Abstract
Cdc7 is a conserved serine/threonine kinase essential for the initiation of DNA replication, likely by activating the MCM DNA helicase at the G1- to S-phase transition. Cdc7 kinase activity requires association with its regulatory subunit Dbf4/activator of S-phase kinase. Cdc7-Dbf4 is also downstream of the conserved Ataxia telangectasia and RAD3-related kinase that responds to stalled replication forks or DNA damage. In this study, we found that Cdc7 protein was very low or undetectable in normal tissues and cell lines but had increased expression in ∼50% of the 62 human tumor cell lines we examined. Most cell lines with increased Cdc7 protein levels also had increased Dbf4 abundance, and some tumor cell lines had extra copies of the DBF4 gene. A high expression of Cdc7 protein was also detected in primary breast, colon, and lung tumors but not in the matched normal tissues. We also found a high correlation between p53 loss and increased CDC7 and DBF4 expression in primary breast cancers (P = 3.6 x 10-9 and 1.8 x 10-10, respectively) and in the cancer cell lines we studied. Therefore, increased Cdc7-Dbf4 abundance may be a common occurrence in human malignancies.
Introduction
The initiation of DNA synthesis requires the assembly of a multi-protein complex at replication origins during G1-phase [1]. These “licensed” replication complexes are activated to initiate DNA synthesis by cyclin-dependent kinases and by the Cdc7-Dbf4 kinase [2]. Cdc7 kinase activity requires a regulatory subunit called Dbf4, which is cyclically expressed during the cell cycle and peaks during S-phase [3,4]. Although first identified in the budding yeast [5,6], orthologs of CDC7 and DBF4 have been found in fission yeast, Aspergillus, Drosophila, Xenopus, mice, and humans, where DBF4 is also called the activator of S-phase kinase (ASK) [7–12]. Here, we will refer to this subunit as Dbf4. In Xenopus and human cells, another Dbf4-related protein (Drf1) can bind to and activate Cdc7 kinase [4,13]. Drf1 is most active during the early embryonic cycles in Xenopus and is absent after gastrulation, suggesting that it may be developmentally regulated in other vertebrate organisms as well [14]. In all organisms, Cdc7 kinase is essential for the initiation of DNA replication likely through its ability to phosphorylate key replication proteins [1]. Recently, it was reported that Cdc7-Dbf4 phosphorylation of Mcm2 is essential for the initiation of DNA replication in mammalian cells [15,16].
The Cdc7-Dbf4 protein kinase is a target of the S-phase checkpoint pathway, and it has an important role in promoting a proper response to DNA damage in multiple organisms [7,17]. Vertebrate Cdc7-Dbf4 is downstream of the Ataxia telangectasia and RAD3-related (ATR) and Chk1 checkpoint kinases in response to UV irradiation [18] where its activity may be inhibited to prevent initiation events. Cdc7 is also a downstream target of ATR and Chk2 after replication fork stalling [19,20], although it is not known if Cdc7 or Dbf4 are direct targets of ATR or Chk1/2. Cdc7-Dbf4 is therefore an essential cell cycle regulator that is also important for genome integrity in the response to DNA damage or replication fork arrest.
Previous studies examining proteins required for the initiation of DNA replication have shown that Cdc6, Mcm2, Mcm5, and Cdt1 are variously up-regulated in cancers of the bladder, colon, cervix, and lung [21–28]. CDC7 mRNA expression is also altered in some cancer cell lines and primary tumors [29], and furthermore, somatic CDC7 mutations were identified in colorectal and gastric carcinomas through comprehensive kinome screens of human tumors [30,31]. These data suggest that alterations in Cdc7-Dbf4 protein abundance or activity may occur during tumorigenesis and have important consequences for cell survival. Indeed, Nambiar et al. [32] characterized Dbf4 as a novel determinant in cutaneous melanoma development with prognostic relevance. They showed that Dbf4 protein is increased in primary melanoma, melanoma metastasis, and melanoma cell lines.
In this study, we compared levels of Cdc7 protein expression in the NCI-60 and additional leukemia cell lines to normal cell lines and tissues. About 50% of 62 tumor cell lines we examined had increased Cdc7 protein abundance. Furthermore, Cdc7-Dbf4 expression levels were correlated in these cancer cell lines. Immunohistochemical analysis of primary tumors showed moderate to intense Cdc7 staining in some breast, colon, and lung cancers but no staining in matched normal tissue. Interestingly, although Cdc7-Dbf4 expression was high in multiple cell lines and primary breast tumors with mutant p53, we found no evidence that p53 expression or induction regulated CDC7 and DBF4 mRNA levels. To investigate whether increased CDC7 or DBF4 copy number might contribute to Cdc7-Dbf4 protein overexpression, we examined the DBF4 and CDC7 genes by fluorescence in situ hybridization (FISH). Although we did not detect CDC7 amplification, multiple copies of DBF4 were detected in 5 of 14 tumor cell lines examined and in 1 primary tumor. Knockdown of Cdc7 expression in tumor cell lines caused growth arrest with or without apoptosis as seen previously [33]. Together, our results indicate that increased Cdc7-Dbf4 protein expression is a common occurrence in human malignancies and suggest that inactivation of the p53 tumor suppressor or increased DBF4 copy number may be contributing mechanisms.
Materials and Methods
Cell Lines
The normal human fetal lung fibroblasts WI-38, the normal human lung fibroblasts IMR-90, and HeLa cells were maintained in minimum essential medium with Earle's salts supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (HI FBS), 2 mM glutamine, 1.5 g/L sodium bicarbonate, 0.1 nM nonessential amino acids, 1 mM sodium pyruvate, and 50 U/ml and 50 µg/ml penicillin/streptomycin, respectively. Tumor cell lines from the NCI-60 set were maintained in RPMI-1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% (vol/vol) HI FBS and 0.05 mg/ml gentamicin. The leukemia cell lines from the NCI-60 set and the five additional leukemia cell lines MOLT-3, Jurkat clone E6-1, A3, GDM-1, and KG-1 were grown in suspension in RPMI-1640 medium supplemented with 10% (vol/vol) HI FBS and 0.05 mg/ml gentamicin.
The HCT-116 p53+/+ and p53-/- cell lines were a gift from B. Vogelstein [34]. They were cultured in McCoy's 5A medium supplemented with 10% (vol/vol) HI FBS.
The bone marrow mononuclear cells were issued from healthy adults and obtained from Cambrex (Walkersville, MD). They were cultured in Iscove's modified Dulbecco's medium, which is a modified Dulbecco's modified Eagle's medium containing high glucose (4500 mg/L), sodium pyruvate, additional amino acids, and HEPES and is supplemented with 15% (vol/vol) HI FBS, 200 nM glutamine, and 2 ng/ml stem cell factor.
Antibodies
Monoclonal antibodies were separately raised against purified GST-HsCdc7 and recombinant HsCdc7-Dbf4 kinase. Antibodies were screened for their ability to immunoblot recombinant HsCdc7 or HsDbf4. We identified multiple monoclonal cell lines that produce anti-Cdc7 and anti-Dbf4 antibodies with a variety of properties, four of which were used in this study: Cdc7-4D9, Cdc7-10B9, Cdc7-2G1, and Dbf4-8H6.
Monoclonal antibodies against the human β-actin and poly(ADP)-ribose polymerase (PARP) proteins were purchased from Sigma (St. Louis, MO) and Cell Signaling Technology (Danvers, MA), respectively. Horseradish peroxidase-conjugated secondary antibodies were obtained from Amersham Biosciences (Piscataway, NJ).
Immunoblot Analysis
Whole-cell extracts were prepared by resuspending cell pellets in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris [pH 8]). The concentrations of proteins in the extracts were determined using the BCA Protein Assay Reagent Kit (Pierce, Rockford, IL) according to the manufacturer's protocol. Equal amounts of proteins were separated in 10% SDS-PAGE gel and were transferred to a nitrocellulose membrane (Millipore Inc., Billerica, MA). Membrane was stained with Ponceau S dye to check for equal loading and homogeneous transfer. The proteins were visualized using the SuperSignalWest Pico Chemiluminescent Substrate kit from Pierce.
