Abstract
The serine-threonine kinase Akt regulates mesangial cell apoptosis, proliferation, and hypertrophy. To define Akt signaling pathways in mesangial cells, we performed a functional proteomic screen for rat mesangial cell proteins phosphorylated by Akt. A group of chaperone proteins, heat shock protein (Hsp) 70, Hsp90α, Hsp90β, Glucose-regulated protein (Grp) Grp78, Grp94, and protein disulfide isomerase (PDI) were identified as potential Akt substrates by two techniques: (a) in vitro phosphorylation of mesangial cell lysate by recombinant active Akt followed by protein separation by SDS-PAGE or 2-DE and phosphoprotein identification by peptide mass fingerprinting using MALDI-MS, or (b) immunoblot analysis of proteins from PDGF-stimulated mesangial cells using an anti-Akt phospho-motif antibody. In vitro kinase reactions using recombinant proteins confirmed that Akt phosphorylates Hsp70, Hsp90α and β, Grp94, and PDI. Immunoprecipitation of Akt from mesangial cell lysate co-precipitated Grp78 and Hsp70. PDGF stimulation of mesangial cells caused an acidic shift in the isoelectric point of Hsp70, Hsp90, and PDI that was dependent on PI-3K activity for Hsp70 and Hsp90. The data suggest that Akt-mediated phosphorylation of stress-induced chaperones represents a mechanism for regulation of chaperone function during mesangial cell responses to physiologic and pathologic stimuli.
Keywords: 2D gel electrophoresis, kinase, phosphorylation, mass spectrometry
INTRODUCTION
Mesangial cells contribute to glomerular injury through their ability to undergo proliferation, hypertrophy, migration, contraction, and apoptosis, and their ability to synthesize extracellular matrix proteins. A number of studies support a role for the serine/threonine kinase, Akt, in these mesangial cell responses. Akt mediates mesangial cell proliferation in response to platelet-derived growth factor (PDGF), arginine vasopressin (AVP), and CXCR3 chemokine receptor activation.1-3 Angiotensin II (ATII)-induced Akt activation participates in mesangial cell hypertrophy.4,5 Akt also mediates insulin, Insulin-like growth factor-1 (IGF-1), and PDGF-induced mesangial cell survival in response to apoptotic stimuli,6-8 and Akt augments Smad 3-stimulated collagen-I expression in response to Transforming Growth Factor-β.9 Culture of mesangial cells in high glucose leads to increased Akt activity, contributing to production of reactive oxygen species and NF-κB activation.10,11
Despite the evidence that Akt plays an important role in mesangial cell responses to glomerular injury, the molecular mechanisms underlying this role remain to be defined. A number of Akt targets have been identified, however, the targets that regulate specific mesangial cell responses are unknown. Targets of Akt identified previously include the forkhead family of transcription factors (FOXO)12, 13 and cyclic AMP responsive element binding protein (CREB),14 which regulate gene transcription. Akt controls protein synthesis through phosphorylation of inhibitors of translation initiation factors such as glycogen synthase kinase-3 (GSK-3).15 Akt promotes cell cycle progression and proliferation through phosphorylation of cyclin kinase inhibitors p2116 and p2717, in addition to mediating transcriptional repression of p27.1 Other targets of Akt include eNOS, caspase 9, Bad, 14−3−3 proteins, and p47phox.8,19-22 Of these targets only Bad and FOXO were reported to be phosphorylated by Akt in mesangial cells.7,13 Recent work by Lewis et al using a proteomic approach to define targets of ERK phosphorylation suggested that a majority of substrates for specific kinases remain to be identified.23 Their study determined that 20 of the 25 ERK substrates found by isoelectric point shift on two dimensional gel electrophoresis (2-DE) gels were previously unknown. Thus, a more complete definition of Akt targets is necessary to understand the molecular basis for Akt participation in glomerular disease.
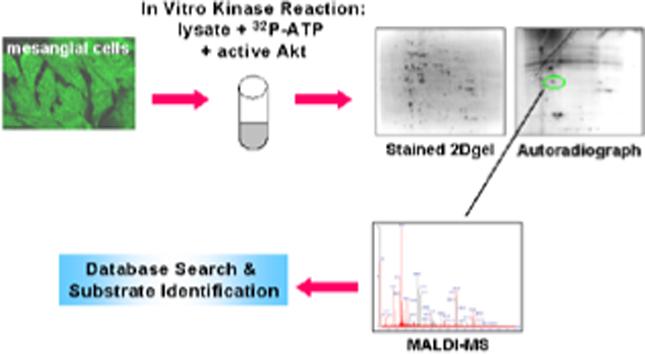
To more completely define Akt signaling pathways in mesangial cells, the present study used two functional proteomic approaches to screen rat mesangial cells. The first consisted of in vitro phosphorylation of mesangial cell lysate by recombinant active Akt followed by electrophoretic protein separation and phosphoprotein identification by mass spectrometry. The second approach involved stimulation of mesangial cells with PDGF, followed by immunoblot analysis of proteins separated by 2-DE using an anti-Akt phospho-motif antibody. A group of chaperone proteins, Heat shock protein 70 (Hsp70), Hsp90, Glucose-regulated protein 78 (Grp78), Grp94, and protein disulfide isomerase (PDI), were identified as Akt substrates. In vitro and in vivo confirmation of these targets suggested that Akt-mediated phosphorylation of molecular chaperones may represent a novel mechanism by which Akt regulates mesangial cell responses to inflammatory and stress stimuli.
METHODS
Culture of Mesangial Cells
Rat mesangial cells immortalized with pSV3-neo were from American type culture collection (ATCC; Manassas, VA). Cells were grown in Dulbecco's Modified Eagle Medium (DMEM) with 15% Fetal Bovine Serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin. Media was replaced with DMEM containing 0.5% FBS 24 hr prior to stimulation with PDGF or cell lysis and protein extraction.
Mesangial Lysate Preparation for Identification of Akt Substrates
To extract proteins, cells were washed with phosphate-buffered saline (PBS) and lysed by a 1 hr incubation in buffer containing 7 M urea, 2 M thiourea, 65 mM CHAPS, 58 mM dithiothreitol (DTT), and 4.5% ampholytes (pH 3−10). Supernatant was collected by centrifugation at 15,000 × g for 15 min. The high urea concentration of the buffer denatured endogenous kinases, but also inactivated exogenously added kinase used for the in vitro kinase reaction.24 Therefore size exclusion chromatography was used to sequentially exchange urea-based lysate into kinase buffer (25 mM HEPES, 25 mM β-glycerophosphate, 10 mM MgCl2, 10 mM MnCl2, 2 mM DTT, 0.1 mM NaVO3, and 65 mM CHAPS) containing a final concentration of 2 M urea.24 Preliminary studies demonstrated that reduction of mesangial lysate urea to 2 M allowed exogenous recombinant active Akt to remain active, while endogenous kinases remained in a denatured state and activity was significantly diminished (data not shown).
Akt Substrate Identification
Kinase reactions with 100 μCi [γ-32P]ATP, 1 μg active recombinant Akt (Upstate, Lake Placid, NY), and 400 μg mesangial cell protein in 2 M urea kinase buffer were carried out at 30°C for 2 hours. 2-DE was performed as previously described.25 Gels of proteins separated by 2-DE were stained with Sypro-Ruby (Bio-Rad) protein stain for 18 hr, while proteins separated by SDS-PAGE alone were stained with coomassie blue. Images of Sypro Ruby-stained gels were acquired using a high resolution 12-bit camera with an ultraviolet light box system (Genomic Solutions), and phosphorylation of all gels was visualized by autoradiography.
For identification of Akt substrates in intact mesangial cells, cells were incubated with or without 10 ng/ml PDGF-BB (Sigma, St. Louis, MO). Cells were lysed in urea-based buffer as described above, and each lysate subject to 2-DE or 10% SDS-PAGE on duplicate gels. For 2-DE immobilized pH gradient (IPG) strips pH 3−10 (Invitrogen, San Diego, CA) were rehydrated overnight with protein samples prepared in rehydration buffer, proteins separated by Isoelectric focusing (IEF) using the ZOOM IPG Runner (Invitrogen) with a maximal voltage of 2000V and 50 μA per gel. Following IEF, IPG strips were applied to 4−12% Bis-Tris gradient gels (Invitrogen) to separate proteins. Following electrophoresis, one gel was stained with colloidal coomassie blue (Genomic solutions), while proteins from the other gel were transferred electrophoretically to nitrocellulose. Membranes were blocked with 5% non-fat milk in Tris-buffered saline supplemented with 0.05% Tween 20 (TTBS), then incubated with anti-phospho-Akt substrate antibody (Cell Signaling, Beverly MA) at a dilution of 1:1000 in 5% BSA in TTBS overnight at 4°C. Immunoblots were developed using horseradish peroxidase-conjugated anti-rabbit IgG, followed by detection with enhanced chemiluminescence.
Candidate phosphoproteins were identified by comparison of stained gels to corresponding autoradiographs or immunoblots.
