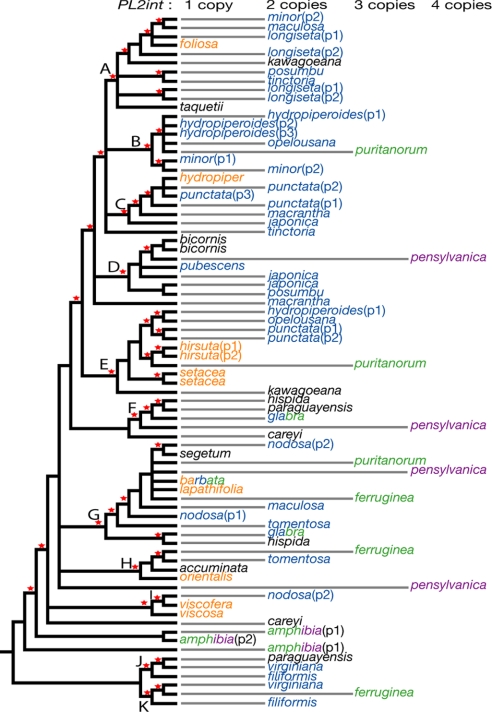
The length of PL2int ranged from 492 bp (P. filiformis.1) to 1054 bp (P. kawagoeana.2), and the GC content ranged from 25.8% (P. posumbu.1) to 31.4% (P. bicornis.1). Twenty-seven accessions showed multiple copies of PL2int (Fig. 1). In general the number of copies corresponds to ploidal level where chromosome numbers have been reported (Fig. 2; Table S1). However, not all surveyed populations of known polyploid species (e.g., P. amphibia, P. hydropiperoides, P. minor, P. nodosa, P. pubescens, and P. punctata) were found to have more than 1 copy, presumably reflecting a failure to amplify additional copies or a loss of redundant copies in the sampled individuals.
Fig. 1.
Fifty percent majority-rule consensus tree from Bayesian inference. Posterior probabilities from Bayesian analyses are above the branch, and bootstrap values from maximum likelihood are below. Colors of taxon names represent ploidy: orange = diploid, blue = tetraploid, green = hexaploid, purple = octaploid, black = chromosome number unknown. See Table S1 for voucher information. P#, population number; .1, .2, .3, .4, clone number.
Fig. 2.
Fifty percent majority-rule consensus tree from Bayesian inference showing the number of copies of PL2int. Posterior probabilities >0.95 are indicated by red stars. Color of taxon names represents ploidy: orange = diploid, blue = tetraploid, green = hexaploid, purple = octaploid, black = chromosome number unknown.
A summary of the aligned dataset is presented in Fig. 1. Various gaps were needed to align the 78-accession matrix, but none of these were coded for phylogenetic analysis, owing to overlaps and inconsistencies. We trimmed the initial 3,166-bp aligned matrix to a 1,875-bp matrix by removing gaps whose sizes were larger than 15 bp and where >90% of the accessions were aligned as gaps.
Phylogenetic Analyses.
A majority-rule consensus tree from our Bayesian analyses is presented in Fig. 1. The clades labeled A through K (Fig. 1) were strongly supported with high posterior probabilities and with moderate to high bootstrap values except for clade D (BT = 57). The monophyly of the clade including A, B, C, and P. tinctoria.2 is strongly supported, and in turn it seems to be closely related to clade D plus P. macrantha.2. A sister group relationship between the A–D clade and clade E is also strongly supported. However, relationships among the A–E clade and clades F, G, H, and I are only weakly supported. As in our previous analyses (36), PL2int analyses indicate that the clade including all Eupersicaria except P. amphibia is very strongly supported. As discussed below, different copies in several species of Eupersicaria clustered with species from the outgroup Tovara (P. virginiana, P. filiformis), indicating hybridization involving more distantly related plants (Fig. 1).
Multiple copies from the same accession were clearly separate in the tree with the exception of the four copies recovered from two populations of P. longiseta, which all nested in clade A. The placements of one or more of the copies from several species (e.g., P. tinctoria, P. macrantha, and P. careyi) are not well resolved. As discussed below (see Fig. 2), clades A, C, E, G, H, and I contain one or two diploid species each; species known to be diploids are not present in clades B, D, and F.
Comparison with Previous Analyses.
