Abstract
Peptide vaccination for cancer immunotherapy requires identification of peptide epitopes derived from antigenic proteins associated with tumors. Heparanase (Hpa) is broadly expressed in various advanced tumors and seems to be an attractive new tumor-associated antigen. The present study was designed to predict and identify HLA-A2-restricted cytotoxic T lymphocyte (CTL) epitopes in the protein of human Hpa. For this purpose, HLA-A2-restricted CTL epitopes were identified using the following four-step procedure: 1) a computer-based epitope prediction from the amino acid sequence of human Hpa, 2) a peptide-binding assay to determine the affinity of the predicted protein with the HLA-A2 molecule, 3) stimulation of the primary T-cell response against the predicted peptides in vitro, and 4) testing of the induced CTLs toward different kinds of carcinoma cells expressing Hpa antigens and/or HLA-A2. The results demonstrated that, of the tested peptides, effectors induced by peptides of human Hpa containing residues 525–533 (PAFSYSFFV, Hpa525), 277–285 (KMLKSFLKA, Hpa277), and 405–413 (WLSLLFKKL, Hpa405) could effectively lyse various tumor cell lines that were Hpa-positive and HLA-A2-matched. We also found that these peptide-specific CTLs could not lyse autologous lymphocytes with low Hpa activity. Further study revealed that Hpa525, Hpa277, and Hpa405 peptides increased the frequency of IFN-γ-producing T cells compared to a negative peptide. Our results suggest that Hpa525, Hpa277, and Hpa405 peptides are new HLA-A2-restricted CTL epitopes capable of inducing Hpa-specific CTLs in vitro. Because Hpa is expressed in most advanced malignant tumors, Hpa525, Hpa277, and Hpa405 peptide-based vaccines may be useful for the immunotherapy for patients with advanced tumors.
Introduction
Malignant tumors are among the most lethal diseases threatening humans. The therapeutic options for the treatment of patients with carcinomas are limited to three fundamental modalities: surgical resection, chemotherapy, and radiation therapy. For especially advanced carcinomas, these modalities do not yield good results. For the past few years, dendritic cell (DC)-based immunotherapy, which has the advantages of strong immunogenicity, weak side effects, and applicability, has become one of the hot topics in studies focused on the therapies for malignant tumors.
One of tactics for tumor immunotherapy is the use of tumor-associated antigen (TAA)-loaded DC. Dozens of TAAs have been described [1]. Unfortunately, the expression of most TAAs is restricted to a few tumor types and to a fraction of patients with these types of tumors, and the appearance of antigen-loss mutations in tumor cells in response to immune pressure is well described [2,3]. To circumvent this issue, a class of TAAs termed universal tumor antigens has been proposed that is hypothesized not only to trigger T cell reactivity against a broad range of tumor types but also to play critical functional roles in tumor growth and development. An ideal universal TAA should have the following characteristics: 1) be expressed by the majority of human cancers but rarely be expressed in normal tissues, 2) be indispensable in the process of tumorigenesis to avoid antigen variation or depletion, 3) include peptide sequences that bind to major histocompatibility complex (MHC) molecules, and 4) be recognized by the T-cell repertoire in an MHC-restricted fashion to elicit specific T-cell response [4,5].
Cytotoxic T lymphocytes (CTLs) are considered to be chief mediators of tumor immunosurveillance through the recognition of TAAs as cognate peptides bound to MHC molecules expressed on the surface of tumor cells. A major achievement in tumor immunology for the last 20 years has been the clear demonstration that CTL epitopes binding to MHC rather than integral TAAs induce CTL reactions. These epitope peptides are usually 8 to 10 amino acids long with 2 to 3 primary anchor residues that interact with the MHC class I molecules and 2 to 3 amino acid residues that bind to the T-cell receptor [6]. Therefore, the identification of CTL epitopes from TAAs has become a critical step in the development of peptide-based immunotherapy for cancer.
Heparanase (Hpa) is the only endogenous endoglycosidase found so far that can degrade the heparan sulfate proteoglycans in the extracellular matrix and basal membrane [7]. Unlike most other TAAs, the expression of Hpa in tumor cells has been linked to tumor invasion and metastasis. Heparanase can be found in almost all metastatic malignant tumor cells. In normal tissue, it is only expressed in leukomonocytes and bone marrow. Inhibition of Hpa can obviously inhibit the proliferation and metastasis of tumor cells [8]. Activation of Hpa is a determinant factor for the occurrence of metastasis, which makes tumor cells break through the extracellular matrix and basal membrane barrier, releases many kinds of cytokines, causes the formation of new vessels, and causes the local permanent planting of tumor cells [5,7–11]. Thus, Hpa is a potential universal TAA for the treatment of advanced stage tumors. Our previous study demonstrated that the DC-loaded full-length Hpa cDNA could induce an Hpa-specific CTL, which showed potent lysis of gastric carcinoma cells that were MHC-matched during Hpa expression, whereas it had no effects on cells that were not MHC-compatible [12]. These results indicate that Hpa can serve as a TAA that could be used for tumor immunotherapy. Conversely, CTL epitopes must exist in the Hpa protein that can induce specific CTL. Recently, Sommerfeldt et al. [13] successfully predicted three epitopes derived from the human Hpa amino acid sequence. Their results demonstrated that these three epitopes could elicit Hpa-specific CTLs capable of lysing breast cancer cells in vitro. Our previous study also demonstrated that effectors induced by peptides of mouse Hpa at residue positions 398–405 (LSLLFKKL, mHpa398) and 519–526 (FSYGFFVI, mHpa519) could lyse three kinds of carcinoma cells expressing both Hpa and H-2Kb (B16 melanoma cells, EL-4 leukoma cells, and Lewis lung cancer cells). In vivo experiments indicated that mHpa398 and mHpa519 peptides offered the possibility not only to immunize against tumors but also to successfully treat tumor-bearing hosts [14].
On the basis of the analysis previously mentioned, the objective of this study was mainly to find other possible HLA-A2-restricted CTL epitopes in human Hpa with the ability to induce an Hpa-specific antitumor immune response. For this purpose, we first predicted candidate epitopes restricted by HLA-A2 in the protein of Hpa using computer algorithms and molecular modeling. We then induced Hpa-specific CTLs from HLA-A2-positive peripheral blood mono-nuclear cells (PBMCs) from five healthy donors with these candidate peptides in vitro to seek CTL epitopes present in the Hpa antigen. We hope to find more Hpa epitopes capable of inducing an Hpa-specific antitumor immune response and provide a foundation for immunotherapy for patients with malignant tumors.
Materials and Methods
Cell Lines
The human TAP-deficient T2 cell line and BB7.2 cell line producing mAb against HLA-A2 were purchased from the American Type Culture Collection (Manassas, VA). The osteogenic sarcoma cell line U2OS (Hpa+,HLA-A2+) was purchased from Beijing Xiehe Medical University (Beijing, China). The gastric cancer cell line KATO-III (Hpa+,HLA-A2+), liver cancer cell line HepG2 (Hpa+,HLA-A2-), breast cancer cell line MCF-7 (Hpa+,HLA-A2+), and colon cancer cell line SW480 (Hpa+,HLA-A2+) were maintained in our laboratory. U2OS cells were cultured in McCoy's 5A medium (Life Technologies, Inc., Gaithersburg, MD; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 µg/ml streptomycin, and 2 mM l-glutamine. BB7.2 cells were maintained in DMEM containing 10% FCS, penicillin (100 U/ml), and streptomycin (100 pg/ml). T2, KATO III, HepG2, MCF-7, and SW480 cells were all cultured in RPMI-1640 medium containing 10% FBS, penicillin (200 U/ml), and streptomycin (100 µg/ml). All cell lines mentioned previously were kept at 37°C in a humidified atmosphere containing 5% CO2.
