Abstract
Human papillomaviruses (HPVs) cause squamous cancers of epithelial surfaces, of which genital cancers are the most common. In this article we have attempted to describe the properties and functions of the viral proteins of HPV type 16, a common cause of genital cancers, and have tried to suggest how their expression may lead to a dysregulated cell which may become malignant. These viruses are attempting to replicate in terminally differentiating keratinocytes and must stimulate G1 to S-phase progression for the replication of their genome. As part of the successful completion of replication and assembly of infectious virus particles, the virus needs at least partial differentiation to occur. Therefore, at the same time as differentiation is occurring, the nuclei of infected cells are in S-phase. While the mechanisms of action of the viral proteins are not completely understood, researchers are making progress and this article strives to bring together the conclusions from some of this work.
Introduction
Human papillomaviruses (HPV) are small double-stranded DNA viruses, which infect stratified epithelium. There are at least 80 different types, which can be divided into those infecting cutaneous surfaces and those infecting mucosal surfaces. The types have traditionally been differentiated by sequence divergence, although it is becoming clear, using antibodies to conformational epitopes on the major and minor capsid proteins, that the virions are serologically different. While the viruses often cause benign proliferative lesions or warts, there are a subgroup of viruses which cause premalignant and malignant lesions. Malignant disease occurs most frequently in the genital tract and carcinoma of the cervix is by far the most common cancer. This article will concentrate on the viruses which infect the genital tract as they are of medical importance and as substantial research has been carried on the biology of these viruses.
Natural history of HPV infection
The viruses enter the epithelium through microlesions and infect the basal epithelial cells where they maintain a copy number of 50–100 genomes per cell. Upon cell division, one daughter cell will remain part of the basal epithelium, while the other daughter cell will migrate up to the next level and start to differentiate. At this stage the viral DNA will segregate with the two daughter cells and replicate to maintain the 50–100 copies per cell. The fact that one daughter cell starts to differentiate is a problem for the virus because it needs the replicative machinery of the cell for viral DNA synthesis and a terminally differentiated cell will contain little or no replicative enzymes. Therefore the virus needs to stimulate G1 to S-phase progression in the face of a cell programmed to terminally differentiate, and so produce the correct environment for DNA replication. However, the virus does require some level of differentiation, since the late promoter, which regulates the mRNA coding for the capsid proteins, is only switched on in partially differentiated cells. The end result, therefore, is that the virus uncouples G1 to S-phase progression from differentiation, and both processes occur in the same cell. As the infected cells move up through the epithelium and partially differentiate the viral DNA is amplified in the granular layer and late gene transcription and translation occur near the top of the epithelium with viral particle assembly taking place in the cornified layer. Infected cells are sloughed off from the top of the epithelium and may be transmitted directly to individuals. Alternatively, infected cells may remain present in the environment before the virus is transmitted to a new epithelial surface. This latter case is true for viruses which infect cutaneous surfaces. Direct transmission of viruses infecting mucosal surfaces can occur by contact during sexual intercourse.
HPV associated diseases
While benign warts are only a cosmetic nuisance, papillomaviruses can also cause a number of more life-threatening diseases (McCance 1998; zur Hausen 1999). A large group of HPV types cause lower genital tract cancers of which cervical cancer is the most common. (Table 1) lists the mucosal viruses involved along with the associated diseases and (Table 2) summarizes the viruses on the basis of whether they are high risk or low risk types. In addition, there is a group of cutaneous viruses that are also involved in squamous cell carcinomas particularly in immunosuppressed allograft recipients and individuals with a rare genetic disease called epidermodylasia verruciformis, which results in partial immunosuppression (McGregor & Rustin 1994). (Table 3) summarizes the viruses associated with cutaneous malignancies. This article will concentrate on the viruses infecting mucosal surfaces, in particular the genital tract, which commonly infect sexually active individuals.
Table 1.
Genital human papillomavirus types and the site of associated lesionsa
| HPV type | Associated lesion | Site |
|---|---|---|
| HPV 6a-f | Condylomata acuminata | Vulva |
| Vagina | ||
| Cervix | ||
| Penis | ||
| shaft | ||
| prepuce | ||
| urethral meatus | ||
| Perianal | ||
| Larynx | ||
| bLSIL/HSIL | Cervix | |
| cVIN I-III | Vulva | |
| dPIN I-III | Penis | |
| 11a, b | Condylomata acuminata | Vulva |
| Cervix | ||
| Perianal | ||
| Larynx | ||
| LSIL/HSIL | Cervix | |
| PIN I-III | Penis | |
| 16 | Condylomata acuminata | Vulva, cervix and penis |
| LSIL/HSIL | Cervix | |
| VIN I-III | Vulva | |
| PIN I-III | Penis | |
| Bowenoid papulosis | Vulva and penis | |
| Malignant carcinoma | Cervix, vulva and penis | |
| 18 | LSIL/HSIL | |
| Malignant carcinoma | Cervix and penis | |
| 31 | LSIL/HSIL | Cervix |
| Malignant carcinoma | ||
| 30 | LSIL/HSIL | Cervix |
| 33 | LSIL/HSIL | Cervix |
| Malignant carcinoma | ||
| 34 | LSIL/HSIL | Cervix |
| 35 | LSIL/HSIL | Cervix |
| Malignant carcinoma | ||
| 39 | LSIL/HSIL | Cervix and penis |
| PIN | ||
| Malignant carcinoma | ||
| 40 | LSIL/HSIL | Cervix and penis |
| 42 | LSIL/HSIL/VIN | Cervix and vulva |
| 43 | LSIL/HSIL | Cervix |
| 44 | LSIL/HSIL | Cervix |
| 45 | LSIL/HSIL | Cervix |
| Malignant carcinoma | ||
| 51 | LSIL/HSIL | Cervix |
| Malignant carcinoma | ||
| 52 | LSIL/HSIL | Cervix |
| Malignant carcinoma | ||
| 53 | Normal cervix | |
| 54 | Condyloma | Penis |
| 55 | Bowenoid papulosis | Penis |
| 56 | LSIL/HSIL | Cervix |
| Malignant carcinoma | ||
| 57 | Intraepithelial | Oral cavity, cervix |
| Neoplasia | ||
| 58 | LSIL/HSIL/ | Cervix |
| Malignant carcinoma | ||
| 59 | VIN | Vulva |
| 61 | VIN | Vulva |
| 66 | LSIL/HSIL | Cervix |
| Malignant carcinoma | ||
| Intraepithelial neoplasia | Cervix | |
| vulva | ||
| 68 | LSIL/HSIL | Cervix |
| Malignant carcinoma | Cervix | |
| 69 | Intraepithelial neoplasia | Cervix |
| 70 | Condyloma | Vulva |
| Malignant carcinoma | Cervix | |
| 71 | VAINe | Vagina |
| 74 | VAIN | vagina |
| 82 | HSIL | Cervix |
Adapted from De Villiers (1989) and updated with the help of Dr C. Wheeler.
Low-grade squamous intraepithelial lesion and high-grade squamous intraepithelial lesion of the cervix.
Vulvar intraepithelial neoplasia.
Penile intraepithelial neoplasia.
Vaginal intraepithelial neoplasia.
Table 2.
Genital HPV and their associated risk of cancer
| Lowrisk | 6, 11, 40, 42–45, 53–55, 57, 59, 61, 67, 69, 71, 74, 82 |
| Highrisk | 16, 18, 31–35, 51–52, 56, 58, 61, 66, 68, 70, 73 |
Table 3.
HPV types isolated from patients with the genetic disease epidermodysplasia verruciformis (EV) and associated diseases:
| HPV type | Lesions |
|---|---|
| 5 | Warts/SCC |
| 8 | SCC |
| 9 | warts |
| 12 | warts |
| 14 | SCC |
| 15 | warts |
| 17 | warts/SCC |
| 19 | warts |
| 20 | warts/SCC |
| 21 | warts |
| 22 | warts |
| 23 | warts |
| 24 | warts |
| 25 | warts |
| 36 | warts |
| 46 | warts |
| 47 | warts |
HPV types 38 (melanoma), 41 (SCC) and 48 (SCC) were isolated from immunosuppressed patients. This table was assembled with the help of Dr C. Wheeler.
Genital tract infections
HPV types cause a variety of lesions in the genital tract of males and females (Schiffman & Brinton 1995). Some types (Table 1) cause benign warts and low-grade premalignant lesions and are not found in malignant tissues. HPV type 6 and 11 (HPV-6, HPV-11) are the most common isolates from these lesions. There is another group of viruses that are found in premalignant and malignant tissues, and HPV-16 and −18 are commonly associated with these lesions, although a large number of other HPV types, which are not so common, have a similar pathogenesis (Table 1).
In adults, transmission is predominantly by sexual contact. However, there is evidence that HPV can be transmitted from mother to neonate, probably during delivery. Laryngeal warts in young children are thought to arise after such episodes of transmission (Kashima et al. 1985; Fredericks et al. 1993; Austen et al. 1996). In addition, genital infection is also possible after mother to neonate transmission (Cason et al. 1999), but it is unclear whether such transmission leads to clinical disease later in life. The viruses cause similar lesions in both males and females, however cancer of the cervix is more common than cancer at other sites. For example, penile cancer is much rarer and occurs mainly in men over 60 years of age (McCance et al. 1986; Malek et al. 1993). Why should cervical cancer be so much more common? The disease arises from the transformation zone on the cervix located between the mature epithelium of the exocervix and the columnar epithelium of the endocervical canal. This zone is immature, metaplastic and hormonally responsive. As such, it appears during puberty and disappears at menopause. It would appear that the transformation zone is more susceptible to external insults such as viral infection resulting in the production of premalignant and possibly malignant cells. The rest of the epithelium in the female and male genital tract is composed of mature epithelium and seems less prone to malignant conversion due to HPV infection. While cancers arising at other sites such as the vaginal wall, vulva, perianal region (both sexes) and the penis are rarer, they are associated with the same high risk HPV types.
Initial infection with the high-risk types causes low- grade disease (low-grade squamous intraepithelial lesions, LSIL), which is manifested by inhibition of the normal differentiation in the lower third of the epithelium. The lesion may remain low-grade, regress, or progress to severe dysplasia or HSIL (high-grade squamous intraepithelial lesions). This latter stage may persist or may start to invade below the basement membrane leading to metastatic disease. The relationship of the HPV genome to the chromosomal DNA may change during progression from premalignant to malignant phases of the disease. Most of the metastatic cells have integrated HPV sequences (∼70%) while the rest have predominantly episomal copies (McCance et al. 1985; McCance et al. 1986; zur Hausen 1999). However, it is not clear if these episomal copies have wild type sequence or have mutations and whether there are small subfragments of the HPV genome integrated. Integration occurs randomly in the cellular chromosomes, but all viral DNA integration events associated with malignant disease, allow for the expression of E6 and E7 proteins. This suggests that the expression of both these proteins is important for the phenotype of the invading cells and only those cells where integration of the viral genome permits the expression of E6 and E7 are selected.
Rates of infection
The number of cases of malignant disease of the cervix is small compared to the number of women infected with HPV. For example, studies have shown that 20–25% of young women (18–30 years of age) are infected with an oncogenic virus, but there are only 15 000 cases of invasive cervical cancer per year in the USA and this occurs most frequently in women in their 4th and 5th decades. Large studies in the USA (Hildesheim et al. 1994) suggest that that the majority of HPV positive women will have a transitory infection and only in a small proportion will infection lead to severe premalignant disease, or HSIL and then onto malignant disease. The transitory infections lead to mild disease, which will spontaneously regress. Therefore, while many individuals are infected, the numbers with severe disease are small. The problem however, is to identify those individuals who are at risk for progressive disease amongst the large number infected.
