Abstract
Metastatic breast cancer cells are characterized by their high propensity to colonize the skeleton and form bone metastases, causing major morbidity and mortality. Identifying key proteins involved in the osteotropic phenotype would represent a major step toward the development of both new prognostic markers and new effective therapies. Cell surface proteins differentially expressed in cancer cells are preferred potential targets for antibody-based targeted therapies. In this study, using cell surface biotinylation and a mass spectrometric approach, we have compared the profile of accessible cell surface proteins between the human breast cancer cell line MDA-MB-231 and its highly osteotropic B02 subclone. This strategy allowed the identification of several proteins either up- or downregulated in the osteotropic cell line, and differential protein expressions were validated using antibody-based techniques. Class I HLAs were down-regulated in the bone metastatic variant, whereas αvβ3 integrins, among others, were consistently up-regulated in this latter cell line. These results show that comprehensive profiling of the cell surface proteome of mother cancerous cell lines and derived organ-specific metastatic cell lines provides an effective approach for the identification of potential accessible marker proteins for both prognosis and antibody-based targeted therapies.
Introduction
Metastases represent the major cause of morbidity for cancer patients, yet combating them represents a challenging task. Breast cancer has a strong predilection for spreading to the skeleton [1], and almost three quarters of breast cancer patients with advanced disease experience bone metastases. These bone metastases are associated with substantial morbidity, including among others bone pain, pathologic fracture, neurologic deficit, and hypercalcemia. Identifying key proteins involved in the osteotropic phenotype would represent a major step toward the development of both new prognostic markers and therapeutic improvements. With recent mass spectrometry-based advances in biomarker discovery, as well as growing knowledge of cellular pathways involved in tumor progression, molecularly targeted therapies now represent an increasing proportion of new drugs entering clinical trials. In the case of breast cancer treatment, humanized antibodies have become a major option. For instance, the humanized monoclonal antibody trastuzumab (Herceptin) represents a successful adjuvant immunotherapy for the membrane receptor Her2/Neu overexpressed in metastatic breast cancers [2,3]. Many other therapeutic monoclonal antibodies were approved for use in oncology target membrane proteins (e.g., CD20, CD33, and CD52, for treatment of lymphomas, acute myelogenous leukemia, and chronic lymphocytic leukemia, respectively). It is noteworthy, yet not surprising, that many clinical biomarkers and therapeutic targets are cell surface proteins. Indeed, membrane proteins play important roles in cell-matrix interactions, cell signaling and adaptation to the environment. Herein, we sought to unveil potential molecular cell surface markers, both on an invasive breast cancer cell line and on its osteotropic metastatic variant. Different bone-seeking clones were previously generated from the now well-described invasive cell line MDA-MB-231 [4–6]. Peyruchaud et al. [6] were able to establish a breast cancer cell line subclone by repeated in vivo passages in bone using the mouse heart injection model: this osteotropic clone, MDA-MB-231-B02, is referred to as “B02” hereafter. On the basis of the comparison of a transcriptomic analyses of MDA-MB-231 and B02 cells, we recently showed that these osteotropic breast cancer cells exhibit an osteomimetic phenotype [7]. However, because mRNA and protein levels may differ [8,9], transcriptomic and proteomic analyses remain complementary. For example, although the αvβ3 integrin expression has been associated with the osteotropic phenotype [10], there was no significant difference in the mRNA transcript for this cell membrane receptor between the B02 clone and the parent cell line [7]. We have therefore undertaken a comparative membrane proteome survey between these two cell lines. Our results showed that class I HLA and several cancer-associated markers were down-regulated in the bone metastatic variant, whereas αv and β3 integrins, among others, were up-regulated. Such differentially expressed proteins could be of major importance in unraveling the mechanisms of “homing” and could eventually represent potential new molecular targets for new compounds aiming at selectively destroying threatening cells (e.g., antibodies bound to bioactive molecules).
Materials and Methods
Cell Culture
The B02 cell line has been established from bone metastases caused by MDA-MB-231. This subclone of MDA-MB-231 has been selected after six in vivo passages in nude mice using a heart injection model and is characterized by its unique predilection for bone metastasis [6,10]. MDA-MB-231 and its derivative cell line B02 were used at early passages until approaching confluency in RPMI 1640 medium (Invitrogen, Merelbeke, Belgium) supplemented with 10% fetal calf serum (ICN, Asse Relegem, Belgium) at 37°C in a humidified 5% CO2 incubator.
Cell Biotinylation
Cells were detached with dispase (Invitrogen). Cells were then washed and resuspended at 106 cells/ml in D-PBS (Invitrogen). Two reagents are of particular interest for the biotinylation of accessible lysines: sulfosuccinimidyl-6-(biotin-amido) hexanoate (EZ-link sulfo-NHS-LC-biotin; Pierce, Erembodegem-Aalst, Belgium) [11–13], and sulfosuccinimidyl-2-(biotinamido) ethyl-1,3-dithiopropionate (EZ-link sulfo-NHS-SS-biotin; Pierce) [11,14]. We found a significant intracellular staining with sulfo-NHS-LC-biotin (as revealed by streptavidin-conjugated to the Alexa 488 fluorescent dye used to visualize the subcellular distribution of labeled proteins), in good agreement with Peirce et al. [11] (data not shown). We therefore used the sulfo-NHS-SS-biotin reagent (0.2 mg/ml for 20 minutes at 4°C), which also allowed the subsequent elution from the streptavidin resin used for affinity purification. Each biotinylation reaction was stopped by an incubation in 50 mM Tris in D-PBS (5 minutes).