Fluorescence In Situ Hybridization
DNA probes recognizing the DBF4 and CDC7 genes are derived from BAC clones RP11-104E6 (location 7q21.12) and RP11-47K11 (location 1p22.2), respectively. They were purchased from BACPAC Resources, Children's Hospital Oakland Research Institute, Oakland, CA. Fluorescence in situ hybridization probes were generated using Vysis SpectrumOrange-conjugated dUTP and the Vysis Nick translation labeling kit following manufacturer's protocol (Abbott Molecular Inc., Des Plaines, IL). On chromosome 1, we used a control probe at the location 1q21.1 (RP11-337C18) labeled with Vysis SpectrumGreen-conjugated dUTP. For the chromosome 7, we used a commercial centromeric probe CEP7 SpectrumGreen (α satellite) that hybridizes to the centromere (band region 7p11.1-q11.1, locus D7Z1; Abbott Molecular Inc.). Fluorescence in situ hybridization experiments were done on cell lines, on metaphase preparations, and frozen tissue sections following standard protocols. Metaphases were prepared following a standard cytogenetic technique. Slides were mounted with VECTASHIELD Mounting Medium with DAPI (VECTOR Laboratories, Burlingame, CA). Fluorescence in situ hybridization control experiments were done on Vysis Normal Male Metaphase Comparative Genomic Hybridization Target Slides made from phytohemagglutinin-stimulated lymphocytes derived from a karyotypically normal male donor (Abbott Molecular Inc.).
Fifty nuclei were analyzed per cell line or per tissue section. Counting was performed, and calculation of ratios was done by dividing 7q21.12 (DBF4) by CEP7 signals and dividing 1p22.2 (CDC7) by 1q21.1 (control) signals in interphase FISH experiments. Ratios that were ≥1.5 were considered overrepresented, and ratios ≥ 2.5 were considered as amplified. Counting was performed with a fluorescence microscope (Axioplan 2 Imaging; Zeiss, Thornwood, NY). The pictures were taken at an original magnification of x63 with a fluorescence microscope (Model BX51; Olympus, Center Valley, PA) using the software Case Data Manager (Version 5.0).
Tissue Samples
Whole human normal tissue homogenates from brain, kidney, liver, lung, mammary gland, ovary, prostate, and small intestine were derived from healthy humans who died of trauma or sudden death (Protein Medley; BD Biosciences Clontech, Palo Alto, CA). Patients who had surgery for invasive beast, lung, and colon cancers were identified through searching the histopathology records available at the Van Andel Research Institute (Grand Rapids, MI). Sections were cut from fresh frozen breast, lung, and colon primary tumors issued from 35 different patients according to standard protocols. For six patients, the matched normal tissues were available. Sections were preserved at -80°C until use.
Immunohistochemistry
Frozen sections were fixed in 70% ethanol for 10 minutes. Immunohistochemistry was performed with the Vectastain ABC Kit (VECTOR Laboratories) according to the manufacturer's protocol. Endogenous peroxidase activity was first blocked by a 30-minute incubation of the sections in 0.03% hydrogen peroxide. After incubations with primary and secondary antibodies, sections were incubated in peroxidase substrate solution until the desired staining intensity developed. We used the diaminobenzidine (DAB) Substrate Kit for Peroxidase (VECTOR Laboratories). Then, the sections were washed for 5 minutes in water. They were counterstained in hematoxylin (VECTOR Laboratories) and were mounted in Cytoseal60 (Richard-Allan Scientific, Kalamazoo, MI). Sections were analyzed using a bright microscope (ECLIPSE E600; Nikon, Melville, NY), and pictures were taken at the original magnifications of x20 and x40 with the SPOT software (Version 4.0.4; Diagnostic Instruments Inc., Sterling Heights, MI).
Gene Expression Analysis
To examine the relationship between CDC7 expression, DBF4 expression, and TP53 gene status, preprocessed gene expression data were obtained from the Gene Expression Omnibus (GSE3494, GSE4922) [35–37]. Expression values for CDC7 and DBF4 were isolated, and expression differences between wild type and mutated TP53 tumors were evaluated using a two-sided Student's t test. For the expression analysis in additional tumors, unprocessed gene expression data derived from more than 1000 tumors were obtained from Gene Expression Omnibus (GSE2109). Gene expression values were preprocessed using the RMA method as implemented in the BioConductor affy package for the R environment [38,39] using updated probe set mappings [40]. For each tumor gene expression value, the median gene expression value of the corresponding nondiseased tissue was subtracted. Tumor samples that could not be associated with the corresponding nondiseased tissue were not evaluated. Relative gene expression levels for CDC7 and DBF4 could be established in 678 tumor samples (breast = 181, cervix = 13, colon = 134, endometrium = 58, kidney = 91, lung = 53, lymph = 6, ovary = 102, pancreas = 5, prostate = 19, stomach = 6, and thyroid = 10). DBF4 expression was evaluated with respect to CDC7 expression and tumor subtype using linear regression. From the regression model, an adjusted R2 and a significance value were computed.
The association between CDC7 protein expression and p53 mutant status in the NCI-60 cell lines was determined using binomial logistic regression where “-” or “+” represented “low” expression and “++”, “+++,” or “++++” represented “high” expression. The model was fit to a combined data set, and to adjust for measurement differences, a term for experimenter was included.
RNA Interference
All siRNA were from Dharmacon (Lafayette, CO). The sequence of the siRNA against CDC7 is AGU AGG ACC UGA AGA GAA A. A “smart pool” against CDC7 gave similar knockdown and induced apoptosis in HeLa cells, but a control nontarget siRNA (AUG AAC GUG AAU UGC UCA A) had no effect on Cdc7 expression. All tumor cell lines were transfected in six-well plates using 100 nM Cdc7 siRNA (or nontarget RNA) with Oligofectamine (Invitrogen, Carlsbad, CA). Subsequent titrations with HeLa cells showed that we could decrease the siRNA concentration to 10 nM with similar results.
Cell viability was determined by Trypan blue staining (0.4%) using standard methods, and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assays were performed according to the manufacturer's instructions (Roche, Indianapolis, IN).
Results
Cdc7-Dbf4 Protein Has Low Abundance in Normal Cell Lines and Tissues But Is Highly Expressed in Many Cancer Cell Lines
We raised monoclonal antibodies against human Cdc7-Dbf4 kinase and screened normal cells, the NCI-60 tumor cell lines plus five additional leukemia cell lines for Cdc7 and Dbf4 protein abundance. (We did not have access to the NCI-60 cell lines RXF-393, LOX IMVI, and MDA-N.)We were unable to detect Cdc7 protein in normal tissues of different origins relative to β-actin (Figure 1A). We also investigated the expression of the Cdc7 protein in whole-cell extracts from normal human fetal lung fibroblasts (WI-38), normal human lung fibroblasts (IMR-90 Figure 1B), and from bone marrow mononuclear cells (data not shown) and only detected a signal for Cdc7 protein in the IMR-90 cells. Because we can detect subnanogram amounts of recombinant Cdc7-Dbf4 proteins using our monoclonal antibodies, these data suggest that Cdc7-Dbf4 protein levels are quite low in normal cell lines and perhaps absent in postmitotic cells.
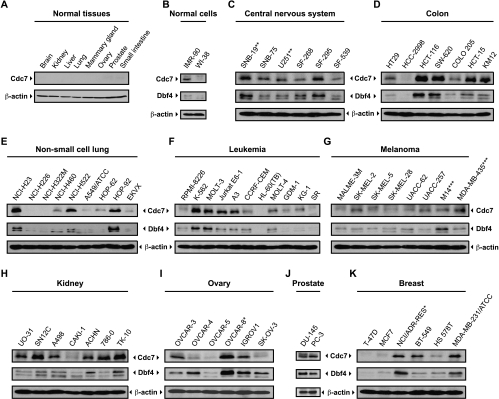
Figure 1.
Abundance of Cdc7 and Dbf4 proteins in normal and tumor cell lines. Twenty micrograms of protein sample from whole-cell extracts was separated on 10% SDS-PAGE gels for normal tissues of different origins (A), and immortalized IMR-90 and WI-38 fibroblast cell lines (B). Panels (C–K) represent tumor cell lines of the central nervous system, colon, non-small cell lung, leukemia, melanoma, kidney, ovary, prostate, and breast. After transfer, membranes were probed with a Cdc7- or Dbf4-specific antibody. Equal loading was confirmed by probing with a β-actin-specific antibody. Asterisk (*) indicates the cell lines that were shown to be derived from the same tumor [41,43].