HPV testing
It has been suggested that HPV infection can lead to cervical cancer. Therefore, testing for the presence of HPV DNA on the cervix may be a better way to diagnose disease than the traditional Pap smear. However, there are problems with DNA typing. Firstly, it is more expensive than the Pap smear and secondly, while the sensitivity is generally greater, this can lead to false positives depending on the type of test. One possible exception in developed countries is the triage of patients with atypical squamous cells of undetermined significance (ASCUS), where DNA testing may help to assign the patient for colposcopy or follow-up (Manos et al. 1999). At present, however, there is no recommendation for this as a standard diagnostic procedure. Regardless of the type of diagnostic test, identifying HPV infection may fail in identifying those women at risk for progressive disease.
Prevention of infection/disease by vaccination
One possible approach to controlling the level of disease would be to prevent infection. Theoretically, this could be achieved by vaccination as with many childhood diseases. At present it is not clear what constitutes a protective immune response in humans but this has not prevented some trials of potential vaccine candidates from taking place. There are two approaches to vaccination. One would be to produce a prophylactic vaccine to prevent infection and the other to produce a therapeutic vaccine to administer, probably to people with invasive disease. The latter types of vaccine contain early proteins such as E6 and E7 and have been shown to produce B and T cells responses in recipients. The prophylactic vaccines contain the major capsid protein L1 in which, when expressed in insect cells, bacteria or yeast forms into an iscohedral structure very similar to the capsid in the virion. These structures are called virus-like particles (VLPs). The rationale behind using capsid proteins stems from work in cattle and dogs, which has shown high levels of protection from experimental infection after vaccination with either BPV-1 or canine papillomavirus L1 proteins in the form of VLPs (Jarrett et al. 1991; Suzich et al. 1995). Phase I clinical trials using HPV-6 and −11 VLPs have been carried out and a robust antibody response was detected with few minor side-effects from vaccination. Whether the responses will protect against infection has yet to be tested. One would expect that a cytotoxic T-cell response would also be necessary for effective protection. While vaccination using VLPs produces a good antibody response, it has yet to be demonstrated in humans that a cytotoxic T-cell response results. Another Phase I trial with HPV-16 and HPV-18 VLPs commences in the autumn of 2000 and it is expected to be as successful as previous trials as far as antibody induction is concerned.
Since L1 and L2 (the minor capsid protein) are highly conserved between many genital HPV types it was hoped that cross-protection might occur. However, when the proteins form into their iscohedral structure there is little or no cross reactivity of antibodies which ‘see’ the conformation epitopes produced by the structure. Antibodies against disrupted VLP, which bind to linear epitopes, do not react with the VLPs or to full virus particles. Therefore, it would appear that individuals will have to be vaccinated with a cocktail of VLPs from different viruses for protection against a range of virus types. Phase II and III trials using VLP are eagerly awaited.
Replication of HPV in stratified epithelium
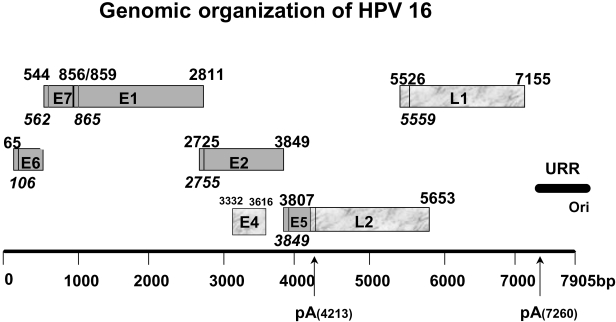
The consensus is that the virus infects the basal epithelial cells through microlesions and replicates to a stable level of 50–100 copies per cell in the basal layer. The cells of the basal layer can divide and one daughter cell remains in the basal layer while the other moves up and starts to terminally differentiate. Differentiated cells contain little or no replicative machinery, therefore the virus will be unable to propagate if cells are permitted to terminally differentiate. Therefore the virus expresses proteins early in infection in the basal and parabasal layers, whose biological properties are consistent with stimulating G1 to S-phase progression. However, at the same time the virus does require a certain level of differentiation because the shift from the early to late promoter, which transcribes capsid protein mRNA, is mediated in differentiated cells. The factors required for this promoter shift have yet to be determined. The fact that some level of differentiation is required is reinforced by the observation that viral DNA amplification and capsid protein synthesis is never observed in basal epithelial cells. The following sections describe possible mechanisms whereby three viral proteins, E6, E7 and E5 (Fig. 1) achieve a cellular environment which is conducive to viral DNA replication.
Figure 1.
Schematic of the HPV-16 genome organization. The early genes are solid grey and the late genes are marble grey. The base pairs of the open reading frames are shown and the ATG start codon is shown in italics. E4 is spliced to the N-terminal of E1 (E1^E4) since it has no ATG of its own. The poly A + signals for the early and late mRNA are shown (pA). The origin (ori) of replication and the upstream regulatory region (URR) or the main early promoter region are indicated.
E6 protein
The HPV-16 E6 protein is a 151 amino acid protein, containing two zinc finger domains. Although small, E6 induces several important changes in the host cell that impact both the normal viral life cycle and the process of immortalization. E6 alone is not capable of immortalizing primary human foreskin keratinocytes (HFK), but can efficiently immortalize human mammary epithelial cells (HMEC) (Kiyono et al. 1998; Liu et al. 1999). However, E6 does act with E7 to immortalize HFK. Expression of E6 as a transgene in mice leads to hyperproliferation of epithelial layers, loss of differentiation in epithelial layers and benign and malignant tumour formation (Song et al. 1999). E6 is one of the earliest expressed genes during an HPV infection. It provides several functions that alter the cellular environment, making it more amenable to production of new viral particles. These include blocking apoptosis through p53 degradation, altering transcription of cellular genes through interaction with p300, and increasing cellular lifespan through increased telomerase activity. These functions serve to facilitate the viral life cycle. These biological activities of E6 are discussed in more detail below.
E6 and cell proliferation
As mentioned above, E6 will act together with E7 to efficiently immortalize HFK. One recent finding is that E6 can up-regulate the activity of the cellular telomerase complex. Telomerase is an enzyme that maintains the telomeric DNA at the ends of linear chromosomes (Greider & Blackburn 1985). It utilizes an RNA molecule (TER) as the template for synthesizing DNA and a reverse transcriptase protein component (TERT) to carry out the synthesis (Greider & Blackburn 1987). Without telomerase, telomeres shorten with each cell division, until they reach a critically short length. Beyond this point further division would induce damage to the coding regions of the chromosome and cell senescence.
It is hypothesized that telomere shortening functions as a sort of molecular clock, controlling the number of cell divisions any cell can undergo and signalling cellular senescence when this limit is reached. Supporting this notion, experiments have shown that expression of exogenous telomerase in primary human fibroblasts effectively immortalizes these cells, allowing them to proliferate beyond 100 population doublings (Bodnar et al. 1998). Additionally, cells that normally proliferate in vivo have detectable telomerase activity, as do most tumour cells, while cells that normally have a limited lifespan in vivo generally do not show telomerase activity (Harle-Bachor & Boukamp 1996; Belair et al. 1997).
Telomerase activity is increased in HFK, HFF and HMEC expressing HPV16 E6 (Kiyono et al. 1998; Klingelhutz et al. 1996; Stoppler et al. 1997). However, the importance of telomerase activation in E6/E7 mediated immortalization remains unclear. It has been shown that E6 alone will immortalize HMEC, and E6 mutants that do not increase telomerase activity do not immortalize (Kiyono et al. 1998). This study also demonstrated that addition of TERT to E7-expressing HFK allows growth and immortalization in serum-free media. HFK typically require both E6 and E7 for efficient immortalization. However, mutants of E6 with altered ability to activate telomerase have not been used in conjunction with E7 in HFK to show directly that immortalization ability is impaired.
Another confounding piece of data is that despite E6-mediated increases in telomerase activity in fibroblasts, telomere lengths do not increase (Filatov et al. 1998). Instead, telomeres are maintained at a short, but apparently sufficient, length to allow for cell survival. However, this paper also demonstrates that there is an increased amount of chromosomal damage occurring in these cells, measured by increased DNA content and increased numbers of chromosomal aberrations as the cells are passaged. Thus, telomerase activation by E6 may be facilitating the transition from a normal to transformed phenotype by telomerase-dependent and –independent pathways.
E6 and apoptosis
It is important that viruses avoid cell death, at least until they have replicated. Apoptosis is a natural death process for cells. The cellular p53 protein is normally induced in response to conditions of cellular stress such as UV irradiation, hypoxia, or viral infection. p53 is a transcription factor that stimulates the expression of genes involved in cell cycle arrest and apoptosis, such as the cyclin dependent kinase inhibitor, p21CIP. E6 has been shown to bind to p53 (Werness et al. 1990) and induce its degradation (Scheffner et al. 1990), a process that is mediated by an ubiquitin ligase called the E6-associated protein (E6-AP) (Huibregtse et al. 1991; Scheffner et al. 1993). Although important for E6 mediated degradation of p53, E6-AP may not be involved in normal regulation of p53 (Talis et al. 1998). In normal cells the mdm2 protein appears to be involved in turnover of p53 but it is not required for E6-mediated degradation (Freedman & Levine 1998). E6 can also retain p53 in the cytoplasm, blocking its translocation to the nucleus and thus inhibiting its function independently of degradation (Mantovani & Banks 1999). The prevention of translocation and degradation of p53 by E6 inhibits the ability of p53 to activate or repress transcription of target genes (Mietz et al. 1992).
Evidence that the interaction of E6 with p53 is important for cell cycle progression is the fact that the p14ARF protein, an alternative gene product of the p16INK4a tumour suppressor locus. p14ARF induces arrest in a manner dependent on p53, causing elevated amounts of p21CIP, resulting in either a G1 or G2 arrest (Stott et al. 1998). Expression of E6 in normal human foreskin fibroblasts (HFF) allows for continued proliferation of these cells despite expression of p14ARF. Another study, carried out in HFF, utilized adriamycin treatment to induce a G2 arrest, and concluded that the ability of E6 to bypass this arrest signal was through a p53 dependent mechanism involving repression of the cyclin B and cdc2 promoters (Passalaris et al. 1999).
In addition to arrest, E6 can bypass apoptosis induced by p53. In response to serum and calcium induced differentiation of HFK, E6 expression decreases the number of apoptotic cells as compared to normal HFK, without blocking differentiation of these cells, as measured by expression of differentiation-specific markers (Alfandari et al. 1999).
Work by Pan and Griep (Pan & Griep 1994; Pan & Griep 1995), using transgenic mice with targeted expression of E6, E7, or both, in the lens of the eye, showed that E6 can block apoptosis in both p53 dependent and independent ways. Another recent study (Jackson & Storey 2000) using E6 genes from mucosal and cutaneous subtypes of HPV demonstrated that all were capable of blocking apoptosis induced by ultraviolet (UV) radiation. However, HPV 18 E6, the only E6 utilised that degraded p53, inhibited apoptosis at the highest UV dosage, suggesting that modulation of p53 plays some role in this inhibition, but does not account for all of the inhibition (Jackson & Storey, 2000).