Preparation of Membrane Fractions and Purification of Biotinylated Proteins
Biotinylated cells were subjected to a freezing-thawing step, then lysed in hypotonic CLB buffer (10 mM HEPES pH 7.5, 5 mM EDTA, 10 mM NaCl, 1 mM KH2PO4, 5 mM NaHCO3, 1 mM CaCl2, 0.5 mM MgCl2, 0.5 mM oxidized glutathione, and protease inhibitors by 50 strokes of a tight-fitting Dounce homogenizer). The homogenate was clarified by centrifugation (3000 rpm for 3 minutes at 4°C) to generate a postnuclear supernatant. This latter fraction was centrifuged again (35,000g for 30 minutes at 4°C) to yield a membrane-enriched pellet. The efficacy of each step was checked using light microscopy by mixing 5 µl of methyl green to 5 µl of each fraction. The membrane-enriched pellet was solubilized in 1% Triton X-100 + 0.2% SDS in PBS and heated at 60°C for 10 minutes. Streptavidin-sepharose slurry (150 µl/mg of total proteins; Amersham Biosciences, Diegem, Belgium) was equilibrated by three washes in buffer A (1% NP-40 and 0.1% SDS in PBS), and binding of biotinylated membrane proteins was allowed overnight in a rotating mixer at 4°C. The resin was then washed twice with buffer A, twice with buffer B (0.1% NP-40, 1 M NaCl in PBS), twice with buffer C (0.1 M sodium carbonate in PBS, pH 11), and once with PBS.
Binding of proteins biotinylated using EZ-Link sulfo-NHS-LC-biotin to the resin and washing efficiencies were checked by SDS-PAGE and were further checked either by Coomassie blue staining or by blot analysis for the subsequent detection of biotin by streptavidinhorseradish peroxidase (data not shown).
Elution of biotinylated proteins was performed for twice for 30 minutes each at 58°C with 100 mMDTT in 1% SDS pH 7.0. Eluted proteins were alkylated with 150 mM iodoacetamide (Sigma, Merelbeke, Belgium) 30 minutes at RT in the dark. The eluate was then filtrated through a sintered glass filter (10-µm pores; Pierce). After trichloroacetic acid precipitation and acetone washing, the pellet was dissolved in 0.2% Rapigest (Waters, Zeek, Belgium) in 50 mM NH4HCO3. Digestion with trypsin (1:100 trypsin-protein ratio; Promega, Leiden, The Netherlands) was performed overnight at 37°C. Rapigest was then cleaved by the addition of highly pure trifluoroacetic acid following manufacturer's instructions.
Mass Spectrometry
Peptide separation was performed by reverse-phase liquid chromatography on an Ultimate LC system (LC Packings, Amsterdam, The Netherlands). The sample (5 µg in 20 µl at 0.25 µg/µl 0.1% formic acid) was first trapped on an SCX micro precolumn [500 µm in internal diameter (i.d.), 15 mm in length, packed with MCA50 bioX-SCX 5 µm; LC Packings] at a flow rate of 30 µl/min followed by a micro precolumn cartridge (300 µm in i.d., 5 mm in length, packed with 5 µmC18 PepMap100; LC Packings). After 5 minutes, the precolumn was connected with the separating nanoflow column at 200 nl/min (75 µm in i.d., 15 cm in length, packed with 3 µm C18 PepMap100; LC Packings) equilibrated in mobile phase A (0.1% formic acid in 2:98 of acetonitrile-degassed milliQ water). A linear elution gradient was applied with mobile phase B (0.1% formic acid in 80:20 of acetonitrile-degassed milliQ water) spanning from 10% to 40% in 95 minutes. The outlet of the LC system was directly connected to the nanoelectrospray source of an EsquireHCTion trapmass spectrometer (Bruker Daltonics, Germany), controlled by Esquire Control v5.2 and Hystar v3.0 (from the Bruker Compass software bundle). For each mass scan, a data-dependent scheme picked the three most intense doubly or triply charged ions to be selectively isolated and fragmented in the trap. The resulting fragments were analyzed using the Ultra Scan mode (m/z range of 50–3000 at 26,000 m/z per second). SCX-trapped peptides were stepwise eluted with five salt concentrations (10, 20, 40, 80, and 200 mM), each followed by the same gradient of mobile phase B.
Data Processing and mgf File Generation
Raw spectra were formatted in DataAnalysis software (v3.4 build 150; Bruker Daltonics). The portions of the chromatogram-containing signal were processed to extract and deconvolute MS/MS spectra, without smoothing or background subtraction. Both deconvoluted and undeconvoluted data were incorporated in the mgf file.