Epitope Prediction and Synthesizing
Human Hpa peptides with HLA-A2-binding motifs were predicted by computer analysis as described previously [15,16]. Five nonapeptides derived from the human Hpa amino acid sequence and one nonapeptide from HIV virus [HIVpol(476–484) (ILLEPVHGV)], which served as a positive control in MHC peptide-binding assay and a negative control in inducing Hpa-specific CTL reactions in vitro, were synthesized by the Beijing Scilight Biotechnology Ltd. Co. (Beijing, China) with a purity >90% as determined by HPLC (Delta 600 Wasters Company). The molecular weight of the peptides was validated by mass spectrum (API2000; PE, Waltham, MA). Lyophilized peptides were dissolved in DMSO (Sigma) and stored at -20°C.
Molecular Modeling and Dynamics Calculation
Models of nonapeptides binding with HLA-A2.1 molecules were established from the crystal structures of the MHC-I-peptide complex in the Brookhaven Protein Data Bank: 3HLA for HLA-A2.1 and 3HSA for the nonapeptides. The HLA-A2.1 model was simplified by using only α1 and α2 domains with 18 water molecules bound to them. Molecular mechanic and dynamic calculations were performed using the Discover 3.0 package. The force field parameters used in this study were those of the consistent valence force field. During the molecular dynamics and minimization calculation, a dielectric constant of 1.0 was used. A 9-Å cutoff distance was applied to calculate the nonbinding interaction. The peptide ligand was first relaxed by 2000 steps of conjugate gradient energy minimization while maintaining the fixed protein. It was then submitted to a 100-ps molecular dynamics calculation at 300 K. During these 100 ps, no protein atoms were allowed to move. The last conformation was then solvated in a 10-Å-thick TIP3P water shell. Energy minimization of the water shell followed by a 200-ps molecular dynamics simulation of the fully solvated HLA-A2/ligand pair was performed at 300 K. Finally, we obtained each of the candidate peptide-MHC complex binding parameters, including nonbond energy, hydrogen bond number, and anchor residue distance.
HLA Stabilization Analysis
The synthesized peptides were used in an MHC stabilization assay using T2 cells as described previously [17,18]. Briefly, T2 cells (2 x 105) were incubated with 200 µl of RPMI-1640 containing 0.1% FCS, 5 x 10-5 M β-mercaptoethanol, and each of the peptides at a concentration of 10 µg/ml for 15 hours. After the incubation, surface HLA-A2 molecules were stained with anti-HLA-A2 mAb (derived from BB7.2 hybridoma culture supernatant) for 30 minutes at 4°C. T2 cells were washed twice with PBS and stained with fluorescein isothiocyanate-conjugated IgG antibodies for 30 minutes. The cells were then rinsed three times with PBS and analyzed with a FACScan (Becton Dickinson, San Jose, CA). The relative binding affinity of the respective peptides was calculated from the mean fluorescence intensities (MFIs) as follows: MFI(peptide) - MFI(unloaded cells)/MFI(unloaded cells). Relative binding affinities >1.5 were considered strong; 1.5 to 1.0, intermediate; and <1.0, low.
Dendritic Cell Generation from Human Peripheral Blood Precursors
Dendritic cells from PBMCs were generated using the procedure described by Romani et al. [19]. Briefly, PBMCs were isolated from five healthy HLA-A2+ donors by Ficoll-Hypaque density gradient centrifugation and then seeded into culture flasks in RPMI-1640 medium supplemented with penicillin (100 U/ml), streptomycin (100 µg/ml), and 10% FBS. After monocytes adhered (incubation for 2 hours), the nonadherent cells were collected and frozen in freeze medium (60% RPMI-1640 and 30% FBS, 10% DMSO) for later use in CTL assays. The adherent cells were cultured for 5 days in RPMI-1640 containing 1000 U/ml of granulocyte-macrophage colony-stimulating factor (R&D Systems, Inc., Minneapolis, MN) and interleukin-4 (IL-4; R&D Systems, Inc.) and were the cultured for an additional 2 days in the presence of 1000 U/ml of tumor necrosis factor α (R&D Systems, Inc.) to induce final maturation. After 7 days of culture, the mature DCs were harvested and analyzed for DC typical phenotypes by FACS analysis.
Reverse Transcription-Polymerase Chain Reaction
Total RNA of target cells (KATO-III gastric cancer cells, U2OS osteogenic sarcoma cells, SW480 colonic cancer cells, MCF-7 breast cancer cell line, MCF-7 cells transduced with full-length cDNA of Hpa, HepG2 liver cancer cells) was isolated from cell extracts using the Tripure RNA isolation kit (Roche, Basel, Switzerland) following the manufacturer's instructions. One microgram of total RNA was reverse-transcribed, and the cDNA was amplified with Taq polymerase (Takara, Dalian, China) using specific primers for Hpa (sense primer: 5′-GAATGGCCCTACCAGGAGCA-3′ antisense primer: 5′-ACGCATTTAGGCCAAAGATCAAG-3′) or β-actin (sense primer: 5′-GTTGCGTTACACCCTTTCTTGACA-3′, antisense primer: 5′-GCACGAAGGCTCATCATTCAAAA-3′). After initial denaturation for 5 minutes at 94°C, the polymerase chain reaction thermal cycle profile was 45 seconds at 94°C, 60 seconds at 66°C, and 60 seconds at 72°C for 30 cycles, followed by a final extension for 5 minutes at 72°C. The products of Reverse Transcription-Polymerase Chain Reaction (RT-PCR) were electrophoresed in a 1.5% agarose gel and visualized by UVP after ethidium bromide staining.
Immunohistochemistry Staining
The previously mentioned target cells were cultured in six-well culture dishes and fixed with 3.7% paraformaldehyde for 7 minutes at room temperature. The dishes were then washed with buffer containing 0.1 M Tris (pH 7.5), 1.5 M NaCl, and 1% BSA and were permeabilized in the presence of 2% Triton X-100. Next, cells were incubated with an Hpa antibody (0.5 µg/ml; Santa Cruz Biotechnology, Santa Cruz, CA) for 1 hour at room temperature, washed, and further incubated with secondary antibody. Finally, the cells were incubated for 15 minutes with an avidin-biotin enzyme reagent. Slides were immersed in DAB/H2O2 solution to develop the staining. Phosphate-buffered saline was used as a negative control in place of the primary antibody.