Trypsin Digestion and Mass Spectrometry
Stained protein spots corresponding to phosphorylated proteins on autoradiographs or immunoblots were excised from gels, subjected to in-gel trypsin digestion as described in detail by Thongboonkerd et al.26
Trypsin-generated peptides were applied by a thin film spotting procedure for matrix-assisted laser desorption and ionization mass spectrometry (MALDI-MS) analysis using α-cyanohydroxycinnamic acid as the matrix on stainless steel targets, as previously described.26 Mass spectral data were obtained using a TOF-Spec 2E (Micromass, Milford, MA) and a 337-nm N2 laser at 20−35% power in the reflector mode. Spectral data were obtained by averaging 10 spectra, each of which was the composite of 10 laser firings. Mass axis calibrations were accomplished using peaks from trypsin autolysis. Peptide masses obtained by MALDI-MS were analyzed by peptide mass fingerprinting using the mascot search engine (www.matrixscience.com) by comparing to theoretical masses in the NCBInr protein database. The search parameters were Rattus for taxonomy and no missed cleavages for trypsin were allowed. Carbamidomethylation was chosen as a fixed modification and methionine oxidation as a variable modification. A maximal tolerance of 150 ppm was allowed for monoisotopic masses. A probability-based MOWSE (MOlecular Weight SEarch) score greater than 58 indicated a significant match that was not a random effect.
In Vitro Kinase Reaction with Recombinant Proteins
Recombinant active Akt (400 ng), 1 μg recombinant substrate protein (Hsp70, Hsp90α, PDI, Grp94 and Grp78 (Stressgen, Victoria, B.C., Canada); Hsp90β (Medical and Biological Laboratories, Woburn, MA)) and 10 μCi [γ-32P]ATP in 30 μl of kinase buffer (20 mM HEPES, 10 mM MgCl2, 10 mM MnCl2) were incubated for 45 minutes at 25°C.21,27
Immunoprecipitation
Untreated or PDGF-stimulated cells were lysed in buffer containing 1% Triton X-100, 1% NP40, 10% glycerol, 137 mM NaCl, 20 mM Tris HCl, 1 μg/μl Aprotinin, 1 μg/μl Leupeptin, 4 mM PMSF, 20 mM NaF, and 1 mM Na3VO4. Lysate was cleared by centrifugation at 15,000 × g for 15 min at 4°C, and the supernatant collected. Mesangial lysate (100 μg) was incubated overnight at 4°C with anti-Akt PH domain agarose-conjugated monoclonal antibodies (Upstate) or normal mouse IgG. The latter were incubated with 10 μl of 50% slurry protein-A agarose beads for 1 hr at 4°C. Beads were washed three times with lysis buffer and proteins eluted by addition of 2× laemmli buffer and boiling for 5 minutes.
Ex Vivo Phosphorylation
Confluent mesangial cells were treated with the PI-3K inhibitor LY294002 (25 μM; Calbiochem, La Jolla, CA) or vehicle (dimethyl sulfoxide) for 1 hr, prior to stimulation with PDGF. Lysates were prepared by addition of urea/thiourea buffer as described above. Separate aliquots of lysate were exchanged to kinase buffer containing 2 M urea, as described above, and these aliquots were incubated with 0.5 Units protein phosphatase 2-A (PP2A; Upstate) for 1.5 hr at 30°C. Proteins were separated by 2-DE using IPG strips of pH 4−7 or 4.5−5.5, and specific proteins were identified by immunoblot analysis.
Microsomal Isolation
Mesangial cell fractionation for isolation of microsomes was performed as described by Rao et al.28
RESULTS
Identification of Akt Substrates
Rat mesangial cell lysate was screened for Akt substrates by two methods. The first approach was by incubation of recombinant active Akt and [γ-32P]ATP with proteins extracted from mesangial cells with an urea/thiourea-based buffer. Previous studies validated the ability to identify kinase substrates using this approach.25,29 Proteins were separated by 2-DE or SDS-PAGE, and potential Akt substrates were identified by comparing autoradiographs to Sypro Ruby or coomassie-stained gels. Proteins were identified by peptide mass fingerprinting using MALDI-MS using the Mascot search engine. Incubation of mesangial lysate in 2 M urea kinase buffer with [γ-P32]ATP alone resulted in minimal phosphorylation of mesangial proteins separated by SDS-PAGE (figure 1), indicating that activity of endogenous kinases was significantly diminished. Initial experiments were conducted to determine the concentration of urea in which endogenous kinases would remain denatured and inactive. The urea concentration of mesangial lysate prepared in 7M urea was sequentially decreased in 1M increments by size exchange chromatography and endogenous kinase activity monitored by incubation of lysate with [γ-P32] ATP for 1 hr, separation of proteins by SDS-PAGE, and visualization of phosphorylated proteins by autoradiography. We found that a final concentration of 2M urea was sufficient to maintain endogenous kinases in a denatured, inactive state, indicated by minimal phosphorylation of cellular proteins, yet allow exogenously added active recombinant Akt to remain active. A total of 20 phosphoproteins were demonstrated by autoradiography after SDS-PAGE separation of mesangial lysate incubated with active recombinant Akt. Nine proteins were phosphorylated only in the presence of active recombinant Akt, while an additional 11 exhibited increased phosphorylation with the addition of Akt. Nine of these 20 phosphorylated proteins could be matched to a protein band on the coomassie blue-stained gel by aligning images of stained gels and autoradiographs by location of molecular weight markers in Microsoft PowerPoint. Six candidate Akt substrates were identified, including Hsp86 and 84 (murine homologs of Hsp90α and β, respectively), enolase, lamin A, annexin I, and a previously identified substrate of Akt, Hsp27.27
Figure 1. Phosphorylation of Mesangial Lysate by Recombinant Active Akt.
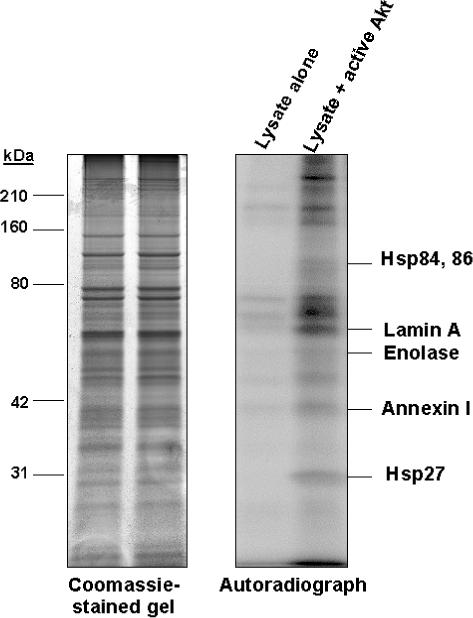
Candidate Akt substrates were identified by phosphorylation of mesangial lysate with recombinant active Akt in the presence of [γ-P32] ATP. Proteins were separated by SDS-PAGE and phosphoproteins detected by autoradiography. Minimal endogenous kinase activity was observed in mesangial lysate incubated in the presence of [γ-P32] ATP without exogenous kinase, while incubation of lysate with active recombinant Akt resulted in phosphorylation of twenty proteins. Phosphoproteins were identified by comparing autoradiographs with corresponding coomassie-stained gels. Five candidate Akt substrates identified by peptide mass fingerprinting of trypsin-digested phosphoproteins using MALDI-MS are labeled on the autoradiograph.
As less than 50% of phosphorylation events corresponded to a protein band and only 25% of phosphoproteins were identified, proteins from an in vitro kinase reaction were also resolved by 2-DE. Figure 2 shows that an additional 5 proteins, Grp78, actin, vimentin, tropomyosin, and the β-subunit of the mitochondrial F1 ATP synthase, a previously identified Akt target,30 were identified as candidate Akt substrates from 2D gels. Only 15 of nearly 60 proteins phosphorylated in the presence of Akt and observed on autoradiographs corresponded to visible protein spots on Sypro-Ruby-stained 2D gels. In addition, inadequate mass spectra were obtained from 10 of the 15 proteins that were excised and subjected to in-gel trypsin digestion. This may be due to incomplete digestion of proteins with trypsin, poor extraction of trypsin-generated peptides from gel pieces, or phosphorylation of proteins of low biological abundance.
Figure 2. Phosphoryaltion of Mesangial Lysate by Recombinant Active Akt.
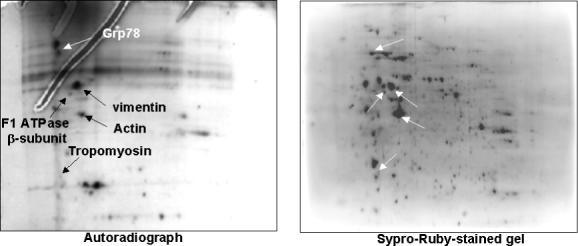
Candidate Akt substrates were identified by phosphorylation of mesangial lysate with recombinant active Akt in the presence of [γ-P32]ATP. Proteins were separated by 2-DE, and phosphoproteins were detected by autoradiography. Phosphoproteins were identified by comparing autoradiographs with corresponding Sypro-Ruby-stained gels. Five candidate Akt substrates identified by peptide mass fingerprinting of trypsin-digested phosphoproteins using MALDI-MS are indicated by arrows.