In addition to interacting with p53, E6 may impact apoptosis through p53-independent pathways. There is some evidence that E6 can interact with Bak, a pro-apoptotic protein expressed at high levels in the upper layers of differentiating epithelium (Krajewski et al. 1996). Thomas and Banks have shown that HPV 18 E6 can physically interact with Bak and mediate its degradation by ubiquitination, through interaction with E6AP, thus reducing apoptosis (Thomas & Banks 1998). They suggest that Bak may be a natural target of the E6-AP, the first identified. More recent work from the same group shows that HPV 16 and 11 E6 proteins can also interact with Bak, but that the interaction is weaker with the nononcogenic subtype, HPV-11, than with HPV-16 (Thomas & Banks 1999). This interaction probably contributes to the p53 independent inhibition of apoptosis.
The ability of E6 to prevent p53 mediated arrest and apoptosis may be especially important in light of two facts. First, HPV E7, when expressed alone, has been shown to increase the amount of p53 present in cells (Demers et al. 1994). Second, the HPV E2 protein, which is involved in viral replication and transcription of viral genes, was recently shown to induce apoptosis through a p53 dependent mechanism, when expressed alone in HPV positive and negative cell lines (Webster et al. 2000). If left unchecked, the increase in p53 and induction of p53 dependent apoptosis would most likely kill an infected cell before viral replication could take place. Thus, the ability of E6 to modulate p53 levels may be integral during productive infection.
E6 and alteration of gene transcription
Several years ago it was shown that HPV 16 E6 was capable of altering transcription of target promoters in a positive and negative manner. E6 expression was shown to transactivate the HPV early promoter in the URR (Fig. 1) (Gius et al. 1988) and the adenovirus E2 promoter (Desaintes et al. 1992). It was later shown to repress the Moloney murine leukaemia virus long-terminal repeat and the cytomegalovirus immediate early promoter (Etscheid et al. 1994). The mechanism for this transcriptional alteration was unclear, but it was not dependent on p53.
The adenovirus E1A protein is capable of interacting with p300 and CBP, transcriptional coactivators that bring together transcription factors and the basal transcription machinery at gene promoters (Eckner et al. 1994; Arany et al. 1995). CBP and p300 also have intrinsic histone acetyltransferase activity, allowing them to acetylate histone core proteins and alter chromatin structure. The E1A-p300/CBP interaction can block transcriptional activation (Chakravarti et al. 1999; Hamamori et al. 1999). Recently, the same activity was described for HPV 16 E6, with the same functional consequence, namely inhibition of p300/CBP mediated transcriptional activation (Patel et al. 1999; Zimmermann et al. 1999). Therefore, an explanation of the earlier work on the repression by E6 may be the interaction of E6 with the coactivators CBP and p300.
E6-CBP/p300 binding probably impacts transcription of other, as yet unidentified, genes. For example, adenovirus E1A, the human herpesvirus 8 vIRF protein and the Epstein-Barr virus EBNA2 protein can increase transcription from the c-myc gene promoter in a manner dependent on transcriptional coactivators p300, CBP or P/CAF (Jayachandra et al. 1999). HPV E6 has also been shown to transactivate the c-myc promoter (Kinoshita et al. 1997) and this may be dependent on the interaction with CBP/p300. Another possible set of target genes are those involved in cytokine production and immune signalling. Control of the IL-6 and IL-8 promoters is largely dependent on the NF-κB transcription factor, which is coactivated by CBP/p300 (Sheppard et al. 1999). HPV-16 E6 has been shown to inhibit NF-κB mediated transcription, therefore the interaction of HPV E6 with p300 or CBP might modulate transcription of these cytokines, effectively inhibiting these signals to the immune system (Patel et al. 1999). Other transcriptional targets affected by the E6-p300/CBP interaction have yet to be identified.
E6 interactions with other proteins
E6 has also been reported to interact physically with several other proteins. These include the E6BP protein (also known as ERC-55), the human homologue of the Drosophila discs large protein and the c-myc protein. The functional significance of these interactions is still largely unknown.
E6BP was identified in a yeast two-hybrid screen for E6 interacting proteins (Chen et al. 1995). It was shown in this study to interact with E6 in vitro and in vivo. E6BP is a calcium binding protein, leading to speculation that this interaction might affect calcium induced differentiation of E6 expressing cells. However, E6 expression does not impede calcium dependent differentiation, at least as measured by expression of differentiation specific markers (Alfandari et al. 1999). Mutational analysis of E6 in HMEC cells revealed two relevant points. First, E6 bp binding and E6-AP binding were correlated, meaning that mutants that were unable to bind E6-AP were also unable to interact with E6 bp. Second, most of these mutants were still able to immortalize HMEC cells, though at slightly reduced efficiency relative to wild type E6 (Liu et al. 1999). Thus, the importance of E6 bp in immortalization by E6 has not been established.
The hDLG protein was also identified as interacting with E6 in a yeast two-hybrid screen and shown to interact with E6 in vitro (Kiyono et al. 1997). The hDLG protein is associated with cell junctions and has also been shown to interact with the APC gene, a known tumour suppressor. It has recently been demonstrated that E6 can target hDLG for degradation, independently of E6–AP interaction (Pim et al. 2000), suggesting a potential role for this interaction in tumour formation. However, additional studies need to be undertaken to clarify the role of hDLG in HPV mediated immortalization.
Finally, E6 has been shown to stimulate the degradation of the c-Myc and N-Myc proteins in vitro and in vivo, decreasing the half life of these proteins significantly (Gross-Mesilaty et al. 1998). These interactions were identified following the discovery that E6-AP could mediate c-myc and N-myc degradation. Again, the importance of this in the normal viral life cycle or during transformation is not clear.
E7 protein
The E7 open reading frame of HPV encodes the major transforming activity. HPV16 E7 is a nuclear protein of 98 amino acids, with casein kinase II phosphorylation sites at serine residues 31 and 32. The protein is divided into domains based upon homology to adenovirus E1A with CR1 consisting of amino acids 1–20, CR2 containing residues 21–39, and CR3 composed of amino acids 40–98. E7 dimerizes via a zinc-finger motif in the CR3 region that is essential for proper protein folding.
Numerous interactions between E7 and cellular proteins have been described. Many of these interactions involve factors that regulate cell growth, especially the transition from the G1 of the mitotic cycle into S-phase. E7 has been demonstrated to associate with the retinoblastoma tumour suppressor protein family (Rb, p107, p130), histone deacetylases (HDAC), AP-1 transcription factors, cyclins, cyclin-dependent kinases (cdks), and cdk inhibitors. These associations likely contribute to the ability of E7 to induce cellular proliferation, immortalization and transformation. The effects of E7 on cell growth are discussed in detail below.
Effects of E7 on cell growth
E7 has been demonstrated to alter growth phenotypes in a variety of cell types, the most biologically relevant of which is primary human foreskin keratinocytes (HFK). Immortalization of HFK correlates with oncogenicity of HPVs, with high-risk HPVs (e.g. HPV-16, − 18, − 31) able to efficiently induce immortalization and low-risk HPVs (e.g. HPV-6, − 11) incapable of extending HFK lifespan (McCance et al. 1988; Barbosa & Schlegel 1989; Hudson et al. 1990). Expression of E7 from oncogenic HPV types is sufficient to immortalize HFK, though the immortalizing activity of E7 is more efficient in the context of E6 expression or the entire HPV genome (Barbosa & Schlegel 1989; Munger & Phelps 1993). Indeed, the presence of E7 is essential in HFK immortalization, as mutation of the E7 open reading frame completely abrogates immortalization by full-length HPV16 genomes (Jewers et al. 1992). Expression of E7 also confers extended proliferation to primary rat embyro fibroblasts (REFs) (Munger & Phelps 1993). The CR1 and CR3 regions of E7 appear critical for its immortalizing activity, as mutations in these domains diminish the efficiency of E7-induced immortalization (Banks et al. 1990; Jewers et al. 1992; Takami et al. 1992). Interestingly, the CR2 domain, which contains the high-affinity Rb-binding site (see below), is not necessary for immortalization of HFK in the context of the whole genome, suggesting that interaction with Rb may be dispensable in engendering this phenotype (Jewers et al. 1992).
E7 can also induce cellular transformation in several assays. HPV-16 E7 can induce anchorage-independent growth in NIH3T3 and, in cooperation with an activated ras, induce focus formation in REFs, infant rat kidney cells, and various rodent fibroblast cell lines (Munger & Phelps 1993). Similar to HFK and REF immortalization, E7 transformation of rodent cells correlates with oncogenic potential of HPVs, and chimeric molecules (utilizing domains of E7 from oncogenic HPV-16 and benign HPV-6) indicate that the CR1 and CR2 regions of E7 are essential for this phenotype (Munger et al. 1991; Takami et al. 1992). Mutational analysis has corroborated the importance of the CR1 and CR2 domains (Banks et al. 1990; Heck et al. 1992; Phelps et al. 1992; Sang & Barbosa 1992; Brokaw et al. 1994). In addition, mutation of the zinc-finger (CR3) reduces the transformation potential of E7 (Chesters et al. 1990; Edmonds & Vousden 1989; McIntyre et al. 1993), however, it is not clear whether these mutations disrupt CR3-specific activities necessary for transformation or simply alter the general structure of E7. Therefore, different domains of E7 are essential for rodent cell transformation and HFK immortalization. This suggests that it is important to use the natural host cell to determine functions of E7.
In addition to its immortalizing and transforming activities, E7 also abrogates several growth arrest signals including exposure to transforming growth factor-β, DNA damage, serum deprivation, anchorage-independent suspension, and suprabasal differentiation (Banks et al. 1990; Demers et al. 1996; Schulze et al. 1998). Although the precise mechanisms by which these signals lead to arrest are not completely defined, many of the cellular factors and pathways involved have been delineated. Interestingly, E7 interacts with several of such factors. In the subsequent sections, the interactions of E7 with specific cellular factors that govern cellular growth will be discussed.
Effects of E7 on cellular transcription
It is becoming increasingly clear that E7 mediates many of its effect via the modulation of gene transcription. Consequently, E7 has been found to interact with several proteins believed to affect the transcription of genes involved in cell cycle progression and/or cellular differentiation. The significance of these interactions is discussed below.
Binding to retinoblastoma protein family members
Perhaps the most well characterized property of E7 is its ability to bind to the retinoblastoma tumour suppressor protein (Rb) (Dyson et al. 1989; Munger et al. 1989). The fact that E7 shares this property with other viral oncoproteins, such as the adenovirus E1A and simian virus 40 large T antigen (Munger 1995), suggests that tumour viruses possess evolutionary conserved attributes, and underscores the importance of Rb binding in the natural history of virus infection. The retinoblastoma protein family members play a central role in the regulation of the eukaryotic cell cycle. Specifically, in its hyphophosphorylated state, Rb can bind to transcription factors such as the E2F family members, and repress the transcription of particular genes. As cells progress from G0 through G1 and into S phase, Rb family members become progressively hyperphosphorylated by G1 cyclin-cyclin dependent kinases, releasing the transcription factor E2F, which in turn activates genes involved in DNA synthesis and cell cycle progression (Dyson 1998). Since E7 is able to bind to hypo-phosphorylated Rb, it is believed that E7 can prematurely induce cells into S phase by disrupting Rb-E2F complexes (Huang et al. 1993; Wu et al. 1993; Patrick et al. 1994).