Although there are some differences in the placement of individual species, the strongly supported clades in our PL2int tree largely correspond to clades found in our previous cpDNA and nrITS analyses (see ref. 31 and Fig. S1). Clades corresponding to clade A in our PL2int tree were recovered in both our cpDNA and nrITS trees. Clades corresponding to B, C, F, and I were found only in our nrITS trees, whereas clades corresponding to E and G appeared only in our cpDNA trees. Clades D and H were not found in our analyses of cpDNA or nrITS sequences. However, movement of the P. bicornis/P. pensylvanica.1 clade from clade D to the vicinity of P. hirsuta and P. setacea would yield a clade supported by nrITS. This result was obtained in some of our analyses and is almost as likely.
Discussion
Allopolyploid Speciation in Eupersicaria.
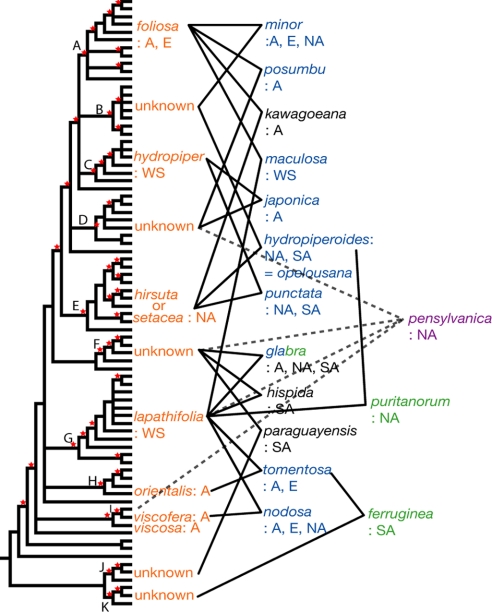
We find that the number of PL2int copies generally corresponds to ploidy level where this has been reported (Table S1). Multiple copies from polyploid species are separately placed within different strongly supported clades (Figs. 1 and 2), which supports the allopolyploid origin of these species after hybridization between possible diploid parents (known or unknown) in these clades. Diploid species, from which we obtained just one copy of PL2int, are rather evenly dispersed among the major clades. As noted, several clades lack known diploid species, either because diploid species are missing from our sample or because these have become extinct. Our results, therefore, suggest that each of the known diploid species, or in some cases “missing” diploids, served as parents for derived allopolyploid species that share PL2int copies with these diploids. For example, one PL2int copy from two accessions of the tetraploid P. punctata (P1 and P2) is most closely related to the copy present in the diploid P. hydropiper in clade C, whereas the other copy is nested in clade E close to the diploids P. hirsuta and P. setacea. This indicates that P. punctata is an allotetraploid species that originated from hybridization between P. hydropiper and probably P. hirsuta or P. setacea. This strongly supports our previous hypothesis on the origin of P. punctata, which was based solely on tree conflict (see ref. 31 and Fig. S1). In that analysis, P. punctata was linked with P. hydropiper on the basis of nrITS, whereas it was united with P. hirsuta and P. setacea on the basis of cpDNA. A battery of incongruence tests indicated that this conflict was not due to stochastic error (31). Our further survey of ITS polymorphism revealed a second copy of ITS in P. punctata that strongly clustered with P. hirsuta and P. setacea. Persicaria punctata shows morphologic intermediacy because it shares distinct glands on the tepals and relatively glabrous stems and leaves with P. hydropiper and relatively long inflorescences with P. hirsuta and P. setacea (31). These lines of evidence jointly support diploid P. hydropiper as the paternal lineage (pollen parent) and diploid P. hirsuta or P. setacea as the maternal lineage (seed parent) for the allotetraploid P. punctata (Fig. 3). Using similar reasoning, our PL2int analyses suggest a total of 15 cases of allopolyploid speciation, including 2 hexaploids and an octaploid (Fig. 3).
Fig. 3.
Hypothesized allopolyploid speciation events, with each polyploid connected to its suggested parental species by solid lines. Dotted lines indicate uncertainty in the case of the sole octaploid, P. pensylvanica. Color of taxon names represents ploidy: orange = diploid, blue = tetraploid, green; = hexaploid, purple = octaploid, black = chromosome number unknown. Abbreviation for geographic range: A, Asia; E, Europe; NA, North America; SA, South America; WS, widespread, present in all regions.
nrITS Underestimates Allopolyploidy.