In contrast, the Cdc7 protein was overexpressed in ∼50% of 62 tumor cell lines relative to β-actin, which has a relatively constant abundance between the different cell lines (Figure 1, C–K). The leukemia cell lines showed a wide range of Cdc7 protein expression (Figure 1F). K-562 (chronic myelogenous leukemia), MOLT-3 (acute lymphocytic leukemia), A3 and Jurkat E6-1 (two Jurkat, acute T-cell leukemia), and MOLT-4 (acute lymphocytic leukemia) exhibited a two- to fivefold increase of Cdc7 protein expression relative to the leukemia lines with lower Cdc7 expression. The higher Cdc7 expression of these cells lines did not correlate with a faster growth rate. Similarly, different levels of Cdc7 protein expression were observed for the central nervous system, colon, lung, melanoma, kidney, ovary, prostate, and breast cancer cell lines (Figure 1). Cdc7 protein was also very abundant in the two prostate cancer cell lines, namely, DU-145 and PC-3 (Figure 1J). We also confirmed that CDC7 and DBF4 mRNA expression was induced in various leukemia cell lines as reported previously [29] (data not shown).
In most cases, Dbf4 protein expression paralleled Cdc7 expression. When Cdc7 protein was overexpressed, Dbf4 protein was either overexpressed or present. When Cdc7 protein expression was very low or absent, Dbf4 protein was also expressed at very low levels. The exceptions were two ovarian cancer cell lines OVCAR-4 and SK-OV-3 that had low levels of Cdc7 but significant Dbf4 expression. We note that the NCI/ADR-RES cell line was originally thought to be an adriamycin-resistant derivative of MCF7 but was recently determined to derive from OVCAR-8 (human ovarian carcinoma cells) [41]. Consistently, we saw that Cdc7 and Dbf4 were overexpressed in both OVCAR-8 and NCI/ADR-RES (Figure 1, I and K).
High Level of Cdc7-Dbf4 Expression Is Correlated with p53 Loss
In Table 1, we list the doubling time, ploidy [42], relative Cdc7 protein levels (Figure 1), and the TP53 gene status [43] for the 57 cell lines from the NCI-60 set of tumor cell lines. Cdc7 protein was scored as undetectable (-), weakly expressed (+), or overexpressed (++ to ++++) from two independent experiments (the second measurements are summarized in Table W1). We found a significant association between Cdc7 protein expression and p53 mutational status (P = .000324). Approximately 90% of the cells overexpressing Cdc7 in our two data sets were mutant for p53. However, there was no correlation with Cdc7 (or Dbf4) protein expression and doubling time of the cells (P = .271), indicating that Cdc7-Dbf4 expression is not correlated with growth rate.
Table 1.
Doubling Time, Ploidy, Cdc7 Expression Level, and TP53 Gene Status in the 57 Cell Lines from the NCI-60 Set.
| Panel Name | Cell Name | Doubling Time* | Ploidy† | Cdc7 Band Intensity‡ | TP53 Gene Status |
|---|---|---|---|---|---|
| Central nervous system cancer | SNB-19§ | 34.6 | 3 | ++++ | m |
| SNB-75 | 62.8 | 2 | ++ | m | |
| U251§ | 23.8 | 2 | ++ | m | |
| SF-268 | 33.1 | 2 | + | m | |
| SF-295 | 29.5 | 5 | +++ | m | |
| SF-539 | 35.4 | 4 | + | m | |
| Colon cancer | HT29 | 19.5 | 3 | ++ | m |
| HCC-2998 | 31.5 | 2 | - | m | |
| HCT-116 | 17.4 | 2 | ++++ | wt | |
| SW-620 | 20.4 | 2 | ++++ | m | |
| COLO 205 | 23.8 | 3 | + | m | |
| HCT-15 | 20.6 | 2 | ++++ | m | |
| KM12 | 23.7 | 2 | ++++ | m | |
| Non-small cell lung cancer | NCI-H23 | 33.4 | 2 | ++++ | m |
| NCI-H226 | 61 | 3 | - | wt | |
| NCI-H322M | 35.3 | 2 | - | m | |
| NCI-H460 | 17.8 | 2 | + | wt | |
| NCI-H522 | 38.2 | 2 | +++ | m | |
| A549/ATCC | 22.9 | 3 | - | wt | |
| HOP-62 | 39 | 4 | + | m | |
| HOP-92 | 79.5 | 4 | +++ | m | |
| EKVX | 43.6 | 3 | - | m | |
| Leukemia | RPMI-8226 | 33.5 | 3 | - | m |
| K-562 | 19.6 | 3 | ++++ | m | |
| CCRF-CEM | 26.7 | 2 | + | m | |
| HL-60(TB) | 28.6 | 2 | - | m | |
| MOLT-4 | 27.9 | 4 | ++ | m | |
| SR | 28.7 | 2 | - | wt | |
| Melanoma | MALME-3M | 46.2 | 4 | - | wt |
| SK-MEL-2 | 45.5 | 4 | ++ | m | |
| SK-MEL-5 | 25.2 | 4 | - | wt | |
| SK-MEL-28 | 35.1 | 4 | - | m | |
| UACC-62 | 31.3 | 3 | - | wt | |
| UACC-257 | 38.5 | 3 | ++ | wt | |
| M14¶ | 26.3 | 3 | ++ | m | |
| MDA-MB-435¶ | 25.8 | 2 | +++ | m | |
| Kidney cancer | UO-31 | 41.7 | 2 | +++ | wt |
| SN12C | 29.5 | 3 | ++++ | m | |
| A498 | 66.8 | 3 | ++ | wt | |
| CAKI-1 | 39 | 3 | - | wt | |
| ACHN | 27.5 | 2 | ++ | wt | |
| 786-0 | 22.4 | 4 | ++++ | m | |
| TK-10 | 51.3 | 4 | ++++ | m | |
| Ovary cancer | OVCAR-3 | 34.7 | 3 | ++ | m |
| OVCAR-4 | 41.4 | 3 | + | m | |
| OVCAR-5 | 48.8 | 2 | - | wt | |
| OVCAR-8¶ | 26.1 | 2 | ++++ | m | |
| IGROV1 | 31 | 4 | ++ | m | |
| SK-OV-3 | 48.7 | 4 | - | m | |
| Prostate cancer | DU-145 | 32.3 | 3 | ++++ | m |
| PC-3 | 27.1 | 4 | ++++ | m | |
| Breast cancer | T-47D | 45.5 | 2 | - | m |
| MCF7 | 25.4 | 3 | - | wt | |
| NCI/ADR-RES$ | 34 | 2 | ++ | m | |
| BT-549 | 53.9 | 3 | ++ | m | |
| HS 578T | 53.8 | 3 | + | m | |
| MDA-MB-231/ATCC | 41.9 | 2 | ++ | m |
Doubling time in hours (see http://dtp.nci.nih.gov/docs/misc/common_files/cell_list.html).
For the ploidy, 2 means diploid, numbers greater than 2 indicate an increase of the chromosome number.
Cdc7 protein expression level was scored from the immunoblot analysis presented in Figure 1 as follows: “-” indicates no Cdc7 expression; “+,” low Cdc7 expression; “≥++,” Cdc7 overexpression.
Indicate cell lines that were shown to be derived from the same tumor.
Indicate cell lines that were shown to be derived from the same tumor.
Indicate cell lines that were shown to be derived from the same tumor.
m, indicates mutant; wt, wild type.
To determine whether there is a correlation between TP53 status and CDC7 or DBF4 mRNA expression in primary tumors, we analyzed the gene expression data from a breast cancer study in which the TP53 gene status was determined using the Oncomine database (www.ocomine.com). The study by Miller et al. [37] included Affymetrix data from 251 breast cancers, that is, 193 wild type and 58 mutant for TP53. By comparing the normalized CDC7 expression, we found that CDC7 expression was significantly higher in breast cancer with mutant TP53 (P = 3.6 x 10-9). Similarly, DBF4 expression was significantly higher in this data set for tumors with mutant TP53 (P = 1.8 x 10-10). These data suggest that the increased CDC7 and DBF4 expression in breast tumors and in cancer cell lines from diverse tissues can be related, at least in part, to the TP53 gene status.
