Abstract
Trichomonas vaginalis is a parasitic protozoan purine auxotroph possessing a unique purine salvage pathway consisting of a bacterial type purine nucleoside phosphorylase (PNP) and a purine nucleoside kinase. Thus, T. vaginalis PNP (TvPNP) functions in the reverse direction relative to PNPs in other organisms. Immucillin-A (ImmA) and DADMe-Immucillin-A (DADMe-ImmA) are transition state mimics of adenosine with geometric and electrostatic features that resemble early and late transition states of adenosine at the transition state stabilized by TvPNP. ImmA demonstrates slow-onset tight-binding inhibition with TvPNP, to give an equilibrium dissociation constant of 87 pM, an inhibitor release half-time of 17.2 min and a Km/Kd ratio of 70,100. DADMe-ImmA resembles a late ribooxacarbenium ion transition state for TvPNP to give a dissociation constant of 30 pM, an inhibitor release half-time of 64 min and a Km/Kd ratio of 203,300. Tight binding of DADMe-ImmA supports a late SN1 transition state. Despite their tight binding to TvPNP, ImmA and DADMe-ImmA are weak inhibitors of human and P. falciparum PNPs. The crystal structures of the TvPNP•ImmA•PO4 and TvPNP•DADMe-ImmA•PO4 ternary complexes differ from previous structures with substrate analogues. The tight binding with DADMe-ImmA is in part due to a 2.7 Å ionic interaction between a PO4 oxygen and the N1’ cation of the hydroxypyrrolidine and is weaker in the TvPNP•ImmA•PO4 structure at 3.5 Å. However, the TvPNP•ImmA•PO4 structure includes hydrogen bonds between the 2’-hydroxyl and the protein that are not present in TvPNP•DADMe-ImmA•PO4. These structures explain why DADMe-ImmA binds tighter than ImmA. Immucillin-H is a 12 nM inhibitor of TvPNP but a 56 pM inhibitor of human PNP. And this difference is explained by isotope-edited difference infrared spectroscopy with [6-18O]ImmH to establish that O6 is the keto tautomer in TvPNP•ImmH•PO4, causing an unfavorable leaving-group interaction.
Introduction
Trichomonas vaginalis is a highly prevalent protozoan human parasite (1) and the causative agent of trichomoniasis (2). Although the drug metronidazole is available for treatment of infections (3), it is poorly tolerated and drug resistance has developed (4). Purine salvage is an essential function for all obligate parasitic protozoa (5). T. vaginalis is unusual in that it lacks a hypoxanthine-guanine phosphoribosyltransferase (HGPRT). Instead, it relies on the nucleoside synthetic reaction of an unusual purine nucleoside phosphorylase (PNP) with broad substrate specificity to form nucleosides that are converted to nucleotides by purine nucleoside kinases (6-8). The purine auxotrophy of T. vaginalis makes PNP an attractive chemotherapeutic target. Adenine, hypoxanthine and guanine are salvaged to their corresponding nucleosides by PNP and subsequently phosphorylated to form AMP, IMP and GMP, respectively. PNPs catalyze the reversible phosphorolytic cleavage of the glycosidic bond of purine nucleosides and deoxynucleotides (9). PNP from mammalian sources are specific for 6-oxypurines whereas PNP from certain bacterial and parasitic sources will also use adenine as a substrate (10). TvPNP is unusual in having phosphorolytic catalytic activity with inosine, adenosine and guanosine, thus permitting salvage of all purine bases. Of these, adenine and hypoxanthine are the most favored. A common feature of all known PNPs is the oxacarbenium ion character of the transition state, where cleavage of the C-N bond occurs through an SN1-like mechanism.
Dissociative transition states can be achieved by following one of several reaction pathways (10). The first involves activation of the leaving group, as in the case of acid catalyzed solvolysis of purine nucleosides (10). The second occurs as a result of interactions between the catalytic site and the ribosyl moiety to stabilize the oxacarbenium ion charge and geometry (10). Other mechanisms involve ionization of the 2’-hydroxyl or (3’-hydroxyl) to favor generation of the oxacarbenium ion or activating the attacking phosphate nucleophile (11).
p-Nitrophenyl β-D-ribofuranoside (pNPR) can be used as a substrate to distinguish between these mechanisms. If pNPR is a good substrate, catalytic site formation of a ribooxacarbenium ion or activation of the phosphate nucleophile dominate transition state formation. Alternatively, catalytic site ionization of a ribosyl hydroxyl will also facilitate the departure of the p-nitrophenolate anion (12). If transition state formation is dominated by leaving group activation, involving mono- or di-protonation of the purine base, pNPR will be a poor substrate since the p-nitrophenylate anion is already active as a leaving group and cannot be further activated by protonation. As pNPR is an O-riboside, protonation of the β-oxygen bridge is also possible but unlikely in an enzyme designed for N-riboside phosphorolysis (13).
Transition state analogue mimics for mammalian and protozoan PNPs have been synthesized and were shown to have high binding affinities and specificity towards their cognate PNPs (Figure 1). Human and malarial PNPs are specific for 6-oxypurines and Immucillin-H (ImmH) [5] and Immucillin-G (ImmG) [4] inhibit with dissociation constants in the pM range, but ImmA [2] is a poor inhibitor since adenosine is not a substrate for these PNPs (14). Adenosine is the primary precursor of the purine salvage pathway in T. vaginalis and is a preferred substrate for TvPNP (15). We hypothesized that Immucillins based on the structure of adenosine would be powerful inhibitors of TvPNP. The most powerful inhibitors of TvPNP reported previously are adenosine analogues, which include Formycin-A (Ki of 2.3 μM; (16)) and a subversive substrate 2-fluoro-2’-deoxyadenosine (F-dAdo, with an IC50 of 106 nM against T. vaginalis in vitro (3)). DADMe-Immucillin-A (DADMe-ImmA) [1] shows slow-onset, tight binding inhibition and a remarkable dissociation constant of 30 pM, binding 76,700 times tighter than Formycin-A, and 203,300 times tighter than adenosine.
Figure 1.
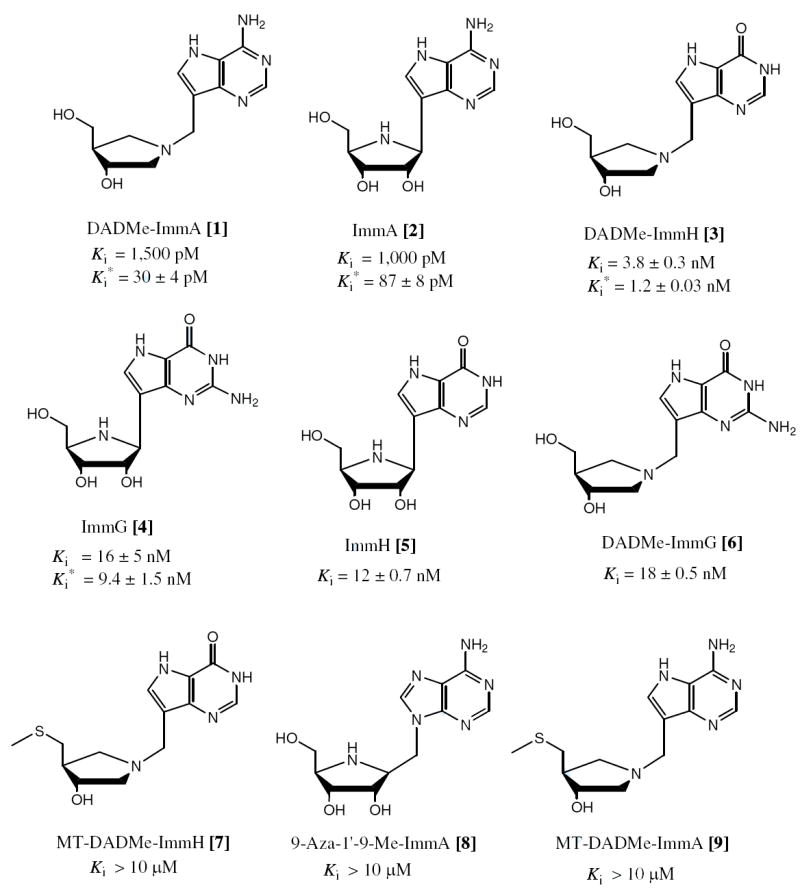
The inhibition constants for Immucillins and DADMe-Immucillins as competitive inhibitors of TvPNP. Inhibition constants for the initial rate (Ki) and the slow onset rate (Ki*) are indicated when slow onset was observed. When no slow onset occurred, only Ki is given. Compounds indicated with Ki > 10 μM showed no inhibition at 150 μM inhibitor under assay conditions listed in the methods.