Another approach identified Akt substrates from intact mesangial cells by stimulating cells with 10 ng/ml PDGF and screening lysate by immunoblot analysis using phospho-Akt substrate antibody. This method was previously utilized to identify ATP citrate lyase and the mitochondrial F1 ATP synthase β-subunit as Akt substrates.31,30 Seven of the 20 proteins stained by anti-phospho-Akt substrate antibody following 10% SDS-PAGE protein separation were identified, including Grp78, Grp94, 14−3−3ε, and filamin (Figure 3A). Approximately 40 candidate Akt substrates were detected on immunoblots following 2-DE protein separation of which 5 were identified, including Hsp70, vimentin, tubulin, actin, and PDI (Figure 3B). Thus, a total of 20 candidate Akt substrates were identified by these proteomic approaches, of which 18 were not previously shown to be targets of Akt. These 20 proteins represent about 20% of the phosphorylation events detected. Table 1 lists the 20 candidate Akt targets, the methods by which they were identified, and categorizes them based on known cellular functions.
Figure 3. Identification of Candidate Akt Substrates in Intact Mesangial Cells.
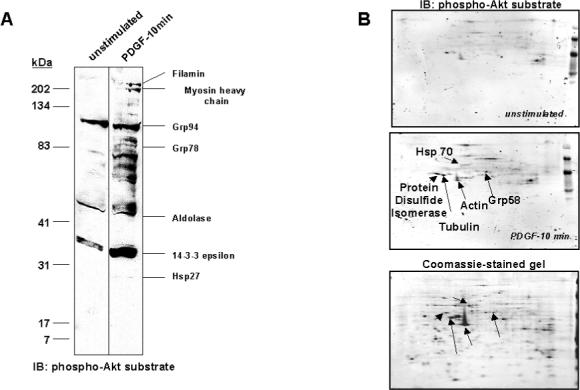
Mesangial cells were cultured in 0.5% FBS/DMEM for 24 hr, prior to incubation with 10 ng/ml PDGF-BB for 10 min, or were left untreated. Lysates were prepared in urea/thiourea buffer and proteins separated by SDS-PAGE (A) or 2-DE (B), on duplicate gels. One of each duplicate gel was stained with Coomassie blue. Proteins from the other gel were transferred to nitrocellulose and immunoblotted with anti-phospho-Akt substrate antibody. Candidate Akt substrates were identified by excision of protein spots/bands from gels, which corresponded to proteins appearing on respective immunoblots, followed by trypsin digestion and identification using peptide mass fingerprinting. Candidate substrates identified are labeled on each figure.
Table 1.
List of candidate Akt Substrates identified by incubation of mesangial cell lysate with active recombinant Akt in an in vitro kinase assay or immunoblot (IB) analysis of lysate from PDGF-stimulated cells.
|
Method of Separation |
||||
|---|---|---|---|---|
| Functional Category | Protein | gi | SDS-PAGE | 2-DE |
| Chaperone | Hsp84 | 51890229 | Kinase Assay | |
| Hsp86 | 54673763 | Kinase Assay | ||
| Hsp70 | 56385 | IB | ||
| Hsp27 | 14010865 | Kinase Assay & IB | ||
| Grp94 | 34862435 | IB | ||
| Grp78 | 25742763 | IB | Kinase Assay | |
| Grp58 | 8393322 | IB | ||
| PDI | 6981324 | IB | ||
| Cytoskeletal | actin, beta | 71620 | Kinase Assay & IB | |
| Vimentin | 14389299 | Kinase Assay & IB | ||
| tropomyosin 5 | 9653293 | Kinase Assay | ||
| tubulin, beta chain | 92930 | IB | ||
| Similar to FLJ00343 protein (filamin) | 34881882 | IB | ||
| myosin heavy chain | 6981236 | IB | ||
| lamin A | 1072002 | Kinase Assay | ||
| Glycolytic | Enolase | 6978809 | Kinase Assay | |
| Aldolase A | 6978487 | IB | ||
| Phospholipid Binding | Annexin I | 6978501 | Kinase Assay | |
| ATP Synthesis | F1-ATP synthase β-subunit | 92350 | Kinase Assay | |
| Adaptor Protein | 14−3−3 ε | 13928824 | IB | |
The identification of chaperone proteins, Hsp70 and Hsp90, and the endoplasmic reticulum (ER) homologs of these proteins, Grp78 and Grp94, as well as PDI, as candidate Akt substrates suggested a previously unidentified mechanism of Akt activity, as these proteins participate in cellular response to inflammatory and oxidative stress32 by regulating protein folding, transport, and degradation. Thus, additional studies were performed to confirm that these chaperones were true substrates of Akt, as the proteomic approaches used may lead to identification of false positives for several reasons. First, extraction of proteins by cell lysis disrupts the spatio-temporal relationship of signaling proteins and protein-protein interactions that provide specificity of target phosphorylation in intact cells, which may permit access of exogenous kinases to proteins with restricted localization in intact cells. In addition, phosphorylation sites not accessible in properly folded proteins may be exposed after urea denaturation. The phospho-Akt susbtrate antibody demonstrates cross-reactivity with motifs different from the Akt phosphorylation motif, and kinases other than Akt can phosphorylate the Akt consensus sequence. Finally, protein spots or bands on gels may contain more than one protein, leading to identification of false substrates.
Table 2 shows mass spectra of peptides obtained by MALDI-MS used to identify the chaperone proteins. Using the search criteria listed in the methods section, Hsp90α and Hsp90β were identified from one protein band as indicated in figure 1. Grp78 was identified by both of our proteomic approaches as indicated in table 1. Grp78 was the only protein identified with a significant MOWSE score from the protein spot corresponding to a phosphoprotein following protein separation by 2D gel electrophoresis. On the other hand, the protein band on SDS-PAGE that contained Grp78, as indicated in figure 3A, revealed a mixture of proteins with significant MOWSE scores and close correlation of protein mass. The mixture of proteins included Moesin (NCBI entry # 13540689; Mass 67868 Da) with 15 matched peptides and a mascot score of 118, Ezrin (NCBI entry # 40804381; Mass 69479) with 10 matched peptides and a macot score of 70, and Radixin (NCBI entry # 40804379; Mass 68700) with 9 matched peptides and a mascot score of 62. Only one peptide from Ezrin and Radixin were matched to Grp78. Three peptides were in common between Grp78 and moesin. No protein mixtures were present in the identification of Hsp70, Grp94, and PDI by peptide mass fingerprinting using our search criteria.
Table 2.
Identification of HSP90α, HSP90β, HSP70, GRP78, GRP94, and PDI as Akt Substrates by peptide mass fingerprinting.