One of the PL2int copies in a putative allopolyploid species is expected to be closely related to the copy in the maternal lineage, whereas the other copy should be closely related to the copy derived from the paternal lineage. The identity of the maternal contributor can be determined by reference to the cpDNA tree, because chloroplasts are maternally inherited in most angiosperms, including Polygonaceae (37). For nrITS there may be three different fates after a hybridization event, which are not mutually exclusive: (i) homogenization to the maternal or to the paternal type, (ii) maintenance of the two types, or (iii) formation of a chimeric mixture of ITS types (12, 13). Only when nrITS is homogenized to the paternal type is topological disagreement between cpDNA and ITS trees likely to be seen. If nrITS is homogenized to the maternal type then, in theory, conflict between cpDNA and nrITS trees will not be seen, and hybridization will be underestimated. The advantage in using a low-copy nuclear region is to avoid the susceptibility of nrITS to concerted evolution (16).
Our PL2int results reveal four tetraploid species to be allopolyploids that had shown no significant conflict between cpDNA and nrITS trees. In the case of P. maculosa, for example, the maternal lineage is traced to the diploid P. foliosa, and the paternal lineage seems to be P. lapathifolia of clade C (Fig. 3). This is consistent with previous work based on isozymes (24). The lack of conflict between cpDNA and nrITS in this case is presumably due to nrITS homogenization to the maternal parent. On the basis of similar arguments P. tomentosa, P. posumbu, and P. pubescens are also probably allopolyploids (Fig. 3). Persicaria tomentosa seems to be derived from diploid P. lapathifolia of clade G and P. orientalis of clade H. In P. posumbu the maternal lineage is traced to the diploid P. foliosa, and the paternal parent is an unknown diploid from clade D. From P. pubescens we recovered only one copy of PL2int (Figs. 1 and 2), despite its being reported as tetraploid (Table S1). It strongly clustered with P. hydropiper in both our cpDNA and nrITS trees. As above, this might be explained by concerted evolution of the nrITS toward the maternal lineage. The diploid paternal parent is unclear, however, because no diploids are yet known from clade D.
Nuclear ITS continues to be widely used (38), and we expect many more such cases of homogenization to be revealed through the use of low-copy nuclear markers. However, to date only a few cases have been reported from the mustard clade Cardamine (17) and from Paeonia (39).
Parentage of Hexaploid and Octaploid Species.
Our results also indicate that 2 hexaploids, P. puritanorum and P. ferruginea, are most likely to be allopolyploids derived in each case by hybridization between a tetraploid and diploid. Two PL2int copies in these hexaploids are placed in clades that include the presumptive diploid parents of the tetraploid species: P. hydropiperoides in the case of P. puritanorum and P. tomentosa in the case of P. ferruginea. The third copy is clustered with the candidate diploid parent: P. lapathifolia in the case of P. puritanorum and a missing diploid in clade K in the case of P. ferruginea (Fig. 3). In neither of these hexaploids was the third lineage detected in our previous cpDNA and nrITS analyses.
The parental lineage for the octaploid, P. pensylvanica, could not be determined precisely owing to the ambiguous placement of 1 copy (Figs. 1 and 2). However, the finding that 2 copies are nested in clades F and G suggests that P. glabra or P. hispida (whose chromosome numbers are not known or are ambiguous; Table S1) might be involved (Figs. 2 and 3).
Allopolyploidy vs. Autopolyploidy.
In contrast to the many suggested cases of allopolyploidy, we find little evidence for autopolyploidy in Persicaria. This finding is consistent with the long-standing view that allopolyploidy is prevalent in polyploid speciation (40, 41), although the role of autopolyploidy may have been underestimated in the past (42). Persicaria longiseta (P1 and P2) and P. bicornis may be autopolyploids, because their two PL2int copies are closely linked within clades A and D, respectively (Fig. 1). Several species (P. tinctoria, P. macrantha, and P. careyi) whose two PL2int copies are not confidently placed in our analyses, could emerge as either auto- or allopolyploids with extended sampling and the use of additional markers. Our finding of significant sequence variation within several species (e.g., the diploid P. setacea and the tetraploid P. japonica showed significant length variation) highlights the potential value of expanded sampling at the population level.
Geographic Location of Inferred Hybridization Events.
Because hybridization requires physical proximity, it is pertinent to assess each proposed event from a geographic perspective. This analysis is complicated, however, by the fact that a number of these species have been moved through human activity. For example, P. lapathifolia and P. hydropiper are of Eurasian origin but are now cosmopolitan in temperate regions (21, 43). Likewise, P. longiseta [= Polygonum caespitosum var. longisetum (Bruijn) Steward] presumably originated in Southeast Asia and was accidentally introduced to eastern North America in the early 1900s, spreading from there across the continent (44, 45).