The structures of TvPNP•DADMe-ImmA•PO4 and TvPNP•ImmA•PO4 show that the carboxyl group of Asp204 hydrogen bonds to N7H and N6H of the 9-deazaadenine base with distances of 3.0 and 2.8 Å. Other TvPNP inhibitor complexes have Asp204 pointing away from the base or have interactions that are significantly weaker (3). Phosphate is held tightly in the TvPNP•DADMe-ImmA•PO4 complex and forms a close ion pair between the incipient phosphate nucleophile and the N1’ cation of bound DADMe-ImmA. In the ImmA complex the ion pair is weaker. Thus, leaving group interactions and an ion pair mimic of the transition state provide the binding energy for the Immucillins.
Materials and Methods
TvPNP Assay
TvPNP was prepared as described earlier (3). The inosine phosphorolytic activity of TvPNP was by monitored by following the conversion of hypoxanthine to uric acid (ε293 = 12.9 mM-1 cm-1) (17) at 25 °C in a coupled assay containing 60 milliunits of xanthine oxidase, variable inosine concentrations (1 μM to 3 mM) in a final volume of 1 mL 50 mM KH2PO4, pH 7.4. The hydrolysis of pNPR was observed by liberation of p-nitrophenol (ε 405 = 12.5 mM-1 cm-1) in 50 mM KH2PO4, Ph 7.4. Reactions were initiated by the addition of enzyme, typically at 2 nM final concentration.
Slow-Onset Inhibition
Slow onset of inhibition was measured following the addition of enzyme to complete assay mixtures at 1 mM inosine and various inhibitor concentrations (18). Inhibitor concentrations were determined spectrophotometrically using the published millimolar extinction coefficients of 8.5 at 275 nm, 9.54 at 261 nm, and 8.92 at 269 nm at pH 7 for 9-deazaadenosine (ImmA based inhibitors), 9-deazainosine (ImmH based inhibitors), and 9-deazaguanosine (ImmG based inhibitors), respectively (19, 20). Enzyme (0.5-5 nM final concentration) was added to assay mixtures followed by monitoring of product formation. Rates were monitored for 1-2 h to determine both the initial reaction rate and to determine if a second reaction rate occurred as a result of slow-onset inhibition. The inhibition constant (Ki) for the initial rate was determined from its fit to eq 1, which describes the competitive inhibition pattern of the Immucillin based inhibitors (Imm) and inosine as substrate (A). Km is the Michaelis constant for inosine (A), Ki is the dissociation constant for the TvPNP•PO4 complex, assuming competitive inhibition. The dissociation constant for the equilibrium complex after slow-onset inhibition has occurred, Ki*2, was determined from fits of the final slopes to eq 1. This analysis is only valid when enzyme concentration is 10-fold less than inhibitor concentration. When the enzyme concentration is significant it depletes free inhibitor and the effective inhibitor concentration can be corrected by the expression: Ifree = Itotal - (1 - vs/vo) Et where Ifree is the free inhibitor concentration, Itotal is the total added inhibitor, vs is the steady-state inhibited catalytic rate, vo is the uninhibited catalytic rate and Et is the total enzyme concentration.
| (1) |
Immucillin off rates
A time-dependent dissociation is expected for bound inhibitor following dilution of the enzyme inhibitor complex into assay mixtures with no inhibitor (20). The tightly bound E*•I•PO4 complex was formed by incubation of 40 μM enzyme subunits with 34 μM inhibitor in 50 mM phosphate buffer, pH 7.4, a condition known to saturate one-third of the catalytic sites and to cause full inhibition under conditions where most of the inhibitor is bound to the enzyme (20). Rapid serial dilution of the E*•I•PO4 complex into buffer (1:200) followed by dilution into the assay mixture (1:200) demonstrated time-dependent release of the inhibitor (20). At high concentrations of inosine (5 mM) as substrate, the rate of activity regain can be used to estimate k3 and k4 (see footnote 2). The rate k4 is estimated by fitting absorbance at 293 nm (P) versus time (t) to the equation: P = vst + (vo − vs) (1 − e-kt) / k where vs is the rate at time zero, vo is the rate at time t and k = k4. The rate of activity regain can also be used to estimate k3 (see footnote 2).
Crystallization of TvPNP-Immucillin-A complexes
TvPNP in 50 mM Hepes pH 7.2 containing 2 mM DTT and 2 mM MgCl2 was concentrated to 8 mg/mL and incubated on ice for 10 min with 1 mM Immucillin-A [2] or 1 mM DADMe-ImmA [1] and 1 mM KH2PO4. The TvPNP•ImmA•PO4 complex was crystallized by hanging drop vapor diffusion at 18 °C using 80 μL of reservoir containing 0.2 M lithium citrate and 20% (w/v) PEG3350, pH 8.5. The TvPNP•DADMe-ImmA•PO4 was crystallized using the same technique but with 0.17 M sodium acetate, 0.085 M Tris-HCL pH 8.8, 25.5% PEG 4000 and 15% glycerol as the reservoir solution.
Data collection
Crystals were soaked in mother liquor supplemented with 20% glycerol and flash cooled to −178 °C prior to data collection. Diffraction from the TvPNP•ImmA•PO4 crystals was consistent with the space group P62 (a = b = 155 and c = 102 Å) with three molecules in the asymmetric unit. The Matthews coefficient was 4.5 Å3/Da, which corresponds to a solvent content of 72%. The TvPNP•DADMe-ImmA•PO4 exhibited diffraction consistent with the space group P212121 (a = 82, b =114 and c = 195 Å) with six molecules in the asymmetric unit. The Matthews coefficient was 2.96 Å3/Da which corresponds to a solvent content of 58%. Diffraction data were collected to a resolution of 2.7 Å for both complexes at beamline X29A at the National Synchrotron Light Source, Brookhaven National Laboratory using an ADSC Quantum 315 detector. Each frame was exposed for 10 s with an oscillation range of 1°. The HKL2000 suite (21) was used for integration and scaling of the data (see Table 1).
Table 1.
Data processing and refinement statistics.
| TvPNP-ImmA | TvPNP-DADMe-ImmA | |
|---|---|---|
| Wavelength (Å) | 0.979301 | 0.979099 |
| Resolution (Å) | 2.7 | 2.7 |
| Unique Reflections | 37032 | 50207 |
| Completeness (%)a | 99.3 (99.5) | 98.1 (93.8) |
| Multiplicitya | 6.2 | 6.9 |
| Rsym (%)a, b | 6.9 (41.8) | 11.6 (59.4) |
| I/σa | 17 (3) | 11.4 (1.7) |
| No. of protein atoms | 5242 | 10721 |
| No. of water molecules | 73 | 170 |
| R-factor | 17.0 | 19.9 |
| Rfree | 22.5 | 25.1 |
| Average B-factor | 60 | 37.8 |
| R.m.s bond (Å) | 0.02 | 0.01 |
| R.m.s angle (Å) | 1.5 | 1.3 |
| Ramachandran analysis | ||
| Most favored | 83.5% | 84.5% |
| Allowed | 16.0% | 15.2% |
| Disallowed | 0.5% | 0.3% |
Values for the highest resolution shell are given in parentheses.
Rsym= (ΣhklΣi|Ii(hkl) - <I(hkl)>|) / ΣhklΣiIi(hkl) for n independent reflections and observations of a given reflection, <I(hkl)> is the average intensity of the i observation.
Structure determination and refinement
The structure of TvPNP in complex with ImmA [2] and DADMe-ImmA [1] and PO42- were solved by molecular replacement using the monomeric form of TvPNP (Protein Data Bank ID code 1Z33.pdb without water) as a search model. Molecular replacement with MOLREP (22), and refinement with REFMAC5 (23) were performed with the CCP4i package (24). COOT (25) was used for molecular modeling. Weak NCS restraints were applied in the initial stages of refinement to improve the maps and removed in the later stages. Clear ligand density was observed in the Fo – Fc maps contured at 5σ.