Induction of Peptide-Specific CTL with Synthetic Hpa Peptides
The assay was done as described previously [20]. Briefly, DCs were loaded with different Hpa peptides at a final concentration of 100 µg/ml for 4 hours and were then irradiated with 20 Gy, which prevented all outgrowths in the control cultures. Autologous T cells were restimulated every 7 days with the previously mentioned peptide-pulsed DCs to generate peptide-specific CTLs. Recombinant interleukin 2 (IL-2) at a concentration of 20 U/ml was added to the culture medium on day 3 after every stimulation. Cytotoxic T lymphocyte activity was then assessed on day 23 by a 4-hour 51Cr release assay. Effectors generated from negative peptide-pulsed DCs were used as controls.
Cytotoxicity Assay
To evaluate the levels of CTL activity, a standard 4-hour 51Cr release assay was used as previously described [21]. Briefly, target cells were incubated with 51Cr (100 µCi per 1 x 106 cells) for 2 hours in a 37°C water bath. After incubation with 51Cr, target cells were washed three times with PBS, resuspended in RPMI-1640 medium, and mixed with effector cells at a 10:1, 20:1, 40:1, or 80:1 effector-to-target (E/T) ratio. Assays were performed in triplicate for each sample at each ratio in a 96-well round-bottomed plate. After a 4-hour incubation, the supernatants were harvested, and the amount of released 51Cr was measured with a gamma counter. The percent-specific lysis was calculated according to the following formula:
Enzyme-Linked Immunospot Assay
IFN-γ secretion of effectors was assayed by enzyme-linked immunospot (ELISPOT) [22]. Multiscreen 96-well assay plates (Dakewe, Shenzhen, China) were precoated overnight at 4°C with anti-IFN-γ antibody according to the manufacturer's instruction. After washing with PBST (PBS-0.05% Tween 20), plates were blocked for 1 hour at 37°C with PBS/1% BSA. Splenocytes were plated in triplicate wells at a density of 2 x 105/100 µl in RPMI-1640 medium. For restimulation, different peptides or medium alone was added. Plates were cultured overnight, washed extensively with PBST, and incubated with anti-IFN-γ mAb for 1 hour at 37°C. After washing, goat antibiotin antibodies (Dakewe) were added, and the plated were incubated for 1 hour at 37°C. Thirty microliters of activator solution (Dakewe) was added to develop spots, and after 10 to 30 minutes, the plates were washed with distilled water to stop the reaction. After being air-dried, the number of spots in each well was counted using the Bioreader 4000 PRO-X (Bio-Sys; Germany).
Results
Epitope Prediction and Molecular Modeling
The amino acid sequence of human Hpa was initially screened for peptides containing the supermotif for HLA-A2. We chose five peptides consisting of nine amino residues, which were found to contain the supermotif for HLA-A2, as predicted epitopes of human Hpa. These five peptides were all suitable HLA-A2-restricted CTL Hpa epitopes as calculated by the quantitative motif method. The sequences of these peptides were PAFSYSFFV (525–533, Hpa525), PLLSDTFAA (353–361, Hpa353), KMLKSFLKA (277–285, Hpa277), PLPDYWLSL (400–408, Hpa400), and WLSLLFKKL (405–413, Hpa405). Molecular modeling showed that these five potential CTL epitopes from Hpa met the criteria for HLA-A2-restricted CTL epitopes. As shown in Table 1, the peptides bound to the HLA-A2 mode structure possessed a side chain of COOH-terminal anchor residues oriented into the binding groove with different distances, ranging from 17 to 20 Å.
Table 1.
Characteristics of Hpa Candidate Epitope Bound to the Modeled HLA-A2.
| Peptide | Sequence | Score | Distance (Å) | H-bond Number | Nonbond Energy | SAS of Anchor Residue (Å) | |
| P2 | P9 | ||||||
| Hpa(525–533) | PAFSYSFFV | 428.78 | 17.62 | 4 | -17,158.9 | 3.6 | 4.9 |
| Hpa(353–361) | PLLSDTFAA | 359.37 | 18.23 | 3 | -16,144.5 | 15.6 | 18.7 |
| Hpa(277–285) | KMLKSFLKA | 140.81 | 17.81 | 6 | -19,237.5 | 5.4 | 6.98 |
| Hpa(400–408) | PLPDYWLSL | 91.21 | 19.58 | 4 | -16,276.2 | 2.9 | 8.7 |
| Hpa(405–413) | WLSLLFKKL | 61.29 | 18.94 | 5 | -18,324.7 | 3.7 | 4.6 |
Major Histocompatibility Complex Peptide-Binding Assay
The binding affinity of the previously mentioned five nonapeptides to HLA-A2 was determined using antigen processing-deficient T2 cells due to their enhanced HLA-A2 expression when exposed to exogenous HLA-A2-binding peptides. An octapeptide mHpa(519–526) (FSYGFFVI) derived from mouse Hpa and a nonapeptide HIVpol (476–484) (ILLEPVHGV) derived from HIV served as negative and positive controls, respectively. As shown in Table 2, all predicted peptides were capable of up-regulating HLA-A2 molecular expression and showed high affinity for HLA-A2.
Table 2.
Analysis of HLA-A2-Binding Affinity of Hpa-Derived Peptides.
| Name | Sequence | Position in Hpa | Mean Fluorescence Intensity | Fluorescence Index*† |
| HIVpol‡ | ILLEPVHGV | 476–484 | 912 ± 24.11 | 8.2 |
| Hpa525 | PAFSYSFFV | 525–533 | 885 ± 9.17 | 7.1 |
| Hpa353 | PLLSDTFAA | 353–361 | 895 ± 48.21 | 7.2 |
| Hpa277 | KMLKSFLKA | 277–285 | 837 ± 88.39 | 6.67 |
| Hpa400 | PLPDYWLSL | 400–408 | 762 ± 40.92 | 5.98 |
| Hpa405 | WLSLLFKKL | 405–413 | 850 ± 47.57 | 6.79 |
| mHpa519§ | FSYGFFVI | 519–526 | 115 ± 5.51 | — |
Fluorescence intensity was calculatedwith the following formula: FI = [MFI(peptide) - MFI(unloaded cells)]/MFI(unloaded cells).
Fluorescence intensity was determined as high (FI > 1.5), intermediate (1.5 > FI > 1.0), or weak (FI < 1.0).
HIVpol served as positive control.
mHpa519 served as negative control.
Expression of Hpa and HLA-A2 in Target Cells
The expression of Hpa mRNA in all cell lines in this study was analyzed by RT-PCR. The results demonstrated that Hpa mRNA was detected in KATO-III gastric cancer cells, U2OS osteogenic sarcoma cells, SW480 colonic cancer cells, and HepG2 liver cancer cells. Meanwhile, Hpa mRNA could not be detected in MCF-7 breast cancer cells but could be detected in MCF-7 cells transduced with the full-length cDNA of Hpa. Moreover, immunohistochemistry indicated the expression of Hpa protein in the cytoplasm of all the mentioned target cells except MCF-7 cells (Figure 1).
Figure 1.
Expression of Hpa in various target cells. (a) Expression of Hpa mRNA in various target cells. 1 indicates DNA marker; 2, KATO-III; 3, SW480; 4, U2OS; 5, HepG2; 6, MCF-7; 7, MCF-7/Hpa. (b) Expression of Hpa protein in various target cells. A indicates KATO-III; B, SW480; C, U2OS; D, HepG2; E, MCF-7; F, MCF-7/Hpa.