Interaction with Rb is primarily mediated through amino acid sequences contained in the conserved amino terminal or CR2 region of E7 (Barbosa et al. 1990). The CR2 region of E7 binds to Rb and its family members, p107 and p130, through the sequence motif LXCXE. The LXCXE motif of E7 has been shown to specifically bind to one Rb pocket region, between 649 and 772 aa of Rb (Huang et al. 1990). Moreover, the LXCXE amino acid sequence is found within other viral oncoproteins such as E1A and SV40 LTAg, as well as many cellular Rb binding proteins including cyclin D1, cyclin D2, cyclin D3, BRG1, histone deacetylase 1 (HDAC-1) and HDAC-2 (Brehm et al. 1998; Dowdy et al. 1993). The high level of conservation of this motif among viral and cellular proteins suggests that viral oncoproteins such as E7 can compete with cellular proteins for binding to Rb.
As mentioned previously, low-risk HPV-6 E7 and high-risk HPV-16 E7 display different immortalization and transforming properties. Considering the relative importance of targeting Rb during oncogenesis, it has been suggested that variance in Rb binding affinities might correlate with differences in viral oncogenic potentials. Indeed, HPV-6 E7 has been shown to possess a lower affinity for Rb than HPV-16 E7 (Gage et al. 1990). Moreover, the variance in binding efficiency can be accounted for by a single difference in the sequences next to their respective LXCXE motifs: HPV-6 E7 contains a glycine at amino acid 20, while HPV-16 E7 possesses an aspartic acid at the equivalent position (Heck et al. 1992; Sang & Barbosa 1992). Although the CR2 domain of E7 mediates high affinity binding to Rb, this region alone is unable to displace E2F from Rb (Huang et al. 1993; Wu et al. 1993). More recent studies have demonstrated that the carboxy terminal region of E7 (CR3) can also bind to Rb. CR3 interacts with a region between 803 aa and 841 aa of Rb. Furthermore, in vitro experiments indicate that CR3 alone is capable of displacing E2F from Rb, leading to transcriptional activation (Patrick et al. 1994). These results suggest that both the CR2 and CR3 domains are required for high affinity binding to Rb and the subsequent disruption of Rb-E2F complexes.
Recent developments in the Rb and E2F literature highlight the complexity and importance of different Rb family members. For example, p130/E2F complexes are the most predominant and are present in quiescent or differentiated cells, while p107/E2F and Rb/E2F can be found in cells entering G1 and S phase (Dyson 1998). Moreover, p107 and p130 are required for the regulation of different subsets of genes (Hurford et al. 1997). The CR2 region of E7 can interact with both p107 and p130 (Dyson et al. 1992; Davies et al. 1993). Considering the ability of E7 to interact with all known Rb family members and the latter's involvement in both differentiation and cell proliferation, it is tempting to suggest a paradigm whereby E7 can uncouple the process of differentiation from cell cycle progression by modulating the transcription of different subsets of genes. This in turn would establish an environment that is more conducive to viral replication.
Considerable evidence points to the importance of Rb binding in the ability of E7 to transform cells. More recent data, however, suggests that Rb binding may not be essential for the immortalization capabilities of E7. Within the context of the whole HPV genome, mutations in the CR2 region of E7 which abrogate binding to Rb, do not affect the ability of E7 to immortalize primary human keratinocytes (Jewers et al. 1992). However, mutations in the CR3 zinc finger motif, Cys-X-X-Cys, of E7 led to complete abrogation of immortalization capabilities (Jewers et al. 1992). This has prompted the search for Rb independent interaction, involving E7 and other cellular factors, which may prove to be more essential for cellular immortalization.
Association with histone deacetylase complexes
Chromatin remodelling through histone acetylation is emerging as an important mechanism by which gene transcription is regulated. Actively transcribed genes show a high level of histone acetylation while repressed genes do not (Grunstein 1997). In addition, it has been demonstrated recently that Rb can associate with histone deacetylase-1 (HDAC-1), and that both Rb and HDAC-1 cooperate in repressing transcription from E2F regulated genes (Brehm 1998; Magnaghi-Jaulin et al. 1998). These observations suggest that histone deacetylase complexes play an important role in cell cycle regulation, and are consequently potential targets for viral oncoproteins. In accordance with this rationale, the CR3 zinc finger domain of E7 has been found to interact with HDAC-1 (Brehm 1998; Brehm et al. 1999). Although this association is Rb independent, E7 binding to HDAC-1 occurs indirectly through Mi2β, a component of the NURD histone deacetylase complex (Brehm et al. 1999). Mutations in the zinc finger domain of E7 do not affect Rb binding, but inhibit interaction with Mi2β/HDAC complexes and seem to abrogate the ability of E7 to relieve Rb repression (Brehm et al. 1999). While the significance of these results remains unclear, the targeting of histone deacetylases provides yet another method by which E7 can de-repress gene transcription, and may explain the essential nature of the CR3 domain in activating E2F-regulated genes as well as immortalizing keratinocytes.
Binding to AP-1 transcription factors
Further attempts to identify Rb independent functions of E7 have led to the discovery that E7 can interact with members of the AP-1 family of transcription factors, including c-Jun, JunB, JunD and c-Fos. AP-1 transcription factors appear to mediate early mitogenic effects and are further implicated in keratinocyte and myeloid cell differentiation. Specific mutational analysis and binding data to c-Jun indicate that the E7 zinc finger motif, but not the Rb binding domain, is involved in this interaction (Antinore et al. 1996). Moreover, E7 binds to 224–249 aa of c-Jun and can trans-activate transcription from a Jun responsive promoter. The E7/c–Jun interaction was further demonstrated to be important in E7′s ability to transform REF in the presence of activated ras (Antinore et al. 1996).
The c-Jun promoter itself contains AP-1 sites and appears to be regulated by Rb/c-Jun complexes (Nead et al. 1998). The interaction between Rb and c-Jun is mediated via the leucine zipper of c-Jun and the B pocket and c-terminus of Rb. Through such an interaction, Rb can recruit c-Jun to an AP-1 consensus site and activate transcription from c-Jun responsive promoters (Nead 1998). Interestingly, it was demonstrated that the presence of E7 inhibited Rb activation of c-Jun transcription, and that this effect was mediated through E7′s LXCXE motif, suggesting that transcriptional down-regulation by E7 in this case is Rb dependent. Moreover, hypophosphorylated Rb is complexed to c-Jun in terminally differentiated keratinocytes but not in cycling cells (Nead 1998). Finally, in terminally differentiated keratinocytes, the presence of E7 seems to cause a significant reduction in c-Jun levels.
The data discussed above suggest an intricate mode of action whereby HPV E7 can modulate both the process of cell cycle progression as well as cell differentiation, through interactions with AP-1 factors and Rb. On the one hand, by binding to c-Jun independently of Rb, E7 may potentiate the activation of genes involved in early cell cycle progression and thus promote S phase entry. On the other hand, by associating with both Rb and c-Jun, E7 may de-regulate keratinocyte differentiation via disruption of Rb/c-Jun complexes. As such, the differential targeting of AP-1 factors and Rb provides a potential mechanism used by E7 to uncouple differentiation from cell cycle progression.
Interaction with TBP and TAFs
The interactions described so far implicate the association of E7 with proteins that regulate the transcription of specific genes; namely those involved in cell cycling and differentiation. E7 however, has been found to associate with members of the basal transcriptional machinery. E7 can bind to the TATA-binding protein (TBP) and to TBP-associated factor-110 (TAF-110) (Mazzarelli et al. 1995; Massimi et al. 1996; Phillips & Vousden 1997). Binding to TBP seems to require three domains of E7: the Rb binding domain, the CKII domain and the CR3 region (Phillips & Vousden 1997). The importance of these interactions is debatable, but they suggest that E7 may mediate its effects on gene transcription by targeting more general transcription factors.
Effects of E7 on cyclin-CDKs
As previously discussed, viral genome amplification occurs in the differentiating epithelium, and, as such, HPV must force the cell to provide DNA replication components and activities in the absence of growth signals. The G1/S cyclins and cyclin-dependent kinases (cyclin d-cdk4, cyclin E-cdk2, and cyclin A-cdk2) are essential regulators of progression from the first gap phase through DNA synthesis (Sherr 1993; Sherr & Roberts 1999). In a proliferating cell, mitogenic stimulation signals the synthesis and assembly of cyclin d-cdk4 which contributes to inactivation of Rb via phosphorylation, leads to expression of cyclin E, and sequesters inhibitors of cdk2 (Cip/Kip inhibitors). Subsequently, cyclin E-cdk2 continues to inactivate Rb and phosphorylates other substrates essential for initiation of DNA replication and S phase entry. During S phase, cyclin A-cdk2 is assembled and remains active through the second gap phase. As integral components of the G1/S and S/G2 transitions, cyclin-cdks are logical targets of HPV. The effect of E7 on each of these cyclin-cdks is discussed below.
Cyclin E-cdk2 is essential for initiation of DNA synthesis and activates cell cycle progression in the absence of cyclin d-cdk4 (Geng et al. 1999; Ohtsubo et al. 1995; Krude et al. 1997), underlining this complex as a potential target by which E7 propels cell cycle reentry. Interestingly, several groups have demonstrated E7-mediated increases in cyclin E-cdk2 activity in asynchronous HFK (Funk et al. 1997; Jones et al. 1997; Ruesch & Laimins 1997; Ruesch & Laimins 1998). In addition, E7-expressing cells maintain cyclin E-cdk2 activity in the presence of growth arrest signals such as epithelial differentiation, serum deprivation, and anchorage-independent growth (Ruesch & Laimins 1998; Schulze et al. 1998). This phenotype is abrogated by mutation of the CR1 region of E7, which is vital for cellular immortalization and transformation (see above). The mechanism by which E7 affects cyclin E-cdk2 activity in vivo has not been clearly defined, though several models have been proposed. The presence of E7 dysregulates cyclin E expression at the transcriptional (via relief of E2F-dependent repression) and post-transcriptional levels, suggesting that E7 may, in part, affect cyclin E-cdk2 activity via increased synthesis of cyclin E (Zerfass et al. 1995; Botz et al. 1996; Martin et al. 1998). However, the contribution of elevated cyclin E expression is, alone, unlikely to be sufficient to increase cyclin E-cdk2 activity, because E7-expressing cells also exhibit dramatically elevated total and cdk2-associated levels of p21CIP, a potent inhibitor of cdk2 (Jones et al. 1997). High exogenous or endogenous cyclin E expression has been shown to be unable to maintain cyclin E-cdk2 activity in the presence of high levels of the Cip/Kip family of cdk inhibitors (p21CIP and p27KIP) (Sewing et al. 1997; Alevizopoulos et al. 1998).
E7 has been detected in complex with p107-cyclin E-cdk2, suggesting that E7 may affect cyclin E-cdk2 through physical interaction (McIntyre et al. 1996). Indeed, recent evidence also suggests that E7 interacts with p21CIP and p27KIP in vitro and in vivo; in vitro experiments have shown that E7 can bind to and derepress the kinase activity of p21CIP-cyclin E-cdk2 complexes (Funk et al. 1997; Jones et al. 1997). However, it is not clear whether active E7-p21CIP-cyclin E-cdk2 complexes exist in vivo. Other reports refute the interaction between E7 and p21CIP (Hickman et al. 1997; Ruesch & Laimins 1997). Therefore, further examination of the mechanism by which E7 affects cyclin E-cdk2 is required. In addition, further studies are necessary to determine the effects of E7-mediated cyclin E-cdk2 dysregulation on viral DNA replication and on long-term phenotypes such as cellular immortalization and transformation.