In the TvPNP•ImmA•PO4 complex residues 206-223 were not well resolved in two of the three monomers in the asymmetric unit (A and C) and these residues were not included in the final model of the TvPNP•ImmA complex. However, interpretable electron density was present for this loop region in the third monomer (B). After the protein and ligands were refined, water molecules were included in the structure. The final structure had R-factor and Rfree of 17.0% and 22.5% respectively.
The six independent molecules in the TvPNP•DADMe-ImmA•PO4 structure also lacked density for the loop region missing in the TvPNP•ImmA•PO4 complex. The C-terminal His tag was partially built in monomer A, B, C, D and F. The final structure of the DADMe•ImmA•PO4 complex had an R-factor of 19.9% with an R-free of 25.5%. All figures were made using PYMOL (26).
[6-18O]ImmH binding in the TvPNP complex
Free TvPNP was concentrated to 3 mM against 0.2 M NaCl using a 0.5 mL 10 K molecular weight retention centrifugal filter (Millipore). [6-18O]ImmH [1] and phosphate buffer, pH 7.4, both at 3 mM were incubated with TvPNP. Free inhibitor and phosphate were washed away with 0.2 M NaCl in order to generate a 3 mM TvPNP•[6-18O]ImmH•PO4 complex for IR studies. The same procedure was used in matched samples to generate the same complex with [6-16O]ImmH. FTIR spectroscopy was performed on a Magna 760 Fourier transform spectrometer (Nicolet Instrument Corp., WI) using a MCT detector. A CaF2 window with a 25 μm Teflon spacer was used. Spectra were collected in the range of 900-4000 cm-1 with 2 cm-1 resolution. A Blackman-Harris three-term apodization and a Happ-Genzel apodization were applied, respectively. Omnic 4.1a (Nicolet Instruments, Corp.) software was used for data collection and analysis.
Results and Discussion
Substrate Specificity of TvPNP
Adenosine is the preferred substrate for the phosphorolysis reaction of TvPNP with a catalytic efficiency of 2.8 × 105 M-1 s-1 and a relatively low Km of 6.1 μM (Table 2). Inosine has higher kcat and Km values for a similar kcat/Km of 7.2 × 104 M-1 s-1. The enzyme is less efficient with guanosine as a substrate by approximately an order of magnitude. Ribooxacarbenium ion stabilization by ribohydrolases can be tested by ribosyl derivatives with good leaving groups and pNPR has been used for this purpose (10). TvPNP had a Km of 90 μM and a kcat of 10-4 s-1 for pNPR (Table 2) to give a kcat/Km for this compound of 1.1. This is 2.5 × 105 fold lower than the kcat/Km for TvPNP with adenosine (16). Catalytic efficiency of TvPNP for adenosine is similar to that observed for bovine and human PNPs with inosine, although the mammalian enzymes do not accept adenosine as a substrate. pNPR is a poor substrate for bovine PNP with a kcat/Km of 0.89 M-1 s-1 (10) similar to the catalytic efficiency with TvPNP. Human PNP had no significant activity with pNPR (level of detection was < 0.1 M-1 s-1, result not shown). Poor substrate function for pNPR suggests a strong requirement for leaving group activation for both mammalian PNPs and TvPNP. Protonation at the N1 and N7 nitrogen(s) of the purine ring and hydrogen-bonds to the 6-exocyclic oxygen are established mechanisms for activation of inosine/hypoxanthine in mammalian PNPs. Specifically, Glu201 of mammalian PNP interacts at N1 and Asp243 spans a protonated N7H and O6 for transition state stabilization in complexes with Immucillin-H [5]. These interactions differ in TvPNP where Asp204 replaces Asn243. In order to accept both adenine and hypoxanthine bases for efficient catalysis in TvPNP, it is likely that the protonation pattern of the carboxylate changes to accommodate the change of H-bond donor/acceptor at C6’ (from amino in adenine to a carbonyl oxygen in hypoxanthine).
Table 2.
Kinetic Constants for TvPNP with Various Substrates
| Substrate | Km (μM) | kcat (s-1) | kcat/Km (M-1 s-1) |
|---|---|---|---|
| Adenosine | 6.1 ± 0.5 | 1.7 ± 0.2 | 2.78 × 105 |
| Guanosine | 59.7 ± 8.1 | 1.1 ± 0.1 | 1.8 × 104 |
| Inosine | 25 ± 1 | 1.8 ± 0.1 | 7.2 × 104 |
| pNPR | 90 ± 42 | 1.0 ± 0.1 × 10-3 | 1.1 |
Slow-Onset Inhibition of TvPNP
Immucillin-A [2] is a slow-onset tight-binding transition state analogue, with a Ki of 1,000 pM and a Ki* of 87 pM (Table 3 and Figure 1). In contrast, ImmA binds to the P. falciparum and human PNPs with Ki* values of greater than 10 μM (27) and 2.6 μM for bovine PNP (28). DADMe-ImmA [1] is a more powerful inhibitor of TvPNP and binds tightly with a Ki* of 30 pM. Some hexameric PNPs, like that from Plasmodium falciparum use 5’-methylthioinosine as substrates (29). In those cases 5’-methylthio-substituted Immucillins are powerful inhibitors (14). For TvPNP, MT-DADMe-ImmA [9] has a dissociation constant of > 200,000 times that for DADMe-ImmA, thus the 5’-methylthio group prevents binding (Figure 1). Because TvPNP does not accept 5’-methylthio groups, it is unlikely to be involved in 5’-methylthioadenosine metabolism in T. vaginalis. ImmG [4] has a dissociation constant of 42 pM for trimeric human PNP, 900 pM for hexameric P. falciparum PNP and relatively weak binding to TvPNP (Ki* = 9.4 nM). DADMe-ImmG [6] is a powerful inhibitor of human PNP with a Ki* of 7 pM, however the Ki* for TvPNP is 18 nM. TvPNP, with its preference for adenosine, prefers DADMe-ImmA as a picomolar transition state analogue while human PNP, with deoxyguanosine as a critical substrate, prefers DADMe-ImmG as a transition state analogue.
Table 3.
Inhibition Constants (Ki and Ki*) for Immucillin Based Inhibitors of TvPNP
| Inhibitor | Ki (pM) | Ki* (pM) |
|---|---|---|
| DADMe-Immucillin-A [1] | 1,500 ± 130 | 30 ± 4 |
|
| ||
| Immucillin-A [2] | 1,000 ± 90 | 87 ± 8 |
| DADMe-Immucillin-H [3] | 3,800 ± 300 | 1,200 ± 30 |
| Immucillin-G [4] | 16,000 ± 5,000 | 9,400 ± 1,500 |
| Immucillin-H [5] | 12,000 ± 700 | naa |
| DADMe-Immucillin-G [6] | 18,000 ± 500 | naa |
| MT-DADMe-Immucillin-H [7] | > 10,000,000 | naa |
| 9-Aza-1’-9-Me- Immucillin-A [8] | > 10,000,000 | naa |
| MT-DADMe-Immucillin-A [9] | > 10,000,000 | naa |
No slow onset observed.
ImmH [5] binds to TvPNP with a Ki of 12 nM, a greatly reduced affinity relative to human and bovine PNPs (dissociation constants of 56 pM and 23 pM respectively). DADMe-ImmH [3] has a dissociation constant of 1.2 nM for TvPNP, an affinity decreased by 40-fold relative to DADMe-ImmA [1] (5). Relatively tight binding of both inosine and adenosine transition state mimics to TvPNP distinguishes this enzyme from all previously reported PNPs. Leaving group interactions at PNP catalytic sites are usually specific for 6-amino or 6-oxy substituents but TvPNP accepts both and this broad specificity is reflected in its transition state analogue specificity. DADMe-ImmA [1] (5) binds 2.2 kcal/mol more favorably than DADMe-ImmH [3] (2). This difference reflects interactions of the 6-amino and 6-carbonyl groups of the Immucillins at the catalytic site and is characterized by the structural and spectroscopic analyses described below.
