Abstract
Streptococcus pneumoniae 5′-methylthioadenosine/S-adenosylhomocysteine hydrolase (MTAN) catalyzes the hydrolytic deadenylation of its substrates to form adenine and 5-methylthioribose or S-ribosylhomocysteine (SRH). MTAN is not found in mammals but is involved in bacterial quorum sensing. MTAN gene disruption affects growth and pathogenicity of bacteria, making it a target for antibiotic design. Kinetic isotope effects and computational studies have established a dissociative SN1 transition state for E. coli MTAN and transition state analogues resembling the transition state are powerful inhibitors of the enzyme (Singh, V., Lee, J. L., Núñez, S., Howell, P. L. and Schramm, V. L. (2005) Biochemistry 44, 11647-11659). The MTAN from S. pneumoniae has 40% sequence identity to E. coli MTAN, but exhibits remarkably distinct kinetic and inhibitory properties. 5′-Methylthio-Immucillin-A (MT-ImmA) is a transition state analogue resembling an early SN1 transition state. It is a weak inhibitor of S. pneumoniae MTAN with a Ki of 1.0 μM. The X-ray structure of S. pneumoniae MTAN with MT-ImmA indicates a dimer with the methylthio group in a flexible hydrophobic pocket. Replacing the methyl group with phenyl (PhT-ImmA), tolyl (p-TolT-ImmA) or ethyl (EtT-ImmA) groups increases the affinity to give Ki values of 335 nM, 60 nM and 40 nM, respectively. DADMe-Immucillins are geometric and electrostatic mimics of a fully-dissociated transition state and bind more tightly than Immucillins. MT-DADMe-Immucillin-A inhibits with a Ki value of 24 nM and replacing the 5′-methyl group with p-Cl-phenyl (p-Cl-PhT-DADMe-ImmA) gave a Ki* value of 0.36 nM. The inhibitory potential of DADMe-Immucillins relative to the Immucillins supports a fully dissociated transition state structure for S. pneumoniae MTAN. Comparison of active site contacts in the X-ray crystal structures of E. coli and S. pneumoniae MTAN with MT-ImmA would predict equal binding, yet most analogues bind 103 to 104 fold more tightly to the E. coli enzyme. Catalytic site efficiency is primarily responsible for this difference since kcat/Km for S. pneumoniae MTAN is <10-2 that of E. coli MTAN.
Keywords: 5′-methylthioadenosine, 5′-methylthioadenosine nucleosidase, quorum sensing, nucleoside hydrolase, transition state, transition state analogue inhibitors, polyamines, MTAN structure, catalytic efficiency
5′-Methylthioadenosine/S-adenosylhomocysteine nucleosidase (MTAN1) is a bacterial enzyme encoded by the pfs gene and catalyzes hydrolytic depurination of 5′-methylthioadenosine (MTA) to form 5-methylthioribose (MTR) and S-adenosylhomocysteine (SAH) to S-ribosylhomocysteine (SRH). Adenine is a product of both reactions (1-2). MTAN is involved in bacterial pathways related to polyamine biosynthesis, quorum sensing, methyl transfer reactions and adenine and methionine salvage (Figure 1; 3-6). The substrates of MTAN are structurally related to S-adenosylmethionine (SAM) and function as product inhibitors in SAM-requiring reactions (7). In bacteria, accumulation of these metabolites is avoided through the function of the pfs gene. Deletion of pfs in E. coli causes a severe growth defect (8). Quorum sensing is important in pathogenicity since deletion of the LuxS quorum sensing gene in S. pneumoniae results in attenuation of pneumococcal infections (9). LuxS functions immediately downstream of pfs and deletion of pfs is expected to have a similar phenotype. Schauder et. al. (10) reported that purified pfs and luxS enzymes are necessary and sufficient for in vitro production of autoinducers-2 (AI2) quorum sensing molecules, using SAH as a substrate. SAH is converted to SRH by MTAN and SRH is a precursor for synthesis of tetrahydrofurans (AI2 molecules). AI2 quorum-sensing molecules are involved in expression of the enzymes for biofilm formation, exotoxin synthesis and antibiotic resistance factors (11-14). A second group of quorum sensing molecules are the N-acylhomoserine lactones (AHL). AHLs are used by gram negative bacteria for intra-species communication and are collectively known as autoinducers-1 (AI1). They are synthesized from S-adenosylmethionine and acylated-acyl carrier protein by AHL synthase. MTA is a product of the AHL synthase reaction and is known to inhibit AHL synthase activity (15). Because of these roles, inhibition of MTAN may have anti-pathogenicity properties through disruption of both AI1 and AI2 quorum sensing pathways.
Figure 1.