Nevertheless, it is still possible to identify the likely location of several hybridization events. For example, the currently widespread, weedy allotetraploid P. maculosa seems to have originated from hybridization between Eurasian P. foliosa and the widespread P. lapathifolia. This hybridization most likely occurred in Asia or Europe, where both of these species are native, after which P. maculosa was transported to the New World, presumably as a crop seed contaminant (25). Similarly, a member of the diploid P. hirsuta/P. setacea lineage may have hybridized in North America with the widespread P. hydropiper to produce the allotetraploid P. punctata, which since then seems to have spread from North to South America (43, 45). The ecologically narrow hexaploid P. puritanorum evidently originated from hybridization between tetraploid P. hydropiperoides (native to North America) and diploid P. lapathifolia (native to Eurasia but naturalized in North America). Unlike the previous examples, this event was not followed by geographic spread; P. puritanorum is geographically restricted to a small area on Cape Cod, Massachusetts, and one site in Nova Scotia (31, 46).
Patterns in the Frequency of Allopolyploid Speciation.
One striking result is that the parents of allopolyploids do not seem to be randomly distributed across the phylogeny. With only two exceptions (involving the parentage of P. maculosa and P. puritanorum), hybridizations seem to have been successful between diploids in the well supported clade that includes clades A–E, or between diploids that fall outside of this clade (Fig. 3). Note that hybridization events outside of the A–E clade even seem to have involved unidentified species outside of Eupersicaria, related to P. virginiana and P. filiformis of the Tovara clade. The pattern of allopolyploidy mostly involving species of clades A–E or species outside of this clade shows no obvious relationship to the geographic ranges of species on either side of this phylogenetic divide, and may instead reflect a biochemical or genetic compatibility barrier, such as specific pollen germination or fertilization cues. Perhaps some change occurred in the origin of the A–E clade that reduced the likelihood of hybridization with species outside of this clade. There is no obvious morphological trait that marks the A–E clade, but the pattern suggests that there may be a genetic difference that largely prevents hybridization between members of the A–E clade and species from other Persicaria lineages. More extensive sampling is needed to confirm this pattern, and studies of the 2 exceptions, P. maculosa and P. puritanorum, may shed light on this issue.
A second noteworthy pattern is the involvement of P. lapathifolia in at least 6 cases of allopolyploid speciation in Eupersicaria. This may in part reflect the cosmopolitan geographic distribution of this species, which may simply have provided more chances for hybridization. However, the only other geographically widespread diploid, P. hydropiper, seems to have been involved only in two cases (Fig. 3). In addition to geographic range, other factors that may influence hybridization potential include number of flowers and their longevity; floral attractiveness to potential pollen vectors; and duration of flowering period. The frequency of successful hybridizations involving P. lapathifolia may partly result from its high flower production (dense floral fascicles containing approximately 8 flowers each), greater floral apparency due to both taller shoot systems and long, “nodding” inflorescences, and lengthy, indeterminate reproductive period, all of which would enhance opportunities for cross-pollination by generalist floral visitors. In contrast, the equally widespread and indeterminately flowering P. hydropiper produces upright, less conspicuous, and less densely arranged inflorescences with only 1 to 2 flowers per fascicle.
A species' ecologic breadth will also influence its likelihood of involvement in hybridization events. Within a given geographic area, gametes are more likely to move between individuals of species that occur in the same habitat. Ecological generalists that occupy diverse habitat types will cooccur with a greater number of potential hybrid partners, promoting an enhanced role for such taxa as hybrid parents. The pattern of hybrid parentage confirms this prediction: P. lapathifolia occurs in a broad range of environmental conditions and is thus ecologically and geographically widespread, whereas P. hydropiper is restricted to a single habitat type in both its native and introduced range (47).
Potential Implications of Allopolyploidy for Invasiveness.
If allopolyploids are characterized by broadly adaptive phenotypic plasticity, they are likely to be ecologic generalists and hence potentially invasive (48, 49). Hybridization creates new gene combinations and epistatic interactions that can expand the range of phenotypic expression, and consequently environmental tolerance, beyond that of parental taxa (41, 50–52). Polyploidy, too, can promote increasingly complex regulatory networks due to subfunctionalization and neofunctionalization of duplicated genes, and concerted epigenetic changes, leading to greater repertoires of plasticity (53). Although polyploidy per se is not always associated with broader ecologic amplitude (54), the joint effects of hybridization and genome doubling may in some cases produce highly plastic new taxa capable of rapid colonization across diverse habitats.