We also detected HLA-A2 expression in all target cells in the study by flow cytometry. The results indicated that the expression of HLA-A2 in KATO-III gastric cancer cells, U2OS osteogenic sarcoma cells, SW480 colonic cancer cells, or MCF-7 breast cancer cells was 90.0%, 86.8%, 73.8%, or 84.9%, respectively. Meanwhile, HLA-A2 expression in HepG2 liver cancer cells was only 3.1%. After being transfected with the plasmid of HLA-A2, the expression of HLA-A2 in HepG2/HLA-A2 cells was increased to 31.3%.
Identification of Hpa-Specific HLA-A2-Restricted CTL Epitopes by Cytotoxicity Assay
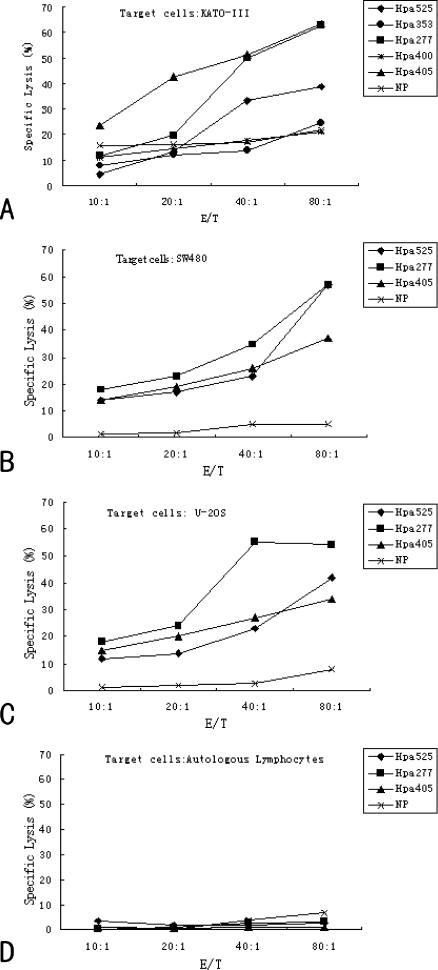
To detect whether the predicted peptides could generate Hpa-specific CTL in vitro, we used a standard 4-hour 51Cr release assay. As shown in Figure 2, the three peptides (Hpa525, Hpa277, and Hpa405) generated a highly specific 51Cr release at an E/T ratio from 40:1 to 80:1. These Hpa-specific CTLs caused greater than 40% lysis of KATO-III gastric cancer cells, which were both hTERT- and HLA-A2-positive at an E/T ratio of 80:1. However, the induced effectors generated from Hpa353 and Hpa400 could not lyse the mentioned target cells. Even at the highest E/T ratio, the lysis rate was approximately 20%. We further assessed U2OS osteogenic sarcoma cells and SW480 colonic cancer cells as target cells, both of which also expressed Hpa and HLA-A2. The results demonstrated that the CTLs induced by predicted peptides Hpa525, Hpa277, and Hpa405 had significant killing effects on U2OS and SW480 cells. Taken together, these results indicate that the predicted peptides Hpa353, Hpa277, and Hpa405 could induce Hpa-specific CTL responses in vitro and may be CTL epitopes of Hpa.
Figure 2.
Specific lysis of CTLs generated from different Hpa-derived peptides against various target cells. Effector-to-target ratios are presented on the x-axis, whereas the y-axis represents the percent of specific lysis. Cytotoxic T lymphocytes generated from HIV virus, HIVpol(476-484) (ILLEPVHGV), served as a negative peptide (NP). (A) Specific lysis of CTLs generated from Hpa525, Hpa353, Hpa277, Hpa400, and Hpa405 peptides against KATO-III. (B, C, and D) Specific lysis of CTLs generated from Hpa525, Hpa277, and Hpa405 peptides against SW480 (B), U2OS (C), and autologous lymphocytes (D).
Specificity of CTLs Directed against Hpa
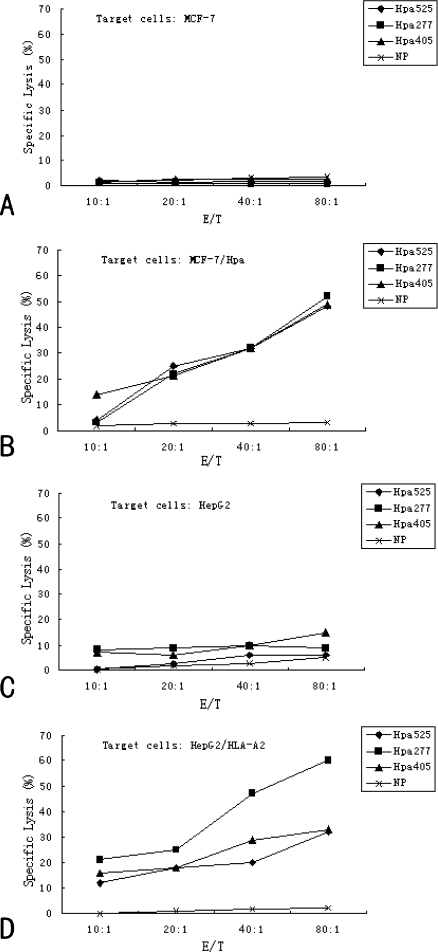
To further confirm the Hpa specificity of the CTLs, we took the advantage of the HLA-A2-positive, Hpa-negative breast cancer cell line, MCF-7 [13,23]. MCF-7 cells were transduced with pIRES2-EGFP-Hpa plasmid, a eukaryotic fluorescent expression vector containing the full-length cDNA of Hpa, by the DOTAP lipofection method according to the manufacturer's protocol [24]. After 24 hours of transfection, 400 µg/ml G418 was added to the RPMI-1640 medium. After G418 selection for 4 weeks, drug-resistant individual clones were randomly collected from the transfected cultures. The selected clone was named MCF-7/Hpa. Reverse transcription-polymerase chain reaction and immunohistochemistry demonstrated that Hpa mRNA and protein was highly expressed in MCF-7/Hpa cells but not in MCF-7 cells (Figure 1). Heparanase peptide-specific CTLs were generated using peptide-pulsed DCs from four normal HLA-A2-positive PBMC samples. After three stimulations, the cytotoxic activities of CTLs induced by Hpa525, Hpa277, and Hpa405 were determined against MCF-7 and MCF-7/Hpa cells at various E/T ratios by the 51Cr release assay. As shown in Figure 3, these Hpa-specific CTLs could lyse MCF-7/Hpa, whereas no obvious lysis of MCF-7 was detected even at the highest E/T ratio. These results clearly demonstrate that most CTLs were specifically targeted against Hpa peptides that were presented in the context of HLA-A2.
Figure 3.