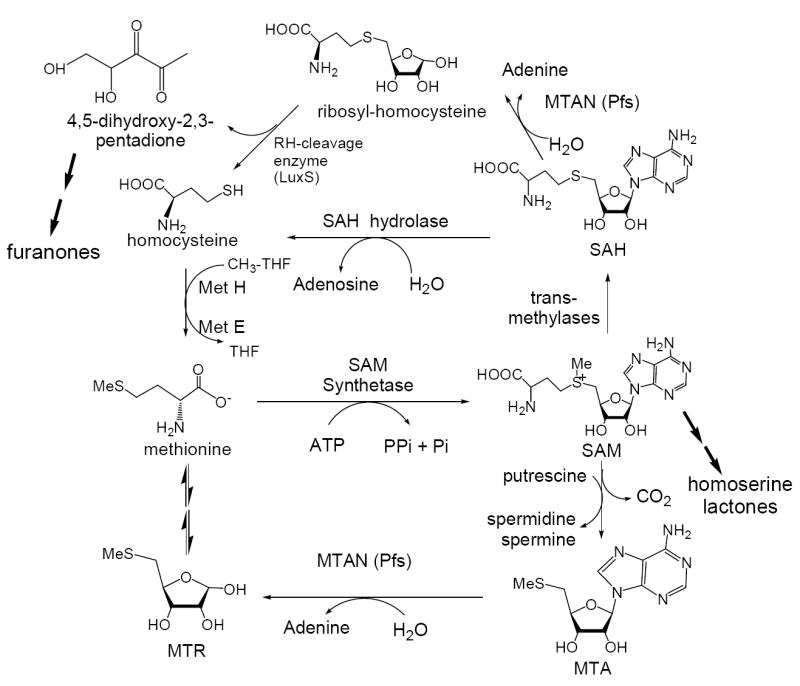
Pathways at the intersection of polyamine synthesis, adenine and methionine salvage and furanone synthesis in bacteria. The furanones are precursors for the synthesis of autoinducers-2, signaling molecules for quorum sensing. MTAN is encoded by the pfs gene in bacteria and catalyzes two reactions in this cycle. This diagram is derived from the summary in reference 11.
MTAN is also involved in the polyamine biosynthetic pathway via adenine and methionine salvage. MTA is a product of both spermidine and spermine synthases and these enzymes are sensitive to MTA accumulation (16, 17). Adenine is recycled to the adenine nucleotide pool by adenine phosphoribosyltransferase (18) and 5-methylthio-d-ribose is converted into methionine (19). Disruption of MTAN catalytic activity is therefore expected to affect adenine and methionine salvage, polyamine biosynthesis and quorum sensing.
Transition states of nucleoside and nucleotide N-ribosyl hydrolases involve formation of ribosyl oxacarbenium ions, where the N-ribosidic bond is cleaved to variable extents without significant bond order to the attacking water nucleophile (20-22). The oxacarbenium ion transition state structures of N-ribosyl transferases can be grouped into early transition states that have significant bond order to the leaving group and late transition states with little or no bond order to the leaving group. Kinetic isotope effect measurements for S. pneumoniae MTAN support a dissociated ribooxacarbenium ion2 transition state related to those recently established for E. coli MTAN and for human and malarial purine nucleoside phosphorylases (Figure 2; 23,24). The Immucillins are transition state analogue inhibitors which mimic the properties of early N-ribosyl transferase transition states found in bovine purine nucleoside phosphorylase (25) and IU-nucleoside hydrolase (20) while the second-generation DADMe-Immucillins resemble highly-dissociated N-ribosyltransferase transition states like those found for E. coli MTAN, human PNP and ricin A-chain (21, 23, 24).
Figure 2.
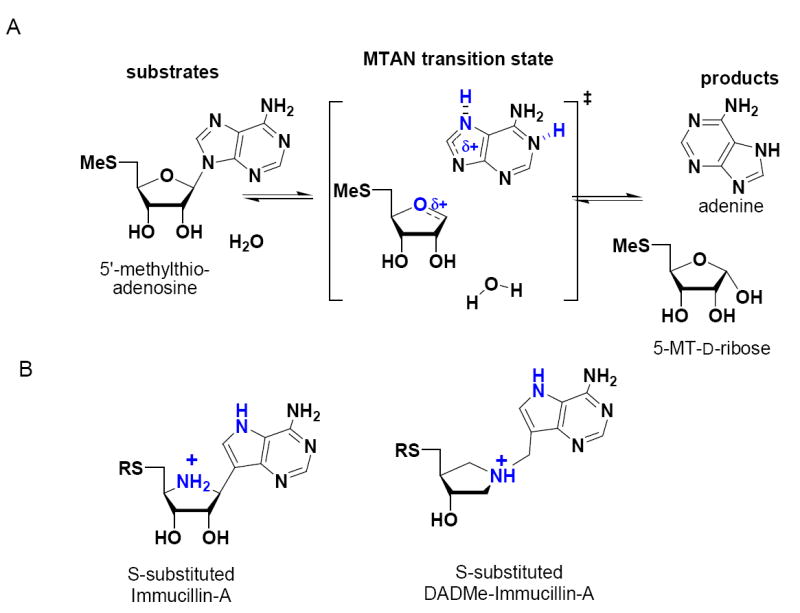
MTAN-catalyzed hydrolysis of MTA and the proposed transition state of the reaction (A). 5′-Thio substituted-Immucillin-A molecules mimic an early transition state where the bond between the ribosyl and adenine groups retains partial bond order and 5′-thio-substituted-DADMe-Immucillin-A molecules mimic a late transition state where the ribosyl cation is fully dissociated from the adenine leaving group, a distance of approximately 3 Å (B). In fully dissociated ribosyl transition states, the site of carbocation formation is at C1′, and the N1′ mimics this geometry. The methylene bridge positions the leaving group at the appropriate distance from the ribooxacarbenium ion site, 9-deazaadenine provides a carbon-carbon bridge for chemical stability and to increase the pKa at N1, N3 and N7. The transition state in (A) is shown to be H-bonded (partially protonated) at N1 and N7 suggested by structure (Figure 7) and KIE studies2.