Rate constants for interaction of DADMe-ImmA and ImmA
Slow-onset tight-binding inhibitors interact through rapid, weak interactions followed by an inhibitor-induced conformational change that causes tight binding (Figure 2). For TvPNP inhibition by DADMe-ImmA [1], a short lived (t1/2 of 79 sec) initial interaction with a 1,500 pM Ki is followed by slow-onset inhibition. The affinity increases 50-fold to give a Ki* of 30 pM after the slow-onset step has equilibrated (after 60 min). The slow-onset inhibition pattern with ImmA [2] is similar with the binding at equilibrium giving a Ki* of 87 (Figure 2). Dilution of inhibited enzyme into excess substrate gave an inhibitor t1/2 off-rate of 17.2 min for ImmA [2] and 64 min for DADMe-ImmA [1] (Table 4). By comparison, this rate is 72 min for the 23 pM interaction of ImmH [5] with bovine PNP (20).
Figure 2.
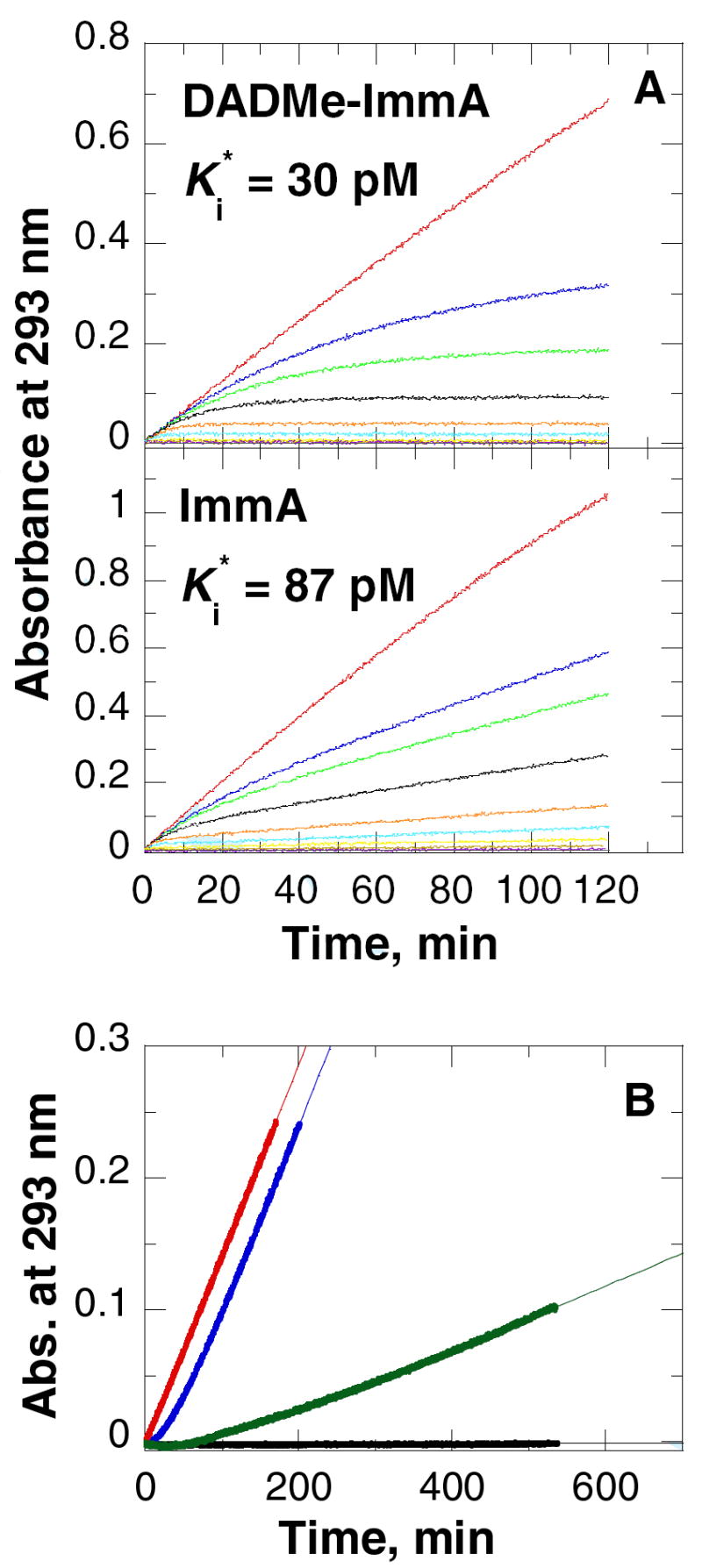
Immucillin binding to and release from TvPNP. (A) Slow-onset nhibition of TvPNP with various concentrations of DADMe-Immucillin-A [1] (upper panel) and Immucillin-A [2] (lower panel), in 50 mM KH2PO4, pH 7.4 at 25 °C. The reactions were initiated by the addition of enzyme (~5 nM) into 1 ml reactions mixtures containing 1 mM inosine as substrate, 60 milliunits of xanthine oxidase, and varied inhibitor concentrations. In the upper panel (A), DADMe-ImmA concentrations are 0 (red), 8 nM (blue), 16 nM (green), 32 nM (black), 80 nM (orange), 160 nM (cyan) and 400 nM (yellow), 800 nM (brown) and 1,600 nM (purple). In the lower panel (A), ImmA concentrations are 0 (red), 6.6 nM (blue), 13.2 nM (green), 26.4 nM (black), 66 nM (orange), 132 nM (cyan) and 330 nM (yellow), 660 nM (brown) and 1,320 nM (purple). Gray curves are negative controls with no enzyme. The UV change at 293 nm due to the formation of uric acid were monitored for 120 min in the coupled xanthine oxidase assays. (B) Dissociation of ImmA (blue curve) and DADMe-ImmA (green curve) from TvPNP. Enzyme (40 μM) and Immucillins (40 μM) were incubated for 1 h at 30 °C followed by 40,000-fold dilution into xanthine oxidase coupling assay mixtures containing 5 mM of inosine and 50 mM KH2PO4, pH 7.4. In the positive control assay (red curve), an equivalent enzyme sample was treated in the same manner except that incubation was with buffer. The background (black curve) shows the spectrophotometer response when no PNP is added to the reaction mixture. Data were fit to the equation for egain of enzyme activity as indicated in the methods.
Table 4.
Immucillin Binding and Release Constants
Crystal structure of TvPNP•ImmA•PO4 and the TvPNP•DADMe-ImmA•PO4
The structures of TvPNP•ImmA•PO4 and TvPNP•DADMe-ImmA•PO4 were determined by X–ray crystallography for comparison with structures of other inhibitors (3). The TvPNP•ImmA•PO4 structure has 3 monomers in the asymmetric unit. The application of a crystallographic 2-fold axis generates the functional hexameric structure which is a trimer of dimers (Figure 3a). The six active sites are composed of residues contributed from a pair of monomers. Each monomer of TvPNP has a β-sheet core flanked by seven α-helices. The TvPNP•DADMe-ImmA•PO4 structure has six monomers in the asymmetric unit which corresponds to the functional hexamer. The structures with bound Immucillins show similarity to TvPNP without ligands at the catalytic site (1Z33.pdb) and to E. coli PNP in complex with 2-F-adenosine and phosphate (1PK9.pdb), but also reveal differences as discussed below (3, 30).
Figure 3.