Database Searching
Proteins were identified using the minimally redundant SwissProt human protein database [15] (release 54.5, SIB; Switzerland, 17,741 human entries), through the MS/MS ion search algorithm of the Mascot search engine (Mascot and Mascot Daemon v2.1) [16] running on a local four-processor computer cluster. The mass tolerances of precursor and fragmented ions were set at 0.6 and 0.3, respectively; allowed modifications were partial oxidization of methionines, carbamidomethyl cysteines, one misscut, and the ions' score cutoff was set to 30. The absolute probability (P) was set to .05. Interesting proteins reported in Table 1 identified by only one peptide were manually inspected using previously described criteria [17]. With the above-mentioned parameters, 522 proteins were found in the MDA cell line, whereas 526 were found in the B02 subclone. The full protein list contained 691 proteins, of which 357 (68%) were found in both cell lines.
Table 1.
Selection of Membrane Proteins Identified in the MDA-MB-231 Cell Line (MDA, n = 4) and MDA-MB-231-B02 Cell Line (B02, n = 4).
| Name | MDA | SC | BO2 | SC | Biologic Process | Cell. Component |
| CD51 (integrin αv) | 1 | 3 | 3 | 4 | Cell comm., Signal transd. | PM |
| CD9 (MRP-1) | 1 | 1 | 4 | 5 | Immune resp. | PM |
| Plasma cell membrane glycoprotein PC1 | 1 | 1 | 4 | 13 | Metabolism energy pathways | PM |
| Kinectin | 1 | 4 | 4 | 19 | Cell growth and/or maintenance | ER, L |
| Prohibitin | 1 | 7 | 4 | 27 | Cell comm., Signal transd. | MC, PM, N, EC, C |
| Moesin | 3 | 35 | 3 | 26 | Cell growth and/or maintenance | PM, C, N |
| EGF receptor | 4 | 12 | 4 | 26 | Cell comm., Signal transd. | PM, C |
| CD49b (integrin α2) | 4 | 19 | 4 | 15 | Cell comm., Signal transd. | PM, C |
| Integrin β6 | 4 | 22 | 4 | 13 | Cell comm., Signal transd. | PM |
| Clathrin, heavy polypeptide | 4 | 47 | 4 | 22 | Cell growth and/or maintenance | C, PM, G |
| CD44 | 4 | 61 | 4 | 29 | Cell comm., Signal transd. | - |
| CD98 | 4 | 68 | 4 | 68 | Transport | PM |
| CD29 (integrin β1) | 4 | 162 | 4 | 60 | Cell comm., Immune cell migr. | PM |
| HLA-A | 4 | 600 | 4 | 51 | Immune resp. | PM, ER, EC |
| CD49c (Integrin α3) | 4 | 24 | 3 | 4 | Cell comm., Signal transd. | PM, C |
| CD155 (PVR) | 1 | 1 | 0 | 0 | Cell comm., Signal transd. | PM, EC |
| CD104 (integrin β4) | 1 | 7 | 0 | 0 | Cell comm., Signal transd., Cell migration, Cell adhesion | ITM |
| CD107b (LAMP2) | 2 | 2 | 1 | 1 | Cell growth and/or maintenance | EC, ER, PM, G |
| CD49e (integrin α5) | 3 | 8 | 1 | 1 | Cell comm., Signal transd. | PM |
| CD97 | 3 | 9 | 1 | 1 | Cell comm., Signal transd. | - |
| CD49f (integrin α6) | 4 | 21 | 1 | 1 | Cell comm., Signal transd. | PM |
| HLA-C | 4 | 374 | 0 | 0 | Immune resp. | PM |
| HLA-B | 4 | 431 | 1 | 19 | Immune resp. | PM, EC |
The proteins are identified from the SwissProt database with Mascot. The numbers in bold indicate in how many runs each protein was detected. Function and location of the proteins are determined using the Human Protein Reference Database (www.hprd.org).
C indicates cytoplasm; Cell comm., cell communication; EC, extracellular; ER, endoplasmic reticulum; G, Golgi; Immune cell migr., immune cell migration; Immune resp., immune response; L, lysosome; MC, mitochondrion; N, nuclear; PM, plasma membrane; SC, spectral counting (italicized numbers represent the number of redundant spectra identifying the same protein or group of protein, e.g. HLA-A, -B, and -C serotypes); Signal transd., signal transduction.
The false-positive rate was estimated, for each sample, by dividing the number of peptides found in the randomized SwissProt database by the number of identified peptides from the normal SwissProt database, according to following formula: fp = nrandom/nnormal, where fp is the estimated false-positive rate, nrandom is the number of peptides identified (queries after filtering) from the random SwissProt database, and nnormal is the number of peptides identified (queries after filtering) from the normal SwissProt database. Considering the eight MS analyses performed for both MDA and B02, fp is equal to 1.58 ± 0.30% (mean ± SEM).