Study of the specificity and restriction of CTLs generated from Hpa525, Hpa277, and Hpa405 epitopes. Effector-to-target ratios are presented on the x-axis, whereas the y-axis represents the percent of specific lysis. Cytotoxic T lymphocytes generated from HIV virus, HIVpol(476-484) (ILLEPVHGV), served as a negative peptide (NP). (A and B) Study of the specificity of CTLs generated from Hpa525, Hpa277, and Hpa405 epitopes. MCF-7 cells, which were HLA-A2-positive but Hpa-negative, were transduced with pIRES2-EGFP-Hpa plasmid by the lipofection method. After 24 hours of transfection and 4 weeks of selection with G418, one drug-resistant individual clone was randomly collected and was named MCF-7/Hpa. The cytotoxic activity of CTLs induced by Hpa525, Hpa277, and Hpa405 was determined against MCF-7 cells (A) and MCF-7/Hpa cells (B) at various E/T ratios using the 51Cr release assay. (C and D) Study of the restriction of CTLs generated from Hpa525, Hpa277, and Hpa405 epitopes. HepG2 cells, which were Hpa-positive but HLA-A2-negative, were transduced with a plasmid containing full-length cDNA of HLA-A2 by the lipofection method. After 24 hours of transfection and 4 weeks of selection with G418, one drug-resistant individual clone was randomly collected and was named HepG2/HLA-A2. The cytotoxic activity of CTLs induced by Hpa525, Hpa277 and Hpa405 was determined against HepG2 cells (C) and HepG2/HLA-A2 cells (D) at various E/T ratios using the 51Cr release assay.
Analysis of HLA-A2 Restriction
To further confirm that these CTL epitopes were restricted by HLA-A2, we took advantage of the Hpa-positive, HLA-A2-negative liver cancer cell line, HepG2 [25]. HepG2 cells were transduced with a eukaryotic vector containing the full-length cDNA of HLA-A2 (given to us by Dr. Wan Y.) by the DOTAP lipofection method according to the manufacturer's protocol. After 24 hours of transduction, 400 µg/ml G418 was added to the RPMI-1640 medium. After G418 selection for 4 weeks, drug-resistant individual clones were randomly collected from the transduced cultures. The selected clone was named HepG2/HLA-A2. HLA-A2 expression was 31.3% in HepG2/HLA-A2, whereas it was 3.1% in HepG2 by flow cytometry (data not shown). The 51Cr release assay showed that CTLs generated from Hpa525, Hpa277, or Hpa405 peptide-pulsed DCs could lyse HepG2/HLA-A2. However, the previously mentioned induced effectors could not lyse HepG2 cells, even at the highest E/T ratio (Figure 3). These results clearly demonstrate that most CTLs are restricted by HLA-A2.
Antibody Inhibition Assay
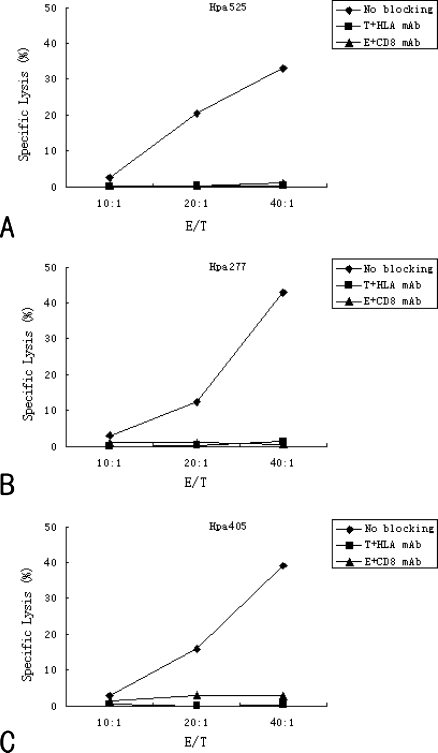
To further determine whether predicted peptide Hpa525-, Hpa277-, and Hpa405-induced effectors recognized Hpa-positive target tumor cells in an HLA-A2-restricted manner and whether the effectors were derived from CD8+ T lymphocytes, mAbs against HLA-A2 were used to block recognition of target cells and mAbs against CD8 were used to block recognition of effectors. In addition, mAbs against HLA-B0702 and mAbs against CD4 served as negative controls. Results showed that after blocking the HLA-A2 sites on the surface of target cells with HLA-A2 mAb or blocking the CD8 molecules on the surface of effector cells with CD8 mAb, the specific killing effects of CTLs could be significantly eliminated (Figure 4). However, mAbs of negative controls could not eliminate the killing effect of Hpa peptide-specific CTLs (data not shown). These results further indicate that the effectors are HLA-A2.1-restricted. Moreover, these Hpa-specific CTLs are mainly raised from CD8+ T lymphocytes.
Figure 4.
Inhibited recognition of induced cells by anti-HLA-A2 or anti-CD8+ antibody. KATO-III target cells were incubated with or without anti-HLA-A2 antibody from BB7.2 cells for 1 hour at 4°C. Moreover, effectors induced by different Hpa-derived peptides were also incubated with or without anti-CD8+ antibody for 1 hour at 4°C. The cytotoxic activities of CTLs induced by Hpa525 (A), Hpa277 (B) and Hpa405 (C) were determined against these KATO-III cells at various E/T ratios using 51Cr release assay. T = target cells; E = effectors.
Killing Effect of Hpa-Specific CTLs on Autologous Lymphocytes
While expressed in malignant tissues, Hpa could be also detected at a low level in some normal cell and tissues. It was reported that Hpa could be expressed in immunologically competent cells, natural killer cells, and inflammatory cells such as neutrophils, granulocytes, and activated T and B cells [12,14,26]. Theoretically, immunotherapy aimed at Hpa may elicit adverse effects on the immune system. To investigate the effect of Hpa-specific CTLs on immunologically activated lymphocytes, CTLs induced by Hpa-specific peptides were also used to lyse autologous lymphocytes and DCs. These results indicate that Hpa peptide vaccinations had no detectable lysis effect on autologous lymphocytes (Figure 2D).
IFN-γ Secretion by ELISPOT
Because CTLs are known to produce the TH1 cytokine IFN-γ, peptide-specific T cells were enumerated by measuring IFN-γ-producing cells by ELISPOT assay. As shown in Figure 5, Hpa525, Hpa277, and Hpa405 peptides were found to generate strong peptide-specific T-cell responses by virtue of their ability to induce increased frequencies of IFN-γ-producing T cells compared to negative peptide (P < .05). These results suggest that Hpa peptide vaccines could increase IFN-γ secretion by effectors and enhance the TH1 immune response.
Figure 5.
IFN-γ-producing cells were enumerated by ELISPOT assay. Negative peptide (NP), HIVpol-derived from HIV virus, served as a negative control. Phytohemagglutinin (PHA) served as a positive control. Column indicates mean; bars, SE. **Statistically significant values at P < 0.01 using a paired Student's t test were assessed to compared with NP group.
Discussion
For the past few years, the analysis of spontaneous immune responses to autologous tumors in cancer patients has allowed for the identification of several categories of TAAs that can be targeted by tumor-specific immune responses based on the recognition of tumor antigens by CTL in an MHC-class I/peptide complex-restricted manner [27,28]. Therefore, cancer-specific immunotherapy has become a very attractive therapeutic approach against carcinomas. Among these approaches, one of the most relevant for the development of tumor immunotherapy is a peptide-based, cancer-specific immunotherapy using universal TAAs, which are expressed by tumor cells but not by most somatic adult tissues. Therefore, the identification of T-cell epitopes from these antigens has become a critical step in the development of peptide-based immunotherapy for cancers.