The effects of E7 on cyclin A-cdk2 (which regulates the progression of S phase) are similar to those observed for cyclin E-cdk2. Cyclin A-cdk2 activity is elevated in asynchronous HFK expressing E7 (Martin et al. 1998). Deregulated expression of cyclin A may contribute to this phenotype, as the presence of E7 leads to constitutive expression of cyclin A throughout the cell cycle (Martin et al. 1998). In concordance, E7-expressing fibroblasts synthesize cyclin A in the presence of growth arrest signals such as anchorage-independent suspension and serum starvation (Zerfass et al. 1995; Schulze et al. 1998). E7 has also been demonstrated to bind cyclin A-cdk2 in a complex involving E2F and p107 (Arroyo et al. 1993; Pagano et al. 1992). The effect of E7 on the activities of the cyclin A-cdk2-E2F-p107 complex has not been elucidated. However, it is noteworthy that cyclin A-cdk2 inhibits the DNA-binding activity of E2F complexes as cells undergo DNA replication, and the expression of mutant E2F proteins lacking the cdk-binding site generates stable DNA-binding E2F complexes and arrests cells in S phase (Krek et al. 1995). Because HPV requires the cellular DNA replication machinery for its own DNA synthesis, prolonging the time during which the terminally differentiating host cell is competent to synthesize DNA would benefit the replicative requirements of the virus. In concert, the effects of E7 on dysregulation of cyclin E-cdk2 (which regulates initiation of S phase) and cyclin A-cdk2 (which regulates the progression of S phase) may contribute to extending the duration of ‘replicative competence’ in the host cell.
E5 protein
The HPV-16 E5 protein is a small (84 aa) hydrophobic membrane protein and is located just downstream of the E2 open reading frame. The gene is not well conserved at the DNA level between HPVs, or animal viruses, although there is a conservation of the physico-chemical properties in that the proteins are all highly hydrophobic and membrane bound (Burkhardt et al. 1989; Conrad et al. 1993). The proteins from both animal and human sources can transform mammalian cells with varying degrees of efficiency. As mentioned above, the protein is hydrophobic with three very hydrophobic peaks and less hydrophobic troughs (Bubb et al. 1988). It has been reported to dimerize even in reducing conditions (Kell et al. 1994). The nature of the protein means that it is membrane bound and distributed predominantly in the endoplasmic reticulum and Golgi, but also in the cytoplasmic membrane (Burkhardt et al. 1989; Conrad et al. 1993; Sparkowski et al. 1995).
The hydrophobic nature of E5 also means that purification is difficult because the protein is insoluble and good antibody reagents directed against E5 are limited. Anti-peptide antibodies directed against N-and C-terminal have been used successfully (Hwang et al. 1995), and addition of peptides as tags to the N-terminal of the protein have also been used (Conrad et al. 1993), taking advantage of the commercially available antibodies to the peptide tags.
Interactions of E5 with growth factor receptors and vacuolar ATPase
Unlike the E6 and E7 proteins which abrogate the effects of negative cellular regulators, the major biological function of the E5 proteins appears to be through cellular growth factor receptors, whose activity it upregulates. The original E5/receptor interaction was described for bovine papillomavirus type 1 (BPV-1) E5, where the viral protein was found to stabilize the epidermal growth factor receptor (EGFR) in the presence of the ligand, EGF, with the result that the complex was not broken down and a hyper-stimulating signal was observed (Martin et al. 1989). Subsequent to this study, BPV-1 E5 was found in a complex with the platelet derived growth factor receptor (PDGFR) (Petti et al. 1991).
It is important to be aware of the kinetics of the interaction of both receptors and their respective ligands. When the ligand binds to the receptors it causes dimerization and the receptors autophosphorylate each other through a kinase domain in the cytoplasmic tail and are rapidly endocytosed. For instance, in the case of the EGFR, 90% of receptors are internalized by 10 min of the addition of EGF. The phosphorylated receptors bind to adaptor proteins, which in turn activate signalling pathways through ras, phospatidylinositol 3-kinase, or phospholipase C. The endosomes containing the receptor/ligand complex are then acidified and become part of the lysosomal compartment of the cell and within 90 min all of the receptors are degraded, resulting in a short lived stimulatory signal.
It was also found that BPV-1 E5 could activate the PDGFR in the absence of the ligand, platelet derived growth factor (PDGF) (Petti et al. 1991). How this is achieved is not clear but it may be caused by the ability of the E5 to dimerize (Burkhardt et al. 1989) with itself and to bind to PDGFR and so in effect cause dimerization of the receptor and its subsequent autophosphorylation (Goldstein et al. 1992). However, the PDGFR is not found on human keratinocytes, where the major growth factor receptor is the epidermal growth factor receptor (EGFR), so most of the work on HPV-16 E5 has been carried with this latter receptor.
Several studies have shown a co–operative interaction between E5 and EGF in the transformation of rodent fibroblasts (Chen & Mounts 1990; Leechanachi et al. 1992; Pim et al. 1992; Straight et al. 1993), for both the benign (HPV-6 E5a) and oncogenic types (HPV-16 E5). There is conflicting evidence on whether E5 binds directly to the EGFR. In one study, HPV-6 E5 was shown to bind to the EGFR when both were expressed in COS-1 cells, but HPV-16 E5 was not observed to bind (Conrad et al. 1994), while in another study there was evidence of binding to EGFR using the same cell type (Hwang et al. 1995). In the former study, both E5 proteins were epitope-tagged, and antitagged antibodies were used to coimmunoprecipitate the proteins, while in the latter, E5 was not tagged and anti-E5 peptide antibodies were used. It remains to be determined why there is this discrepancy in the binding results, but the fact that one set of E5 proteins was tagged with a hydrophilic peptide may have resulted in the protein being inefficiently immunoprecipitated due to masking by the hydrophobic protein.
The E5 proteins from both HPV-16 and BPV-1 also bind to the 16 kDa subunit of the vacuolar ATPase (Conrad et al. 1993; Valle & Banks 1995). The ATPase is a large protein complex made up of a number of subunits of which the 16 kDa forms the pore through which H + ions pass to acidify the contents of endosomes. The effect on the acidification of endosomes has only been tested with HPV-16 E5 in human keratinocytes (Straight et al. 1995). E5 was shown to inhibit acidification of endosomes, resulting in the prolongation of the active signal from the EGFR. The inhibition of pH change results in the retention of the receptor in the endosomal compartment and recycling of 40% of the receptors back to the surface in the absence of the ligand. This probably accounts for the increased numbers of EGFR observed on E5-expressing keratinocytes. The fact that the ligand is removed from the receptor is a consequence of the dissociation pH being 6.5, a level reached even in the presence of E5. The inhibition of acidification is probably not dependent on the EGFR, although this has not been tested. Instead it is likely that the acidification of any endosome will be effected by the presence of HPV-16 E5. The direct testing of the binding of E5 to the 16 kDa subunit has not been shown in human keratinocytes due to the low levels of E5 protein and its very hydrophobic nature. However, coexpression of both E5 and the bovine vacuolar ATPase subunit in COS-1 cells has shown a complex containing both E5 and the 16 kDa protein (Conrad et al. 1993). It is assumed, although not proven, that this is a direct interaction and that there is not a third protein involved. Recent work on E5 has shown that while binding to the 16 kDa subunit is essential for biological activity, it is not sufficient, and other properties of E5 are required for the inhibition of v-ATPase function (Adam et al. 2000).
There are down-stream effects of E5 expression in rodent cells. For instance, there is an increase in c-Fos and c-Jun expression in cells especially in the presence of EGF (Leechanachi 1992; Bouvard et al. 1994). The upstream regulatory region (URR) of most HPV types contain an AP-1 binding site and in the presence of E5 there is an increase in transcription from this promoter in E5-containing NIH 3T3 cells (Bouvard et al. 1994). This is a potentially important observation because one of the earliest mRNAs produced by HPV-16 DNA in infected tissues potentially codes for E5, so this protein may be important for the production of early messages coding for E6 and E7. There is also an increase in the MAP kinase kinase and MAP kinase activity (Gu & Matlashewski 1995) in cells stimulated by EGF and this increase is enhanced and prolonged in the presence of E5. Therefore it appears that the various pathways activated by the interaction of EGF with the EGF receptor are enhanced and prolonged in the presence of HPV-16 E5. It is, however, not clear at the moment whether all of this can be accounted for by the inhibition of acidification of the endosomal contents leading to prolonged signalling by the EGF/EGFR complex, or whether E5 can act on the pathways independent of the effects on endosomal pH.
Since EGF is a growth requirement for human keratinocytes and is necessary through part of the G1 phase of the cell cycle, the activity of E5 suggests that it acts to stimulate cells through this phase of the cell cycle and into S-phase.
Transformation and immortalization properties of E5
As mentioned above, HPV-16 E5 can transform immortalized rodent fibroblasts, and the frequency of transformation is increased in the presence of EGF, but not PDGF. The transformation assays have all been carried out using soft agar to measure growth in suspension. HPV-16 E5 is unable to form transformed foci on cell monolayers and therefore is considered to have only moderate transforming activity, unlike the BPV-1 E5 gene, which can form transformed foci on rodent monolayer cultures. It should be remembered that the E5 protein of BPV-1 is the major transforming protein unlike the situation with HPVs, where E7 has the major transforming role.
HPV-16 E5 is unable to immortalize primary human keratinocytes (Straight et al. 1993; Straight et al. 1995), however there are reports of extended life span of cells expressing E5 from an exogenous promoter (Straight et al. 1995). In addition, the full length HPV-16 genome with a premature stop codon in the E5 gene is only able to immortalize keratinocytes at 10% of the frequency of the wild type genome, suggesting that E5 plays a complementary role in the immortalization process (Stoppler et al. 1996). Therefore, in the life cycle of the virus E5 plays an important role along with E6 and E7 proteins.
E1 and E2 proteins
Both E1 and E12 have been shown to be essential for the replication of the origin of various HPV types (Ustav & Stenlund 1991). As well as binding to specific sequences at the origin of replication, both E1 and E2 proteins bind each other. E2 binds to palindromic sequence ACCN6GGT (Spalholz et al. 1987) of which there are three sets surrounding the origin. The E1 binding site is AT rich region with more variability than the E2 binding site and lies between E2 sites (Frattini & Laimins 1994a; Sun et al. 1996). As mentioned above, both proteins are required for episomal replication of the genome in human keratinocytes and when either open reading is disrupted the genome integrates into the host chromosome.
E2 binds DNA as a dimer through a domain in the c-terminal part of the protein. E1 probably binds as a hexamer and the binding domain is in the c-terminal half of the protein (Sedman & Stenlund 1998). E1 binds weakly to DNA, but the affinity is increased in the presence of E2. The episomal replication of HPV-31 in keratinocytes has been shown to be controlled by the levels of E1 and E2 and increasing the expression of either increases the copy number of the HPV-31 genome per cell (Frattini & Laimins 1994b). Therefore copy number can be controlled by E1 and E2, but the transcriptional control of these proteins has not been elucidated. One of the difficulties of this type of experiment is that the control of E1 and E2 transcription is probably dictated by the differentiation state of the cell.
Why do some HPV types cause cancer?
Both benign and malignant viruses have to stimulate G1 progression and S-phase entry for successful replication of their genomes. They probably achieve this through overlapping, but not identical pathways. The pathways disrupted by the oncogenic viruses must lead to a greater potential for chromosomal accidents, which can lead to malignant conversion, whereas pathways modulated by the nononcogenic types result in the virus achieving its objective with minimal disruption to the epithelial cell. Examples of the differences in biological properties of HPV proteins from oncogenic and nononcogenic viruses are the interaction of the E7 proteins with the Rb proteins, where the affinity of binding of the oncogenic E7 is greater than the nononcogenic protein, or the fact that the E6 protein from oncogenic viruses can bind and degrade p53 more efficiently than the E6 protein from benign viruses. Therefore, a better understanding of the functions of each of the HPV proteins will help determine the reasons why infection with the different viruses has quite distinct consequences for the host. In addition, the knowledge gained on the functions of the HPV proteins will elucidate many of the cellular pathways that are essential for keratinocyte differentiation and cell cycle progression.