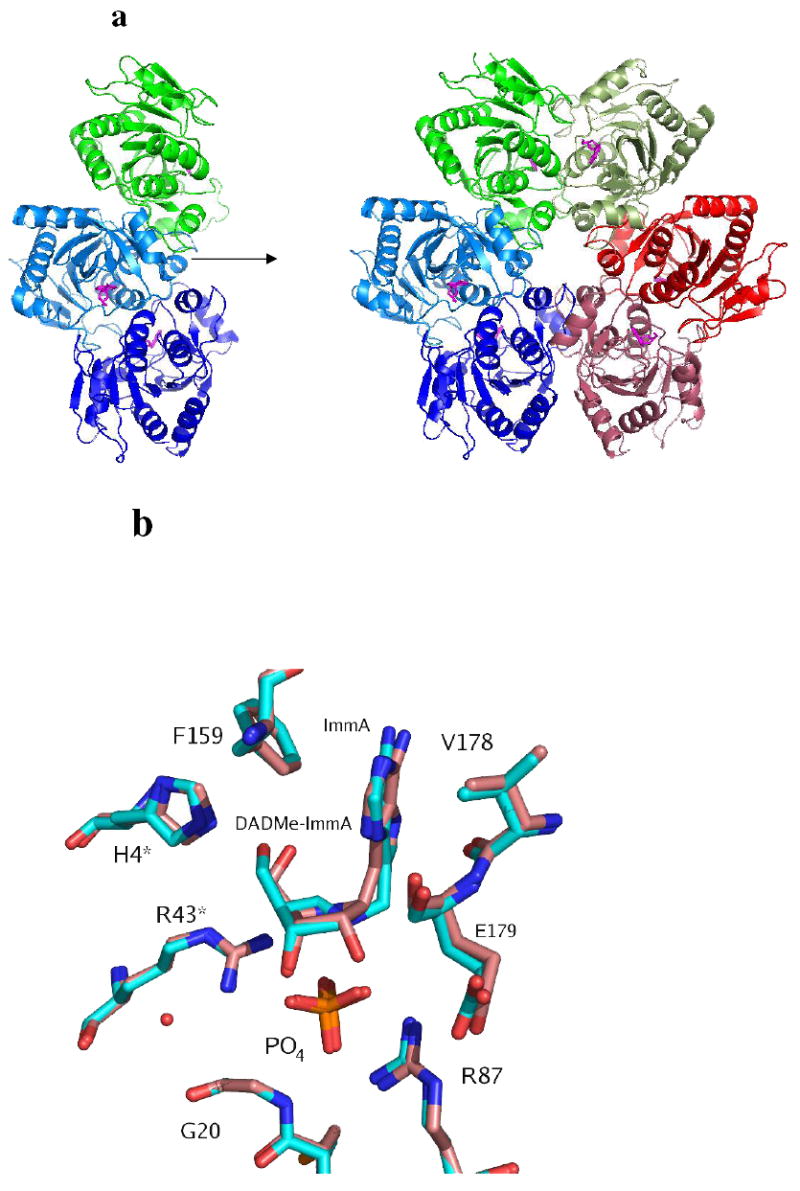
X-ray crystal structure of TvPNP with ImmA [2] and PO4 bound in the active site. In a) the molecules in the asymmetric unit generate the hexamer by applying two-fold crystallographic symmetry. Each pair of monomers is represented with similar colors. The crystal structure of TvPNP in complex with DADMe-ImmA and PO4 bound in the active sites was solved with a hexamer in the asymmeric unit. b) Superpositions of the Immucillins at the active sites of the two TvPNP structures. The complex with ImmA [2] is shown in pink and the complex with DADMe-ImmA [1] is shown in cyan.
Active site of TvPNP
The structures of TvPNP•ImmA•PO4 and TvPNP•DADMe-ImmA•PO4 show the active site located at the interface between each pair of monomers. Clear electron density was observed for the inhibitor and for bound phosphate. The ligands in monomers A and C in the ImmA•PO4 complex have higher B-factors (80 Å2) than those in monomer B (60 Å2), suggesting slightly higher ligand occupancy or structural order in monomer B compared to monomer A and C. The three dimers of the hexamer in the asymmetric unit of the TvPNP•DADMe-ImmA•PO4 complex show similar B-factor patterns to those of the ImmA complex. Differential B-factors may relate to crystallographic differences or the negative cooperativity known to be induced by inhibitor binding in mammalian PNPs (20).
The two TvPNP-Immucillin-A complexes are similar to each other and superimpose well with an r.m.s. deviation of 0.3 Å comparing 236 residues (Figure 3b). The active site can be divided into the purine base binding site, the sugar binding site and the phosphate binding site. The purine base is recognized by four hydrophobic residues Phe159, Val178, Met180 and Ile206 where Phe159 and Val178 make hydrophobic interactions with the base (Figures 4a). The side chains of Ile206 and Met180 form hydrophobic van der Waals interactions near the 6-amino group and the N3 of the 9-deazahypoxanthine ring of bound ImmA and DADMe-ImmA.
Figure 4.
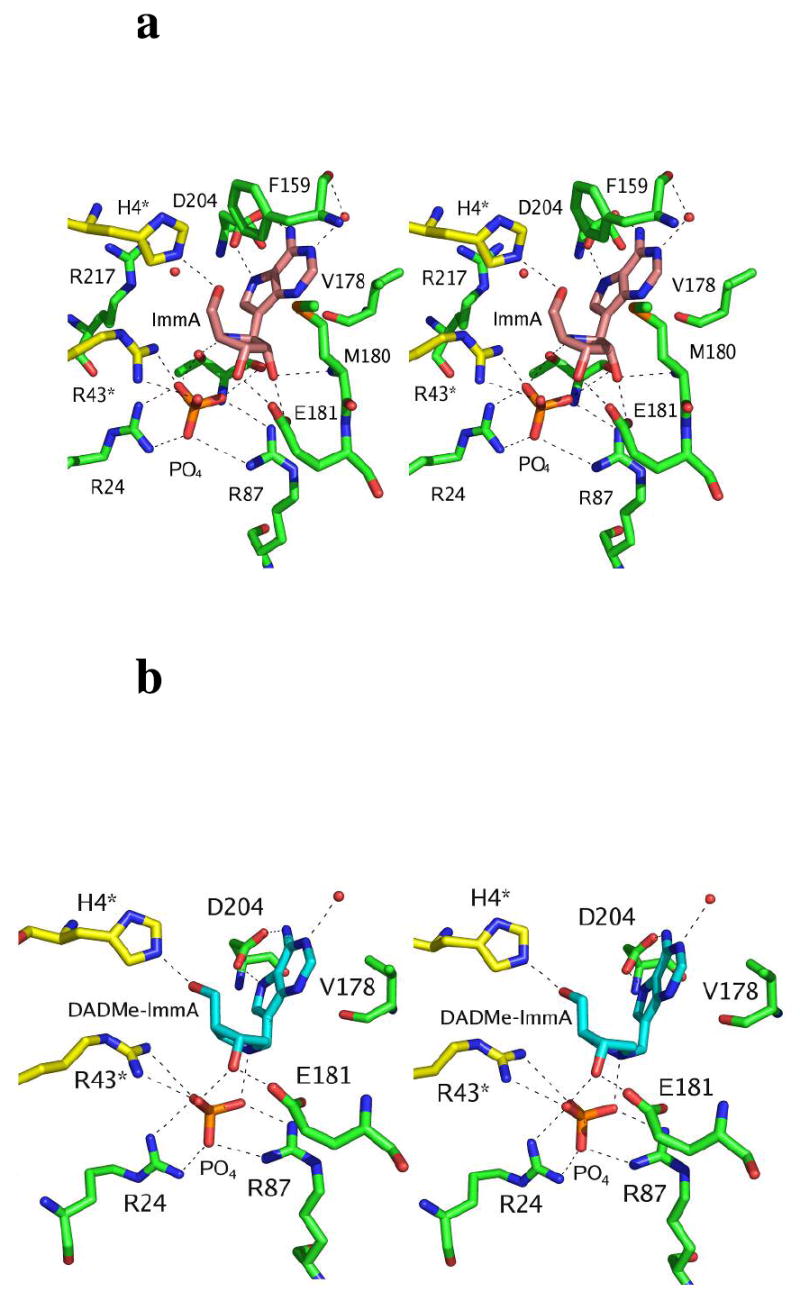
Stereoviews of the catalytic site contacts in TvPNP with the transition state analogue inhibitors ImmA [2] and DADMe-ImmA [1] and PO4 in the catalytic sites. In a) the stereo view of the active site of TvPNP in complex with ImmA- and PO4 is shown and is compared in b) to the stereo view of the active site of TvPNP in complex with DADMe-ImmA and PO4.
N1 of the deazapurine base accepts a hydrogen bond from a structurally conserved water that also hydrogen bonds to the carbonyl oxygen of Phe159 (distances of 2.7-2.9 Å). OD2 and OD1 of the catalytically important Asp204 make hydrogen bond interactions to N7 and N6 of the base with distances of 2.7 and 3.0 Å respectively (Figure 5). The close interaction of Asp204 and the base suggest formation of the transition state by Asp204 acting as the general acid proton donor to N7 of the adenine leaving group. Proton donation from Asp204 to form the transition state requires that Asp204 be protonated prior to reaching the transition state. Thus, the pKa of this group, prior to transition state formation must have a sufficiently high pKa to permit formation of acidic Asp204 at neutral pH.