A recent study suggested that several genes involved in DNA replication (including CDC7 and DBF4) may be repressed by p53 [44]. Therefore, we compared CDC7 and DBF4 mRNA expression using reverse transcription-polymerase chain reaction (RT-PCR) in isogenic HCT-116 p53+/+ and p53-/- cells [34]. The parental HCT-116 colon tumor cells have a relatively normal diploid DNA content and have intact DNA damage checkpoints. We found that CDC7 and DBF4 expression were indistinguishable in both cell lines (Figure W1A) and we saw no difference in Cdc7-Dbf4 protein expression (data not shown). To test whether the induction of p53 after exposure to DNA damage repressed CDC7 and DBF4 expression, we also measured expression of these genes in HCT-116 p53+/+ cells after exposure to bleomycin. Once again, we saw no evidence for p53 regulation of CDC7 or DBF4 expression (Figure W1B) after exposure to DNA damage. Thus, our data suggest that p53 does not directly regulate expression of the CDC7-DBF4 genes in HCT-116 cells.
DBF4 Copy Number Is Increased in Some Cancer Cell Lines
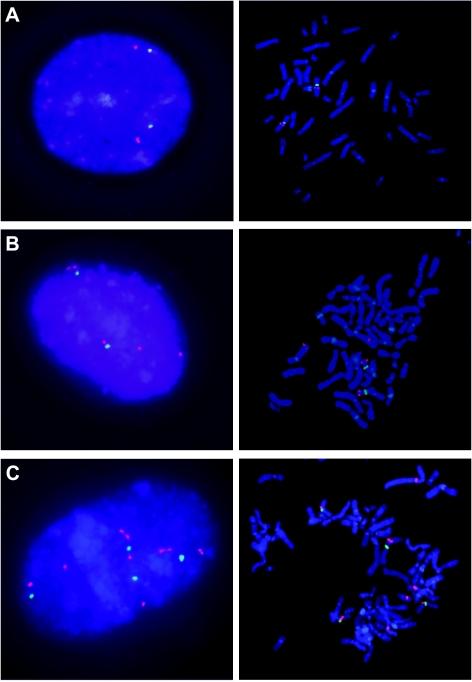
To identify a mechanism that might contribute to Cdc7 and Dbf4 overexpression, we examined CDC7 and DBF4 gene copy number. DBF4 is located at 7q21.12 and CDC7 at 1p22.2. We performed DBF4 FISH on the breast, ovarian, and prostate cancer cell lines (Figure 2 and Table 2). As a control, we performed FISH on phytohemagglutinin-stimulated lymphocytes derived from a karyotypically normal male donor (Figure 2A). Fluorescence in situ hybridization patterns observed in the interphase and metaphase spreads of the cancer cell lines OVCAR-8 and OVCAR-4 suggest that gene dosage is one of the mechanisms by which Dbf4 protein level is altered. For these two ovary cancer cell lines, we can easily visualize the presence of two and four extra copies of the DBF4 gene for OVCAR-8 and OVCAR-4, respectively (Figure 2, B and C).
Figure 2.
Increased DBF4 copy number in cancer cell lines. Fluorescence in situ hybridization experiments were done with a centromeric probe specific to the chromosome 7 (CEP7; SpectrumGreen-labeled) versus the DBF4-specific probe (SpectrumOrange-labeled) on interphase nuclei and on metaphase spreads. (A) Control FISH experiment was done on phytohemagglutinin-stimulated lymphocytes derived from a karyotypically normal male donor. Pictures are also shown for two ovary cancer cell lines OVCAR-8 and OVCAR-4 (B and C), respectively.
Table 2.
DBF4 Gene Dosage in Ovary, Prostate, and Breast Cancer Cell Lines.
| Cancer Cell Lines | DBF4 | DBF4/CEP7 Ratio | |
|---|---|---|---|
| Range | Mean | ||
| Ovary | |||
| OVCAR-3 | 2–13 | 3.78 | 1.55 |
| OVCAR-4 | 4–13 | 6.46 | 1.55 |
| OVCAR-5 | 1–3 | 2.06 | 0.64 |
| OVCAR-8* | 3–9 | 4.28 | 1.95 |
| IGROV1 | 3–9 | 4.04 | 1.24 |
| SK-OV-3 | 2–3 | 2.46 | 1.02 |
| Prostate | |||
| DU-145 | 3–6 | 3.62 | 1.02 |
| PC-3 | 5–14 | 6.16 | 1 |
| Breast | |||
| T-47D | 2–4 | 2.28 | 0.54 |
| MCF7 | 4–6 | 5.4 | 2.57 |
| NCI/ADR-RES* | 3–5 | 4.18 | 2.07 |
| BT-549 | 1–6 | 2.98 | 0.97 |
| HS 578T | 1–6 | 2.72 | 0.97 |
| MDA-MB-231/ATCC | 2–6 | 2.78 | 1.00 |
Range denotes the range of signals per cell counted in 50 nuclei for each cell line, and mean denotes the mean number of signals per cell. Gene overrepresentation is noted if the ratio of DBF4/CEP7 is ≥1.5 and gene amplification is noted if the ratio is ≥2.5.
Indicates cell lines that were shown to be derived from the same tumor.
Fluorescence in situ hybridization analyses on interphase nuclei were used to quantify the number of DBF4 gene copies per CEP7 copies per cell in the cell lines. A ratio ≥ 1.5 for DBF4 (orange) to CEP7 (green) was observed in three (OVCAR-3, OVCAR-4, and OVCAR-8) of six ovary cell lines (Table 2). These results are consistent with increased Dbf4 protein levels in these three cell lines (Figure 1I). We also found a DBF4/CEP7 ratio of 0.64 for OVCAR-5, which correlated with our inability to detect Dbf4 protein in this cell line (Table 2 and Figure 1I). We also examined relative DBF4 copy number in the two prostate cancer cell lines, namely, DU-145 and PC-3. Although the calculated ratios were 1 for both cell lines (Table 2), we observed more DBF4 gene copies, that is, four for DU-145 and six for PC-3, due to increased copies of chromosome 7 (Table 2). Among the breast cancer cell lines, NCI/ADR-RES and MCF7 showed an increased DBF4/CEP7 ratio (Table 2). For the NCI/ADR-RES, we obtained a ratio of 2, consistent with the ratio calculated for the OVCAR-8 cell line, from which NCI/ADR-RES is derived [41]. For the MCF7 cells, we calculated a ratio of 2.57; however, this cell line did not have increased Dbf4 protein levels (Figure 1K). The lower ratio obtained for the T-47D (0.54) once again correlated with the absence of detectable Dbf4 protein (Figure 1K).
We also examined CDC7 copy number using FISH (Spectrum Orange-labeled) for the six breast cancer cell lines relative to a control probe on the 1q arm (SpectrumGreen-labeled) but found no evidence of CDC7 gene amplification on interphase nuclei (data not shown).
Cdc7 Protein Is Highly Expressed in Some Primary Tumors
We examined 20 breast carcinomas, 10 colorectal cancers, and 5 non-small lung cancers from 35 different patients by immunohistochemical staining, using a combination of two monoclonal antibodies against Cdc7 protein. As expected, the Cdc7 staining in each case was nuclear as determined by the hematoxylin counterstaining of the nuclear structures. In each case, the negative controls with either no primary or secondary antibody showed no staining (data not shown). Cdc7 protein was scored as (-) when absent, (+) when the staining was weak, and (≥++) when the staining was intense. We were able to detect weak Cdc7 protein in 5 of 20 breast carcinomas and intense staining in 15 of 20 tumors (Table W2). Interestingly, 6 of 15 breast tumors overexpressing Cdc7 protein were metastatic but 0 of 5 tumors that exhibited weak staining were metastatic. We obtained the same trend for the colon cancers: all stained for Cdc7 protein, 2 of 10 had low levels and 8 of 10 were intensely stained for Cdc7 protein (Table W2). For the lung cancers, three of five sections showed no Cdc7 staining but two of five revealed Cdc7 overexpression (Table W2). In summary, 71% (25/35) of the primary tumors we tested had significantly elevated Cdc7 protein levels.