Comparative data on adaptive plasticity in Persicaria are consistent with the idea that genomes of alloploid origin can be extremely phenotypically plastic. Genotypes of the tetraploid P. maculosa (= Polygonum persicaria L.) express broadly adaptive norms of reaction for physiologic rates, tissue allocation, spatial root deployment, fitness components, and offspring traits, compared with individuals of its somewhat more ecologically restricted putative diploid parent P. lapathifolia (= Polygonum lapathifolium L.) and other close relatives (e.g., 55, 56). The native octaploid P. pensylvanica (= Polygonum pensylvanicum L.) is also a highly plastic, invasive ecological generalist. However, allopolyploidy does not always produce such adaptively plastic, generalist taxa: P. puritinorum, a hexaploid of hybrid origin, is restricted to a single habitat type and narrow geographic range (31). The effects of allopolyploidy on phenotypic expression and environmental tolerance seem to be genome dependent (54).
An important anthropogenic effect on plant evolution is that human-mediated introduction of nonnative species creates new hybridization opportunities by bringing previously separated taxa into contact (57). To the extent that allopolyploid genomes have particularly broad repertoires of environmental response, this points to a potentially disruptive evolutionary synergy in which species introductions lead to the generation of new taxa that are especially likely to be invasive. A well documented example of this scenario is the case of Spartina anglica in Great Britain, the highly invasive allopolyploid product of two diploid species, one native and one inadvertently introduced by shipping ballast (58). Our results indicate that P. punctata also may exemplify this synergy: this very common allopolyploid species is likely to have originated fairly recently in North America and subsequently spread to South America, whereas its putative diploid parents (P. hydropiper and P. hirsuta or P. setacea) have failed to spread. Comparative studies of plasticity and invasiveness in taxa with contrasting genetic architectures will further illuminate this potentially important aspect of allopolyploid speciation.
Materials and Methods
Taxon Sampling.
Information on the accessions used in this study is presented in Table S1. Forty-six accessions represent 37 species of Persicaria, 35 of which belong to Eupersicaria [= Persicaria sect. Persicaria: (36)]. Living samples were collected from fieldwork during 2002–2005 in North America, China, and South Korea. Twelve accessions were sampled from herbarium specimens in the Yale University Herbarium or borrowed from the Harvard University Herbaria and the University of New Hampshire Herbarium. Two species of the most closely related group, Tovara, were included for rooting purposes (36).
DNA Extraction, Amplification, and Sequencing.
Total genomic DNA was extracted from fresh or dried leaf samples, using a DNeasy Plant Mini Kit (Qiagen) with the addition of proteinase K (20 mg/liter per reaction) and 2-mercaptoethanol, especially for herbarium samples. To amplify the PL2int region 2 degenerate primers, LFsxl-2 and LFtxr (32), were initially used and more specific primers for Persicaria, PLFY-F3 (5′-CTT GAT TAC TTG TTC CAC C-3′) and PLFY-R7 (5′-CCY GCY TTC TTT GCR TAC-3′), were designed in conserved regions of the second and third exons. PCR was carried out by using a step-down annealing temperature of 3°C from 68°C to 47°C. All PCR products showing single bands were directly sequenced, but samples showing multiple bands or polymorphism in sequences were cloned by using a TOPO TA cloning kit (Invitrogen). At least 8 colonies were picked and sequenced to survey sequence variations in multiple copies.
Sequence Alignment and Phylogenetic Analyses.
Alignment of PL2int sequences was conducted by using CLUSTAL X (59) and MUSCLE (60), with manual adjustment; partial coding regions in exons 2 and 3 were maintained during alignment. Alignment required numerous gaps of varying size; single gaps larger than 15 bp were removed for phylogenetic analyses.
Bayesian inferences were conducted in MrBayes 3.12 (61), using the general time reversible model with Gamma distributed rate heterogeneity (GTR+G), as suggested by MODELTEST 3.06 (62). Parameters relating sequence evolution and likelihood probabilities were estimated by 5 × 106 generations, using Markov Chain Monte Carlo (MCMC). Trees were sampled every 100th generation and analyzed with TRACER v1.3 (http://evolve.zoo.ox.ac.uk/beast/) to determine the “burn-in.” A 50% majority rule consensus tree was calculated to generate a posterior probability for each node after removing 25% of the generations (12,500 sampled trees).