Transition state analogue inhibitors bind to their enzymes tighter than the substrate by a factor related to the catalytic rate of acceleration imposed by the enzyme (26-28). Therefore the binding potential of transition state analogue inhibitors is related to catalytic potential. Transition state analogues were recently reported for E. coli MTAN with binding affinity to 47 fM (29). The MTAN from S. pneumoniae characterized here has a lower kcat and greater Km for MTA than E. coli MTAN for a kcat/km decrease of 845-fold (30). Therefore, the same transition state analogues would be expected to bind 845-fold less tightly to S. pneumoniae MTAN than to E. coli MTAN. Surprisingly, both partially and fully-dissociated SN1 transition state analogue mimics (29, 31, 32) of MTA bind 103 to 104 less well to the S. pneumoniae enzyme (29). Differences in transition state efficiency account for most of this binding energy, but weaker enzymatic interactions must account for remaining differences.
The crystal structure of MTAN from S. pneumoniae was solved with MT-ImmA bound to the catalytic sites and is compared to previous crystal structures of the E. coli enzyme. It is remarkable that MTAN isozymes with 40% sequence identity and near-identity at the catalytic sites reveal up to 13,000-fold differences in binding transition state analogues. Crystal structures, catalytic reactivity and inhibitor binding indicate that transition state stabilization is primarily responsible for transition state binding differences between E. coli and S. pneumoniae MTAN isozymes, but that these differences must lie in features of protein structure or dynamics beyond the highly similar catalytic sites.
Material and Methods
Streptococcus pneumoniae Methylthioadenosine Nucleosidase
Purification of S. pneumoniae chromosomal DNA was performed as previously described (33). The pfs gene, encoding 5′-methylthioadenosine nucleosidase, was obtained by PCR amplification of the gene from genomic DNA of Streptococcus pneumoniae using Pfu DNA polymerase and the primers (5′-ATTCCATATGAAAATAGGAATTATTGCTGC-3; 5′-CCCTCGAGTCAATGATGATGATGATGATGATCTAAAGCCTTCAAAAAGGC-3′) containing Nde I and Xho I restriction sites (underlined), respectively and a His6 affinity tag. The PCR product was cloned into the PCR-Blunt vector (Invitrogen) and transformed into TOP 10 competent cells. After purifying the plasmid DNA from TOP 10 cells, the pfs gene was cleaved with Nde I and Xho I restriction enzymes from the recombinant PCR-Blunt plasmid and ligated into an Nde I- and Xho I-digested pET23a(+) plasmid (Novagen).
The plasmid containing the pfs gene was expressed in the E. coli strain BL21 (DE3). The BL21 (DE3) cells containing pET23a(+):pfs or mutant genes were grown at 37°C to an A600 of 0.5 in LB medium containing 50 μg/mL carbenicillin. Cultures were induced with 0.5 mM IPTG and growth was continued for an additional 4 hours at 37°C.
Purification and Analysis of the Recombinant Methylthioadenosine Nucleosidase
Cells (~10 grams) were suspended in 60 mL of 40 mM TEA-HCl, pH 7.8, containing protease inhibitors (Boehringer Mannheim) and 200 μg/mL of lysozyme, 5 mM MgCl2, and 100 μg/mL of DNase I. After sonication, cells debris was removed by centrifugation for 45 min at 11 000 rpm at 4°C. The supernatant was dialyzed against 20 mM TEA-HCl containing 300 mM NaCl, pH 7.8, for 2 hours at 4°C. After centrifugation, the supernatant was applied to a 50 mL Ni2+-NTA His-Bind affinity column (Novagen), and proteins were eluted with a linear gradient (25 to 250 mM) of imidazole at 1 mL/min. Active fractions were pooled, concentrated to 4 mL, and loaded onto a 1.6 × 70 cm Superdex 200 gel filtration column. The enzyme was eluted with 20 mM TEA-HCl containing 300 mM NaCl, pH 7.8, at 0.5 mL/min and the active fractions were pooled and concentrated.
Protein concentrations were measured using a Bio-Rad protein assay kit with bovine serum albumin as a standard. The purity of MTAN was determined by SDS-polyacrylamide gel electrophoresis according to the method of Laemmli (34).
Crystallization of MTAN•MT-ImmA
Recombinant S. pneumoniae MTAN was co-crystallized with a 1:1.5 molar ratio of MT-ImmA (31) by hanging-drop vapor diffusion at 18 °C. MTAN protein (2 μL of 7.8 mg/ml) containing MT-ImmA was mixed with 2.0 μL of the reservoir solution containing 100 mM Na acetate, pH 4.6, 20 mM of CaCl2 and 30% 2-methyl-2,4-pentanediol (Hampton), and equilibrated against 1.0 mL of the reservoir solution. Plate-shaped crystals appeared in approximately 45 days and grew to maximum size of 0.1 × 0.1 × 0.05 mm3. Diffraction from the crystals was consistent with the space group P21 (a = 80.6 Å, b = 139.0 Å, c = 84.8 Å, β = 117.9 °). Assuming there are six molecules in the asymmetric unit, the Matthews coefficient Vm = 2.83 Å3/Da with 56 % solvent content.