Figure 5.
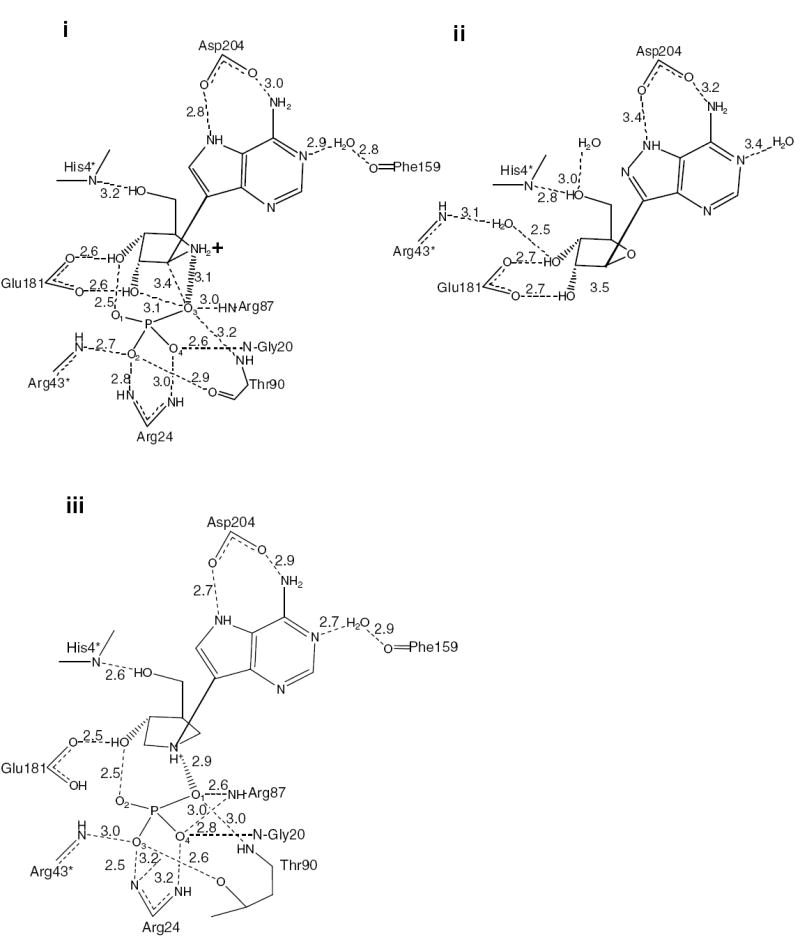
A stereoview overlay of the active site complexes of TvPNP•ImmA•PO4 in grey (deposited to the PDB with ID number 2I4T), TvPNP•DADMe-ImmA•PO4 in cyan (deposited with PDBID number 2ISC), TvPNP•Formycin-A (1z36.pdb) in yellow and TvPNP•2-fluoroadenosine (1z35.pdb) (3) in yellow. Asp204 has the closer interactions with DADMe-ImmA and ImmA than to the other two TvPNP complexes.
The ribose sugar is recognized by His4* (* indicates a contact from the adjacent subunit), Glu181 and Thr90. The 5′-OH group of the sugar makes hydrogen bonds to NE2 of His4* (2.6-3.2 Å). The O2’ and O3’ of the sugar participate in several hydrogen bonds with OE1 and OE2 of Glu181 (2.6 Å and 2.6 Å) and to O1 and O3 of the phosphate group (2.5 Å and 3.1 Å) in the TvPNP-ImmA complex. DADMe-ImmA lacks the O2’ group and is missing the hydrogen bonds associated with this group (Figure 5). Instead the N1’ of the ribose ring participates in a 2.7 Å ion pair with the O3 of the phosphate group. Ion pair formation in a relatively hydrophobic catalytic site contributes to the high affinity of DADMe-ImmA for TvPNP.
The anionic phosphate binding site contains cations contributed by Arg24, Arg87 and Arg43* and H-bond donors from the main chain amino group of Gly20. Another cation is provided by the bound Immucillins. For ImmA the cationic imino nitrogen N4’ of ImmA is near phosphate as is the N1’ cation from bound DADMe-ImmA. The PO4 moiety O2 also hydrogen-bonds with the carbonyl oxygen of Thr90, while O3 and O1 interact with the 2′-OH (3.1 Å) and 3′-OH (2.5 Å) of ImmA, respectively. Phosphate O2 makes hydrogen bond or ion pair interactions with NH2 of Arg24 (2.8 Å), NH2 of Arg43* (2.7 Å) and to the OG1 of Thr90 (2.7-2.9 Å). Phosphate O4 interacts with NH1 of Arg24 (3.0 Å) and the amino group of Gly20 (2.6 Å). Phosphate O3 interacts with the NH2 of Arg87 (2.7-3.0 Å), the amino group of Thr90 (3.0-3.2 Å) and the N4’ cation of ImmA (3.1 Å). The N1’ of the carbocation in the DADMe-ImmA complex forms an ion pair with the same oxygen of the PO4 (2.9 Å). The apparent nucleophilic oxygen is O3 of the phosphate and the distance between the nucleophile, C1′ and the cationic N4’ of the ImmA is 3.4 and 3.1 Å, respectively. DADMe-ImmA has a 2.9 Å distance between the ribocation mimic and the 9-deazaadenine leaving group to resemble the geometry of a fully developed SN1 transition state. Transition state analysis of human, bovine and P. falciparum PNPs have placed the attacking phosphate oxygen nucleophile approximately 3.0 Å from the C1′ carbocation at these transition states (31, 32). The 3.0 Å distance between N4′ of bound ImmA and the anionic O3 of phosphate provides both geometric and ionic mimicry of these transition states. However, when the ImmA transition state analogue is bound, its N4’ cation is slightly more distant and the cationic charge is moved from the position of the cation in the actual transition state.
Comparison of TvPNP•ImmA•PO4 with homologous structures
The catalytic mechanism proposed for TvPNP involves Asp204 as the proton donor to N7 of the leaving group. In the crystal structure of TvPNP•ImmA•PO4, Asp204 accepts hydrogen bonds from NH26 and NH7 of the ImmA base. In the DADMe-ImmA complex these hydrogen bonds are even shorter in agreement with its tighter binding. Most previously reported TvPNP complexes (3, 27) have the Asp204 pointing away from the base (Figure 6). Only TvPNP in complex with Formycin-A (1z36.pdb) (3) has Asp204 in the proposed active conformation with hydrogen bonds to the N7 and NH26 of the base, similar to Immucillin interactions (Figure 6, 7). Formycin-A is a 2.3 μM inhibitor of TvPNP and the interactions to Asp204 in the TvPNP•Formycin•A structure (3) are weaker, (3.4 and 3.2 Å, respectively), than those seen in the TvPNP•ImmA•PO4 structure (3.0 and 2.8 Å, respectively) and in the DADMe-ImmA complex (2.9 and 2.7 Å respectively). Although these are near the limits of experimental uncertainty, distances systematically become shorter with the more tightly-bound inhibitors. The favorable hydrogen bonds between NH7 and NH26 and Asp204 with bound ImmA or DADMe-ImmA support transition state stabilization of adenosine by these interactions.
Figure 6.
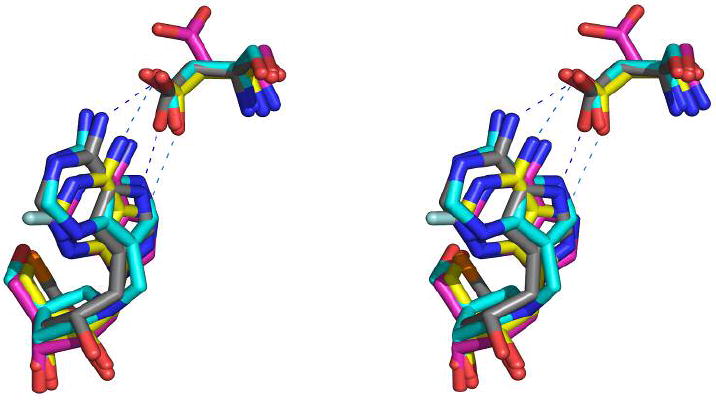
Distance maps of (i) TvPNP•ImmA•PO4 compared to (ii) TvPNP•Formycin-A (from 1z36.pdb, 3) and (iii) TvPNP•DADMe-ImmA•PO4.