Western Blot Analysis
MDA and B02 cells were lysed in RIPA extraction buffer (1% NP-40, 0.5% deoxycholate, 0.1% SDS in 50 mM Tris pH 7.5, 150 mM NaCl) containing an anti-protease cocktail (Complete, Roche, Vilvoorde, Belgium). Proteins were separated on 12.5% or 7.5% polyacrylamide gels under reducing conditions. Transfer was performed overnight at 30 V. Membranes were blocked in TBS-Tween (0.1% v/v) with 5% nonfat dried milk, and primary antibodies were incubated overnight at 4°C.
Equal protein loading and transfer were assessed by the subsequent use of an anti-alpha-tubulin antibody (clone B-5-1-2; Sigma) applied on stripped membranes. Bound antibodies were visualized by incubation of the membranes with ECL chemiluminescent substrate (Amersham Biosciences) and exposure to x-ray films (Fuji, Dusseldorf, Germany).
Antibodies
Mouse monoclonal anti-HLA class I antibody was from Sigma. The antibodies anti-CD107b, anti-αv, and anti-β3 integrins were from BD Biosciences (Erembodegem, Belgium).
Confocal Microscopy
MDA-MB-231 cells were biotinylated as described above or left unlabeled, washed twice, and allowed to adhere to glass adhesion slides. Adherent cells were fixed (3% paraformaldehyde for 10 minutes at RT), rinsed, blocked (1 mg/ml BSA in PBS), and incubated with streptavidin-Alexa (1:4000 for 1 hour at RT in the dark; Invitrogen/Molecular Probes, Merelbeke, Belgium). Cells were visualized using a TCS SP laser scanning confocal microscope (Leica, Groot Bijgaarden, Belgium).
FACS Analyses
Cells were washed twice in PBS and incubated with mouse immunoglobulin (IgG, 1 µl per 106 cells for 1 hour at 4°C). Cells were then incubated (at 1:1000 dilution for 1 hour at 4°C) with fluorescein isothiocyanate-conjugated monoclonal secondary antibodies. Cells were acquired using a FACSCalibur (Becton Dickinson) flow cytometer and data analyzed using CellQuest software (BD Biosciences) and FlowJo software (Tree Star, Inc., Ashland, OR).
Results
Membrane Biotinylation
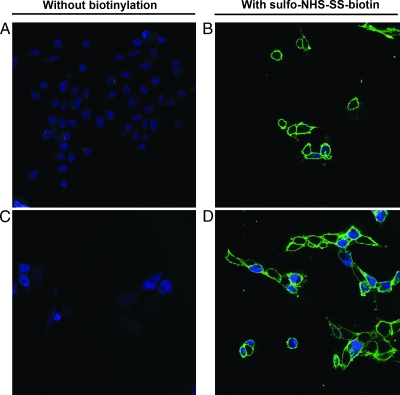
To identify differentially expressed proteins between a well-known parental invasive breast cancer cell line (MDA-MB-231) and its osteotropic cell derivative (B02), we combined membrane protein labeling and cell fractionation. Cell surface biotinylation provides an effective tool for the detection and purification of proteins and relies on biotin labeling of solvent-exposed primary amines residues (i.e., lysines) on the membrane proteins [18]. To test for endogenous and/or nonspecific binding of streptavidin-conjugated proteins, we omitted the biotinylation step for both MDA (Figure 1A) and B02 (Figure 1C). In these conditions, we did not detect any binding of streptavidin-Alexa. Representative pictures of biotinylated membranes of the parental cells and the osteotropic cells are shown in Figure 1, B and D, respectively. After permeabilization, some intracellular staining patterns appeared but were restricted to few cells.
Figure 1.
Biotinylation of plasma membranes using sulfo-NHS-SS-biotin. MDA-MB-231 and MDA-MB-231-B02 were left unlabeled (A and C, respectively), or biotinylated using sulfo-NHS-SS-biotin (B and D, respectively). Biotin labeling was revealed using streptavidin-Alexa 488. Nuclei were stained with TO-PRO-3 fluorescent dye. Confocal images were generated using a Leica TCS SP at an original magnification of x630. Data shown are representative pictures of repeated experiments.