| Protein | NCBInr Entry | Matched Peptidesa | Coverage (%)b | Mascot Score | Matched Massc | Startd | Ende | Peptide Sequencef |
|---|---|---|---|---|---|---|---|---|
| Hsp90β | 51890229 | 16/36 | 27 | 178 | 1274.64 | 42 | 53 | ELISNASDALDK |
| 2255.01 | 149 | 168 | HNDDEQYAWESSAGGSFTVR | |||||
| 950.46 | 169 | 177 | ADHGEPIGR | |||||
| 1310.56 | 187 | 196 | EDQTEYLEER | |||||
| 1150.55 | 276 | 284 | YIDQEELNK | |||||
| 900.52 | 285 | 291 | TKPIWTR | |||||
| 1526.74 | 307 | 319 | SLTNDWEDHLAVK | |||||
| 1347.66 | 320 | 330 | HFSVEGQLEFR | |||||
| 828.52 | 331 | 337 | ALLFIPR | |||||
| 1512.78 | 379 | 392 | GVVDSEDLPLNISR | |||||
| 1140.55 | 439 | 448 | LGIHEDSTNR | |||||
| 2175.94 | 457 | 475 | YHTSQSGDEMTSLSEYVSR | |||||
| 1248.61 | 492 | 502 | EQVANSAFVER | |||||
| 2431.1 | 584 | 604 | LVSSPCCIVTSTYGWTANMER | |||||
| 1263.49 | 613 | 623 | DNSTMGYMMAK | |||||
| 1781.94 | 625 | 639 | HLEINPDHPIVETLR | |||||
| Peptides not matched: 662.42, 663.41, 706.56, 714.38, 732.52, 742.42, 768.46 1168.65, 1194.68, 1208.65, 1236.68, 1364.78, 1515.84, 1541.78, 1778.91, 1786.99, 1790.95, 1911.07, 2374.21, 2447.15 | ||||||||
| Hsp90α | 54673763 | 12/36 | 18 | 109 | 2254.95 | 154 | 173 | HNDDEQYAWESSAGGSFTVR |
| 1310.56 | 192 | 201 | EDQTEYLEER | |||||
| 1777.94 | 210 | 224 | HSQFIGYPITLFVEK | |||||
| 1150.55 | 285 | 293 | YIDQEELNK | |||||
| 900.52 | 294 | 300 | TKPIWTR | |||||
| 1540.75 | 316 | 328 | SLTNDWEEHLAVK | |||||
| 1347.66 | 329 | 339 | HFSVEGQLEFR | |||||
| 1512.78 | 388 | 401 | GVVDSEDLPLNISR | |||||
| 1167.56 | 448 | 457 | LGIHEDSQNR | |||||
| 713.28 | 480 | 484 | DYCTR | |||||
| 1207.62 | 491 | 500 | HIYFITGETK | |||||
| 1785.94 | 634 | 648 | HLEINPDHSIIETLR | |||||
| Peptides not matched: 662.42, 663.41, 706.56, 732.52, 742.42, 768.46, 829.55, 951.53, 1141.61, 1194.68, 1236.68, 1249.67, 1264.67, 1275.74, 1364.78, 1515.84, 1527.84, 1782.98, 1790.95, 1911.07, 2177.00, 2374.21, 2432.18, 2447.15 | ||||||||
| Grp78 | 25742763 | 20/48 | 38 | 233 | 1227.62 | 50 | 60 | VEIIANDQGNR |
| 1565.77 | 61 | 74 | ITPSYVAFTPEGER | |||||
| 1676.8 | 82 | 96 | NQLTSNPENTVFDAK | |||||
| 1603.86 | 124 | 138 | TKPYIQVDIGGGQTK | |||||
| 1551.79 | 139 | 152 | TFAPEEISAMVLTK | |||||
| 1886.96 | 165 | 181 | VTHAVVTVPAYFNDAQR | |||||
| 1232.62 | 186 | 197 | DAGTIAGLNVMR | |||||
| 1658.89 | 198 | 213 | IINEPTAAAIAYGLDK | |||||
| 918.46 | 262 | 268 | VMEHFIK | |||||
| 996.51 | 298 | 306 | ALSSQHQAR | |||||
| 2147.99 | 307 | 324 | IEIESFFEGEDFSETLTR | |||||
| 917.47 | 345 | 352 | VLEDSDLK | |||||
| 1459.75 | 354 | 367 | SDIDEIVLVGGSTR | |||||
| 1835.93 | 448 | 464 | SQIFSTASDNQPTVTIK | |||||
| 1190.63 | 465 | 474 | VYEGERPLTK | |||||
| 1933.01 | 475 | 492 | DNHLLGTFDLTGIPPAPR | |||||
| 1073.55 | 524 | 532 | ITITNDQNR | |||||
| 985.51 | 533 | 540 | LTPEEIER | |||||
| 1315.63 | 563 | 573 | NELESYAYSLK | |||||
| 1396.78 | 622 | 633 | ELEEIVQPIISK | |||||
| Peptides not matched:728.34, 743.26, 753.34, 788.34, 856.45, 870.46, 887.43, 894.41, 959.46, 976.44, 1104.50, 1182.57, 1211.55, 1265.61, 1359.69, 1481.78, 1488.72, 1512.78, 1528.71, 1588.85, 1693.78, 1703.89, 1719.87, 1815.97, 2016.07, 2082.03, 2163.06, 2839.38 | ||||||||
| PDI | 6981324 | 16/38 | 32 | 194 | 861.46 | 59 | 66 | ALAPEYAK |
| 1779.83 | 83 | 98 | VDATEESDLAQQYGVR | |||||
| 695.32 | 116 | 121 | EYTAGR | |||||
| 1050.5 | 215 | 223 | NNFEGEITK | |||||
| 1964.04 | 232 | 248 | HNQLPLVIEFTEQTAPK | |||||
| 1080.67 | 256 | 264 | THILLFLPK | |||||
| 869.38 | 265 | 272 | SVSDYDGK | |||||
| 1832.91 | 287 | 301 | ILFIFIDSDHTDNQR | |||||
| 965.56 | 302 | 309 | ILEFFGLK | |||||
| 859.39 | 311 | 317 | EECPAVR | |||||
| 1221.62 | 318 | 327 | LITLEEEMTK | |||||
| 1408.67 | 328 | 339 | YKPESDELTAEK | |||||
| 1515.73 | 340 | 351 | ITQFCHHFLEGK | |||||
| 1226.55 | 377 | 386 | NFEEVAFDEK | |||||
| 927.52 | 438 | 445 | VHSFPTLK | |||||
| 909.43 | 446 | 453 | FFPASADR | |||||
| Peptides not matched: 751.35, 758.33, 774.33, 808.40, 988.48, 994.12, 1002.53, 1016.52, 1067.50, 1118.50, 1179.59, 1199.65, 1234.58, 1243.66, 1287.70, 1308.64, 1424.70, 1475.74, 1486.71, 1493.73, 1729.87, 1794.83 | ||||||||
| Grp94 | 34862435 | 25/46 | 33 | 277 | 960.45 | 44 | 51 | TDDEVVQR |
| 1784.89 | 52 | 67 | EEEAIQLDGLNASQIR | |||||
| 1080.54 | 76 | 84 | FAFQAEVNR | |||||
| 962.58 | 88 | 95 | LIINSLYK | |||||
| 676.39 | 98 | 102 | EIFLR | |||||
| 1274.64 | 103 | 114 | ELISNASDALDK | |||||
| 2029.06 | 117 | 135 | LISLTDENALAGNEELTVK | |||||
| 1512.77 | 143 | 156 | NLLHVTDTGVGMTR | |||||
| 944.59 | 244 | 252 | GTTITLVLK | |||||
| 1524.72 | 253 | 265 | EEASDYLELDTIK | |||||
| 1186.67 | 385 | 395 | SILFVPTSAPR | |||||
| 1014.47 | 396 | 404 | GLFDEYGSK | |||||
| 782.43 | 429 | 434 | YLNFVK | |||||
| 1484.75 | 435 | 448 | GVVDSDDLPLNVSR | |||||
| 882.46 | 449 | 455 | ETLQQHK | |||||
| 1138.57 | 494 | 503 | LGVIEDHSNR | |||||
| 2249.03 | 512 | 530 | FQSSHHSTDITSLDQYVER | |||||
| 1030.49 | 538 | 546 | IYFMAGSSR | |||||
| 1149.52 | 548 | 557 | EAESSPFVER | |||||
| 753.32 | 598 | 603 | FDESEK | |||||
| 658.38 | 634 | 639 | AVVSQR | |||||
| 2355.14 | 640 | 660 | LTESPCALVASQYGWSGNMER | |||||
| 865.38 | 664 | 671 | AQAYQTGK | |||||
| 1288.59 | 672 | 682 | DISTNYYASQK | |||||
| 875.45 | 684 | 690 | TFEINPR | |||||
| Peptides not matched: 856.45, 870.46, 890.42, 987.46, 1004.48, 1045.53, 1278.62, 1378.66, 1413.65, 1417.71, 1574.84, 1602.77, 1663.88, 1734.74, 1799.94, 2044.09, 2200.11, 2225.14, 2300.15, 2839.41 | ||||||||
| Hsp70 | 56385 | 10/19 | 22 | 96 | 1227.62 | 26 | 36 | VEIIANDQGNR |
| 1486.69 | 37 | 49 | TTPSYVAFTDTER | |||||
| 1664.78 | 57 | 71 | NQVAMNPTNTVFDAK | |||||
| 1980.99 | 138 | 155 | TVTNAVVTVPAYFNDSQR | |||||
| 1198.67 | 160 | 171 | DAGTIAGLNVLR | |||||
| 1658.89 | 172 | 187 | IINEPTAAAIAYGLDK | |||||
| 1690.71 | 221 | 236 | STAGDTHLGGEDFDNR | |||||
| 1252.61 | 302 | 311 | FEELNADLFR | |||||
| 2773.32 | 424 | 447 | QTQTFTTYSDNQPGVLIQVYEGER | |||||
| 1196.66 | 459 | 469 | FELTGIPPAPR | |||||
| Peptides not matched: 709.34, 1029.67, 1057.69, 1082.55, 1145.64 1211.71, 1457.86, 1593.03, 2169.09 | ||||||||
number of trypsin-generated peptides matched to identified protein
Percentage of protein covered by matched peptide
mass of matched peptide
start and end positions of matched peptide, respectively
start and end positions of matched peptide, respectively
corresponding peptide sequence
In Vitro Phosphorylation of Chaperone Proteins by Akt
To confirm the ability of Akt to directly phosphorylate chaperone proteins, recombinant active Akt was incubated with recombinant chaperone proteins and [γ-P32]ATP in an in vitro kinase reaction. Proteins were separated by SDS-PAGE and phosphorylated proteins visualized by autoradiography. Akt phosphorylated recombinant human Hsp90α and Hsp90β, recombinant canine Grp94, and purified PDI (Figure 4A). Because active recombinant Akt, which is autophosphorylated, and Hsp70 and Grp78 migrate in close proximity on 10% SDS gels, phosphorylation of these chaperone proteins could not be detected. Thus, proteins from kinase reactions containing Hsp70 and Grp78 were separated on 4−12% gradient gels. Figure 4B shows that Akt phosphorylated recombinant rat Hsp70, but in vitro Akt phosphorylation of Grp78 was not detected.