References
- Adam JA, Briggs MW, McCance DJ. A mutagenic analysis of the E5 protein of human papillomavirus type 16 reveals that E5 binding to the vacuolar H+-ATPase is not sufficient for biological actiity, using mammalian and yeast expression systems. Virology. 2000;272:315–325. doi: 10.1006/viro.2000.0376. [DOI] [PubMed] [Google Scholar]
- Alevizopoulos K, Catarin B, Vlach J, Amati B. A novel function of adenovirus E1A is required to overcome growth arrest by the CDK2 inhibitor p27 (Kip1) EMBO J. 1998;17:5987–5997. doi: 10.1093/emboj/17.20.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfandari J, Shnitman Magal S, Jackman A, Schlegel R, Gonen P, Sherman L. HPV16, E6 oncoprotein inhibits apoptosis induced during serum-calcium differentiation of foreskin human keratinocytes. Virology. 1999;257:383–396. doi: 10.1006/viro.1999.9675. [DOI] [PubMed] [Google Scholar]
- Antinore MJ, Birrer MJ, Patel D, Nader L, McCance DJ. The human papillomavirus type 16, E7 gene product interacts with and trans-activates the AP1 family of transcription factors. EMBO J. 1996;15:1950–1960. [PMC free article] [PubMed] [Google Scholar]
- Arany Z, Newsome D, Oldread E, Livingston DM, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- Arroyo M, Bagchi S, Raychaudhuri P. Association of the human papillomavirus type 16, E7 protein with the S-phase-specific E2F-cyclin A complex. Mol. Cell. Biol. 1993;13:6537–6546. doi: 10.1128/mcb.13.10.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austen DF, Reynolds P, Schottenfeld D, Fraumeni JF.J. Laryngeal Cancer. New York: Oxford University Press Inc; 1996. [Google Scholar]
- Banks L, Edmonds C, Vousden KH. Ability of the HPV16, E7 protein to bind RB and induce DNA synthesis is not sufficient for efficient transforming activity in NIH3T3 cells. Oncogene. 1990;5:1383–1389. [PubMed] [Google Scholar]
- Barbosa MS, Edmonds C, Fisher C, Schiller JT, Lowy DR, Vousden KH. The region of the HPV E7 oncoprotein homologous to adenovirus E1a and Sv40 large T antigen contains separate domains for Rb binding and casein kinase II phosphorylation. EMBO J. 1990;9:153–160. doi: 10.1002/j.1460-2075.1990.tb08091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa MS, Schlegel R. The E6 and E7 genes of HPV-18 are sufficient for inducing two-stage in vitro transformation of human keratinocytes. Oncogene. 1989;4:1529–1532. [PubMed] [Google Scholar]
- Belair CD, Yeager TR, Lopez PM, Reznikoff CA. Telomerase activity: a biomarker of cell proliferation, not malignant transformation. ProcNatlAcadSciUSA. 1997;94:13677–13682. doi: 10.1073/pnas.94.25.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, et al. Extension of life-span by introduction of telomerase into normal human cells [see comments] Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Botz J, Zerfass-Thome K, Spitkovsky D, et al. Cell cycle regulation of the murine cyclin E gene depends on an E2F binding site in the promoter. MolCell Biol. 1996;16:3401–3409. doi: 10.1128/mcb.16.7.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvard V, Matlashewski G, Gu ZM, Storey A, Banks L. The human papillomavirus type 16, E5 gene cooperates with the E7 gene to stimulate proliferation of primary cells and increases viral gene expression. Virology. 1994;203:73–80. doi: 10.1006/viro.1994.1456. [DOI] [PubMed] [Google Scholar]
- Brehm A, Miskka EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastome protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- Brehm A, Nielsen SJ, Miska EA, et al. The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J. 1999;18:2449–2458. doi: 10.1093/emboj/18.9.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokaw JL, Yee CL, Munger K. A mutational analysis of the amino terminal domain of the human papillomavirus type 16, E7 oncoprotein. Virology. 1994;205:603–607. doi: 10.1006/viro.1994.1688. [DOI] [PubMed] [Google Scholar]
- Bubb V, McCance DJ, Schlegel R. DNA sequence of the HPV-16 E5 ORF and the structural conservation of its encoded protein. Virology. 1988;163:243–246. doi: 10.1016/0042-6822(88)90259-0. [DOI] [PubMed] [Google Scholar]
- Burkhardt A, Willingham M, Gay C, Jeang KT, Schlegel R. The E5 oncoprotein of bovine papillomavirus is oriented asymmetrically in Golgi and plasma membranes. Virology. 1989;170:334–339. doi: 10.1016/0042-6822(89)90391-7. [DOI] [PubMed] [Google Scholar]
- Cason J, Best JM, Raju KS. Vertical transmission of human papillomaviruses [letter; comment]. American Journal of Obstetrics &. Gynecology. 1999;180:774–775. doi: 10.1016/s0002-9378(99)70293-0. [DOI] [PubMed] [Google Scholar]
- Chakravarti D, Ogryzko V, Kao HY, et al. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- Chen JJ, Reid CE, Band V, Androphy EJ. Interaction of papillomavirus E6 oncoproteins with a putative calcium-binding protein. Science. 1995;269:529–531. doi: 10.1126/science.7624774. [DOI] [PubMed] [Google Scholar]
- Chen SL, Mounts P. Transforming activity of E5a protein of human papillomavirus type 6 in NIH 3T3 and C127 cells. JVirol. 1990;64:3226–3233. doi: 10.1128/jvi.64.7.3226-3233.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesters PM, Vousden KH, Edmonds C, McCance DJ. Analysis of human papillomavirus type 16 open reading frame E7 immortalizing function in rat embryo fibroblast cells. JGeneral Virology. 1990;71:449–453. doi: 10.1099/0022-1317-71-2-449. [DOI] [PubMed] [Google Scholar]
- Conrad M, Bubb VJ, Schlegel R. The human papillomavirus type 6 and 16, E5 proteins are membrane-associated proteins which associate with the 16-kilodalton pore-forming protein. JVirol. 1993;67:6170–6178. doi: 10.1128/jvi.67.10.6170-6178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M, Goldstein D, Andresson T, Schlegel R. The E5 protein of HPV-6, but not HPV-16, associates efficiently with cellular growth factor receptors. Virology. 1994;200:796–800. doi: 10.1006/viro.1994.1244. [DOI] [PubMed] [Google Scholar]
- Davies R, Hicks R, Crook T, Morris J, Vousden K. Human papillomavirus type 16, E7 associates with a histone H1 kinase and with p107 through sequences necessary for transformation. JVirol. 1993;67:2521–2528. doi: 10.1128/jvi.67.5.2521-2528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demers GW, Espling E, Harry JB, Etscheid BG, Galloway DA. Abrogation of growth arrest signals by human papillomavirus type 16, E7 is mediated by sequences required for transformation. JVirol. 1996;70:6862–6869. doi: 10.1128/jvi.70.10.6862-6869.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demers GW, Halbert CL, Galloway DA. Elevated wild-type p53 protein levels in human epithelial cell lines immortalized by the human papillomavirus type 16, E7 gene. Virology. 1994;198:169–174. doi: 10.1006/viro.1994.1019. [DOI] [PubMed] [Google Scholar]
- Desaintes C, Hallez S, Van Alphen P, Burny A. Transcriptional activation of several heterologous promoters by the E6 protein of human papillomavirus type 16. JVirol. 1992;66:325–333. doi: 10.1128/jvi.66.1.325-333.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Villiers EM. Heterogeneity of the human papillomavirus group. JVirol. 1989;63:4898–4903. doi: 10.1128/jvi.63.11.4898-4903.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdy SF, Hinds PW, Louie K, Reed SI, Arnold A, Weinberg RA. Physical interaction of the retinoblastoma protein with human D cyclins. Cell. 1993;73:499–511. doi: 10.1016/0092-8674(93)90137-f. [DOI] [PubMed] [Google Scholar]
- Dyson N. The regulation of E2F by Rb-family proteins. Genes & Development. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- Dyson N, Guida P, Munger K, Harlow E. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. JVirol. 1992;66:6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N, Howley PM, Munger K, Harlow E. The human papilloma virus-16, E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- Eckner R, Ewen ME, Newsome D, et al. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes & Development. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- Edmonds C, Vousden KH. A point mutational analysis of human papillomavirus type 16, E7 protein. JVirol. 1989;63:2650–2656. doi: 10.1128/jvi.63.6.2650-2656.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etscheid BG, Foster SA, Galloway DA. The E6 protein of human papillomavirus type 16 functions as a transcriptional repressor in a mechanism independent of the tumor suppressor protein, p53. Virology. 1994;205:583–585. doi: 10.1006/viro.1994.1684. [DOI] [PubMed] [Google Scholar]
- Filatov L, Golubovskaya V, Hurt JC, Byrd LL, Phillips JM, Kaufmann WK. Chromosomal instability is correlated with telomere erosion and inactivation of G2 checkpoint function in human fibroblasts expressing human papillomavirus type 16, E6 oncoprotein. Oncogene. 1998;16:1825–1838. doi: 10.1038/sj.onc.1201711. [DOI] [PubMed] [Google Scholar]
- Frattini MG, Laimins LA. Binding of the human papillomavirus E1 origin-recognition protein is regulated through complex formation with the E2 enhancer-binding protein. ProcNatlAcadSciUSA. 1994a;91:12398–1240. doi: 10.1073/pnas.91.26.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frattini MG, Laimins LA. The role of the E1 and E2 proteins in the replication of human papillomavirus type 31b. Virology. 1994b;204:799–804. doi: 10.1006/viro.1994.1596. [DOI] [PubMed] [Google Scholar]
- Fredericks BD, Balkin A, Daniel HW, Schonrock J, Ward B, Frazer IH. Transmission of human papillomavirus from mother to child. Aust. NZ J. Obstetrics Gynaecol. 1993;33:30–32. doi: 10.1111/j.1479-828x.1993.tb02047.x. [DOI] [PubMed] [Google Scholar]
- Freedman DA, Levine AJ. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol. Cell. Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk JO, Waga S, Harry JB, Espling E, Stillman B, Galloway DA. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes &. Development. 1997;11:2090–2100. doi: 10.1101/gad.11.16.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage JR, Meyers C, Wettstein FO. The E7 proteins of the nononcogenic human papillomavirus type 6b (HPV-6b) and of the oncogenic HPV-16 differ in retinoblastoma protein binding and other properties. JVirol. 1990;64:723–730. doi: 10.1128/jvi.64.2.723-730.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Whoriskey W, Park MY, et al. Rescue of cyclin D1 deficiency by knockin cyclin E. Cell. 1999;97:767–777. doi: 10.1016/s0092-8674(00)80788-6. [DOI] [PubMed] [Google Scholar]