Data collection and processing
An MTAN crystal was flash cooled in a N2 stream at −178 °C and X-ray diffraction data were collected at a wavelength of 0.98 Å on a Mar165 CCD detector using synchrotron radiation at beamline X9A at the National Synchrotron Light Source (Brookhaven National Laboratory). Data were reduced with the HKL package (35) and were 88.8% complete to 1.60 Å resolution with Rsym of 4.8%.
Structure determination and refinement
Based on the analysis of solvent content, 4-10 molecules are possible in the asymmetric unit of S. pneumoniae MTAN crystals. Six molecules in the asymmetric unit seemed likely, considering that these crystals demonstrate medium diffraction strength and the related E. coli MTAN forms tight dimers in its crystal structure (36). The structure of S. pneumoniae MTAN•MT-ImmA was solved by the molecular replacement method using the E. coli MTAN dimer as a search model. Two pairs of dimers were located using the program AmoRe, and these dimer pairs share the same orientation and are related by a translation (37). Further analysis by the AmoRe routine failed to yield any additional molecules. Analysis of the packing in the crystal lattice revealed that the MTAN molecules pack tightly in layers that are approximately 20 Å apart and a third pair of dimer was expected to sit between these layers. The third pair of dimers was located using program the EPMR by fixing the two pairs of known dimers as a partial structure (38). Rigid-body refinement yielded an initial Rcryst and Rfree of 33.9% and 35.8% using 8.0 - 4.0 Å data. The structure of MTAN•MT-ImmA was refined with the CNS program (39) and manual intervention using program O (40). The final model includes residues 1-230, one MT-ImmA molecule for each monomer and a total of 592 water molecules. The structure gave Rcrys and Rfree values of 19.3% and 21.2%, respectively. The model displays excellent stereochemistry as determined by PROCHECK (41), with 92.7% of the residues in the most favored region, 7.1% in the additionally allowed region, 0.2% in the generously allowed region, and none in the disallowed region of Ramachandran plot. Data collection and refinement statistics are shown in Table 1.
Table 1.
Data collection and refinement statistics.
| Data Collection | |
| Space group | P21 |
| Unit cell: a (Å) | 80.571 |
| b (Å) | 138.958 |
| c (Å) | 84.755 |
| β (°) | 117.921 |
| Resolution limits (Å) | 20-1.60 (1.66-1.60)* |
| Completeness (%) | 88.8 (78.2) |
| Rsym (%) | 4.8 (25.3) |
| Number of reflections | |
| Unique | 193,068 |
| Total | 809,358 |
| Structure Refinement | |
| Rcryts (%) | 19.3 |
| Rfree (%) | 21.1 |
| Number of amino acids | 1380 |
| Number of waters | 592 |
| Number of ligands | 6 MT-ImmA |
| RMS deviations, bond (Å) | 0.005 |
| RMS deviations, angle (deg.) | 1.2 |
Values in parentheses are for the highest resolution shell
Synthesis of Immucillins and DADMe-Immucillins
Synthesis of the MT-Immucillin and MT-DADMe-Immucillin inhibitors and their analogues was accomplished by methods described previously (42-45).
Determination of inhibition constants
The activity of S. pneumoniae MTAN was measured by monitoring the formation of 2,8-dihydroxyadenine at 293 nm in a coupled reaction where adenine is oxidized by xanthine oxidase. Under the assay conditions, the Δε for conversion of MTA to 2,8-dihydroxyadenine is 15.2 mM-1 cm-1 in 0.1 M Hepes buffer pH 7.0. The concentrations of most inhibitors were determined by ultraviolet absorption using the extinction coefficient of 9-deazaadenine of 8.5 mM-1 cm-1 at 275 nm, pH 7.0. Solutions of p-Cl-PhT-DADMe-Indole, MT-DADMe-3-deaza-ImmA, BnT-Pz-DADMe-ImmA and MT-Pz-ImmA were made from sample weight. Concentration of MT-ImmH was determined with the extinction coefficient of 9-deazahypoxanthine of 9.54 mM-1 cm-1 at 261 nm (46). The reactions for measuring the inhibition constants for Immucillins and DADMe-Immucillins were initiated by adding the enzyme (1-5 nM) to the reaction mixture, typically containing 0.3 to 3 mM MTA, and 100 mM Hepes pH 7.5, 50 mM KCl, 0.5 unit/mL xanthine oxidase and varying inhibitor concentrations in 1 mL reaction volume at 25 °C. Controls having no inhibitor and no enzyme were included in all experiments. The Ki values for inhibitors were obtained by fitting the initial rate and inhibitor concentration to the expression for competitive inhibition. This expression is valid only under the condition where the inhibitor concentration is >10 times the enzyme concentration. However when the inhibitor concentration was <10 fold greater than the enzyme concentration, the effective inhibitor concentration was obtained by the expression; , where I’ is the effective inhibitor concentration, and v0 are the initial rate in the presence and absence of inhibitor, and Et is the total enzyme concentration. Certain transition state analogue inhibitors displayed a second linear reaction rate following slow-onset tight-binding of the complex, indicating that a second thermodynamic equilibrium had been achieved. The final equilibrium dissociation constant Ki* was obtained by fitting the rates to the equation for competitive inhibition for the final reaction rates (46).