Peptide vaccination for cancer immunotherapy requires identification of peptide epitopes derived from antigenic proteins associated with the tumor. Such peptides can bind to MHC proteins (MHC molecules) on the tumor cell surface, with the potential to initiate a host immune response against the tumor. A breakthrough in the identification of peptide epitopes was the elucidation that ligands of a certain MHC molecule carry chemically related amino acids in certain positions, leading to the definition of a peptide motif for every MHC allele [29]. This knowledge was rapidly used to predict potential epitopes from various antigens and initiated the so-called reverse immunology, which has been the most successful strategy for the identification of T-cell epitopes [30–32]. This approach included a four-step procedure: 1) computer-based epitope prediction from the amino acid sequence of a candidate antigen, 2) peptide-binding assays to determine the affinity of the predicted peptide for the MHC molecule, 3) the stimulation of the primary T-cell response against predicted peptides in vitro, and 4) testing of the resultant CTLs against target cells endogenously expressing the antigen [12]. Using this approach, a number of T-cell epitopes have been identified from several antigens such as hTERT [33,34], MAGE [35,36], NY-ESO-1 [37], MUC-1 [38], and Ebola virus [16]. Moreover, with the elucidation of the crystal structures of human MHC class I molecule HLA-A2, molecular modeling of HLA-A2-restricted epitopes became possible. Lim et al. [39] found that the peptides bound to the HLA-A2 mode structure possess a side chain of a COOH-terminal anchor residue oriented into the binding groove exhibiting a specific distance, ranging from 15 to 21 Å, between the two anchor residues.
It has been reported that most patients with advanced tumors express Hpa. Specific inhibition of Hpa should reduce tumor angiogenesis and invasion [7,8,12,13]. More importantly, if a tumor develops immune escape variants against Hpa-specific CTLs by down-regulating Hpa expression, such variants might have reduced angiogenic and invasive capacity. The high immunogenicity of Hpa and its broad expression make this protein a very promising target for tumor-specific vaccination strategies. If Hpa epitopes were presented by different tumor cells and recognized by CTL, vaccines designed to boost CTL responses against Hpa epitopes would be useful for clinical therapy for advanced tumors. Although three HLA-A2-restricted epitopes have been reported by Sommerfeldt et al. [13], we do not know if there are other new HLA-A2-restricted epitopes existing in the full-length amino acid sequence of Hpa.
In the present study, we first predicted candidate epitopes from human Hpa antigen based on computer algorithms. Computational methods combined with in vitro/in vivo studies have proven to be very useful in the identification of immunogenic T cell epitopes from defined antigens and pathogens [16,40]. In this study, 30 HLA-A2-restricted epitopes were predicted within the structural proteins of human Hpa by supermotif combined with quantitative motif. The five epitopes with the highest scores, Hpa(525–533) (PAFSYSFFV), Hpa(353–361) (PLLSDTFAA), Hpa(277–285) (KMLKSFLKA), Hpa(400–408) (PLPDYWLSL), and Hpa(405–413) (WLSLLFKKL), were selected in this study. Interestingly, two epitopes discovered by Sommerfeldt et al. [Hpa8–16(ALPPPLMLL), Hpa16–24(LLLGPLGPL)] were also included in our predicted peptides. Another epitope discovered by Sommerfeldt et al., Hpa183–191 (DLIFGLNAL), is located very close to our predicted epitope Hpa184–192 (LIFGLNALL) (data not shown). These results suggest that the computer-based epitope prediction used in the present study is effective and feasible. Second, we characterized the above five predicted candidates by molecular modeling and found that the above five peptides were suitable HLA-A2-restricted CTL epitopes. Third, the peptide-binding assay was used to determine the affinity of every epitope with HLA-A2 and showed that the five epitopes had high affinity for the HLA-A2 molecule. Finally, we used a standard 4-hour 51Cr release assay to determine the induction of CTLs by every epitope. We found that only Hpa525, Hpa277, and Hpa405 could induce Hpa-specific immune responses. Further study demonstrated that the antitumor immunity to KATO III gastric cancer cells, MCF-7 breast cancer cells, SW480 colon cancer cells, U2OS osteogenic sarcoma cells, and HepG2 liver cancer cells induced by the above three epitopes was Hpa-specific and HLA-A2-restricted. Thus, our present study indicates that peptide Hpa(525–533) (PAFSYSFFV), Hpa(277–285) (KMLKSFLKA), and Hpa(405–413) (WLSLLFKKL) are new HLA-A2-restricted CTL epitopes capable of inducing Hpa-specific CTLs in vitro. Because Hpa is expressed in most advanced malignant tumors, the above three Hpa epitopes could contribute to the design of epitope-based vaccines for patients with advanced cancers.
For the development of cancer vaccines, safety concerns still limit their use in the future. Although Hpa expression is largely restricted to cancer cells, it has been detected in activated immune cells, including T and B cells, DCs, macrophages, neutrophils, and mast cells, mediating extravasation and traffic to inflammatory sites [8,12,41,42]. Consequently, any Hpa-based cancer vaccine therapy will require assessment of potential adverse effects associated with autoimmunity to cells and organs that are Hpa-positive. Our previous study showed that CTLs induced by DCs loaded with Hpa full-length cDNA could not lyse autologous lymphocytes in vitro [12]. In this study, to investigate the effects of Hpa peptide-specific CTLs on immunologically activated lymphocytes, Hpa peptide-specific CTLs were also used to lyse autologous lymphocytes. The results revealed that Hpa peptide vaccination did not remarkably lyse these lymphocytes. It is conceivable that the level of Hpa expression in normal cells is below the threshold needed for recognition by these Hpa peptide-specific CTL populations [9].
In conclusion, our results suggest that Hpa(525–533) (PAFSYSFFV), Hpa(277–285) (KMLKSFLKA), and Hpa(405–413) (WLSLLFKKL) derived from human Hpa might be capable of inducing HLA-A2-restricted CD8+ CTL, which would be a powerful weapon in cancer-specific immunotherapy. Of course, several studies have demonstrated that transduction of DCs with the entire TAA gene could elicit more powerful immune responses than peptide-pulsed DCs. This is because DCs transduced with the entire TAA gene may present multiple epitopes including previously unknown epitopes associated with different MHC class I molecules [21,43]. To overcome this shortcoming, we are now planning to evaluate immune responses generated by several mixed epitopes of Hpa in vivo and in vitro.
Acknowledgments
The authors thank Christopher R. Parish and Mark D. Hulett (Division of Immunology and Genetics, John Curtin School of Medical Research, The Australian National University) for generously providing plasmid pcDNA3 containing the full-length cDNA of Hpa.
Abbreviations
- Hpa
heparanase
- TAA
tumor-associated antigen
- DC
dendritic cell
- CTLs
cytotoxic T lymphocytes
- E/T
effector-to-target
- MFI
mean fluorescence intensity
- FI
fluorescence index
Footnotes
This work was supported by the grants from the National Nature Science Foundation of China (30200123 and 30570841) and the key project of science and technology of Chongqing (CSTC, 2008AB5002).