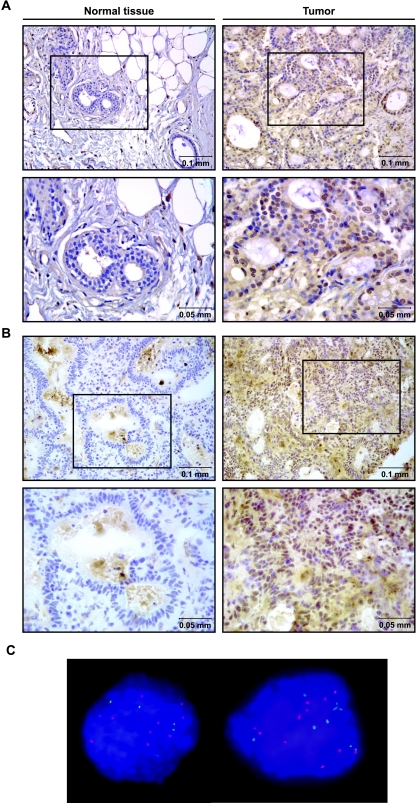
For six patient samples, we performed the immunohistochemistry on tumor sections and matched normal tissues. We observed Cdc7 nuclear staining for primary tumors but no staining in the adjacent normal tissues in all six patients. Representative pictures are shown for normal breast tissue and breast carcinoma (Figure 3A) and for normal colon tissue and colon carcinoma (Figure 3B). These results correlate very well with the immunoblot analysis results described previously, which detected Cdc7 expression in some cancer cell lines but not in normal cells or tissues.
Figure 3.
Immunostaining of Cdc7 in primary breast and colon tumors. (A) Cdc7 protein staining of normal breast tissue (left) and the corresponding breast carcinoma (right) shown at low and high (below) magnifications. (B) Normal colon tissue (left) and tumor (right) stained for Cdc7 protein at low and high (below) magnifications. (C) Fluorescence in situ hybridization experiments were done as described in Figure 2. A representative picture of nuclei from a breast primary tumor showing increased DBF4 gene copy number is shown.
DBF4 Gene Status in Human Primary Tumors Overexpressing Cdc7 Protein
To determine if increased copies of the DBF4 or CDC7 genes occurred in primary tumors, we performed DBF4 and CDC7 FISH on seven breast and three colon primary tumors with high Cdc7 protein expression. Interestingly, we found one breast carcinoma that had increased DBF4 copy number with ∼10 copies of the DBF4 gene and 5 copies of the CEP7 control (counting data not shown; Figure 3C). We saw no evidence for CDC7 duplication or amplification. Consistent with the results seen in the cancer cell lines, increased DBF4 gene copy number may also occur in some primary tumors.
CDC7 and DBF4 mRNA Expression Is Increased in Multiple Primary Tumors of Diverse Origin
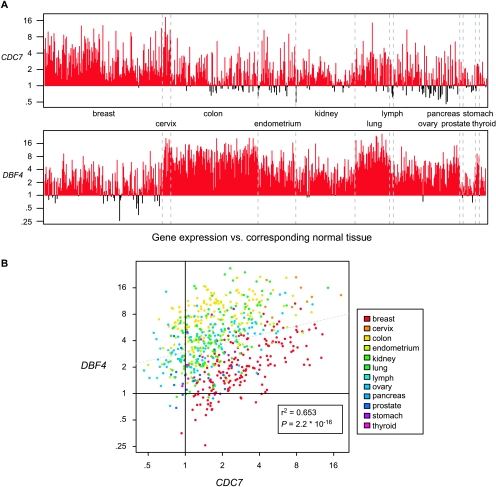
Finally, we examined CDC7 and DBF4 gene expression in 678 tumors of diverse origin relative to the expression in the normal tissues. These data were obtained from the Gene Expression Omnibus (GSE2109). Unprocessed gene expression data derived from more than 1000 tumors were preprocessed using the RMA method as described in the Materials and Methods section. For each tumor gene expression value, the median gene expression value of the corresponding normal tissue was subtracted, which was possible for 678 tumor samples. As shown in Figure 4A, both CDC7 and DBF4 expressions were significantly increased relative to normal controls in most of the 12 tumor types in this data set. DBF4 expression was consistently high in tumors of the colon, lung, and ovary, whereas CDC7 was consistently high in tumors of the breast and lung. When we examined the expression of both genes in individual tumor samples, we found a significant correlation (P ≤ 2.2 x 10-16) for their coordinate expression (Figure 4B). These data agree well with our survey of Cdc7 and Dbf4 protein expression in cancer cell lines and primary tumors and argue that high CDC7 and DBF4 expression is a common occurrence in human tumors.
Figure 4.
Overexpression of CDC7 and DBF4 in human cancers. (A) The gene expression level of CDC7 and DBF4 in tumor samples compared with corresponding normal tissue (see the Materials and Methods section). Plotted is the fold increase or fold decrease in gene expression in each tumor sample (n = 678). Red bars highlight the tumors that have increased gene expression. (B) CDC7 and DBF4 expression data described in (A) as a scatter plot with correlation coefficient, significance value (boxed), and best-fit line generated using linear regression.
siRNA-Mediated Knockdown of CDC7 Causes Growth Arrest and Apoptosis in Tumor Cell Lines
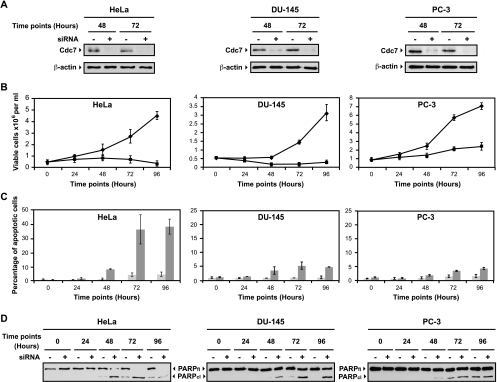
Cdc7 has been shown to be essential in mammalian cells by antibody microinjection experiments [10], a targeted knockout of the murine CDC7 gene [45], and siRNA against CDC7 expression in human cells [33].We independently found that siRNA-mediated knockdown of Cdc7 in HeLa cells caused growth arrest followed by apoptosis (Figure 5 and Figure W2) but that Cdc7 knockdown in the normal immortalized fibroblast cell line WI-38 caused growth arrest with high viability but with no apoptotic induction (not shown). As a control, non-target siRNA-transfected HeLa cells behaved similarly to the mock-transfected cells in that they retained Cdc7 protein expression and did not significantly undergo an apoptotic response (Figure W2). We show blots for Cdc7 protein at 48 and 72 hours after transfection (Figure 5A), but we typically see that Cdc7 protein is undetectable at 24 hours after transfection and remains absent through the 96-hour time point (Figure W2). We further examined the result of CDC7 knockdown in the two prostate cell lines of the NCI-60 panel, namely, DU-145 and PC-3 cells. Both of these cell lines express high levels of Cdc7 and Dbf4 protein (Figure 1J). Cdc7 knockdown in these two cells also causes growth arrest but only a small fraction of cells undergo apoptosis, evidenced by the TUNEL assay (Figure 5, B and C) and PARP blots (Figure 5D). Both cell lines maintain a high viability by Trypan blue staining after Cdc7 knockdown (data not shown). Because both of these cell lines are mutant for p53 and are capable of undergoing apoptosis under various conditions [46,47], the large difference in apoptotic response with HeLa cells suggests some other underlying genetic difference. PC-3 cells are known to express high levels of the antiapoptotic regulator Bcl-2 [46,47], so this might contribute to their failure to undergo significant apoptosis after Cdc7 knockdown.
Figure 5.
siRNA knockdown of CDC7 in tumor cell lines causes growth arrest and apoptosis. (A) Cdc7 immunoblots at 48 and 72 hours after transfection for mock and Cdc7 siRNA-transfected HeLa, DU-145, and PC-3 cells. Actin loading control is shown below. (B) Graphs showing cell number for control-transfected (◆) and Cdc7 knockdown cells (▪) over time. Data represent the mean of at least two experiments ± SD. (C) The percentage of TUNEL-positive cells are shown for each cell line for control transfected (light gray bars) and Cdc7 knockdown cells (dark gray bars) +/- SD. (D) Immunoblots of PARP full length (FL) and cleavage product (CL) are shown for the transfection time course.