Sample Processing
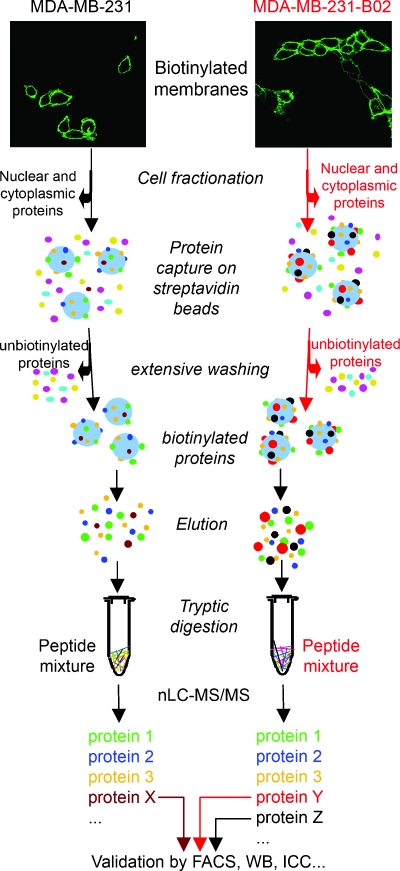
Figure 2 schematically depicts the method used in this study for the identification of membrane proteins. After cell fractionation, biotinylated proteins were captured on streptavidin resin, and after thorough washes to remove abundant cytosolic proteins with both high-salt and high-pH buffers [19], the proteins were eluted thanks to the disulfide bonds between the lysines of labeled proteins and the biotin moieties. Proteins were then submitted to proteolytic digestion. Expression profiles of biotinylated proteins in the two human breast carcinoma cell lines were then determined using the Multidimensional Protein Identification Technology technique, based on a two-dimensional separation of tryptic peptide digest using nanoflow liquid chromatography coupled to electrospray tandem mass spectrometry. To assess the specificity of the streptavidin column, nonbiotinylated breast cancer cells were processed identically to the biotinylated ones and analyzed by MS. The number of identified proteins was much less in nonbiotinylated breast cancer cell samples than in biotinylated samples. Proteins found in nonbiotinylated samples (approximately 10 proteins, data not shown) were most likely “sticky” proteins that unspecifically bound to the streptavidin beads.
Figure 2.
Schematic description of the method used in this study for the identification of membrane proteins in the invasive human breast cancer cell line MDA-MB-231 and its bone metastatic variant MDA-MB-231-B02. Biotinylation was carried out by incubating the cells in a solution containing a reactive ester that links biotin to the free primary amines of membrane proteins. After cell fractionation, biotinylated proteins are captured on streptavidin resin and, after thorough washes, are eluted thanks to the disulfide bond between the lysines and the biotin. Proteins are then submitted to proteolytic digestion. Biotinylated proteins are then sequenced by shotgun mass spectrometry using nLC-ESI MS/MS. Identified proteins differentially expressed in the cells are validated by fluorescent-activated cell sorting (FACS; Figure 3) or Western blot analysis (WB; Figure 4).
Table 1 shows a selection of membrane proteins identified with high confidence from four different analyses of biotinylated MDA samples and four different analyses of biotinylated B02 samples (see Table W1 for sequences, scores, and total number of peptides identifying these proteins of interest). The rationale used for protein inclusion in Table 1 was the relevance of the proteins based on the following: 1) the regulation of the proteins; 2) the actual presence of the protein in the membrane fraction, as determined by checking in the Human Protein Reference Database [20] (a database based on experimental evidences manually extracted from the literature and not on literature mining programs); and 3) the literature mining for a most probable relevance of the proteins in cancer- or metastasisrelated events.
Regulation was assessed by both “on-off” and spectral counting methods: for each analysis, a given protein was present or absent (“on” or “off”). Whenever present, we used the associated spectral counting (i.e., the number of redundant spectra of the same peptide) as a quantification mean [21] to have an idea of the relative protein abundance in the samples.
Down-regulation of HLAs
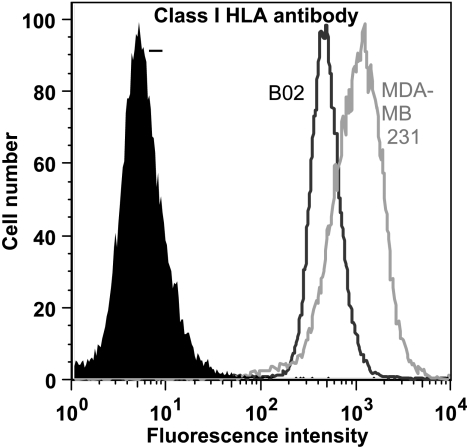
The most striking difference between the two human breast cancer cell lines was clearly the down-regulation or absence of numerous HLA (class I) proteins from the cell surface of the bone metastatic variant B02 (Figure W1 shows a representative MS/MS fragmentation spectra of the peptide FIAVGYVDDTQFVR, identifying class I HLAs). HLA-A are found in both cell lines, but the spectral counting shows a huge down-regulation in B02. This down-regulation is also true for HLA-B, and HLA-C was simply not found in the B02 MS analyses. We sought to confirm this differential expression by FACS (Figure 3) and Western blot analysis (Figure 4A) using an antibody recognizing all class I HLAs. FACS analysis clearly shows that the HLA proteins are down-regulated in B02, confirming the data obtained from mass spectrometry. The Western blot unambiguously also shows that a huge difference actually exists between the two cell lines, with a prominent HLA expression in MDA.
Figure 3.
FACS analysis of class I HLA proteins in MDA and B02 cell lines. MDA and B02 cells were labeled either with class I HLA antibody and analyzed with a FACSCalibur (BD Biosciences). Negative controls (-) are shown in black, B02 in dark gray, and MDA in light gray.
Figure 4.