References
- 1.Novellino L, Castelli C, Parmiani G. A listing of human tumor antigens recognized by T cells: March 2004 update. Cancer Immunol Immunother. 2005;54:187–207. doi: 10.1007/s00262-004-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jager E, Ringhoffer M, Karbach J, Arand M, Oesch F, Knuth A. Inverse relationship of melanocyte differentiation antigen expression in melanoma tissues and CD8+ cytotoxic-T-cell responses: evidence for immunoselection of antigen-loss variants in vivo. Int J Cancer. 1996;66:470–476. doi: 10.1002/(SICI)1097-0215(19960516)66:4<470::AID-IJC10>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 3.Yee C, Thompson JA, Roche P, Byrd DR, Lee PP, Piepkorn M, Kenyon K, Davis MM, Riddell SR, Greenberg PD. Melanocyte destruction after antigen-specific immunotherapy of melanoma: direct evidence of T cell-mediated vitiligo. J Exp Med. 2000;192:1637–1644. doi: 10.1084/jem.192.11.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schultze JL, Maecker B, von Bergwelt-Baildon MS, Anderson KS, Vonderheide RH. Tumour immunotherapy: new tools, new treatment modalities and new T-cell antigens. Vox Sang. 2001;80:81–89. doi: 10.1046/j.1423-0410.2001.00014.x. [DOI] [PubMed] [Google Scholar]
- 5.Vonderheide RH. Prospects and challenges of building a cancer vaccine targeing telomerase. Biochimie. 2008;90:173–180. doi: 10.1016/j.biochi.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rammensee HG, Falk K, Rotzschke O. Peptides naturally presented by MHC class I molecules. Annu Rev Immunol. 1993;11:213–244. doi: 10.1146/annurev.iy.11.040193.001241. [DOI] [PubMed] [Google Scholar]
- 7.Vlodavsky I, Ilan N, Naggi A, Casu B. Heparanase: structure, biological functions, and inhibition by heparin-derived mimetics of heparan sulfate. Curr Pharm Des. 2007;13:2057–2073. doi: 10.2174/138161207781039742. [DOI] [PubMed] [Google Scholar]
- 8.McKenzie EA. Heparanase: a target for drug discovery in cancer and inflammation. Br J Pharmacol. 2007;151:1–14. doi: 10.1038/sj.bjp.0707182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theodoro TR, de Matos LL, Sant Anna AV, Fonseca FL, Semedo P, Martins LC, Nader HB, Del Giglio A, da Silva Pinhal MA. Heparanase expression in circulating lymphocytes of breast cancer patients depends on the presence of the primary tumor and/or systemic metastasis. Neoplasia. 2007;9:504–510. doi: 10.1593/neo.07241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shafat I, Barak AB, Postovsky S, Elhasid R, Ilan N, Vlovdavsky I, Arush MW. Heparanase levels are elevated in the plasma of pediatric cancer patients and correlate with response to anti-cancer treatment. Neoplasia. 2007;9:909–916. doi: 10.1593/neo.07673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shafat I, Pode D, Tamar P, Ilan N, Vlodavsky I, Nisman B. Clinical significance of urine heparanase in bladder cancer progression. Neoplasia. 2008;10:125–130. doi: 10.1593/neo.07875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai YG, Fang DC, Chen L, Tang XD, Chen T, Yu ST, Luo YH, Xiong Z, Wang DX, Yang SM. Dendritic cells reconstituted with a human heparanase gene induce potent cytotoxic T-cell responses against gastric tumor cells in vitro. Tumor Biol. 2007;28:238–246. doi: 10.1159/000107584. [DOI] [PubMed] [Google Scholar]
- 13.Sommerfeldt N, Beckhove P, Ge Y, Schutz F, Choi C, Bucur M, Domschke C, Sohn C, Schneeweis A, Rom J, et al. Heparanase: a new metastasis-associated antigen recognized in breast cancer patients by spontaneously induced memory T lymphocytes. Cancer Res. 2006;66:7716–7723. doi: 10.1158/0008-5472.CAN-05-2363. [DOI] [PubMed] [Google Scholar]
- 14.Tang XD, Wan Y, Chen L, Chen T, Yu ST, Xiong Z, Fang DC, Liang GP, Yang SM. Immunogenic H-2Kb-restricted CTL epitopes derived from mouse heparanase that elicit antitumor immune response in vivo. Cancer Res. 2008;68:1529–1537. doi: 10.1158/0008-5472.CAN-07-5965. [DOI] [PubMed] [Google Scholar]
- 15.Schirle M, Weinschenk T, Stevanovic S. Combining computer algorithms with experimental approaches permits the rapid and accurate identification of T cell epitopes from defined antigens. J Immunol Methods. 2001;257:1–16. doi: 10.1016/s0022-1759(01)00459-8. [DOI] [PubMed] [Google Scholar]
- 16.Sundar K, Boesen A, Coico R. Computational prediction and identification of HLA-A2.1-specific Ebola virus CTL epitopes. Virology. 2007;360:257–263. doi: 10.1016/j.virol.2006.09.042. [DOI] [PubMed] [Google Scholar]
- 17.Nijman HW, Houbiers JG, Vierboom MP, van der Burg SH, Drifhout JW, D'Amaro J, Kenemans P, Melief CJ, Kast WM. Identification of peptide sequences that potentially trigger HLA-A2.1-restricted cytotoxic T lymphocytes. Eur J Immunol. 1993;23:215–1219. doi: 10.1002/eji.1830230603. [DOI] [PubMed] [Google Scholar]
- 18.Nakao M, Shichijo S, Imazumi T, Inoue Y, Matsunaga K, Yamada A, Kikuchi M, Tsuda N, Ohta K, Takamori S, et al. Identification of a gene coding for a new squamous cell carcinoma antigen recognized by the CTL. J Immunol. 2000;164:2565–2574. doi: 10.4049/jimmunol.164.5.2565. [DOI] [PubMed] [Google Scholar]
- 19.Romani N, Gruner S, Brang D, Kampgen E, Lenz A, Trochenbacher B, Konwalinka G, Fritsch PO, Steinman RM, Schuler G. Proliferating dendritic cell progenitors in human blood. J Exp Med. 1994;180:83–93. doi: 10.1084/jem.180.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beckhove P, Feuerer M, Dolenc M, Schuetz F, Choi C, Sommerfeldt N, Schwendemann J, Ehlert K, Altevogt P, Bastert G, et al. Specifically activated memory T cell subsets from cancer patients recognize and reject xenotransplanted autologous tumors. J Clin Invest. 2004;114:67–76. doi: 10.1172/JCI20278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho HI, Kim HJ, Oh ST, Kim TG. In vitro induction of carcino-embryonic antigen (CEA)-specific cytotoxic T lymphocytes by dendritic cells transduced with recombinant adenoviruses. Vaccine. 2003;22(2):224–236. doi: 10.1016/s0264-410x(03)00569-3. [DOI] [PubMed] [Google Scholar]