Figure 4. In Vitro Phosphorylation of Recombinant Hsp70, Hsp90α and β, Grp78, Grp94, and PDI by Recombinant Active Akt.
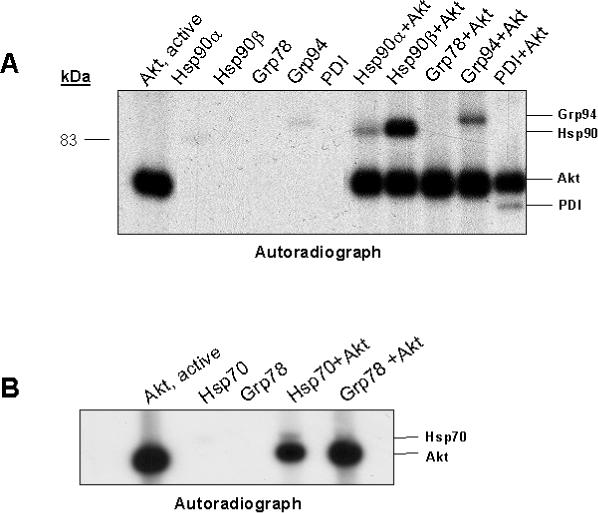
1μg of recombinant human Hsp90α and β, rat Hsp70, canine Grp94, hamster Grp78, and purified PDI was incubated alone or with recombinant active Akt (400ng), in the presence of [γ-P32]ATP, at 25oC for 45 minutes. Reactions were terminated by addition of Laemmli buffer and proteins were separated by 10% SDS-PAGE (A) or 4−12% gradient gel (B). Phosphorylated proteins were visualized by autoradiography. A, autoradiograph demonstrating autophosphorylation of recombinant active Akt, alone, and minimal phosphorylation of recombinant Hsp90α and Grp94 when incubated alone in the presence of [γ-P32] ATP. Incubation of recombinant chaperone proteins with recombinant active Akt resulted in phosphorylation of Hsp90α and β, Grp94, and PDI (A) and recombinant Hsp70 (B), however, recombinant Grp78 was not phosphorylated by Akt in the in vitro kinase reaction (B).
Ex Vivo Phosphorylation of Chaperones
All of the chaperones identified have been reported to exist as phosphorylated proteins in vivo.33-37 Additionally, a number of kinases have been identified that are capable of phosphorylating Hsp90, Grp78, Grp94 and PDI, including casein kinase II, dsDNA-activated kinase, src, and sphingosine-dependent kinases.38-41 To determine if Akt phosphorylates chaperones in intact mesangial cells, each chaperone was evaluated for a shift in isoelectric point consistent with phosphorylation, following PDGF stimulation. To determine the role of Akt in this phosphorylation, mesangial cells were pre-treated with 25 μM of the PI-3K inhibitor LY294002 prior to PDGF stimulation. This concentration was shown to block PDGF-stimulated Akt activity (data not shown). To predict the shift in isoelectric point associated with phosphorylation, theoretical changes in chaperone pI based on the number of phosphorylation sites were estimated using the Scansite website. Based on a maximum of 8 phosphorylation sites, the predicted maximal shift in the isoelectric point of Hsp90 was from 5.05 to 4.92, of Grp94 was 4.99 to 4.76, of Grp78 was 5.1 to 4.92, of PDI was 4.87 to 4.65, and of Hsp70 was 5.56 to 5.13. Based on these predictions, lysates were separated by 2-DE using pH 4−7 or 4.5−5.5 IPG strips, followed by immunoblotting for specific substrates. The protein spots on the immunoblots were subject to densitometric analysis using Progenesis Discovery software (Nonlinear Dynamics, Durham, NC) to determine the relative abundance of acidic and basic species of each protein. Boundaries were formed around each spot on 2-DE immunoblots, as indicated in figure 5, and spots were matched between different experimental conditions (figure 5B). In the cases where protein spots were not sufficiently separated by IEF, the protein was equally divided into two species, following alignment of 2D immunoblots from the basic end of the gel. Spot pixel values were subject to Progenesis background correction of average on the boundary and no normalization to total immunoblot pixel values was carried out. Figure 5A demonstrates a redistribution of Hsp90 toward the acidic end of its isoelectric point after 30 minutes of PDGF stimulation. This shift was partially inhibited by PI-3K inhibition with LY294002. Treatment of lysate from cells stimulated with PDGF for 30 min with the phosphatase PP2A returned Hsp90 to basal distribution on 2-DE gels. In unstimulated cells, 40% of total Hsp90 was the acidic species, which increased to 56% following 10 minutes of PDGF stimulation and to 61% following 30 minutes of PDGF stimulation. Treatment of mesangial cells with LY294002 and treatment of lysate with PP2A resulted in a reduction of the acidic species to 47% and 40% of total Hsp90, respectively. Progenesis analysis of immunoblots for Hsp70 indicated that the most acidic species increased from 8% to 28% of total Hsp70 detected following 10 minutes of PDGF stimulation, and decreased to 5% of total Hsp70 following 30 minutes of stimulation. Treatment of mesangial cells with LY294002 prior to stimulation with PDGF for 10 min and treatment of lysate from cells stimulated for 10 min with PP2A each reduced the acidic species to 5% of total Hsp70. Analysis of Grp78 2-DE immunoblots did not detect a change in abundance of the acidic species following PDGF treatment in the presence or absence of LY294002 (figure 5C). In all conditions the acidic species of Grp78 accounted for about 30% of total Grp78 detected. Stimulation of mesangial cells with PDGF resulted in a minimal acidic shift in the pI of PDI at 30 minutes from 5% to 7% of total PDI detected (figure 5D), and this percent was not altered by LY294002 pretreatment. 2-DE immunoblot analysis of Grp94 was not possible due to inadequate focusing of Grp94 on narrow-range IPG strips. The results indicate that Hsp70 and Hsp90 undergo PI-3K-dependent phosphorylation in mesangial cells stimulated by PDGF.
Figure 5. Phosphorylation of Hsp70, Hsp90, Grp78, and PDI in Mesangial Cells.
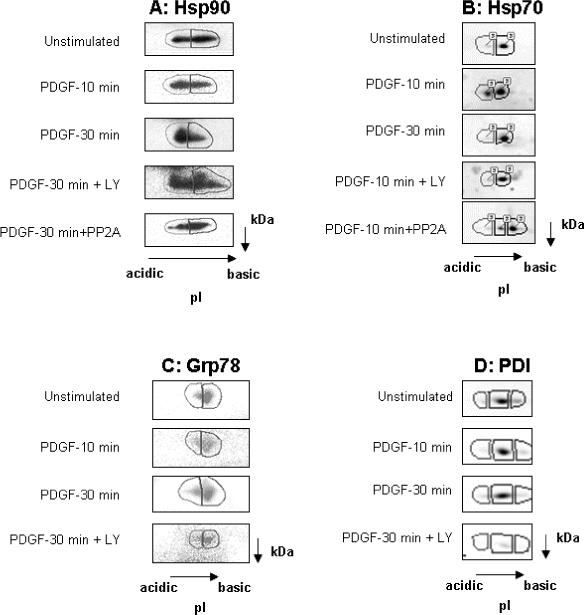
Mesangial cells were treated with the PI-3K inhibitor LY294002 (25μM) or vehicle for 1 hr, prior to stimulation with PDGF. Lysate were prepared in urea/thiourea buffer and exchanged to 2 M urea kinase buffer. Separate aliquots of PDGF-stimulated lysate were incubated with 0.5 Units protein phosphatase 2-A (PP2A) for 1.5 hr at 30°C. Proteins were separated by 2-DE using IPG strips pH 4−7 (Hsp90, Hsp70, and Grp78; panel A, B, and C) or pH 4.5−5.5 (PDI; panelD), and specific proteins identified by immunoblot analysis. Panel A, 30 min of PDGF stimulation increased abundance of more acidic Hsp90 species. Both LY294002 and phosphatase treatment reversed the PDGF-induced acidic shift in Hsp90. Panel B, PDGF induced an acidic shift in Hsp70 by 10 min of stimulation and this effect was diminished by 30 min. Treatment with LY294002 and PP2A prevented and reversed, respectively, the acidic shift in Hsp70 pI induced by PDGF. Panels C & D, 30 min of PDGF stimulation increased abundance of the most acidic species of Grp 78 and PDI and this effect was inhibited by pre-treatment with LY294002.
Association of Akt with Chaperone Proteins
As Akt has been reported to exist in a complex with Hsp7042 and Hsp9043, the ability of Akt to co-precipitate with chaperones in mesangial cells under basal and PDGF-stimulated conditions was determined. Akt was immunoprecipitated from untreated and PDGF-stimulated mesangial cell lysate, and the presence of chaperones examined by immunoblot analysis. Figure 6 shows that Grp78 and Hsp70 associated with Akt in unstimulated cells, and PDGF stimulation enhanced this association. PDI, Hsp90, and Grp94 were not detected in Akt immunoprecipitates under basal or stimulated conditions.