Western blot analyses. The different proteins revealed by mass spectrometry were probed with their respective antibodies after Western blot analysis: expression of class I HLA (A), CD107b (B), CD51 (integrins αv; C), and CD61 (integrin β3; D) are shown, and the amount of proteins loaded (20 µg) was normalized using tubulin alpha (E).
We also checked the expression of some other proteins to validate the MS data. We investigated the expression levels of three CD markers, CD107b, CD51, and CD61, all three proteins being involved in tumorigenesis and/or metastasis. The expression levels were assessed by Western blot analysis, as shown in Figure 4, B, C, and D, for CD107b, CD51, and CD61, respectively. The actual expression of these proteins was validated for each CD marker, confirming that the MS analyses were able to depict correctly global protein expression patterns of the parental versus the osteotropic cell line.
Discussion
The ability of cancer cells to grow in a specific site depends not only on features that are inherent to cancer cell but also on features that are inherent to the organ and on the interactions between cancer cells and the metastatic site. These interactions are most probably essential in determining whether the cancer cells have a high or low probability of growing. In this regard, the up- or down-regulation of membrane-associated proteins involved in the cross talk between circulating invasive cells and the organs might be important for the establishment of metastases. Using cell surface labeling and a high-throughput proteomic method, we have identified differentially expressed proteins between two cell lines that could be of special interest in characterizing the osteotropic phenotype. We analyzed the membrane proteome of the bone-seeking B02 cell line, selected by multiple in vivo passages from the parental breast cancer cell line MDA-MB-231. The methodology used enabled us to discover several differentially expressed proteins. This shotgun approach permits fast and accurate identification of small but complex biologic sample mixtures, overcoming many limitations of two-dimensional gel electrophoresis for proteome analysis [22]. A quite similar method was already used to compare MCF7 and BT474 breast cancer cell lines but did not include cell surface biotinylation [23]. It might be also useful to benefit from direct protein quantitation using, for instance, stable isotopic labeling with amino acids in cell culture. However, we showed herein that Multidimensional Protein Identification Technology, a technique now well established in many MS laboratories, is sufficient for identifying differentially expressed membrane proteins and even estimate the protein abundance [21]. More difficult to set up and also more expensive technologies such as stable isotopic labeling with amino acids are not absolutely needed yet can provide interesting additional data [24].
Our approach led to the discovery of proteins that are either upregulated or down-regulated in the B02 cell line. For instance, B02 cells overexpressed the αvβ3 integrins when compared to the parental cell line. This was an expected result knowing the unanimously recognized role of these integrins during bone metastasis development: αvβ3 integrins confer to breast tumor cells a greater propensity to metastasize to bone [10], can even promote spontaneous metastasis of breast cancer cells to bone [25,26], and represents a therapeutic target for the treatment of experimental bone metastases [26]. Because this overexpression was not revealed at the transcriptomic level [7], these data confirm the additional and complementary value of the proteomic approach to the transcriptomic analysis. Three other interesting proteins were found up-regulated in the B02 clone following MS analyses. The first one is prohibitin, a multifunctional membrane-associated protein that was interestingly found up-regulated in superinvasive cancer cells [27]. Its molecular function as a transmembrane signaling receptor is still elusive [28]. This protein would deserve further investigations in bone metastases, because direct intratumoral injection of prohibitin 3′UTR RNA can cause complete regression of tumors and metastases [29]. The second protein up-regulated in B02 is MRP-1, a cell surface glycoprotein that is known to complex with integrins [30]. This protein can modulate key events, such as cell adhesion, migration, proliferation, and growth, and might be of special interest in the bone tropism. The third protein up-regulated in the B02 cell line is kinectin, an evolutionary conserved integral membrane protein [31]. Less is known about kinectin in cancer, but kinectin was at least found to be alternatively spliced in hepatocellular carcinoma [32]. All these proteins would deserve further investigations.
Other proteins were found down-regulated in the B02 cells. The most important finding is the strong down-regulation of MHC (HLA) class I in the bone-seeking clone. It has already been shown that escape caused by deficient expression of MHC class I antigens on tumor cells may support homing or survival of disseminated tumor cells in lymphoid tissue [33]. Similarly, reduced MHC class I expression was found in metastatic cells from the bone marrow of patients with squamous cell carcinoma of the head and neck region [34]. It has been suggested that the lack of MHC class I expression is a potential mechanism to escape from HLA class I-restricted lysis by cytotoxic T cells. This escape from immunity could therefore explain at least in part the establishment of B02 clones in the bone environment.
Others proteins were found to be down-regulated in the B02 cell line, such as CD107b, also known as lysosomal-associated membrane protein 2. This protein is a heavily glycosylated transmembrane protein, whose up-regulation on the surface of tumor cell lines has been associated with an enhanced metastatic potential by increasing adhesion to extracellular matrix and endothelium [35].