Discussion
Cdc7 and Dbf4 proteins form a complex in vivo [3] to generate an active protein kinase in all organisms tested (reviewed by Sclafani [2]). Human Cdc7-Dbf4 kinase activity is essential for cell proliferation by activating DNA replication [10,32,33] (and data not shown). Although Cdc7 abundance is constant through the cell cycle, its kinase activity is strictly dependent on Dbf4, which significantly increases in abundance before S-phase entry.
The NCI-60 cell lines represent the most common forms of cancer in the United States [48] and are widely used by the research community. Unfortunately, neither CDC7 nor either of the DBF4-related genes were represented in the published microarray data for the NCI-60 cell lines [49–51]. In this study, we showed that there was a wide range of Cdc7 protein expression among the NCI-60 cell lines and five additional leukemia cell lines. In contrast, we consistently observed low or undetectable Cdc7 protein in normal tissues and cell lines. Our results support and significantly extend a previous examination of CDC7 mRNA expression in cell lines and different primary tumors versus normal tissues [29]. Hess et al. [29] found a significant number (15/48) of the tumor samples expressing CDC7 mRNA at higher levels than those seen in the normal tissue. We determined that ∼50% of tumor cell lines we tested expressed high levels of Cdc7 and Dbf4 protein, including many leukemia cell lines. These data suggest that Cdc7-Dbf4 protein has a relatively low abundance in normal cells and is perhaps absent in nonproliferating tissues but has increased expression in some cancer cell lines derived from diverse tissues.
We also defined a significant correlation between Cdc7 protein expression level and the TP53 gene status. On the basis of published mutational data in tumor suppressor or oncogenes in the NCI-60 cell line set [43], we established that Cdc7 overexpression is frequently associated with the presence of mutation(s) in the TP53 gene. We also found that CDC7 and DBF4 expression levels were highly correlated with TP53 status in primary breast tumors, suggesting either that p53 directly represses the CDC7 and DBF4 genes [44] or its loss indirectly activates CDC7 and DBF4 expression. However, we saw no difference in CDC7 and DBF4 expression in isogenic HCT-116 cell lines that differ only by deletion of TP53. Also, induction of p53 in HCT-116 cells after exposure to bleomycin did not alter CDC7 or DBF4 mRNA expression. Because we also found some p53 mutant cell lines that had barely detectable Cdc7-Dbf4 protein expression, together these data indicate that p53 inactivation alone does not determine whether the protein kinase is highly expressed. It may be that p53 loss and an additional genetic alteration that frequently occurs during tumorigenesis give rise to increased Cdc7-Dbf4 expression.
Because Cdc7-Dbf4 is a two-subunit kinase, it was of interest to investigate whether Cdc7 and Dbf4 protein expressions were correlated in primary tumors and cancer cell lines. We analyzed the Dbf4 expression in 62 cancer cell lines with varying Cdc7 expression levels and found a tight correlation between the expression levels of both proteins. Dbf4 protein is frequently coexpressed with Cdc7 and low levels of Dbf4 correspond with low or undetectable levels of Cdc7 protein. Therefore, in the cancer cell lines we analyzed, the expression of the two partners is highly correlated. Expression of the two genes was also highly correlated in 678 tumors of diverse origin (P ≤ 2.2 x 10-16).
A comparative genomic hybridization analysis of 38 breast cancer cell lines showed that the 7q21–22 region, containing the DBF4 gene, is an amplification site (11% of the cases) [52]. Moreover, the regional gene activation of this region was also found as a potential mechanism for acquired drug resistance, with or without changes in gene dosage and in breast and ovary cancer cell lines [53,54]. We naturally wondered if gene amplification could explain the higher Dbf4 protein expression seen in a subset of tumor cell lines. Seven of 14 ovarian, breast, and prostate tumor cell lines had an increased copy number of the DBF4 gene (by FISH analysis) resulting from unbalanced chromosome translocations or increased chromosome 7 copy numbers. With the exception of MCF7, these cell lines also had increased Dbf4 protein levels. In contrast, FISH analysis revealed no evidence for increased CDC7 copy number, indicating that the increased Cdc7 protein expression was independent of 1p22 amplification in the cell lines we examined. Other mechanisms, such as epigenetic modification and chromatin remodeling, may contribute to the regional gene activation in this region. Recently, a group identified that melanoma cell lines with defects in the DNA damage G1 checkpoint displayed an enhanced expression of proliferation-associated genes such as CDC7 [55].
The results obtained by immunohistochemistry on primary tumors paralleled our immunoblot analyses on cell lines. We showed that Cdc7 protein could be detected directly in primary tumors but not in the normal tissues. Of 35 tumors from breast, colon, and lung, 25 exhibited high-level Cdc7 expression. Of particular interest was the finding that 15 of 20 breast carcinomas showed intense Cdc7 staining suggesting that increased Cdc7 expression may be a common feature of breast cancer. Because six metastatic breast carcinomas showed intense Cdc7 staining, increased Cdc7 expression occurs in more aggressive tumors as well. We further showed that most breast cancers surveyed increased CDC7 expression over normal controls (162/181 ≥ 1.5-fold and 123/181 ≥ 2-fold increase; Figure 4A).
We found that DBF4 expression was increased in almost all 678 tumors we surveyed, especially tumors of the colon, lung, and ovary. Recently, it was shown that Dbf4 protein was not only associated with a higher relative change in the transition from nevi to cutaneous melanoma but was also up-regulated in a high number of primary melanoma and melanoma metastasis patients [32]. On the basis of our data, it is possible that increased DBF4 copy number or p53 loss might contribute to Dbf4 protein overexpression. The Cdc6 and Mcm5 replication initiation proteins are also overexpressed in cervical and bladder carcinomas [26,28] suggesting that increased expression of particular replication initiation proteins may be a common feature of certain tumors. Taken together, these data suggest that increased Cdc7 and Dbf4 expression may be selected during tumorigenesis in some tissue types.
It will be of interest to determine the functional consequences of altered Cdc7 and Dbf4 protein expression levels across multiple cell lines. Previous studies have shown that knockdown of Cdc7 in the HeLa cervical cancer cell line (but not normal cells) results in an apoptotic response [33]. We saw the same effect but note that in two prostate cancer cell lines, loss of Cdc7 did not result in significant apoptosis. Because Cdc7-Dbf4 is required for entry into S-phase, higher levels of Cdc7-Dbf4 kinase might aid the proliferative capacity of tumor cells. However, increased Cdc7 expression was not correlated with the proliferative status of the cells (P = .271). Therefore, increased Cdc7 expression is apparently not required for a high proliferation rate. In fact, transient overexpression of the hamster Cdc7 and/or Dbf4 caused a cell cycle arrest [56], and in human cells, overexpression of both human Cdc7 and Dbf4 does not cause significant effects on cell cycle progression [57]. Because Cdc7-Dbf4 has been implicated in the response to stalled forks or various forms of DNA damage [7,17–20], increased Cdc7-Dbf4 kinase may instead aid recovery or repair of stalled replication forks to enhance survival of some tumor cells. Together, these observations suggest that increased Cdc7-Dbf4 expression may be a common and/or important step during tumorigenesis.
Supplemental Materials and Methods
Bleomycin Treatment
Bleomycin Sigma-Aldrich was resuspended in water at 20 mg/ml. A final concentration of 5 µg/ml was added the culture medium for 2 and 4 hours.
Reverse Transcription-Polymerase Chain Reaction
Total RNA was extracted from the cells using Trizol (Invitrogen) according to the manufacturer's instructions. Approximately 0.5 µg of total DNase-treated RNA from each cell line was reverse-transcribed to cDNA and amplified using SuperScript One-Step RT-PCR with Platinum Taq (Invitrogen) in a final volume of 50 µl. The cDNA synthesis was done at 52°C for 30 minutes then the cDNA was amplified following different conditions depending on the mRNA, in duplicate. The following oligonucleotide primer sets were used for the reactions with expected product size in parentheses: CDC7 (681 bp) 5′-GGCAAGATAATGTCATGGGA-3′ (sense) and 5′-TCCTCATCACAGCACTATTC-3′ (antisense); DBF4 (515 bp) 5′-GCATATACTGCAGAAACCACT-3′ (sense) and 5′-GAGGTTCCACCATACTTATC-3′ (antisense); β-actin (353 bp) 5′-GCTCGTCGTCGACAACGGCTC-3′ (sense) and 5′-CAAACATGATCTGGGTCATCTTCTC-3′ (antisense). The primers were designed to span adjacent exons to avoid genomic DNA contamination. Polymerase chain reactions were carried out for 2 minutes at 94°C followed by 27 (for β-actin), 28 (for CDC7), and 33 cycles (for DBF4) of 15 seconds at 94°C, 30 seconds at 52°C, and 1 minute at 68°C, followed by a 5-minute extension at 68°C. The products were separated by electrophoresis on 1.2% agarose gels in 1x TAE (Trisacetate-EDTA) buffer and were detected by ethidium bromide staining. The β-actin mRNA was used as a control for equal sample loading.