Figure 6. Interaction of Akt with Chaperone Proteins in Mesangial Cells.
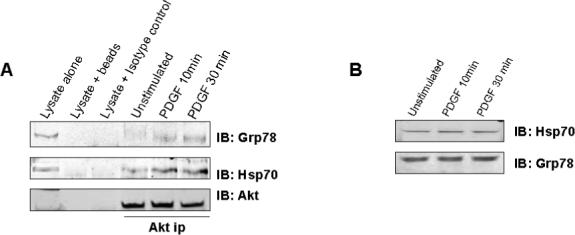
To determine if Akt interacts with chaperone proteins in mesangial cells, agarose-conjugated anti-Akt antibody was used to immunoprecipitate Akt from lysate of mesangial cells that were either unstimulated or stimulated with PDGF for 10 and 30 minutes. The precipitated complex was eluted from the beads, separated by SDS-PAGE and proteins transferred to nitrocellulose. The presence of chaperone proteins in the complex was determined by immunobloting for each specific chaperone. The figure shows that Akt was associated with Grp78 and Hsp70 in unstimulated mesangial cells (A). Stimulation of mesangial cells with PDGF resulted in increased binding of Grp78 and Hps70 (A). PDGF Stimulation of mesangial cells for up to 30 minutes did not alter expression of Grp78 or Hsp70 (B).
Growth Factors are known to induce expression of chaperone proteins,44,45 however, stimulation of mesangial cells with PDGF for up to 30 minutes did not alter expression of HSp70 of Grp78 (figure 6B). These results indicate that some chaperone proteins associate with the Akt signaling complex, and this association is regulated by the activation state of mesangial cells.
Akt and Grp78 are Present in Both Cytosol and Microsomes of Mesangial Cells
Akt is known to be located in the cytoplasm, nuclei, and mitochondria,46,30 and Akt was localized to ER-containing microsomes in adipocytes.47 Several of the proteins identified in the present study, including PDI, Grp78, and Grp94, are abundant in the endoplasmic reticulum (ER). Therefore, the presence of Akt in the microsomal fraction of mesangial cells was examined under basal and PDGF-stimulated conditions, by immunoblot analysis. Microsomal and cytosolic fractionation was confirmed by immunoblotting for caspase 12, a resident ER caspase. Figure 7A demonstrates that uncleaved caspase 12 was only present in the microsomal fraction. Figure 7B shows that Akt was present in both cytosol and microsomal fraction under basal and PDGF-stimulated conditions.
Figure 7. Akt associates with the microsomal fraction in mesangial cells.
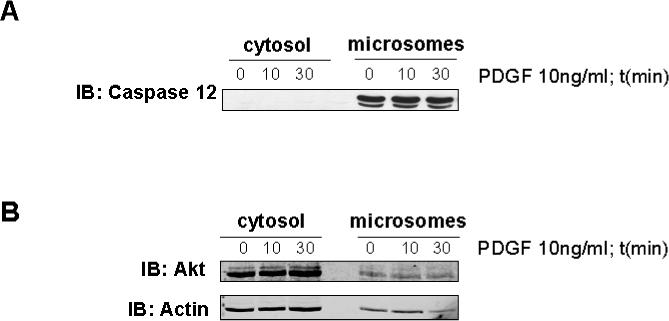
Unstimulated and 10 and 30 min PDGF-stimulated mesangial cells were fractionated into cytosolic and ER-containing microsomal fractions, and the presence of Akt in the two compartments was determined by immunoblot analysis. Panel A is an immunoblot showing for caspase 12 confirming cytosolic and microsomal separation. Procaspase 12, ∼ 55 kDa, is only present in the microsomal fraction. Panel B is an immunoblot of cytosolic and microsomal fractions for Akt showing that Akt is present in both the cytosolic and microsomal fractions and the amount of Akt does not change with PDGF stimulation.
DISCUSSION
The serine/threonine kinase, Akt, mediates mesangial cell responses critical to the development of glomerulonephritis and diabetic nephropathy.1,3 The signaling pathways by which Akt regulates these functions are unknown, and defining these pathways requires identification of Akt substrates. The goal of the present study was to identify additional targets of Akt in mesangial cells that may mediate Akt-induced mesangial cell responses in disease. The novel finding of the current study is that Akt phosphorylated chaperone proteins of the cytosolic Hsp70/Hsp90 folding system, the ER homolog of Hsp90, Grp94, and the ER-abundant folding catalyst, PDI, in mesangial cells. These chaperone proteins participate in cellular response to inflammatory mediator- and oxidant-induced stress, proliferation, and protein synthesis, by regulating folding, transport, and degradation of proteins. Expression of molecular chaperones is increased in the kidney during glomerulonephritis, transplantation-induced hypoxia, and ischemia-reperfusion injury.48 The results of the present study suggest that Akt phosphorylation of stress-induced chaperones may contribute to mesangial cell proliferation or hypertrophy during glomerular injury.
The proteomic approaches used in this study to identify substrates following SDS-PAGE or 2-DE of in vitro kinase reactions involving addition of exogenous active kinase to cell lysates were previously validated by our laboratory.25,29 These techniques eliminate the need for addition of pharmacological kinase inhibitors or the genetic introduction of mutant kinases into intact cells. The second approach utilized an antibody capable of recognizing proteins phosphorylated in the consensus Akt phosphorylation motif to screen for candidate Akt substrates among proteins extracted from PDGF-stimulated mesangial cells, as previously reported.30, 31 The validity of these approaches in mesangial cells was supported by identification of two known Akt substrates, Hsp27 and mitochondrial F1 ATP synthase β-subunit. We reported previously that Akt phosphorylates 14−3−3ζ on Ser-58.21 As 14−3−3ε contains the consensus sequence around Ser-59 similar to that at Ser-58 of 14−3−3ζ, this 14−3−3 isoform is a likely Akt substrate. The sensitivity of these gel-based approaches, however, is low. No more than 20% of Akt targets were identified by the combination of approaches used in this study. Thus, more sensitive techniques will be required to fully identify the range of Akt targets in mesangial cells.
The chaperone proteins identified as Akt targets in the present study may mediate renal cell responses to inflammatory and toxic stimuli. In mesangial cells, Hsp70 expression is induced by treatment with cyclopentanone prostaglandins, and heat shock induction of Hsp70 protects mesangial cells against xanthine-induced oxidant stress.49,50 Increased expression of Hsp90 is required for mesangial cell proliferation in rat mesangioproliferative anti-Thy1.1 glomerulonephritis.51 Based on the potential role for these chaperones in glomerular disease, additional experiments were performed to confirm that these candidates were true Akt substrates. These confirmatory studies were necessary due to several limitations of the proteomic approaches. First, extraction of proteins by cell lysis disrupts the spatio-temporal relationship of signaling proteins and the protein-protein interactions that provide specificity of target phosphorylation in intact cells. In addition, phosphorylation sites not accessible in properly folded proteins may be exposed after urea denaturation. The phospho-Akt substrate antibody demonstrates cross-reactivity with motifs different from the Akt phosphorylation motif, and kinases other than Akt phosphorylate the Akt consensus sequence.
Initial confirmatory studies examined the ability of active recombinant Akt to phosphorylate recombinant chaperone proteins in vitro. These studies showed that Hsp70, Hsp90α and β, Grp94, and purified PDI, but not Grp78, were phosphorylated in vitro. Next, ex vivo phosphorylation of these chaperone proteins was examined by a shift in pI on 2-DE. PDGF stimulation of mesangial cells induced Hsp90 and Hsp70 to redistribute toward more acidic forms of the protein in a PI 3K-dependent manner, suggesting these proteins were phosphorylated, while a shift in PDI was more subtle. The ability of phosphatase treatment to reverse the PDGF-induced shift in pI of Hsp90 and Hsp70 confirmed that the pI shift observed was due to phosphorylation following PDGF stimulation. The effect of phosphorylation on the isoelectric point of a protein varies, depending on the buffering capacity of the amino acid composition of the protein and its size. The Scansite online website (http://Scansite.mit.edu) was utilized to theoretically predict phosphorylation-induced changes in protein isoelectric points. Based on a maximum of eight phosphorylation sites, the isoelectric point was predicted to decrease by ∼ 0.2 pH units for all chaperones except Hsp70, which was predicted to shift by 0.4 pH units. The observed changes in chaperone isoelectric point upon PDGF stimulation in the current study were consistent with the small predicted shift in pI. While Akt was unable to directly phosphorylate recombinant Grp78 in vitro, and determination of an acidic shift in Grp78 with PDGF stimulation ex vivo was inconclusive, a potential role for this kinase in regulating Grp78 function may exist. This possibility is supported by the co-immunoprecipitation of Grp78 with Akt from mesangial cell lysates. Additionally, localization of Grp78 and PDI in the ER may hinder the amount phosphorylated by Akt, such that phosphorylation-induced changes in isoelectric point cannot be detected. While Hsp70 co-immunoprecipitated with Akt, Hsp90 could not be detected in Akt immunoprecipitates. Previous reports, using deletion mutants of Akt, showed that amino acid residues 229−309 on Akt bind Hsp90.43 The absence of Hsp90 in Akt immunoprecipitates in the current study cannot be explained at present. Despite this finding, the data indicate that Akt phosphorylates the chaperone proteins of the Hsp70/Hsp90 system.