To summarize, we searched for proteins that could play a role in the establishment of circulating cancer cells in the bone environment, by comparing the membrane proteome of osteotropic cells versus the parental cell line. The down-regulation of class I HLAs contribute most likely to the establishment of B02 cells in the bone environment by preventing immunity responses against these cancer cells. On the other side, up-regulation of membrane proteins from circulating metastatic cells can constitute potentially important targets for antibody-based anticancer treatments.
Supplementary Material
Acknowledgments
The authors are grateful to Rowan Dobson for revising and editing the manuscript. The authors thank Gabriel Mazzuchelli for expert assistance in mass spectrometry and Frédéric Rosu for the generation of the random database.
Abbreviations
- NHS
N-hydroxy-succinimid
- RT
room temperature
Footnotes
This work was supported by the EU FP6 framework programs Selective Targeting of Angiogenesis and of Tumor Stroma and Molecular Mechanisms of Organ-Specific Metastatic Growth Processes in Breast cancer. Grant sponsors of this work also include the National Fund for Scientific Research (Belgium), Télévie, the Centre Anti-Cancéreux près l'Université de Liège, the Léon Frédéricq Foundation, and Tournesol. The Esquire HCT device belongs to the GIGA proteomic platform. P.K. is a research fellow of the Belgian National Fund for Scientific Research.
This article refers to a supplementary materials, which are designated by Figure W1 and Table W1 and are available online at www.neoplasia.com.
References
- 1.Coleman RE, Rubens RD. The clinical course of bone metastases from breast cancer. Br J Cancer. 1987;55:61–66. doi: 10.1038/bjc.1987.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schaefer NG, Pestalozzi BC, Knuth A, Renner C. Potential use of humanized antibodies in the treatment of breast cancer. Expert Rev Anticancer Ther. 2006;6:1065–1074. doi: 10.1586/14737140.6.7.1065. [DOI] [PubMed] [Google Scholar]
- 3.Adams GP, Weiner LM. Monoclonal antibody therapy of cancer. Nat Biotechnol. 2005;23:1147–1157. doi: 10.1038/nbt1137. [DOI] [PubMed] [Google Scholar]
- 4.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 5.Yoneda T, Williams PJ, Hiraga T, Niewolna M, Nishimura R. A bone-seeking clone exhibits different biological properties from the MDAMB-231 parental human breast cancer cells and a brain-seeking clone in vivo and in vitro. J Bone Miner Res. 2001;16:1486–1495. doi: 10.1359/jbmr.2001.16.8.1486. [DOI] [PubMed] [Google Scholar]
- 6.Peyruchaud O, Winding B, Pecheur I, Serre CM, Delmas P, Clezardin P. Early detection of bone metastases in a murine model using fluorescent human breast cancer cells: application to the use of the bisphosphonate zoledronic acid in the treatment of osteolytic lesions. J Bone Miner Res. 2001;16:2027–2034. doi: 10.1359/jbmr.2001.16.11.2027. [DOI] [PubMed] [Google Scholar]
- 7.Bellahcene A, Bachelier R, Detry C, Lidereau R, Clezardin P, Castronovo V. Transcriptome analysis reveals an osteoblast-like phenotype for human osteotropic breast cancer cells. Breast Cancer Res Treat. 2007;101:135–148. doi: 10.1007/s10549-006-9279-8. [DOI] [PubMed] [Google Scholar]
- 8.Anderson L, Seilhamer J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis. 1997;18:533–537. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- 9.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pecheur I, Peyruchaud O, Serre CM, Guglielmi J, Voland C, Bourre F, Margue C, Cohen-Solal M, Buffet A, Kieffer N, et al. Integrin α(v)β3 expression confers on tumor cells a greater propensity to metastasize to bone. FASEB J. 2002;16:1266–1268. doi: 10.1096/fj.01-0911fje. [DOI] [PubMed] [Google Scholar]
- 11.Peirce MJ, Wait R, Begum S, Saklatvala J, Cope AP. Expression profiling of lymphocyte plasma membrane proteins. Mol Cell Proteomics. 2004;3:56–65. doi: 10.1074/mcp.M300064-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Shin BK, Wang H, Yim AM, Le Naour F, Brichory F, Jang JH, Zhao R, Puravs E, Tra J, Michael CW, et al. Global profiling of the cell surface proteome of cancer cells uncovers an abundance of proteins with chaperone function. J Biol Chem. 2003;278:7607–7616. doi: 10.1074/jbc.M210455200. [DOI] [PubMed] [Google Scholar]
- 13.Sabarth N, Lamer S, Zimny-Arndt U, Jungblut PR, Meyer TF, Bumann D. Identification of surface proteins of Helicobacter pylori by selective biotinylation, affinity purification, and two-dimensional gel electrophoresis. J Biol Chem. 2002;277:27896–27902. doi: 10.1074/jbc.M204473200. [DOI] [PubMed] [Google Scholar]