Maximum likelihood analyses were conducted with GARLI 0.95 (63) (http://www.bio.utexas.edu/faculty/antisense/garli/Garli.html), using the GTR+G+I model (I = proportion of invariable sites). Parameters were estimated by using a genetic algorithm with the default settings and automatic run termination after 10,000 generations without improvement of the topology. To assess node confidences nonparametric bootstrap analysis was conducted, based on 500 analyses using the same options in GARLI, and a 50% majority-rule consensus tree was obtained by using PAUP* 4.0b10 (64).
Supplementary Material
Acknowledgments.
We thank members of the Donoghue laboratory for valuable comments and discussion, and Chang-Le Ma, Ming Deng, and Min-Ha Kim for their help with fieldwork in China and Korea. We are indebted to Harvard University Herbaria, University of New Hampshire Herbarium, and Yale University Herbarium for critical specimens, and especially to Nur Ritter for information on South American species. Bruce Baldwin, Tao Sang, and Doug Soltis provided valuable reviews of the manuscript. This work was partially supported by a John F. Enders Fellowship from Yale University.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU196792–EU196869).
This article contains supporting information online at www.pnas.org/cgi/content/full/0805141105/DCSupplemental.
References
- 1.Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- 2.Grant V. Plant Speciation. New York: Columbia Univ Press; 1981. [Google Scholar]
- 3.Arnold ML. Natural Hybridization and Evolution. Oxford: Oxford Univ Press; 1997. [Google Scholar]
- 4.Rieseberg LH, Willis JH. Plant speciation. Science. 2007;317:910–914. doi: 10.1126/science.1137729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soltis DE, Soltis PS. Polyploidy: Recurrent formation and genome evolution. Trends Ecol Evol. 1999;14:348–352. doi: 10.1016/s0169-5347(99)01638-9. [DOI] [PubMed] [Google Scholar]
- 6.Otto SP, Whitton J. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- 7.Masterson J. Stomatal size in fossil plants: Evidence for polyploidy in majority angiosperms. Science. 1994;264:421–423. doi: 10.1126/science.264.5157.421. [DOI] [PubMed] [Google Scholar]
- 8.De Wet JMJ. In: Polyploidy: Biological Relevance. Lewis WH, editor. New York: Plenum Press; 1980. pp. 3–15. [Google Scholar]
- 9.Soltis DE, et al. Recent and recurrent polyploidy in Tragopogon (Asteraceae): Cytogenetic, genomic and genetic comparisons. Biol J Linn Soc. 2004;82:485–501. [Google Scholar]
- 10.Hughes CE, Bailey CD, Harris SA. Divergent and reticulate species relationships in Leucaena (Fabaceae) inferred from multiple data source: Insight into polyploid origins and nrDNA polymorphism. Am J Bot. 2002;89:1057–1073. doi: 10.3732/ajb.89.7.1057. [DOI] [PubMed] [Google Scholar]
- 11.Sang T, Crawford DJ, Stuessy TF. Documentation of reticulate evolution in Peonies (Paeonia) using internal transcribed spacer sequences of nuclear ribosomal DNA: Implications for biogeography and concerted evolution. Proc Natl Acad Sci USA. 1995;92:6813–6817. doi: 10.1073/pnas.92.15.6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wendel JF, Schnabel A, Seelanan T. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium) Proc Natl Acad Sci USA. 1995;92:280–284. doi: 10.1073/pnas.92.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Álvarez I, Wendel JF. Ribosomal ITS sequences and plant phylogenetic inference. Mol Biol Evol. 2003;29:417–434. doi: 10.1016/s1055-7903(03)00208-2. [DOI] [PubMed] [Google Scholar]
- 14.Lee JY, Mummenhoff K, Bowman JL. Allopolyploidization and evolution of species with reduced floral structures in Lepidium L. (Brassicaceae) Proc Natl Acad Sci USA. 2002;99:16835–16840. doi: 10.1073/pnas.242415399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sang T. Utility of low-copy nuclear gene sequences in plant phylogenetics. Crit Rev Biochem Mol. 2002;37:121–147. doi: 10.1080/10409230290771474. [DOI] [PubMed] [Google Scholar]