RESULTS AND DISCUSSION
Kinetic characterization of S. pneumoniae MTAN
MTAN from S. pneumoniae has dual substrate specificity and hydrolyzes the N-glycosidic bond of 5′-methylthioadenosine (MTA) and S-adenosylhomocysteine (SAH). The kinetic parameters (Km and kcat) for S. pneumoniae MTAN with MTA as substrate are 23 ± 9 μM and 0.25 ± 0.04 s-1, respectively (Table 2). For SAH the Km and kcat values are 13 ± 4 μM and 0.37 ± 0.05 s-1, respectively. The catalytic efficiency of S. pneumoniae MTAN calculated from the kcat/Km ratio was 1.1 ± 0.7 × 104 M-1 s-1 for MTA and 2.8 ± 0.2 × 104 M-1 s-1 for SAH. These values are lower than the catalytic efficiency of E. coli MTAN which has kcat/Km ratio of 9.3 × 106 M-1 s-1 for MTA and 1.6 × 106 M-1 s-1 for SAH (eg. 29). Thus, S. pneumoniae MTAN is less efficient by a factor of 845 relative to E. coli MTAN for MTA. For SAH, E. coli MTAN is 57-fold more efficient that the S. pneumoniae MTAN, and this is reflected in the action of the transition state inhibitor profile (see below). The S. pneumoniae MTAN is specific for the 5’-thio substituents and no kinetic activity was observed with adenosine up to 1 mM. The adenine base is also required as 5′-methylthioinosine did not show significant activity at concentrations to 1 mM.
Table 2.
Kinetic constants for S. pneumoniae and E. coli MTANs.
|
S. pneumoniae MTAN
|
E. coli MTAN
|
|||||
|---|---|---|---|---|---|---|
| Km(μM) | kcat(S-1) | kcat/Km(M-1 S-1) | Km(μM) | kcat(S-1) | kcat/Km(M-1 S-1) | |
| MTA | 23 ± 9 | 0.25 ± 0.04 | 1.1 ± 0.7 × 104 | 0.43 ± 0.2 | 4.0 ± 0.1 | 9.3 ± 3.1 × 106 |
| SAH | 13 ± 4 | 0.37 ± 0.05 | 2.8 ± 0.2 × 104 | 1.3 ± 0.2 | 2.1 ± 0.1 | 1.6 ± 0.8 × 106 |
| a Adenosine | NA | NA | NA | NA | NA | NA |
| b Inosine | NA | NA | NA | NA | NA | NA |
No activity was observed with adenosine and inosine up to 1mM concentration.
Inhibitor Analysis of S. pneumoniae MTAN
Transition state analysis of S. pneumoniae MTAN indicates a fully dissociated SN1 transition state2, similar to that of E. coli MTAN (23). Dissociative (DN*AN) transition states are characterized by a ribooxacarbenium ion with insignificant covalent participation of either the nucleophile or the leaving group (23). Loss of the N-ribosidic bond creates cationic character at the anomeric carbon to induce a partial double bond character at the O4′-C1′ bond and creates an sp2 center at the cationic anomeric carbon. The increased electron density in the leaving group results in an increased pKa at all proton-accepting sites including N1, N3 and N7, with N9 remaining inaccessible to protonation at the transition state because of the nearby C1’ cation, and the requirement that the transition state retains equal probability for return to reactant and for progression to products. The highest pKa in the adenine leaving group can be satisfied by protonation at N1, N3 or N7. Without protonation, the adenine leaving group will be anionic, and will be a poor leaving group compared to neutral adenine. Kinetic isotope effects suggest that adenine is not fully protonated at the transition state2 of the S. pneumoniae MTAN in contrast to N7 protonation found at the transition state for E. coli MTAN (23). Crystallographic comparison (see below) suggests H-bond donations to N1 and N7 of bound transition state analogues, and these contribute to electron withdrawal from the adenine leaving group. The poor leaving-group ability of a partially-anionic adenine is reflected in the poor catalytic efficiency of S. pneumoniae MTAN compared to the E. coli enzyme (Table 2).
Inhibition of S. pneumoniae MTAN by Immucillins
Immucillins mimic the features of early transition states in which significant bond order remains to the leaving group (Figure 1). Immucillins have a nitrogen replacing the ring oxygen. The nitrogen has a pKa of 6.9 and has been shown to be protonated in the active sites of human purine nucleoside phosphorylase and a purine phosphoribosyltransferase (47). The protonated nitrogen cation mimics the ribooxacarbenium ion character at the transition state. A second feature of early transition states provided by Immucillins is a H-bond donor rather than a H-bond acceptor at N7 to mimic the increased pKa of N7 at the transition state. The altered bond conjugation in 9-deazaadenine increases the pKa of N7 to > 10 (29), thus the 9-deazaadenine ring resembles a hydrogen-bonded N7 at the transition state. Immucillins designed for E. coli MTAN also incorporated 5′-alkyl or arylthio groups to resemble the 5′-methylthio or 5′-homocystenyl groups of MTA and SAH (29).