Supplementary Material
Acknowledgments
The authors thank Bert Vogelstein for the HCT-116 cell lines; James Resau and the tissue repository at Spectrum Health for tissue samples; Han-Mo Koo and George Vande Woude for the NCI-60 cell lines; Pam Swiatek and Julie Koeman for preparation of the FISH probes, helpful advice, and use of the fluorescence microscope; Brian Cao and Ping Zhao for raising monoclonal antibodies; and Bart Williams for encouragement and comments on the manuscript.
Footnotes
The authors thank the support of the Van Andel Institute and grants from the Michigan Economic Development Corporation (GR-205/085P1000549) and the American Cancer Society (RSG0506301GMC) for funding this work.
This article refers to supplementary materials, which are designated by Figures W1, W2, and Tables W1, W2 and are available online at www.neoplasia.com.
References
- 1.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 2.Sclafani RA. Cdc7p-Dbf4p becomes famous in the cell cycle. J Cell Sci. 2000;113:2111–2117. doi: 10.1242/jcs.113.12.2111. [DOI] [PubMed] [Google Scholar]
- 3.Kumagai H, Sato N, Yamada M, Mahony D, Seghezzi W, Lees E, Arai K, Masai H. A novel growth- and cell cycle-regulated protein, ASK, activates human Cdc7-related kinase and is essential for G1/S transition in mammalian cells. Mol Cell Biol. 1999;19:5083–5095. doi: 10.1128/mcb.19.7.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montagnoli A, Bosotti R, Villa F, Rialland M, Brotherton D, Mercurio C, Berthelsen J, Santocanale C. Drf1, a novel regulatory subunit for human Cdc7 kinase. EMBO J. 2002;21:3171–3181. doi: 10.1093/emboj/cdf290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartwell LH. Three additional genes required for deoxyribonucleic acid synthesis in Saccharomyces cerevisiae. J Bacteriol. 1973;115:966–974. doi: 10.1128/jb.115.3.966-974.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnston LH, Thomas AP. A further two mutants defective in initiation of the S phase in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1982;186:445–448. doi: 10.1007/BF00729467. [DOI] [PubMed] [Google Scholar]
- 7.Takeda T, Ogino K, Matsui E, Cho MK, Kumagai H, Miyake T, Arai K, Masai H. A fission yeast gene, him1(+)/dfp1(+), encoding a regulatory subunit for Hsk1 kinase, plays essential roles in S-phase initiation as well as in S-phase checkpoint control and recovery from DNA damage. Mol Cell Biol. 1999;19:5535–5547. doi: 10.1128/mcb.19.8.5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faul T, Staib C, Nanda I, Schmid M, Grummt F. Identification and characterization of mouse homologue to yeast Cdc7 protein and chromosomal localization of the cognate mouse gene Cdc7l. Chromosoma. 1999;108:26–31. doi: 10.1007/s004120050348. [DOI] [PubMed] [Google Scholar]
- 9.James SW, Bullock KA, Gygax SE, Kraynack BA, Matura RA, MacLeod JA, McNeal KK, Prasauckas KA, Scacheri PC, Shenefiel HL, et al. nimO, an Aspergillus gene related to budding yeast Dbf4, is required for DNA synthesis and mitotic checkpoint control. J Cell Sci. 1999;112:1313–1324. doi: 10.1242/jcs.112.9.1313. [DOI] [PubMed] [Google Scholar]
- 10.Jiang W, McDonald D, Hope TJ, Hunter T. Mammalian Cdc7-Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J. 1999;18:5703–5713. doi: 10.1093/emboj/18.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landis G, Tower J. The Drosophila chiffon gene is required for chorion gene amplification, and is related to the yeast Dbf4 regulator of DNA replication and cell cycle. Development. 1999;126:4281–4293. doi: 10.1242/dev.126.19.4281. [DOI] [PubMed] [Google Scholar]
- 12.Sato N, Arai K, Masai H. Human and Xenopus cDNAs encoding budding yeast Cdc7-related kinases: in vitro phosphorylation of MCM subunits by a putative human homologue of Cdc7. EMBO J. 1997;16:4340–4351. doi: 10.1093/emboj/16.14.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanow SK, Gold DA, Yoo HY, Dunphy WG. Xenopus Drf1, a regulator of Cdc7, displays checkpoint-dependent accumulation on chromatin during an S-phase arrest. J Biol Chem. 2003;278:41083–41092. doi: 10.1074/jbc.M307144200. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi TS, Walter JC. Cdc7-Drf1 is a developmentally regulated protein kinase required for the initiation of vertebrate DNA replication. Genes Dev. 2005;19:2295–2300. doi: 10.1101/gad.1339805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho WH, Lee YJ, Kong SI, Hurwitz J, Lee JK. CDC7 kinase phosphorylates serine residues adjacent to acidic amino acids in the minichromosome maintenance 2 protein. Proc Natl Acad Sci USA. 2006;103:11521–11526. doi: 10.1073/pnas.0604990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsuji T, Ficarro SB, Jiang W. Essential role of phosphorylation of MCM2 by Cdc7/Dbf4 in the initiation of DNA replication in mammalian cells. Mol Biol Cell. 2006;17:4459–4472. doi: 10.1091/mbc.E06-03-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinreich M, Stillman B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 1999;18:5334–5346. doi: 10.1093/emboj/18.19.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heffernan TP, Unsal-Kacmaz K, Heinloth AN, Simpson DA, Paules RS, Sancar A, Cordeiro-Stone M, Kaufmann WK. Cdc7/Dbf4 and the human S checkpoint response to UVC. J Biol Chem. 2007;282:9458–9468. doi: 10.1074/jbc.M611292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costanzo V, Gautier J. Single-strand DNA gaps trigger an ATR- and Cdc7-dependent checkpoint. Cell Cycle. 2003;2:17. doi: 10.4161/cc.2.1.290. [DOI] [PubMed] [Google Scholar]
- 20.Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell. 2003;11:203–213. doi: 10.1016/s1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- 21.Going JJ, Keith WN, Neilson L, Stoeber K, Stuart RC, Williams GH. Aberrant expression of minichromosome maintenance proteins 2 and 5, and Ki-67 in dysplastic squamous oesophageal epithelium and Barrett's mucosa. Gut. 2002;50:373–377. doi: 10.1136/gut.50.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karakaidos P, Taraviras S, Vassiliou LV, Zacharatos P, Kastrinakis NG, Kougiou D, Kouloukoussa M, Nishitani H, Papavassiliou AG, Lygerou Z, et al. Overexpression of the replication licensing regulators hCdt1 and hCdc6 characterizes a subset of non-small-cell lung carcinomas: synergistic effect with mutant p53 on tumor growth and chromosomal instability—evidence of E2F-1 transcriptional control over hCdt1. Am J Pathol. 2004;165:1351–1365. doi: 10.1016/S0002-9440(10)63393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korkolopoulou P, Givalos N, Saetta A, Goudopoulou A, Gakiopoulou H, Thymara I, Thomas-Tsagli E, Patsouris E. Minichromosome maintenance proteins 2 and 5 expression in muscle-invasive urothelial cancer: a multivariate survival study including proliferation markers and cell cycle regulators. Hum Pathol. 2005;36:899–907. doi: 10.1016/j.humpath.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 24.Murphy N, Ring M, Heffron CC, King B, Killalea AG, Hughes C, Martin CM, McGuinness E, Sheils O, O'Leary JJ. p16INK4A, CDC6, and MCM5: predictive biomarkers in cervical preinvasive neoplasia and cervical cancer. J Clin Pathol. 2005;58:525–534. doi: 10.1136/jcp.2004.018895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy N, Ring M, Heffron CC, Martin CM, McGuinness E, Sheils O, O'Leary JJ. Quantitation of CDC6 and MCM5 mRNA in cervical intraepithelial neoplasia and invasive squamous cell carcinoma of the cervix. Mod Pathol. 2005;18:844–849. doi: 10.1038/modpathol.3800361. [DOI] [PubMed] [Google Scholar]