- Gius D, Grossman S, Bedell MA, Laimins LA. Inducible and constitutive enhancer domains in the noncoding region of human papillomavirus type 18. JVirol. 1988;62:665–672. doi: 10.1128/jvi.62.3.665-672.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DJ, Andresson T, Sparkowski JJ, Schlegel R. The BPV-1 E5 protein, the 16 kDa membrane pore-forming protein and the PDGF receptor exist in a complex that is dependent on hydrophobic transmembrane interactions. EMBO J. 1992;11:4851–4859. doi: 10.1002/j.1460-2075.1992.tb05591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- Gross-Mesilaty S, Reinstein E, Bercovich B, et al. Basal and human papillomavirus E6 oncoprotein-induced degradation of Myc proteins by the ubiquitin pathway. ProcNatlAcadSciUSA. 1998;95:8058–8063. doi: 10.1073/pnas.95.14.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Gu Z, Matlashewski G. Effect of human papillomavirus type 16 oncogenes on MAP kinase activity. JVirol. 1995;69:8051–8056. doi: 10.1128/jvi.69.12.8051-8056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamori Y, Sartorelli V, Ogryzko V. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- Harle-Bachor C, Boukamp P. Telomerase activity in the regenerative basal layer of the epidermis in human skin and in immortal and carcinoma-derived skin keratinocytes. ProcNatlAcadSciUSA. 1996;93:6476–6481. doi: 10.1073/pnas.93.13.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck DV, Yee CL, Howley PM, Munger K. Efficiency of binding the retinoblastoma protein correlates with the transforming capacity of the E7 oncoproteins of the human papillomaviruses. ProcNatlAcadSciUSA. 1992;89:4442–4446. doi: 10.1073/pnas.89.10.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman ES, Bates S, Vousden KH. Pertubation of the p53 response by human papillomavirus type 16, E7. JVirol. 1997;71:3710–3718. doi: 10.1128/jvi.71.5.3710-3718.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildesheim A, Schiffman MH, Gravitt PE, et al. Persistence of type-specific human papillomavirus infection among cytological normal women. JInfectious Diseases. 1994;169:235–240. doi: 10.1093/infdis/169.2.235. [DOI] [PubMed] [Google Scholar]
- Huang PS, Patrick DR, Edwards G, et al. Protein domains governing interactions between E2F, the retinoblastoma gene product, and human papillomavirus type 16, E7 protein. Mol. Cell. Biol. 1993;13:953–960. doi: 10.1128/mcb.13.2.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Wang NP, Tseng BY, Lee WH, Lee EH. Two distinct and frequently mutated regions of retinoblastoma protein are required for binding to SV40 T antigen. EMBO J. 1990;9:1815–1822. doi: 10.1002/j.1460-2075.1990.tb08306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson JB, Bedell MA, McCance DJ, Laiminis LA. Immortalization and altered differentiation of human keratinocytes in vitro by the E6 and E7 open reading frames of human papillomavirus type 18. JVirol. 1990;64:519–526. doi: 10.1128/jvi.64.2.519-526.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse J, Schneffer M, Howley PM. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurford RK, Jr, Cobrinik D, Lee MH, Dyson N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes & Development. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- Hwang ES, Nottoli T, Dimaio D. The HPV16, E5 protein: expression, detection, and stable complex formation with transmembrane proteins in COS cells. Virology. 1995;211:227–233. doi: 10.1006/viro.1995.1395. [DOI] [PubMed] [Google Scholar]
- Jackson S, Storey A. E6 proteins from diverse cutaneous HPV types inhibit apoptosis in response to UV damage. Oncogene. 2000;19:592–598. doi: 10.1038/sj.onc.1203339. [DOI] [PubMed] [Google Scholar]
- Jarrett WF, Smith KT, O'Neil BW, et al. Studies on vaccination against papillomaviruses: prophylactic and therapeutic vaccination with recombinant structural proteins. Virology. 1991;184:33–42. doi: 10.1016/0042-6822(91)90819-w. [DOI] [PubMed] [Google Scholar]
- Jayachandra S, Low KG, Thlick AE, et al. Three unrelated viral transforming proteins (vIRF, EBNA2, and E1A) induce the MYC oncogene through the interferon-responsive PRF element by using different transcription coadaptors. ProcNatlAcadSciUSA. 1999;96:11566–11571. doi: 10.1073/pnas.96.20.11566. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jewers RJ, Hildebrandt P, Ludlow JW, Kell B, McCance DJ. Regions of human papillomavirus type 16, E7 oncoprotein required for immortalization of human keratinocytes. JVirol. 1992;66:1329–1335. doi: 10.1128/jvi.66.3.1329-1335.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DL, Alani RM, Munger K. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes & Development. 1997;11:2101–2111. doi: 10.1101/gad.11.16.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima H, Levanthal B, Mounts P. Scoring System to Assess Severity and Course in Recurrent Repiratory Papillomatosis. New York: Alan R. Liss; 1985. [Google Scholar]
- Kell B, Jewers JR, Cason J, Pkarian F, Kaye JN, Best JM. Detection of E5 oncoprotein in human appillomavirus type 16-positive cervical scrapes using antibodies raised to synthetic peptides. JGeneral Virology. 1994;75:2451–2456. doi: 10.1099/0022-1317-75-9-2451. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Shirasawa H, Shino Y, et al. Transactivation of prothymosin alpha and c-myc promoters by human papillomavirus type 16, E6 protein. Virology. 1997;232:53–61. doi: 10.1006/viro.1997.8536. [DOI] [PubMed] [Google Scholar]
- Kiyono T, Foster SA, Koop JI, McDougall JK, Galloway DA, Klingelhutz AJ. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells [see comments] Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- Kiyono T, Hiraiwa A, Fujita M, Hayashi Y, Akiyama T, Ishibashi M. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. ProcNatlAcadSciUSA. 1997;94:11612–11616. doi: 10.1073/pnas.94.21.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingelhutz AJ, Foster SA, McDougall JK. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature. 1996;380:79–82. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- Krajewski S, Krajewska M, Reed JC. Immunohistochemical analysis of in vivo patterns of Bak expression, a proapoptotic member of the Bcl-2 protein family. Cancer Res. 1996;56:2849–2855. [PubMed] [Google Scholar]
- Krek W, Xu G, Livingston DM. Cyclin A-kinase regulation of E2F-1 DNA binding function underlies suppression of an S phase checkpoint. Cell. 1995;83:1149–1158. doi: 10.1016/0092-8674(95)90141-8. [DOI] [PubMed] [Google Scholar]
- Krude T, Jackman M, Pines J, Laskey RA. Cyclin/cdk-dependent initiation of DNA replication in a human cell-free system. Cell. 1997;88:109–119. doi: 10.1016/s0092-8674(00)81863-2. [DOI] [PubMed] [Google Scholar]
- Leechanachi P, Banks L, Moreau F, Matlashewski G. The E5 gene from human papillomavirus type 16 is an oncogene which enhances growth factor-mediated signal transduction to the nucleus. Oncogene. 1992;7:19–25. [PubMed] [Google Scholar]
- Liu Y, Chen JJ, Gao Q, Dalal S, et al. Multiple functions of human papillomavirus type 16 contribute to the immortalization of mammary epithelial cells. JVirol. 1999;73:7297–7307. doi: 10.1128/jvi.73.9.7297-7307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L, Groisman R, Naguibneva I, et al. Retinoblastoma protein represses transcription by recruiting a histone deacetylase [see comments] Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- Malek RS, Goellner JR, Smith TF, Espy MJ, Cupp MR. Human papillomavirus infection and intraepithelial, in situ, and invasive cancer of penis. Urology. 1993;42:159–170. doi: 10.1016/0090-4295(93)90640-v. [DOI] [PubMed] [Google Scholar]
- Manos MM, Kinney WK, Hurley LO, et al. Identifying women with cervical neoplasia. Using human papillomavirus DNA testing for equivocal Papanicolou results. JAmMedAssoc. 1999;281:1605–1610. doi: 10.1001/jama.281.17.1605. [DOI] [PubMed] [Google Scholar]
- Mantovani F, Banks L. Inhibition of E6 induced degradation of p53 is not sufficient for stabilization of p53 protein in cervical tumour derived cell lines. Oncogene. 1999;18:3309–3315. doi: 10.1038/sj.onc.1202688. [DOI] [PubMed] [Google Scholar]
- Martin LG, Demers GW, Galloway DA. Disruption of the G1/S transition in human papillomavirus type 16, E7-expressing human cells is associated with altered regulation of cyclin E. JVirol. 1998;72:975–985. doi: 10.1128/jvi.72.2.975-985.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P, Vass WC, Schiller JT, Lowy DR, Velu TJ. The bovine papillomavirus E5 transforming protein can stimulate the transforming activity of EGF and CSF-1 receptors. Cell. 1989;59:21–32. doi: 10.1016/0092-8674(89)90866-0. [DOI] [PubMed] [Google Scholar]
- Massimi P, Pim D, Storey A, Banks L. HPV-16 E7 and adenovirus E1a complex formation with TATA box binding protein is enhanced by casein kinase II phosphorylation. Oncogene. 1996;12:2325–2330. [PubMed] [Google Scholar]
- Mazzarelli JM, Atkins GB, Geisberg JV, Ricciardi RP. The viral oncoproteins Ad5 E1A, HPV16, E7 and SV40 TAg bind a common region of the TBP-associated factor-110. Oncogene. 1995;11:1859–1864. [PubMed] [Google Scholar]
- McCance DJ. Human papillomaviruses and cervical cancer [editorial] JMedMicrobiology. 1998;47:371–373. doi: 10.1099/00222615-47-5-371. [DOI] [PubMed] [Google Scholar]
- McCance DJ, Campion MJ, Clarkson PK, et al. Prevalence of human papillomavirus type 16 DNA sequences in cervical intraepithelial neoplasia and invasive carcinoma of the cervix. British Journal of Obstetrics & Gynaecology. 1985;92:1101–1105. doi: 10.1111/j.1471-0528.1985.tb03019.x. [DOI] [PubMed] [Google Scholar]
- McCance DJ, Kalache A, Ashdown K, et al. Human papillomavirus types 16 and 18 in carcinomas of the penis from Brazil. IntJCancer. 1986;37:55–59. doi: 10.1002/ijc.2910370110. [DOI] [PubMed] [Google Scholar]
- McCance DJ, Kopan R, Fuchs E, Laimins LA. Human papillomavirus type 16 alters human epithelial cell differentiation in vitro. ProcNatlAcadSciUSA. 1988;85:7169–7173. doi: 10.1073/pnas.85.19.7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor JM, Rustin MH.A. Human papillomavirus and skin cancer. Postgraduate MedJournal. 1994;70:682–685. doi: 10.1136/pgmj.70.828.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre MC, Frattini MG, Grossman SR, Laimins LA. Human papillomavirus type 18, E7 protein requires intact Cys-X-X-Cys motifs for zinc binding, dimerization, and transformation but not for Rb binding. JVirol. 1993;67:3142–3150. doi: 10.1128/jvi.67.6.3142-3150.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre MC, Ruesch MN, Laimins LA. Human papillomavirus E7 oncoproteins bind a single form of cyclin E in a complex with cdk2 and p107. Virology. 1996;215:73–82. doi: 10.1006/viro.1996.0008. [DOI] [PubMed] [Google Scholar]
- Mietz JA, Unger T, Huibregtse JM, Howley PM. The transcriptional transactivation function of wild-type p53 is inhibited by SV40 large T-antigen and by HPV-16 E6 oncoprotein. EMBO J. 1992;11:5013–5020. doi: 10.1002/j.1460-2075.1992.tb05608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger K. The molecular biology of cervical cancer. JCellular Biochemistry – Supplement. 1995;23:55–60. doi: 10.1002/jcb.240590908. [DOI] [PubMed] [Google Scholar]
- Munger K, Phelps WC. The human papillomavirus E7 protein as a transforming and transactivating factor. Biochimica Biophysica Acta. 1993;1155:111–123. doi: 10.1016/0304-419x(93)90025-8. [DOI] [PubMed] [Google Scholar]