MT-ImmA [11] is the parent compound of the Immucillin series. It binds to S. pneumoniae MTAN with a dissociation constant of 1.0 μM to give a Km/Ki of 23 (Figure 3). The crystal structure of S. pneumoniae MTAN (see below) with MT-ImmA [11] suggests that the methylthio binds in a hydrophobic pocket that could adjust to accommodate other hydrophobic groups, including homocysteine. Longer alkyl groups at the 5′-position increased the binding affinity of Immucillins. Thus, EtT-ImmA [1] has a Ki of 40 nM, binding 25 times more tightly than MT-ImmA. With Km/Ki of 575, EtT-ImmA is the tightest binding inhibitor in the Immucillins series. Incorporation of a polar group as in 2-F-EtT-ImmA [10] increased the Ki of EtT-ImmA [1] from 40 nM to 394 nM. Similarly, incorporation of a hydroxyl group altered the Ki for EtOHT-ImmA [13] to 1.8 μM. The crystal structure of S. pneumoniae MTAN (see below) indicates that aromatic residues including Phe207, Tyr107 and Phe105 surround the methylthio group of MT-ImmA [11]. The PhT-ImmA [8] contains a planar hydrophobic 5′-group and binds with a Ki of 335 nM, a three fold improvement in the binding compared to MT-ImmA. Increasing the size of the aromatic group from phenyl to napthyl marginally improved binding affinity to give a Ki*of 220 nM for NapT-ImmA [7], and this was the only inhibitor of the Immucillin series to show slow-onset inhibition. Incorporating a methyl bridge as in BnT-ImmA [6] improved the dissociation constant to 206 nM. Similar enhancements in affinity were observed in p-Cl-PhT-ImmA [5] and m-Cl-PhT-ImmA [4] with Ki values of 193 nM and 100 nM, respectively. Increasing the hydrophobicity of the aromatic group with toluyl groups improved the affinity with p-TolT-ImmA [2] and m-TolT-ImmA [3] giving Ki values of 60 nM and 77 nM, respectively. The Km/Ki factor for p-TolT-ImmA is 380. The ability of MTAN to accept relatively large substituents in place of the 5′-methylthio group reflects its dual substrate specificity for MTA [19] and SAH [20].
Figure 3.
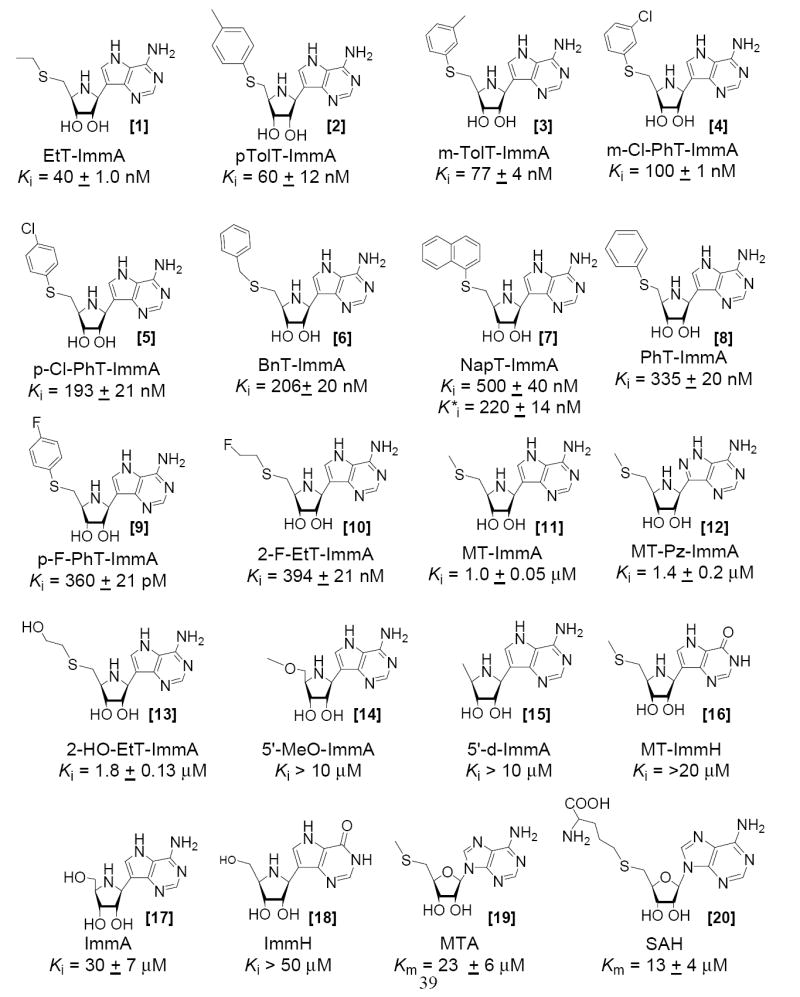
Inhibition constants for MTAN with the 5′-thio-substituted Immucillins are compared to the Km value for MTA [19] and SAH [20].