- 22.Lycke NY, Coico RF. ELISPOT assay for measurement of antigen-specific and polyclonal antibody responses. In: Coligan J, et al., editors. Current Protocols in Immunology. Hoboken, NJ: John Wiley and Sons Inc; 1996. pp. 7.14.1–7.14.7. [DOI] [PubMed] [Google Scholar]
- 23.Halpern M, Zahalka MA, Traub L, Moroz C. Antibodies to placental immunoregulatory ferritin (PLIF) with transfer of polyclonal lymphocytes arrests MCF-7 human breast cancer growth in nude mouse model. Neoplasia. 2007;9:487–494. doi: 10.1593/neo.07259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L, Li ZH, Yang SM, Li JJ, Chai YG, Fang DC, Luo YH, Wang DX. Construction of sense and antisense human heparanase and green fluorescent protein eukaryotic co-expression vectors and expression in hepatic cancer cell lines. Acta Academiae Medicinae Militaris Tertiae. 2006;28:47–49. [Google Scholar]
- 25.Chen L, Liang GP, Tang XD, Chen T, Cai YG, Fang DC, Yu ST, Luo YH, Yang SM. In vitro anti-tumor immune response induced by dendritic cells transfected with hTERTrecombinant adenovirus. Biochem Biophy Res Commun. 2006;351:927–934. doi: 10.1016/j.bbrc.2006.10.165. [DOI] [PubMed] [Google Scholar]
- 26.Parish CR, Freeman C, Hulett MD. Heparanase: a key enzyme involved in cell invasion. Biochim Biophys Acta. 2001;1471:M99–M108. doi: 10.1016/s0304-419x(01)00017-8. [DOI] [PubMed] [Google Scholar]
- 27.Coulie PG, Brichard V, Van Pel A, Wolfel T, Schneider J, Traversari C, Mattei S, De Plaen E, Lurquin C, Szikora JP, et al. A new gene coding for a differentiation antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J Exp Med. 1994;180:35–42. doi: 10.1084/jem.180.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheever MA, Disis ML, Bernhard H, Gralow JR, Hand SL, Huseby ES, Qin HL, Takahashi M, Chen W. Immunity to oncogenic proteins. Immunol Rev. 1995;145:33–59. doi: 10.1111/j.1600-065x.1995.tb00076.x. [DOI] [PubMed] [Google Scholar]
- 29.Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee HG. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 30.Pamer EG, Harty JT, Bevan MJ. Precise prediction of a dominant class I MHC-restricted epitope of Listeria monocytogenes. Nature. 1991;353:852–855. doi: 10.1038/353852a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rotzschke O, Falk K, Stevanovic S, Jung G, Walden P, Rammensee HG. Exact prediction of a natural T cell epitope. Eur J Immunol. 1991;21:2891–2894. doi: 10.1002/eji.1830211136. [DOI] [PubMed] [Google Scholar]
- 32.Celis E, Tsai V, Crimi C, DeMars R, Wentworth PA, Chesnut RW, Grey HM, Sette A, Serra HM. Induction of anti-tumor cytotoxic T lymphocytes in normal humans using primary cultures and synthetic peptide epitopes. Proc Natl Acad Sci USA. 1994;91:2105–2109. doi: 10.1073/pnas.91.6.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scardino A, Gross DA, Alves P, Schultze JL, Graff-Dubois S, Faure O, Tourdot S, Chouaib S, Nadler LM, Lemonnier FA, et al. HER-2/neu and hTERT cryptic epitopes as novel targets for broad spectrum tumor immunotherapy. J Immunol. 2002;168:5900–5906. doi: 10.4049/jimmunol.168.11.5900. [DOI] [PubMed] [Google Scholar]
- 34.Adotevi O, Mollier K, Neuveut C, Cardinaud S, Boulanger E, Mignen B, Fridman WH, Zanetti M, Charneau P, Tartour E, et al. Immunogenic HLA-B0702-restricted epitopes derived from human telomerase reverse transcriptase that elicit antitumor cytotoxic T-cell responses. Clin Cancer Res. 2006;12:3158–3167. doi: 10.1158/1078-0432.CCR-05-2647. [DOI] [PubMed] [Google Scholar]
- 35.Eggert AO, Andersen MH, Voigt H, Schrama D, Kämpgen E, Straten PT, Becker JC. Characterization of mouse MAGE-derived H-2Kb-restricted CTL epitopes. Eur J Immunol. 2004;34(11):3285–3290. doi: 10.1002/eji.200324618. [DOI] [PubMed] [Google Scholar]
- 36.Zhu B, Chen Z, Cheng X, Cheng X, Liu Z, Guo J, Jia Z, Zou L, Wang Z, Hu Y, et al. Identification of HLA-A0201-restricted cytotoxic T lymphocyte epitope from TRAG-3 antigen. Clin Cancer Res. 2003;9:1850–1857. [PubMed] [Google Scholar]
- 37.Yamaguchi H, Tanaka F, Ohta M, Inoue H, Mori M. Identification of HLA-A24-restricted CTL epitope from cancer testis antigen, NY-ESO-1, and induction of a specific antitumor immune response. Clin Cancer Res. 2004;10:890–896. doi: 10.1158/1078-0432.ccr-1086-3. [DOI] [PubMed] [Google Scholar]
- 38.Wierecky J, Mueller M, Brossart P. Dendritic cell-based cancer immunotherapy targeting MUC-1. Cancer Immunol Immunother. 2006;55:63–67. doi: 10.1007/s00262-005-0673-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim JS, Kim S, Lee HG, Lee KY, Kwon TJ, Kim K. Selection of peptides that bind to the HLA-A2.1 molecule by molecular modelling. Mol Immunol. 1996;33:221–230. doi: 10.1016/0161-5890(95)00065-8. [DOI] [PubMed] [Google Scholar]
- 40.Boesen A, Sundar K, Coico R. Lassa fever virus peptides predicted by computational analysis induce epitope-specific cytotoxic-T-lymphocyte responses in HLA-A2.1 transgenic mice. Clin Diagn Lab Immunol. 2005;12:1223–1230. doi: 10.1128/CDLI.12.10.1223-1230.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaday GG, Lider O. Extracellular matrix moieties, cytokines, and enzymes: dynamic effects on immune cell behavior and inflammation. J Leukoc Biol. 2000;67:49–159. doi: 10.1002/jlb.67.2.149. [DOI] [PubMed] [Google Scholar]
- 42.Benhamron S, Nechushtan H, Verbovetski I, Krispin A, Abboud-Jarrous G, Zcharia E, Edovitsky E, Nahari E, Peretz T, Vlodavsky I, et al. Translocation of active heparanase to cell surface regulates degradation of extracellular matrix heparan sulfate upon transmigration of mature monocyte-derived dendritic cells. J Immunol. 2006;176:6417–6424. doi: 10.4049/jimmunol.176.11.6417. [DOI] [PubMed] [Google Scholar]
- 43.Carnevale-Schianca F, Cignetti A, Capaldi A, Vitaggio K, Vallario A, Ricchiardi A, Sperti E, Ferraris R, Gatti M, Grignani G, et al. Allogeneic nonmyeloablative hematopoietic cell transplantation in metastatic colon cancer: tumor-specific T cells directed to a tumor-associated antigen are generated in vivo during GVHD. Blood. 2006;107:3795–3803. doi: 10.1182/blood-2005-10-3945. [DOI] [PubMed] [Google Scholar]