- 16.Small RL, Cronn RC, Wendel JF. Use of nuclear genes for phylogeny reconstruction in plants. Aust Syst Bot. 2004;17:145–170. [Google Scholar]
- 17.Lihova J, Shimizu KK, Marhold K. Allopolyploid origin of Cardamine asarifolia (Brassicaceae): Incongruence between plastid and nuclear ribosomal DNA sequences solved by a single-copy nuclear gene. Mol Phylogenet Evol. 2006;39:759–786. doi: 10.1016/j.ympev.2006.01.027. [DOI] [PubMed] [Google Scholar]
- 18.Brysting AK, et al. Untangling complex histories of genome mergings in high polyploids. Syst Biol. 2007;56:467–476. doi: 10.1080/10635150701424553. [DOI] [PubMed] [Google Scholar]
- 19.Fassett NC. The variation of Polygonum punctatum. Brittonia. 1949;6:369–393. [Google Scholar]
- 20.Greene EL. Leaflets of Botanical Observation and Criticism. Washington, DC: 1904. pp. 17–32. [Google Scholar]
- 21.Mitchell RS, Dean JK. Polygonaceae (Buckwheat Family) of New York State. New York State Mus Bull. 1978;431:1–79. [Google Scholar]
- 22.Stanford EE. Possibilities of hybridism as a cause of variation in Polygonum. Rhodora. 1925;27:81–89. [Google Scholar]
- 23.McDonald CB. A biosystematic study of the Polygonum hydropiperoides (Polygonaceae) complex. Am J Bot. 1980;67:664–670. [Google Scholar]
- 24.Consaul LL, Warwick SI, McNeill J. Allozyme variation in the Polygonum lapathifolium complex. Can J Botany. 1991;69:2261–2270. [Google Scholar]
- 25.Simmonds NW. Polygonum persicaria L. J Ecol. 1945;33:121–131. [Google Scholar]
- 26.Stanford EE. The inflorescence and flower-form in Polygonum, subgenus Persicaria. Rhodora. 1925;27:41–47. [Google Scholar]
- 27.Dalci M. The taxonomy of the section Persicaria (Tourn.) L. in the genus Polygonum (Tourn.) L. (Polygonaceae) in the United States east of the Rocky Mountains. Communications de la Facultae des sciences de L'Universitae d'Ankara. 1974;18:133–153. [Google Scholar]
- 28.Timson J. A study of hybridization in Polygonum section Persicaria. J Linn Soc Bot. 1964;59:155–160. [Google Scholar]
- 29.Sultan SE. Phenotypic plasticity in plants: A case study in ecological development. Evol Dev. 2003;5:25–33. doi: 10.1046/j.1525-142x.2003.03005.x. [DOI] [PubMed] [Google Scholar]
- 30.Sultan SE, Bazzaz FA. Phenotypic plasticity in Polygonum persicaria. III. The evolution of ecological breadth for nutrient environment. Evolution. 1993;47:1050–1071. doi: 10.1111/j.1558-5646.1993.tb02134.x. [DOI] [PubMed] [Google Scholar]
- 31.Kim S-T, Donoghue MJ. Incongruence between cpDNA and nrITS trees indicates extensive hybridization within Eupersicaria (Polygonaceae) Am J Bot. doi: 10.3732/ajb.0700008. in press. [DOI] [PubMed] [Google Scholar]
- 32.Frohlich MW, Meyerowitz EM. The search for flower homeotic gene homologs in basal angiosperms and Gnetales: A potential new source of data on the evolutionary origin of flowers. Int J Plant Sci. 1997;158:S131–S142. [Google Scholar]
- 33.Oh S-H, Potter D. Phylogenetic utility of the second intron of LEAFY in Neilla and Stephanandra (Rosaceae) and implications for the origin of Stephanandra. Mol Phylogenet Evol. 2003;29:203–215. doi: 10.1016/s1055-7903(03)00093-9. [DOI] [PubMed] [Google Scholar]
- 34.Grob GBJ, Gravendeel B, Eurlings MCM. Potential phylogenetic utility of the nuclear FLORICAULA/LEAFY second intron: Comparison with three chloroplast DNA regions in Amorphophallus (Araceae) Mol Phylogenet Evol. 2004;30:13–23. doi: 10.1016/s1055-7903(03)00183-0. [DOI] [PubMed] [Google Scholar]
- 35.Nishimoto Y, Ohnishi O, Hasegawa M. Topological incongruence between nuclear and chloroplast DNA trees suggesting hybridization in the urophyllum group of the genus Fagopyrum (Polygonaceae) Genes Genet Syst. 2003;78:139–153. doi: 10.1266/ggs.78.139. [DOI] [PubMed] [Google Scholar]
- 36.Kim S-T, Donoghue MJ. Molecular phylogeny of Persicaria (Polygonaceae) Syst Bot. 2008;33:77–86. [Google Scholar]
- 37.Hollingsworth ML, Bailey JP, Hollingsworth PM, Ferris C. Chloroplast DNA variation and hybridization between invasive populations of Japanese knotweed and giant knotweed (Fallopia, Polygonaceae) Bot J Linn Soc. 1999;129:139–154. [Google Scholar]