- 26.Stoeber K, Halsall I, Freeman A, Swinn R, Doble A, Morris L, Coleman N, Bullock N, Laskey RA, Hales CN, et al. Immunoassay for urothelial cancers that detects DNA replication protein Mcm5 in urine. Lancet. 1999;354:1524–1525. doi: 10.1016/S0140-6736(99)04265-8. [DOI] [PubMed] [Google Scholar]
- 27.Stoeber K, Swinn R, Prevost AT, de Clive-Lowe P, Halsall I, Dilworth SM, Marr J, Turner WH, Bullock N, Doble A, et al. Diagnosis of genito-urinary tract cancer by detection of minichromosome maintenance 5 protein in urine sediments. J Natl Cancer Inst. 2002;94:1071–1079. doi: 10.1093/jnci/94.14.1071. [DOI] [PubMed] [Google Scholar]
- 28.Williams GH, Romanowski P, Morris L, Madine M, Mills AD, Stoeber K, Marr J, Laskey RA, Coleman N. Improved cervical smear assessment using antibodies against proteins that regulate DNA replication. Proc Natl Acad Sci USA. 1998;95:14932–14937. doi: 10.1073/pnas.95.25.14932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hess GF, Drong RF, Weiland KL, Slightom JL, Sclafani RA, Hollingsworth RE. A human homolog of the yeast CDC7 gene is overexpressed in some tumors and transformed cell lines. Gene. 1998;211:133–140. doi: 10.1016/s0378-1119(98)00094-8. [DOI] [PubMed] [Google Scholar]
- 30.Bardelli A, Parsons DW, Silliman N, Ptak J, Szabo S, Saha S, Markowitz S, Willson JK, Parmigiani G, Kinzler KW, et al. Mutational analysis of the tyrosine kinome in colorectal cancers. Science. 2003;300:949. doi: 10.1126/science.1082596. [DOI] [PubMed] [Google Scholar]
- 31.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nambiar S, Mirmohammadsadegh A, Hassan M, Mota R, Marini A, Alaoui A, Tannapfel A, Hegemann JH, Hengge UR. Identification and functional characterization of ASK/Dbf4, a novel cell survival gene in cutaneous melanoma with prognostic relevance. Carcinogenesis. 2007;28:2501–2510. doi: 10.1093/carcin/bgm197. [DOI] [PubMed] [Google Scholar]
- 33.Montagnoli A, Tenca P, Sola F, Carpani D, Brotherton D, Albanese C, Santocanale C. Cdc7 inhibition reveals a p53-dependent replication checkpoint that is defective in cancer cells. Cancer Res. 2004;64:7110–7116. doi: 10.1158/0008-5472.CAN-04-1547. [DOI] [PubMed] [Google Scholar]
- 34.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 35.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivshina AV, George J, Senko O, Mow B, Putti TC, Smeds J, Lindahl T, Pawitan Y, Hall P, Nordgren H, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 37.Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu ET, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ihaka R, Gentleman RC. A language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 40.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liscovitch M, Ravid D. A case study in misidentification of cancer cell lines: MCF-7/AdrR cells (re-designated NCI/ADR-RES) are derived from OVCAR-8 human ovarian carcinoma cells. Cancer Lett. 2007;245:350–352. doi: 10.1016/j.canlet.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 42.Roschke AV, Tonon G, Gehlhaus KS, McTyre N, Bussey KJ, Lababidi S, Scudiero DA, Weinstein JN, Kirsch IR. Karyotypic complexity of the NCI-60 drug-screening panel. Cancer Res. 2003;63:8634–8647. [PubMed] [Google Scholar]
- 43.Ikediobi ON, Davies H, Bignell G, Edkins S, Stevens C, O'Meara S, Santarius T, Avis T, Barthorpe S, Brackenbury L, et al. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol Cancer Ther. 2006;5:2606–2612. doi: 10.1158/1535-7163.MCT-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spurgers KB, Gold DL, Coombes KR, Bohnenstiehl NL, Mullins B, Meyn RE, Logothetis CJ, McDonnell TJ. Identification of cell cycle regulatory genes as principal targets of p53-mediated transcriptional repression. J Biol Chem. 2006;281:25134–25142. doi: 10.1074/jbc.M513901200. [DOI] [PubMed] [Google Scholar]
- 45.Kim JM, Nakao K, Nakamura K, Saito I, Katsuki M, Arai K, Masai H. Inactivation of Cdc7 kinase in mouse ES cells results in S-phase arrest and p53-dependent cell death. EMBO J. 2002;21:2168–2179. doi: 10.1093/emboj/21.9.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rokhlin OW, Bishop GA, Hostager BS, Waldschmidt TJ, Sidorenko SP, Pavloff N, Kiefer MC, Umansky SR, Glover RA, Cohen MB. Fasmediated apoptosis in human prostatic carcinoma cell lines. Cancer Res. 1997;57:1758–1768. [PubMed] [Google Scholar]
- 47.Tang DG, Li L, Chopra DP, Porter AT. Extended survivability of prostate cancer cells in the absence of trophic factors: increased proliferation, evasion of apoptosis, and the role of apoptosis proteins. Cancer Res. 1998;58:3466–3479. [PubMed] [Google Scholar]
- 48.Monga M, Sausville EA. Developmental therapeutics program at the NCI: molecular target and drug discovery process. Leukemia. 2002;16:520–526. doi: 10.1038/sj.leu.2402464. [DOI] [PubMed] [Google Scholar]
- 49.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 50.Scherf U, Ross DT, Waltham M, Smith LH, Lee JK, Tanabe L, Kohn KW, Reinhold WC, Myers TG, Andrews DT, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 51.Staunton JE, Slonim DK, Coller HA, Tamayo P, Angelo MJ, Park J, Scherf U, Lee JK, Reinhold WO, Weinstein JN, et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci USA. 2001;98:10787–10792. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Forozan F, Mahlamaki EH, Monni O, Chen Y, Veldman R, Jiang Y, Gooden GC, Ethier SP, Kallioniemi A, Kallioniemi OP. Comparative genomic hybridization analysis of 38 breast cancer cell lines: a basis for interpreting complementary DNA microarray data. Cancer Res. 2000;60:4519–4525. [PubMed] [Google Scholar]
- 53.McDonald SL, Stevenson DA, Moir SE, Hutcheon AW, Haites NE, Heys SD, Schofield AC. Genomic changes identified by comparative genomic hybridisation in docetaxel-resistant breast cancer cell lines. Eur J Cancer. 2005;41:1086–1094. doi: 10.1016/j.ejca.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 54.Wang YC, Juric D, Francisco B, Yu RX, Duran GE, Chen GK, Chen X, Sikic BI. Regional activation of chromosomal arm 7q with and without gene amplification in taxane-selected human ovarian cancer cell lines. Genes Chromosomes Cancer. 2006;45:365–374. doi: 10.1002/gcc.20300. [DOI] [PubMed] [Google Scholar]
- 55.Kaufmann WK, Nevis KR, Qu P, Ibrahim JG, Zhou T, Zhou Y, Simpson DA, Helms-Deaton J, Cordeiro-Stone M, Moore DT, et al. Defective cell cycle checkpoint functions in melanoma are associated with altered patterns of gene expression. J Invest Dermatol. 2008;128:175–187. doi: 10.1038/sj.jid.5700935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo B, Romero J, Kim BJ, Lee H. High levels of Cdc7 and Dbf4 proteins can arrest cell-cycle progression. Eur J Cell Biol. 2005;84:927–938. doi: 10.1016/j.ejcb.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 57.Sato N, Sato M, Nakayama M, Saitoh R, Arai K, Masai H. Cell cycle regulation of chromatin binding and nuclear localization of human Cdc7-ASK kinase complex. Genes Cells. 2003;8:451–463. doi: 10.1046/j.1365-2443.2003.00647.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.