Inhibition of S. pneumoniae MTAN by DADMe-Immucillins
DADMe-Immucillins mimic the dissociative SN1 transition states of N-ribosyltransferases (21, 23, 24). Placing the imino nitrogen at the 1’-position in the DADMe-Immucillins locates the carbocation charge in the position to mimic the C1’ carbocation of the transition state. The pKa value of the 1′-pyrrolidine nitrogen is 9.2 (48) compared to a pKa of ~ 6.5 for N4′ in the iminoribitols. At neutral pH, N1’ is >99% protonated in DADMe-Immucillins. The fully-dissociated geometry at the MTAN transition state is well matched with a methylene bridge between the 1′-pyrrolidine nitrogen and the 9-deazaadenine. The 2.5 Å between these groups is similar to the 3 Å distance found in the transition state of E. coli MTAN. The 9-deazaadenine group provides chemical stability and maintains an elevated pKa at N7. MT-DADMe-ImmA [34] is the parent compound of the DADMe-Immucillin series and binds with an inhibition constant of 24 nM, a 40-fold improvement compared to MT-ImmA [11] (Figure 4). DADMe-Immucillins were also modified by incorporating hydrophobic groups to replace the 5′-methyl group and many of the 5′-hydrophobic DADMe-ImmA analogues are slow-onset tight binding inhibitors (Figure 5). The binding affinity of DADMe-Immucillins increased with the hydrophobic nature of the 5′-substituent. EtT-DADMe-ImmA [32] and iPrT-DADMe-ImmA [33] gave Ki values of 10 nM, PrT-DADMe-ImmA [31] has a Ki* of 4.0 nM and BuT-DADMe-ImmA [26] gave a Ki* of 2.0 nM. BuT-DADMe-ImmA [26] is the tightest binding inhibitor of the 5′-alkylthio analogues with a Km/Ki* of 11,500. Incorporation of aromatic groups improved the binding affinity, with PhT-DADMe-ImmA [27], BnT-DADMe-ImmA [28] and p-F-PhT-DADMe-ImmA [30] bind with Ki* values of 2, 2.4 and 3.5 nM, respectively, an order of magnitude improvement relative to MT-DADMe-ImmA. An additional increase in the binding occurred with p-Cl-PhT-DADMe-ImmA [21] to give a Ki* of 360 pM, ~6 fold tighter than PhT-DADMe-ImmA [27]. With a Km/Ki* value of 63,890, it is the tightest binding transition state analogue for S. pneumoniae MTAN. m-Cl-PhT-DADMe-ImmA [29] bound less well and was similar to PhT-DADMe-ImmA [27].
Figure 4.
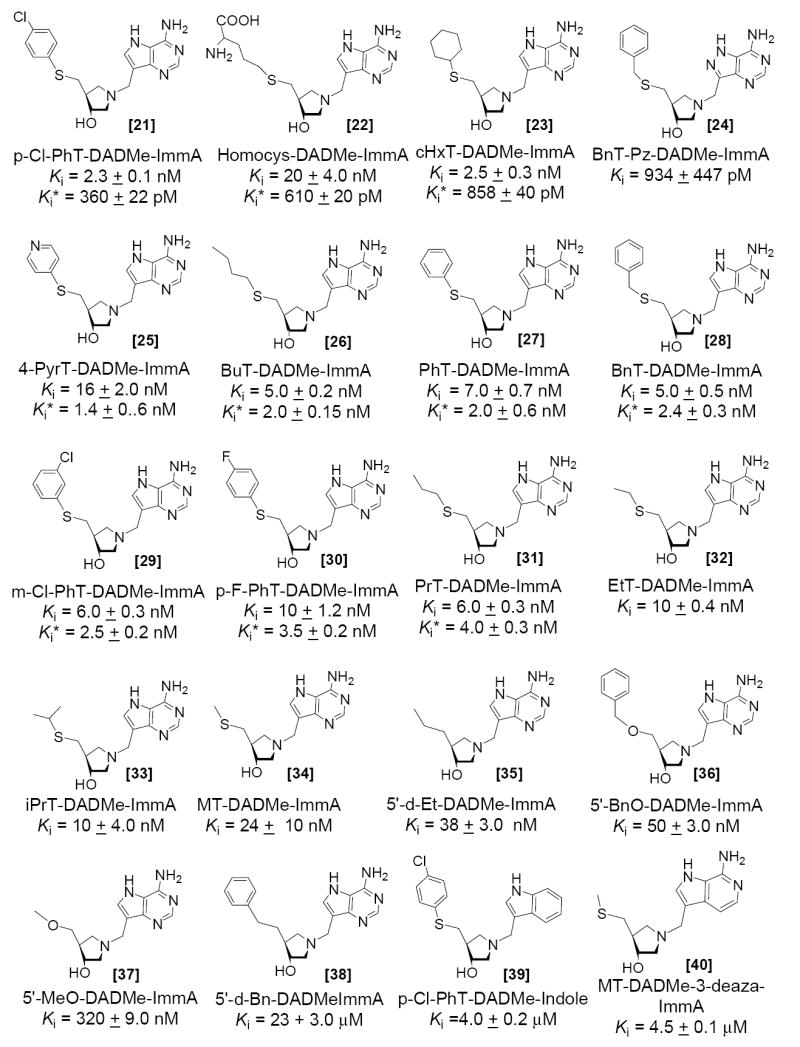
Inhibition constants for MTAN with the 5′-thio-substituted DADMe-Immucillins.