Figure 7.
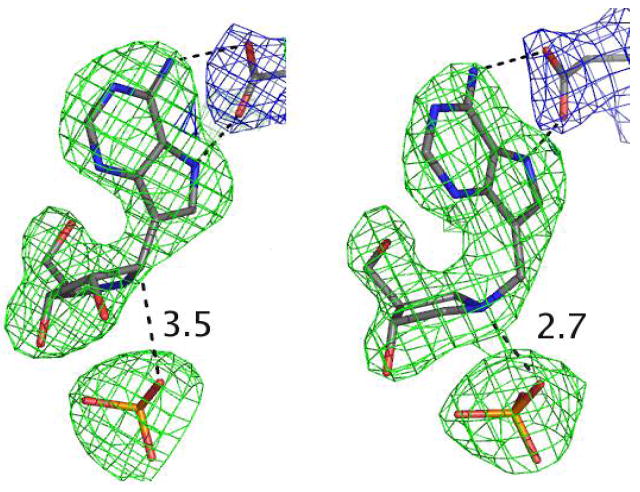
Omit maps for ImmA and phosphate contoured at 3 σ around the inhibitors (green) and the 2FoFc map contoured at 1σ around the Asp204 (in blue) are shown on the left. The right panel is the same analysis for the complex with DADMe-ImmA. The distances between the C1’ of ImmA (left) and the N1’ of DADMe-ImmA (right) and the phosphate groups are shown in angstroms.
ImmH binding in the TvPNP complex
The promiscuous nature of TvPNP with regards to inosine, guanosine and adenosine substrate specificity led us to the hypothesis that a 6-enol tautomer of inosine and guanosine might be required for catalysis. Protonated Asp204 donates its proton to N7 at the transition state of these substrates. Therefore, the carbonyl oxygen (=O) would form an unfavorable interaction with O6 unless it exists at the enol (-OH) form. Then, favorable interactions could occur at both N7 and O6. This hypothesis was tested by the binding of ImmH to form TvPNP•ImmH. The IR difference spectrum between the TvPNP•ImmH and TvPNP•[6-18O]ImmH complexes (Figure 5) contains only vibrational modes from [6-16O/6-18O]ImmH when bound to TvPNP. The positive bands are due to [6-16O]ImmH and the negative bands are a feature of [6-18O]ImmH. The observation of a C6=O stretch frequency at 1672 cm-1 in the ImmH•TvPNP complex suggests that most, if not all bound ImmH exists in the keto form. The positive peak in the difference spectra is shifted from 1667 cm-1 to 1672 cm-1 (a difference of + 6) in the TvPNP•ImmH complex. This frequency change indicates a weaker H-bond between the enzyme and C6=O when ImmH is bound to TvPNP than in solution. In contrast, the binding of ImmH in the complex of human PNP and PO4 reveals a stronger H-bond to C6=O, but not the enol form (not shown). The relatively weak binding interaction (Ki = 12 nM) seen between ImmH and TvPNP is 138-fold weaker (3.0 kcal/mol) than for ImmA, consistent with the loss of one H-bond between the exocyclic group at C6 and Asp204.
Conclusions
The adenosine transition state analogue DADMe-ImmA [1] resembles a fully dissociated adenosyl ribocation transition state and is a powerful, slow-onset tight-binding inhibitor for TvPNP with a Ki* value of 30 pM and a Km/Ki* value of 203,300. Immucillin-A [2] resembles an early SN1 transition state with adenosine partly dissociated at the N-ribosidic bond, and binds with a dissociation constant of 87 pM. This inhibition pattern suggests that TvPNP forms a fully dissociated transition state for the phosphorolysis of adenosine. The 6-amino group is important to transition state analogue binding since DADMe-ImmH [3] and ImmH [5] bind with dissociation constants of 1.2 and 12 nM, respectively. In contrast, human, bovine and P. falciparum PNPs bind ImmH [5] with dissociation constants of 56, 23 and 860 pM, respectively. The interaction of DADMe-ImmA [1] with TvPNP is the tightest-binding enzymatic interaction known for an adenosine-based Immucillin. The relative affinity for Immucillins and DADMe-Immucillins has been shown to be a dependable indication of early and late dissociative (SN1) transition states (31). The transition state formed by TvPNP is therefore likely to have no significant bond order to the leaving group, similar to human PNP. Like TvPNP, human PNP also shows higher affinity for DADMe-Immucillins than for the Immucillins. An exception for TvPNP is the transition state analogues of guanosine. Guanosine has a 10-fold lower catalytic efficiency than adenosine for TvPNP and DADMe-ImmG [6] and ImmG [4] exhibit dissociation constants of 18 and 9.4 nM respectively. This pattern suggests that phosphorolysis of guanosine may have an earlier transition state than that for adenosine.
Poor use of pNPR as a substrate by TvPNP is an indication that much of the catalytic potential for this enzyme arises between TvPNP and leaving group interactions (32). The tight binding of DADMe-ImmA to TvPNP is explained in the crystal structure of TvPNP•DADMe-ImmA•PO4 where Asp204 makes strong hydrogen bonds to the deazaadenine group and a 2.9 Å ion pair is formed between the phosphate anion and the N1’ cation. The unique substrate specificity of TvPNP has permitted the design of a powerful and specific transition state analogue for this unusual and specific isozyme of the PNP family.
Figure 8.
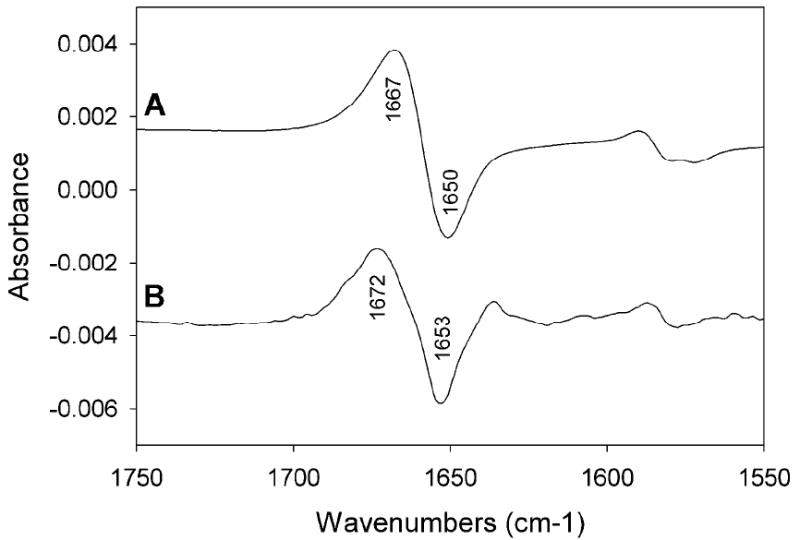
(A) Difference FTIR spectra of [6-O16]ImmH [1] and [6-O18]ImmH in solution (B) difference FTIR spectra of TvPNP•[6-O18]ImmH•PO4 and TvPNP•[6-O16]ImmH•PO4 complexes. Spectra with and without added 50 mM phosphate buffer gave the same result.
Acknowledgments
The authors thank Dr. James Errey for valuable discussions. We thank the staff at NSLS Brookhaven National Lab for technical assistance at X29A. This work was supported by grants from the National Institutes of Health and the Wenner-Gren Foundation.
Footnotes
Supported by NIH Research Grants GM41916 and AI60660
Abbreviations: DADMe-ImmA, 4’-deaza-1’-aza-2’-deoxy-1’-(9-methylene)-Immucillin-A; ImmA, Immucillin-A; DADMe-ImmH, 4’-deaza-1’-aza-2’-deoxy-1’-(9-methylene)-Immucillin-H; PNP, purine nucleoside phosphorylase; TvPNP, Trichomonas vaginalis purine nucleoside phosphorylase; HGPRT, hypoxanthine-guanine phosphoribosyltransferase; pNPR, p-nitrophenyl β-d-ribofuranoside.