The functional consequences of this phosphorylation in mesangial cells remain to be established. The Hsp70/Hsp90 chaperone system is an important regulator of signal transduction pathways for steroid hormone receptors, kinase signaling, and transcription factors.52,53 Both Hsp70 and Hsp90 exist as phosphoproteins in vivo. The phosphorylation status of Hsp70 is altered during heat stress,54 and exercise,55 yet the effect of phosphorylation on Hsp70 function is not known. Hsp90 is required for correct folding and activity of kinases, including Akt, B-raf, and casein kinase II.53 Phosphorylation of Hsp90 has been shown to allow release of client proteins from the Hsp90 complex56 and to stimulate activity of clients such as eIF2-α kinase.57 These data suggest that within the Hsp90 complex, Akt phosphorylation of Hsp90 may induce release of Akt or other Hsp90 clients from the protein complex, or regulate the activity status of Akt.
Identification of chaperones abundant in the ER (Grp94, PDI, and Grp78) as Akt substrates, led us to determine whether Akt was distributed within the ER of mesangial cells. Akt was demonstrated within the ER-containing microsomes. Akt has previously been shown to distribute to ER-containing microsomes of adipocytes, and insulin treatment resulted in different phosphorylation profiles of Akt in the cytosol, microsomes, and plasma membrane.47 Grp78 was present in the cytosolic fraction of mesangial cells in this study. Thus, Akt phosphorylation of Grp78, Grp94 or PDI could take place in the ER or in the cytosol.
The molecular chaperones of ER origin, PDI, Grp94 and Grp78, have also been shown to exist as phosphoproteins in vivo. Thus far, sphingosine-dependent kinases and casein kinase II are the only kinases shown to phosphorylate PDI and Grp94 in vivo.38,41 Grp78 and Grp94 are ER homologs of cytosolic Hsp70 and Hsp90, respectively, and are induced during glucose starvation or accumulation of non-transported proteins. Like chaperones of cytosolic origin, the physiological role of ER chaperone phosphorylation is unclear. Phosphorylation of Grp78 decreases during ER stress,58 and only un-phosphorylated Grp78 is complexed with client proteins.59 Phosphorylation of PDI is differentially regulated by TNFα stimulation60 and ischemia-reperfusion61 and may serve to regulate interaction with client proteins, as with Grp78. Thus, Akt may play a role in the unfolded protein response and ER-associated protein degradation triggered by ER stress, by regulating chaperone interaction with client proteins.
CONCLUSIONS
This study identified molecular chaperones of cytosolic and ER origin as novel Akt substrates. The biological significance of Hsp and Grp phosphorylation by Akt was not examined, but phosphorylation of these proteins may regulate interaction with other chaperones and target proteins. Increased expression of these molecular chaperones promotes cell survival as part of a stress response to reactive oxygen species generation, inflammation, heat shock, and glucose starvation. Thus, Akt phosphorylation of stress-induced chaperones may contribute to mesangial cell proliferation or hypertrophy during glomerular injury.
The combination of two proteomic screens of mesangial cell proteins for Akt substrates identified chaperone proteins of the cytosolic Hsp70/Hsp90 folding system, ER homologs of these proteins Grp78 and Grp94, and the ER-abundant folding catalyst, PDI. These chaperones participate in cellular responses by regulating folding, transport, and degradation of proteins. Akt phosphorylation of chaperones may contribute to mesangial cell responses during glomerular injury.
ACKNOWLEDGMENTS
We thank the technical assistance of Angela Kain and Janice Scherzer. We also thank Dr. Jian Cai for technical assistance in the MALDI-MS analysis.
This research was supported by American Heart Association Ohio Valley Affiliate Post-doctoral Fellowship Grant (to MTB), Department of Veteran Affairs (to KRM and to JBK), the National Institutes of Health R21 DK62389 (to KRM), and the American Heart Association Scientist Development Grant (to MJR).
Abbreviations
- Hsp
heat shock protein
- Grp
glucose-regulated protein
- PDI
Protein Disulfide Isomerase
- PDGF
platelet-derived growth factor
- PI-3K
phosphatidylinositol 3-kinase
REFERENCES
- 1.Choudhury GG. J. Biol. Chem. 2001;276(38):35636–35643. doi: 10.1074/jbc.M100946200. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh PM, Mikhailova M, Bedolla R, Kreisberg JI. Am. J. Physiol. Renal Physiol. 2001;280(6):F972–F979. doi: 10.1152/ajprenal.2001.280.6.F972. [DOI] [PubMed] [Google Scholar]
- 3.Bonacchi A, Romagnani P, Romanelli RG, Efsen E, Annunziato F, Lasagni L, Francalanci M, Serio M, Laffi G, Pinzani M, Gentilini P, Marra F. J. Biol. Chem. 2001;276(13):9945–9954. doi: 10.1074/jbc.M010303200. [DOI] [PubMed] [Google Scholar]
- 4.Gorin Y, Ricono JM, Kim NH, Bhandari B, Choudhury GG, Abboud HE. Am. J. Physiol. Renal Physiol. 2003;285(2):F219–F229. doi: 10.1152/ajprenal.00414.2002. [DOI] [PubMed] [Google Scholar]
- 5.Gorin Y, Kim NH, Feliers D, Bhandari B, Choudhury GG, Abboud HE. FASEB J. 2001;15(11):1909–1920. doi: 10.1096/fj..01-0165com. [DOI] [PubMed] [Google Scholar]
- 6.Hiromura K, Monkawa T, Petermann AT, Durvasula RV, Shankland SJ. Kidney Int. 2002;61(4):1312–1321. doi: 10.1046/j.1523-1755.2002.00257.x. [DOI] [PubMed] [Google Scholar]
- 7.Kang BP, Urbonas A, Baddoo A, Baskin S, Malhotra A, Meggs LG. Am. J. Physiol. Renal Physiol. 2003;285(5):F1013–F1024. doi: 10.1152/ajprenal.00209.2003. [DOI] [PubMed] [Google Scholar]
- 8.Shimamura H, Terada Y, Okado T, Tanaka H, Inoshita S, Sasaki S. J. Am. Soc. Nephrol. 2003;14(6):1427–1434. doi: 10.1097/01.asn.0000066140.99610.32. [DOI] [PubMed] [Google Scholar]
- 9.Runyan CE, Schnaper HW, Poncelet AC. J. Biol. Chem. 2004;279(4):2632–2639. doi: 10.1074/jbc.M310412200. [DOI] [PubMed] [Google Scholar]
- 10.Kang BP, Frencher S, Reddy V, Kessler A, Malhotra A, Meggs LG. Am. J. Physiol. Renal Physiol. 2003;284(3):F455–F466. doi: 10.1152/ajprenal.00137.2002. [DOI] [PubMed] [Google Scholar]
- 11.Sheu ML, Ho FM, Chao KF, Kuo ML, Liu SH. Mol. Pharmacol. 2004;66(1):187–196. doi: 10.1124/mol.66.1.187. [DOI] [PubMed] [Google Scholar]
- 12.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Cell. 1999;96(6):857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 13.Choudhury GG, Lenin M, Calhaun C, Zhang JH, Abboud HE. Cell Signal. 2003;15(2):161–170. doi: 10.1016/s0898-6568(02)00057-8. [DOI] [PubMed] [Google Scholar]
- 14.Du K, Montminy M. J. Biol. Chem. 1998;273(49):32377–32379. doi: 10.1074/jbc.273.49.32377. [DOI] [PubMed] [Google Scholar]
- 15.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Nature. 1995;378(6559):785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 16.Zhou BP, Hung MC. Semin. Oncol. 2002;29(3 suppl 11):62–70. doi: 10.1053/sonc.2002.34057. [DOI] [PubMed] [Google Scholar]