- 14.Scheurer SB, Roesli C, Neri D, Elia G. A comparison of different biotinylation reagents, tryptic digestion procedures, and mass spectrometric techniques for 2-D peptide mapping of membrane proteins. Proteomics. 2005;5:3035–3039. doi: 10.1002/pmic.200402069. [DOI] [PubMed] [Google Scholar]
- 15.Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Mol Cell Proteomics. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 17.Steen H, Mann M. The ABC's (and XYZ's) of peptide sequencing. Nat Rev Mol Cell Biol. 2004;5:699–711. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- 18.Hurley WL, Finkelstein E, Holst BD. Identification of surface proteins on bovine leukocytes by a biotin-avidin protein blotting technique. J Immunol Methods. 1985;85:195–202. doi: 10.1016/0022-1759(85)90287-x. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y, Zhang W, Kho Y, Zhao Y. Proteomic analysis of integral plasma membrane proteins. Anal Chem. 2004;76:1817–1823. doi: 10.1021/ac0354037. [DOI] [PubMed] [Google Scholar]
- 20.Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H, Sadygov RG, Yates JR., III A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 22.Peng J, Gygi SP. Proteomics: the move to mixtures. J Mass Spectrom. 2001;36:1083–1091. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
- 23.Xiang R, Shi Y, Dillon DA, Negin B, Horvath C, Wilkins JA. 2D LC/MS analysis of membrane proteins from breast cancer cell lines MCF7 and BT474. J Proteome Res. 2004;3:1278–1283. doi: 10.1021/pr049852e. [DOI] [PubMed] [Google Scholar]
- 24.Liang X, Zhao J, Hajivandi M, Mu R, Tao J, Amshey JW, Pope RM. Quantification of membrane and membrane-bound proteins in normal and malignant breast cancer cells isolated from the same patient with primary breast carcinoma. J Proteome Res. 2006;5:2632–2641. doi: 10.1021/pr060125o. [DOI] [PubMed] [Google Scholar]
- 25.Sloan EK, Pouliot N, Stanley KL, Chia J, oseley JM, Hards DK, Anderson RL. Tumor-specific expression of αvβ3 integrin promotes spontaneous metastasis of breast cancer to bone. Breast Cancer Res. 2006;8:R20. doi: 10.1186/bcr1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Y, Bachelier R, Treilleux I, Pujuguet P, Peyruchaud O, Baron R, Clément-Lacroix P, Clézardin P. Tumor αvβ3 integrin is a therapeutic target for breast cancer bone metastases. Cancer Res. 2007;67:5821–5830. doi: 10.1158/0008-5472.CAN-06-4499. [DOI] [PubMed] [Google Scholar]
- 27.Dowling P, Meleady P, Dowd A, Henry M, Glynn S, Clynes M. Proteomic analysis of isolated membrane fractions from superinvasive cancer cells. Biochim Biophys Acta. 2007;1774:93–101. doi: 10.1016/j.bbapap.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 28.Terashima M, Kim KM, Adachi T, Nielsen PJ, Reth M, Kohler G, Lamers MC. The IgM antigen receptor of B lymphocytes is associated with prohibitin and a prohibitin-related protein. EMBO J. 1994;13:3782–3792. doi: 10.1002/j.1460-2075.1994.tb06689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jupe ER, Lerner M, Thompson L, Branam D, Manjeshwar S, Farmer D, Taylor B, Hollingsworth A, Brackett D. Therapeutic effects of prohibitin RNA in a metastatic rat mammary tumor model. Mol Ther. 2000;1:162–163. [Google Scholar]
- 30.Berditchevski F. Complexes of tetraspanins with integrins: more than meets the eye. J Cell Sci. 2001;114:4143–4151. doi: 10.1242/jcs.114.23.4143. [DOI] [PubMed] [Google Scholar]
- 31.Futterer A, Kruppa G, Kramer B, Lemke H, Kronke M. Molecular cloning and characterization of human kinectin. Mol Biol Cell. 1995;6:161–170. doi: 10.1091/mbc.6.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang HC, Su YR, Han KJ, Pang XW, Peng JR, Liang B, Wang S, Chen WF. Multiple variants and a differential splicing pattern of kinectin in human hepatocellular carcinoma. Biochem Cell Biol. 2004;82:321–327. doi: 10.1139/o04-003. [DOI] [PubMed] [Google Scholar]
- 33.Passlick B, Pantel K, Kubuschok B, Angstwurm M, Neher A, Thetter O, Schweiberer L, Izbicki JR. Expression of MHC molecules and ICAM-1 on non-small cell lung carcinomas: association with early lymphatic spread of tumour cells. Eur J Cancer. 1996;32A:141–145. doi: 10.1016/0959-8049(95)00551-x. [DOI] [PubMed] [Google Scholar]
- 34.Andratschke M, Pauli C, Stein M, Chaubal S, Wollenberg B. MHC-class I antigen expression on micrometastases in bone marrow of patients with head and neck squamous cell cancer. Anticancer Res. 2003;23:1467–1471. [PubMed] [Google Scholar]
- 35.Saitoh O, Wang WC, Lotan R, Fukuda M. Differential glycosylation and cell surface expression of lysosomal membrane glycoproteins in sublines of a human colon cancer exhibiting distinct metastatic potentials. J Biol Chem. 1992;267:5700–5711. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.