- 38.Feliner G, Rosselló J. Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Mol Phylogenet Evol. 2007;44:911–919. doi: 10.1016/j.ympev.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Sang T, Zhang D. Reconstructing hybrid speciation using sequences of low copy nuclear genes: Hybrid origins of five Paeonia species based on Adh gene phylogenies. Syst Bot. 1999;24:148–163. [Google Scholar]
- 40.Stebbins GL., Jr . Variation and Evolution in Plants. New York: Columbia Univ Press; 1950. [Google Scholar]
- 41.Soltis DE, Soltis PS, Tate JA. Advances in the study of polyploidy since plant speciation. New Phytol. 2004;161:173–191. [Google Scholar]
- 42.Soltis DE, et al. Autopolyploidy in angiosperms: Have we grossly underestimated the number of species? Taxon. 2007;56:13–30. [Google Scholar]
- 43.Gleason HA, Cronquist A. Manual of Vascular Plants of Northeastern United States and Adjacent Canada. Bronx, NY: New York Botanical Garden; 1991. [Google Scholar]
- 44.Paterson AK. Range expansion of Polygonum caespitosum var. longisetum in the United States. Bartonia. 2000;60:57–69. [Google Scholar]
- 45.Cialdella AM. Revision de las especies Argentinas de Polygonum s.l. (Polygonaceae) Darwiniana. 1989;29:179–246. [Google Scholar]
- 46.Fernald ML. A new Polygonum from southeastern Massachusetts. Rhodora. 1919;21:140–142. [Google Scholar]
- 47.Sultan SE, Wilczek AM, Hann SD, Brosi BJ. Contrasting ecological breadth of cooccuring annual Polygonum species. J Ecol. 1998;86:363–383. [Google Scholar]
- 48.Sultan SE. Promising directions in plant phenotypic plasticity. Perspect Plant Ecol. 2004;6:227–233. [Google Scholar]
- 49.Richards CL, et al. Jack of all trades, master of some? On the role of phenotypic plasticity in plant invasions. Ecol Lett. 2006;9:981–993. doi: 10.1111/j.1461-0248.2006.00950.x. [DOI] [PubMed] [Google Scholar]
- 50.Ferguson DM, Sang T. Speciation through homoploid hybridization between allotetraploids in peonies (Paeonia) Proc Natl Acad Sci USA. 2001;98:3915–3919. doi: 10.1073/pnas.061288698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rieseberg LH, et al. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;301:1211–1216. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- 52.Wares JP, Hughes AR, Grosberg RK. In: Species Invasions. Sax DF, Staachowicz JJ, Gaines SD, editors. Sunderland, MA: Sinauer; 2005. pp. 229–257. [Google Scholar]
- 53.Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bretagnolle F, Thompson JD. Phenotypic plasticity in sympatric diploid and autotetraploid Dactylis glomerata. Int J Plant Sci. 2001;162:309–316. [Google Scholar]
- 55.Sultan SE. Phenotypic plasticity for fitness components in Polygonum species of contrasting ecological breadth. Ecology. 2001;82:328–343. [Google Scholar]
- 56.Griffith T, Sultan SE. Shade tolerance plasticity in response to neutral vs. green shade cues in Polygonum species of contrasting ecological breadth. New Phytol. 2005;166:141–148. doi: 10.1111/j.1469-8137.2004.01277.x. [DOI] [PubMed] [Google Scholar]
- 57.Vellend M, et al. Effects of exotic species on evolutionary diversification. Trends Ecol Evol. 2007;22:481–488. doi: 10.1016/j.tree.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 58.Raybould AF, et al. The evolution of Spartina anglica C.E. Hubbard (Gramineae): The origin and genetic variability. Biol J Linn Soc. 1991;43:111–126. [Google Scholar]
- 59.Thompson JD, et al. The clustal X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 62.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 63.Zwickl DJ. Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets Under the Maximum Likelihood Criterion. Austin: The University of Texas; 2006. [Google Scholar]
- 64.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and other Methods) 4th ed. Sunderland, MA: Sinauer; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.