Origin of Ki and Ki* based on the slow-onset tight-binding of Immucillins:
Ki = k2/k1
K1* = Ki (k4/k3+k4)
References
- 1.Radonjic IV, Dzamic AM, Mitrovic SM, Arsic Arsenijevic VS, Popadic DM, Kranjcic Zec IF. Diagnosis of Trichomonas vaginalis infection: The sensitivities and specificities of microscopy, culture and PCR assay. Eur J Obstet Gynecol Reprod Biol. 2005;126:116–120. doi: 10.1016/j.ejogrb.2005.07.033. [DOI] [PubMed] [Google Scholar]
- 2.Gero AM, Kang EW, Harvey JE, Schofield PJ, Clinch K, Furneaux RH. Trichomonas vaginalis: detection of nucleoside hydrolase activity as a potential screening procedure. Exp Parasitol. 2000;94:125–128. doi: 10.1006/expr.1999.4484. [DOI] [PubMed] [Google Scholar]
- 3.Zang Y, Wang W-H, Wu S-W, Ealick SE, Wang CC. Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex. J Biol Chem. 2005;280:22318–22325. doi: 10.1074/jbc.M501843200. [DOI] [PubMed] [Google Scholar]
- 4.Brown DM, Upcroft JA, Dodd HN, Chen N, Upcroft P. Alternative 2-keto acid oxidoreductase activities in Trichomonas vaginalis. Mol Biochem Parasitol. 1999;98:203–214. doi: 10.1016/s0166-6851(98)00169-8. [DOI] [PubMed] [Google Scholar]
- 5.de Koning HP, Bridges DJ, Burchmore RJS. Purine and pyrimidine transport in pathogenic protozoa: From biology to therapy. FEMS Microbiol Rev. 2005;29:987–1020. doi: 10.1016/j.femsre.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Stoeckler JD. Purine nucleoside phosphorylase: a target for chemotherapy. In: Glazer RI, editor. Development in Cancer Chemotherapy. CRC Press, Inc.; Boca Raton, FL: 1984. pp. 35–60. [Google Scholar]
- 7.Montgomery JA. Purine nucleoside phosphorylase: a target for drug design. Med Res Rev. 1993;13:209–228. doi: 10.1002/med.2610130302. [DOI] [PubMed] [Google Scholar]
- 8.Bzowska A, Kulikowska E, Shugar D. Purine nucleoside phosphorylase: properties, functions and clinical aspects. Pharmacol Ther. 2000;88:349–425. doi: 10.1016/s0163-7258(00)00097-8. [DOI] [PubMed] [Google Scholar]
- 9.Koellner G, Bzowska A, Wielgus-Kutrowska B, Luic M, Steiner T, Saenger W, Stepinski J. Open and closed conformation of the E. coli purine nucleoside phosphorylase active center and implications for the catalytic mechanism. J Mol Biol. 2002;315:351–371. doi: 10.1006/jmbi.2001.5211. [DOI] [PubMed] [Google Scholar]
- 10.Mazella LJ, Parkin DW, Tyler PC, Furneaux RH, Schramm VL. Mechanistic diagnosis of N-ribohydrolases and purine nucleoside phosphorylase. J Am Chem Soc. 1996;118:2111–2112. [Google Scholar]
- 11.Singh V, Schramm VL. Transition state structure of human 5’-methylthioadenosine phosphorylase. J Am Chem Soc. 2006;128 doi: 10.1021/ja065419p. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Handlon AL, Xu C, Muller-Steffner HM, Schuber F, Oppenheimer NJ. 2’-Ribose substituent effects on the chemical and enzymatic hydrolysis of NAD+ J Am Chem Soc. 1994;116:12087–12088. [Google Scholar]
- 13.Rosenburg S, Kirsch JF. Oxygen-18 leaving group kinetic isotope effects on the hydrolysis of nitrophenyl glycosides. 2. lysozyme and β-glucosidase: acid and alkaline hydrolysis. Biochemistry. 1981;20:3196–3207. doi: 10.1021/bi00514a032. [DOI] [PubMed] [Google Scholar]
- 14.Lewandowicz A, Taylor Ringia E, Ting LM, Kim K, Tyler PC, Evans GB, Zubkova OV, Mee S, Painter GF, Lenz DH, Furneaux RH, Schramm VL. Energetic mapping of transition state analogue interactions with human and plasmodium falciparum purine nucleoside phosphorylases. J Biol Chem. 2005;280:30320–30328. doi: 10.1074/jbc.M505033200. [DOI] [PubMed] [Google Scholar]
- 15.Mungagala NR, Wang CC. Adenosine is a primary precursor of all purine nucletides in Trichomonas vaginalis. Mol Biochem Parasitol. 2003;127:143–149. doi: 10.1016/s0166-6851(02)00330-4. [DOI] [PubMed] [Google Scholar]
- 16.Mungagala NR, Wang CC. The purine nucleoside phosphorylase from Trichomonas vaginalis is a homologue of the bacterial enzyme. Biochemistry. 2002;41:10382–10389. doi: 10.1021/bi026025n. [DOI] [PubMed] [Google Scholar]
- 17.Kim BK, Cha S, Parks RE., Jr Purine nucleoside phosphorylase from human erythroyctes. II. Kinetic analysis and substrate-binding studies. J Biol Chem. 1968;243:1771–1776. [PubMed] [Google Scholar]
- 18.Kicska GA, Tyler PC, Evans GB, Furneaux RH, Kim K, Schramm VL. Transition state analogue inhibitors of purine nucleoside phosphorylase from Plasmodium falciparum. J Biol Chem. 2002;277:3219–3225. doi: 10.1074/jbc.M105905200. [DOI] [PubMed] [Google Scholar]
- 19.Singh V, Evans GB, Lenz DH, Mason JM, Clinch K, Mee S, Painter GF, Tyler PC, Furneaux RH, Lee JE, Howell PL, Schramm VL. Femtomolar transition state analogue inhibitors of 5’-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Escherichia coli. J Biol Chem. 2005;280:18265–18273. doi: 10.1074/jbc.M414472200. [DOI] [PubMed] [Google Scholar]
- 20.Miles RW, Tyler PC, Furneaux RH, Bagdassarian CK, Schramm VL. One-third-the-sites transition-state inhibitors for purine nucleoside phosphorylase. Biochemistry. 1998;37:8615–8621. doi: 10.1021/bi980658d. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 23.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 24.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Cryst. 2003;D59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Model-Building Tools for Molecular Graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: 2002. [Google Scholar]
- 27.Kicska GA, Tyler PC, Evans GB, Furneaux RH, Schramm VL, Kim K. Purine-less death in Plasmodium falciparum induced by Immucillin-H, a transition state analogue of purine nucleoside phosphorylase. J Biol Chem. 2002;277:3226–3231. doi: 10.1074/jbc.M105906200. [DOI] [PubMed] [Google Scholar]
- 28.Kicska GA, Tyler PC, Evans GB, Furneaux RH, Shi W, Fedorov A, Lewandowicz A, Cahill SM, Almo SC, Schramm VL. Atomic dissection of the hydrogen bond network for transition-state analogue binding to purine nucleoside phosphorylase. Biochemistry. 2002;41:14489–14498. doi: 10.1021/bi026636f. [DOI] [PubMed] [Google Scholar]
- 29.Ting LM, Shi W, Lewandovicz A, Singh V, Mwakingwe A, Birck MR, Ringia EA, Bench G, Madrid DC, Tyler PC, Evans GB, Furneaux RH, Schramm VL. Targetign a novel Plasmodium falciparum purine recycling pathway with specific immucillins. J Biol Chem. 2005;280:9547–9554. doi: 10.1074/jbc.M412693200. [DOI] [PubMed] [Google Scholar]
- 30.Bennett EM, Li C, Allan PW, Parker WB, Ealick SE. Structural basis for substrate specificity of E. coli purine nucleoside phosphorylase. J Biol Chem. 2003;278:47110–47118. doi: 10.1074/jbc.M304622200. [DOI] [PubMed] [Google Scholar]
- 31.Taylor Ringia EA, Tyler PC, Evans GB, Furneaux RH, Murkin AS, Schramm VL. Transition state analogue discrimination by related purine nucleoside phosphorylases. J Am Chem Soc. 2006;128:7126–7127. doi: 10.1021/ja061403n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schramm VL. Enzymatic transition state poise and transition state analogues. Acc Chem Res. 2003;36:588–96. doi: 10.1021/ar0200495. [DOI] [PubMed] [Google Scholar]