- 17.Liang J, Zubovitz J, Petrocelli T, Kotchetkov R, Connor MK, Han K, Lee JH, Ciarallo S, Catzavelos C, Beniston R, Franssen E, Slingerland JM. Nat. Med. 2002;8(10):1153–1160. doi: 10.1038/nm761. [DOI] [PubMed] [Google Scholar]
- 18.Dimmeler S, Fleming I, Fisslthaler B, Hermann C, Busse R, Zeiher AM. Nature. 1999;399(6736):601–605. doi: 10.1038/21224. [DOI] [PubMed] [Google Scholar]
- 19.Cardone MH, Roy N, Stennicke HR, Salvesen GS, Franke TF, Stanbridge E, Frisch S, Reed JC. Science. 1998;282(5392):1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 20.del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Science. 1997;278(5338):687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 21.Powell DW, Rane MJ, Chen Q, Singh S, McLeish KR. J. Biol. Chem. 2002;277(24):21639–1642. doi: 10.1074/jbc.M203167200. [DOI] [PubMed] [Google Scholar]
- 22.Chen Q, Powell DW, Rane MJ, Singh S, Butt W, Klein JB, McLeish KR. J. Immunol. 2003;170(10):5302–5308. doi: 10.4049/jimmunol.170.10.5302. [DOI] [PubMed] [Google Scholar]
- 23.Lewis TS, Hunt JB, Aveline LD, Jonscher KR, Louie DF, Yeh JM, Nahreini TS, Resing KA, Ahn NG. Mol. Cell. 2000;6(6):1343–1354. doi: 10.1016/s1097-2765(00)00132-5. [DOI] [PubMed] [Google Scholar]
- 24.Powell DW, Pierce WM, McLeish KR. Mass Spectrom. Rev. 2005;24:847–864. doi: 10.1002/mas.20044. [DOI] [PubMed] [Google Scholar]
- 25.Powell DW, Rane MJ, Joughin BA, Kalmukova R, Hong JH, Tidor B, Dean WL, Pierce WM, Klein JB, Yaffe MB, McLeish KR. Mol. Cell. Biol. 2003;23(15):5376–5387. doi: 10.1128/MCB.23.15.5376-5387.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thongboonkerd V, Luengpailin J, Cao J, Pierce WM, Cai J, Klein JB, Doyle RJ. J. Biol. Chem. 2002;277(19):16599–16605. doi: 10.1074/jbc.M200746200. [DOI] [PubMed] [Google Scholar]
- 27.Rane MJ, Pan Y, Singh S, Powell DW, Wu R, Cummins T, Chen Q, McLeish KR, Klein JB. J. Biol. Chem. 2003;278(30):27828–27835. doi: 10.1074/jbc.M303417200. [DOI] [PubMed] [Google Scholar]
- 28.Rao RV, Hermel E, Castro-Obregon S, del Rio G, Ellerby LM, Ellerby HM, Bredesen DE. J. Biol. Chem. 2001;276(36):33869–33874. doi: 10.1074/jbc.M102225200. [DOI] [PubMed] [Google Scholar]
- 29.Singh S, Powell DW, Rane MJ, Millard TH, Trent JO, Pierce WM, Klein JB, Machesky LM, McLeish KR. J. Biol. Chem. 2003;278(38):36410–36417. doi: 10.1074/jbc.M306428200. [DOI] [PubMed] [Google Scholar]
- 30.Bijur GN, Jope RS. J. Neurochem. 2003;87(6):1427–1435. doi: 10.1046/j.1471-4159.2003.02113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berwick DC, Hers I, Heesom KJ, Moule SK, Tavare JM. J. Biol. Chem. 2002;277(37):33895–33900. doi: 10.1074/jbc.M204681200. [DOI] [PubMed] [Google Scholar]
- 32.Kregel KC. Appl. Physiol. 2002;92(5):2177–2186. doi: 10.1152/japplphysiol.01267.2001. [DOI] [PubMed] [Google Scholar]
- 33.Garnier C, Lafitte D, Jorgensen TJ, Jensen ON, Briand C, Peyrot V. Eur. J. Biochem. 2001;268(8):2402–2407. doi: 10.1046/j.1432-1327.2001.02121.x. [DOI] [PubMed] [Google Scholar]
- 34.Hernando R, Manso R. Eur. J. Biochem. 1997;243(1−2):460–467. doi: 10.1111/j.1432-1033.1997.0460a.x. [DOI] [PubMed] [Google Scholar]
- 35.Cala SE. Biochim. Biophys. Acta. 2000;1496(2−3):296–310. doi: 10.1016/s0167-4889(00)00028-8. [DOI] [PubMed] [Google Scholar]
- 36.Hendershot LM, Ting J, Lee AS. Mol. Cell. Biol. 1988;8(10):4250–4256. doi: 10.1128/mcb.8.10.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quemeneur E, Guthapfel R, Gueguen P. J. Biol. Chem. 1994;269(8):5485–5488. [PubMed] [Google Scholar]
- 38.Cala SE, Jones LR. J. Biol. Chem. 1994;269(8):5926–5931. [PubMed] [Google Scholar]
- 39.Lees-Miller SP, Anderson CW. J. Biol. Chem. 1989;264(29):17275–17280. [PubMed] [Google Scholar]
- 40.Carlino A, Toledo H, Vidal V, Redfield B, Strassman J, Abdel-Ghany M, Racker E, Weissbach H, Brot N. Biochem. Biophys. Res. Commun. 1994;201(3):1548–1553. doi: 10.1006/bbrc.1994.1880. [DOI] [PubMed] [Google Scholar]
- 41.Megidish T, Takio K, Titani K, Iwabuchi K, Hamaguchi A, Igarashi Y, Hakomori S. Biochemistry. 1999;38(11):3369–3378. doi: 10.1021/bi982548c. [DOI] [PubMed] [Google Scholar]
- 42.Gao T, Newton AC. J. Biol. Chem. 2002;277(35):31585–31592. doi: 10.1074/jbc.M204335200. [DOI] [PubMed] [Google Scholar]
- 43.Sato S, Fujita N, Tsuruo T. Proc. Natl. Acad. Sci. 2000;97(20):10832–10837. doi: 10.1073/pnas.170276797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jerome V, Leger J, Devin J, Baulieu EE, Catelli MG. Growth Factors. 1991;4(4):317–327. doi: 10.3109/08977199109043917. [DOI] [PubMed] [Google Scholar]
- 45.Ting LP, Tu CL, Chou CK. J. Biol. Chem. 1989;264(6):3404–3408. [PubMed] [Google Scholar]
- 46.Camper-Kirby D, Welch S, Walker A, Shiraishi I, Setchell KD, Schaefer E, Kajstura J, Anversa P, Sussman MA. Circ. Res. 2001;88(10):1020–1027. doi: 10.1161/hh1001.090858. [DOI] [PubMed] [Google Scholar]
- 47.Carvalho E, Eliasson B, Wesslau C, Smith U. Diabetologia. 2000;43(9):1107–1115. doi: 10.1007/s001250051501. [DOI] [PubMed] [Google Scholar]
- 48.Borkan SC, Gullans SR. Ann. Rev. Physiol. 2002;64:503–527. doi: 10.1146/annurev.physiol.64.081501.155819. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X, Lu L, Dixon C, Wilmer W, Song H, Chen X, Rovin BH. Kidney Int. 2004;65(3):798–810. doi: 10.1111/j.1523-1755.2004.00454.x. [DOI] [PubMed] [Google Scholar]
- 50.Chen HC, Guh JY, Tsai JH, Lai YH. Kidney Int. 1999;56(4):1270–1273. doi: 10.1046/j.1523-1755.1999.00693.x. [DOI] [PubMed] [Google Scholar]
- 51.Pieper M, Rupprecht HD, Bruch KM, De Heer E, Schocklmann HO. Kidney Int. 2000;58(6):2377–2389. doi: 10.1046/j.1523-1755.2000.00421.x. [DOI] [PubMed] [Google Scholar]
- 52.Wegele H, Muller L, Buchner J. Rev. Physiol. Biochem. Pharmacol. 2004;151:1–44. doi: 10.1007/s10254-003-0021-1. [DOI] [PubMed] [Google Scholar]
- 53.Pratt WB, Toft DO. Exp. Biol. Med. 2003;228(2):111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 54.Cvoro A, Dundjerski J, Trajkovic D, Matic G. Cell. Biol. Int. 1999;23(4):313–320. doi: 10.1006/cbir.1998.0247. [DOI] [PubMed] [Google Scholar]
- 55.Gonzalez B, Manso R. J. Physiol. 2004;556(2):369–385. doi: 10.1113/jphysiol.2003.058420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao YG, Gilmore R, Leone G, Coffey MC, Weber B, Lee PW. J. Biol. Chem. 2001;276(35):32822–32827. doi: 10.1074/jbc.M105562200. [DOI] [PubMed] [Google Scholar]
- 57.Szyszka R, Kramer G, Hardesty B. Biochemistry. 1989;28(4):1435–1438. doi: 10.1021/bi00430a001. [DOI] [PubMed] [Google Scholar]
- 58.Satoh M, Nakai A, Sokawa Y, Hirayoshi K, Nagata K. Exp. Cell. Res. 1993;205(1):76–83. doi: 10.1006/excr.1993.1060. [DOI] [PubMed] [Google Scholar]
- 59.Freiden PJ, Gaut JR, Hendershot LM. EMBO J. 1992;11(1):63–70. doi: 10.1002/j.1460-2075.1992.tb05028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yanagida M, Miura Y, Yagasaki K, Taoka M, Isobe T, Takahashi N. Electrophoresis. 2000;21(9):1890–1898. doi: 10.1002/(SICI)1522-2683(20000501)21:9<1890::AID-ELPS1890>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 61.Sakai J, Ishikawa H, Koji0ma S, Satoh H, Yamamoto S, Kanaoka M. Proteomics. 2003;3(7):1318–1324. doi: 10.1002/pmic.200300432. [DOI] [PubMed] [Google Scholar]