- Munger K, Werness BA, Dyson N, Phelps WC, Harlow E, Howley PM. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger K, Yee CL, Phelps WC, Pietenpol JA, Moses HL, Howley PM. Biochemical and biological differences between E7 oncoproteins of the high- and low-risk human papillomavirus types are determined by amino-terminal sequences. JVirol. 1991;65:3943–3948. doi: 10.1128/jvi.65.7.3943-3948.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nead N, Antinore MJ, Baglia L, Ludlow JW, McCance DJ. Rb binds c-Jun and activates transcription. EMBO J. 1998;17:2342–2352. doi: 10.1093/emboj/17.8.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo M, Theodoras AM, Schumacher J, Roberts JM, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol. Cell. Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano M, Durst M, Joswig S, Draetta G, Jansen-Durr P. Binding of the human E2F transcription factor to the retinoblastoma protein but not to cyclin A is abolished in HPV-16-immortalized cells. Oncogene. 1992;7:1681–1686. [PubMed] [Google Scholar]
- Pan H, Griep AE. Altered cell cycle regulation in the lens of HPV-16 E6 or E7 transgenic mice: implications for tumor suppressor gene function in development. Genes & Development. 1994;8:1285–1299. doi: 10.1101/gad.8.11.1285. [DOI] [PubMed] [Google Scholar]
- Pan H, Griep AE. Temporally distinct patterns of p53-dependent and p53-independent apoptosis during mouse lens development. Genes & Development. 1995;9:2157–2169. doi: 10.1101/gad.9.17.2157. [DOI] [PubMed] [Google Scholar]
- Passalaris TM, Benanti JA, Gewin L, Kiyono T, Galloway DA. The G (2) checkpoint is maintained by redundant pathways. Mol. Cell. Biol. 1999;19:5872–5881. doi: 10.1128/mcb.19.9.5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D, Huang SM, Baglia LA, McCance DJ. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 1999;18:5061–5072. doi: 10.1093/emboj/18.18.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick DR, Oliff A, Heimbrook DC. Identification of a novel retinoblastoma gene product binding site on human papillomavirus type 16, E7 protein. JBiolChemistry. 1994;269:6842–6850. [PubMed] [Google Scholar]
- Petti L, Nilson LA, Dimaio D. Activation of the platelet-derived growth factor receptor by the bovine papillomavirus E5 transforming protein. EMBO J. 1991;10:845–855. doi: 10.1002/j.1460-2075.1991.tb08017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps WC, Munger K, Yee CL, Barnes JA, Howley PM. Structure-function analysis of the human papillomavirus type 16, E7 oncoprotein. JVirol. 1992;66:2418–2427. doi: 10.1128/jvi.66.4.2418-2427.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AC, Vousden KH. Analysis of the interaction between human papillomavirus type 16, E7 and the TATA-binding protein, TBP. JGeneral Virology. 1997;78:905–909. doi: 10.1099/0022-1317-78-4-905. [DOI] [PubMed] [Google Scholar]
- Pim D, Collins M, Banks L. Human papillomavirus type 16, E5 gene stimulates the transforming activity of the epidermal growth factor receptor. Oncogene. 1992;7:27–32. [PubMed] [Google Scholar]
- Pim D, Thomas M, Javier R, Gardiol D, Banks L. HPV E6 targeted degradation of the discs large protein: evidence for the involvement of a novel ubiquitin ligase. Oncogene. 2000;19:719–725. doi: 10.1038/sj.onc.1203374. [DOI] [PubMed] [Google Scholar]
- Ruesch M, Laimins LA. Human papillomavirus oncoproteins alter differentiation-dependent cell cycle exit on suspension in semisolid medium. Virology. 1998;250:19–29. doi: 10.1006/viro.1998.9359. [DOI] [PubMed] [Google Scholar]
- Ruesch MN, Laimins LA. Initiation of DNA synthesis by human papillomavirus E7 oncoproteins is resistant to p21-mediated inhibition of cyclin E-cdk2 activity. JVirol. 1997;71:5570–5578. doi: 10.1128/jvi.71.7.5570-5578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang BC, Barbosa MS. Single amino acid substitutions in ‘low-risk’ human papillomavirus (HPV) type 6, E7 protein enhance features characteristic of the ‘high-risk’ HPV E7 oncoproteins. ProcNatlAcadSciUSA. 1992;89:8063–8067. doi: 10.1073/pnas.89.17.8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- Schiffman MH, Brinton LA. The epidemiology of cervical carcinogenesis. Cancer. 1995;76:1888–1901. doi: 10.1002/1097-0142(19951115)76:10+<1888::aid-cncr2820761305>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Schulze A, Mannhardt B, Zerfass-Thome K, Zwerschke W, Jansen-Durr P. Anchorage-independent transcription of the cyclin A gene induced by the E7 oncoprotein of human papillomavirus type 16. JVirol. 1998;72:2323–2334. doi: 10.1128/jvi.72.3.2323-2334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedman J, Stenlund A. The papillomavirus E1 protein forms a DNA-dependent hexameric complex with ATPase and DNA helicase activities. JVirol. 1998;72:6893–6897. doi: 10.1128/jvi.72.8.6893-6897.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewing A, Wiseman B, Lloyd AC, Land H. High-intensity Raf signal causes cell cycle arrest mediated by p21Cip1. Mol. Cell. Biol. 1997;17:5588–5597. doi: 10.1128/mcb.17.9.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard HM, Corneillie SI, Espiritu C, Gatti A, Liu X. New insights into the mechanism of inhibition of p53 by simian virus 40 large T antigen. Mol. Cell. Biol. 1999;19:2746–2753. doi: 10.1128/mcb.19.4.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. Mammalian G1 cyclins. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes & Development. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Song S, Pitot HC, Lambert PF. The human papillomavirus type 16, E6 gene alone is sufficient to induce carcinomas in transgenic animals. JVirol. 1999;73:5887–5893. doi: 10.1128/jvi.73.7.5887-5893.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalholz BA, Lambert PF, Yee CL, Howley PM. Bovine papillomavirus transcriptional regulation: localization of the E2-responsive elements of the long control region. JVirol. 1987;61:2128–2137. doi: 10.1128/jvi.61.7.2128-2137.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkowski J, Anders J, Schlegel R. E5 oncoprotein retained in the endoplasmic reticulum/cis Golgi still induces PDGF receptor autophosphorylation but does not transform cells. EMBO J. 1995;14:3055–3063. doi: 10.1002/j.1460-2075.1995.tb07308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppler H, Hartmann DP, Sherman L, Schlegel R. The human papillomavirus type 16, E6 and E7 oncoproteins dissociate cellular telomerase activity from the maintenance of telomere length. JBiolChemistry. 1997;272:13332–13337. doi: 10.1074/jbc.272.20.13332. [DOI] [PubMed] [Google Scholar]
- Stoppler MC, Straight SW, Tsao G, Schlegel R, McCance DJ. The E5 gene of HPV-16 enhances keratinocyte immortalization by full-length DNA. Virology. 1996;223:251–254. doi: 10.1006/viro.1996.0475. [DOI] [PubMed] [Google Scholar]
- Stott FJ, Bates S, James MC, et al. The alternative product from the human CDKN2A locus, p14 (ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight SW, Herman B, McCance DJ. The E5 oncoprotein of human papillomavirus type 16 inhibits the acidification of endosomes in human keratinocytes. JVirol. 1995;69:3185–3192. doi: 10.1128/jvi.69.5.3185-3192.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight SW, Hinkle PM, Jewers RJ, McCance DJ. The E5 oncoprotein of human papillomavirus type 16 transforms fibroblasts and effects the downregulation of the epidermal growth factor receptor in keratinocytes. JVirol. 1993;67:4521–4532. doi: 10.1128/jvi.67.8.4521-4532.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YN, Lu JZ, McCance DJ. Mapping of HPV-11 E1 binding site and determination of other important cis elements for replication of the origin. Virology. 1996;216:219–222. doi: 10.1006/viro.1996.0050. [DOI] [PubMed] [Google Scholar]
- Suzich JA, Ghim SJ, Palmer-Hill FJ, et al. Systemic immunization with papillomavirus L1 protein completely prevents the development of viral mucosal papillomas. ProcNatlAcadSciUSA. 1995;92:11553–11557. doi: 10.1073/pnas.92.25.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takami Y, Sasagawa T, Sudiro TM, Yutsudo M, Hakura A. Determination of the functional difference between human papillomavirus type 6 and 16, E7 proteins by their 30 N-terminal amino acid residues. Virology. 1992;186:489–495. doi: 10.1016/0042-6822(92)90014-g. [DOI] [PubMed] [Google Scholar]
- Talis AL, Huibregtse JM, Howley PM. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV) -positive and HPV-negative cells. JBiolChemistry. 1998;273:6439–6445. doi: 10.1074/jbc.273.11.6439. [DOI] [PubMed] [Google Scholar]
- Thomas M, Banks L. Inhibition of Bak-induced apoptosis by HPV-18 E6. Oncogene. 1998;17:2943–2954. doi: 10.1038/sj.onc.1202223. [DOI] [PubMed] [Google Scholar]
- Thomas M, Banks L. Human papillomavirus (HPV) E6 interactions with Bak are conserved amongst E6 proteins from high and low risk HPV types. JGeneral Virology. 1999;80:1513–1517. doi: 10.1099/0022-1317-80-6-1513. [DOI] [PubMed] [Google Scholar]
- Ustav M, Stenlund A. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 1991;10:449–457. doi: 10.1002/j.1460-2075.1991.tb07967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle G, Banks L. The human papillomavirus HPV-6 and HPV-16 E5 proteins co-operate with HPV-16 E7 in the transformation of primary rodent cells. JGeneral Virology. 1995;76:1239–1245. doi: 10.1099/0022-1317-76-5-1239. [DOI] [PubMed] [Google Scholar]
- Webster K, Parish J, Pandya M, Stern PL, Clarke AR, Gaston K. The human papillomavirus (HPV) 16, E2 protein induces apoptosis in the absence of other HPV proteins and via a p53-dependent pathway. JBiolChemistry. 2000;275:87–94. doi: 10.1074/jbc.275.1.87. [DOI] [PubMed] [Google Scholar]
- Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18, E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- Wu EW, Clemens KE, Heck DV, Munger K. The human papillomavirus E7 oncoprotein and the cellular transcription factor E2F bind to separate sites on the retinoblastoma tumor suppressor protein. JVirol. 1993;67:2402–2407. doi: 10.1128/jvi.67.4.2402-2407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerfass K, Schulze A, Spitkovsky D, Friedman V, Henglein B, Jansen-Durr P. Sequential activation of cyclin E and cyclin A gene expression by human papillomavirus type 16, E7 through sequences necessary for transformation. JVirol. 1995;69:6389–6399. doi: 10.1128/jvi.69.10.6389-6399.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann H, Degenkolbe R, Bernard HU, O'Connor MJ. The human papillomavirus type 16, E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. JVirol. 1999;73:6209–6219. doi: 10.1128/jvi.73.8.6209-6219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H. Viruses in human cancers. European JCancer. 1999;35:1878–1885. doi: 10.1016/s0959-8049(99)00291-9. [DOI] [PubMed] [Google Scholar]