Figure 5.
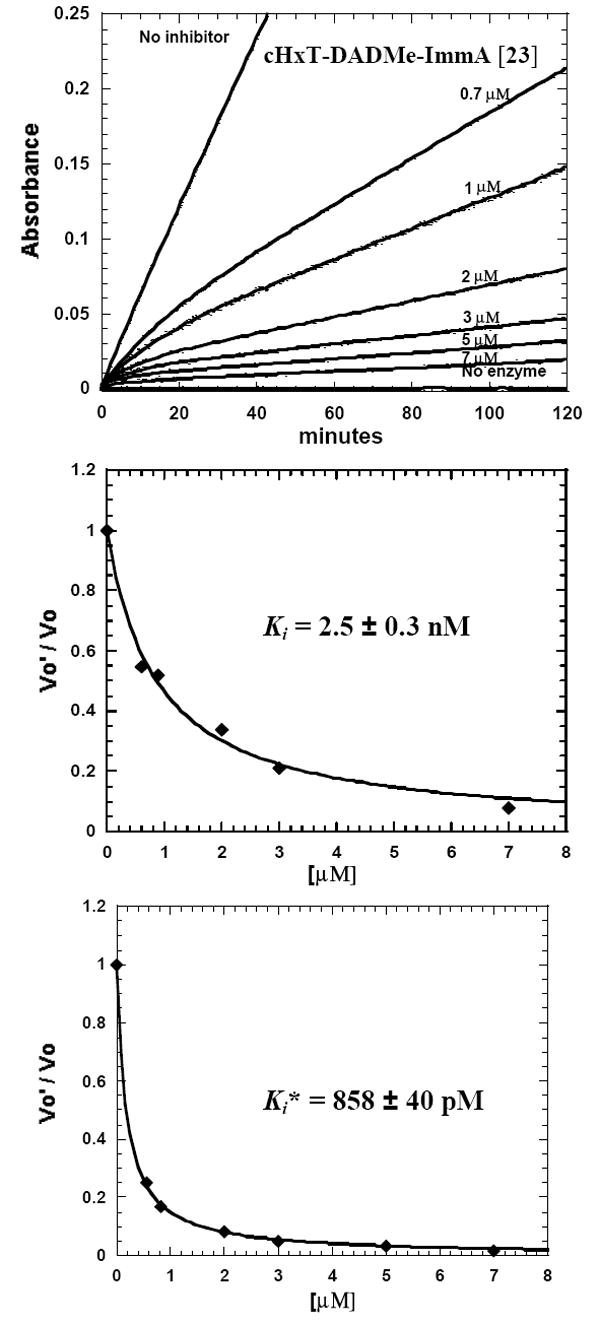
Inhibition of S. pneumoniae MTAN by cHexyl-DADMe-ImmA [23], an example of data analysis for a slow-onset, tight-binding DADMe-Immucillin derivative. The reaction rate and slow-onset inhibition was monitored by the conversion of MTA [17] to 2,8-dihydroxyadenine at 293 nm in a coupled reaction with xanthine oxidase (Upper panel). The coupled assay permits use of high substrate concentration (2.0 mM, 87 × Km for MTA) to compete against these powerful inhibitors. Control experiments demonstrate that Immucillins do not inhibit xanthine oxidase. Values of Ki (middle panel) and Ki* (lower panel) were obtained from the initial (0 to 5 min) and the final rates (25 to 30 min). Similar experiments were used to measure inhibition by slow-onset tight-binding inhibitors shown in Figure 3 and 4.
S. pneumoniae MTAN has dual substrate specificity for MTA [19] and SAH [20] and gives a Km for SAH of 13 μM, almost 2 fold lower than for MTA. This difference is also seen in the transition state analogues, where homocysteinyl-DADMe-ImmA [22] is a slow onset inhibitor with a Ki* of 610 pM, 18-fold increased affinity relative to MT-DADMe-ImmA [34]. Adenosine and methylthioadenosine are not substrates, and accordingly, Immucillin-A [17] is a poor inhibitor with a Ki of 30 μM and MT-Immucillin-H [16] or Immucillin-H [18] are not inhibitors at micromolar concentrations.
The thermodynamic benefit of the DADMe inhibitor geometry can be evaluated by comparing the inhibition constants of Immucillins and DADMe-Immucillins with same 5′-thio substituents. p-Cl-PhT-ImmA [5] and p-Cl-PhT-DADMe-ImmA [21] gave dissociation constants of 193 nM and 360 pM, a factor of 536 tighter for the DADMe-Immucillin inhibitor. Similar values were observed in comparing p-F-PhT-ImmA [9] and p-F-PhT-DADMe-ImmA [30] where the DADMe-Immucillin binds 102 fold more tightly. Likewise, PhT-DADMe-ImmA [27] binds 167 fold more tightly than PhT-ImmA [8]. Finally, the BnT- group achieved an extra 100 fold affinity from being in the DADMe context. The methylene bridge and DADMe features improve binding affinity by 2 to 3 kcal/mol, supportive of a fully dissociated transition state for S. pneumoniae MTAN.
Substitutions in the 9-deazaadenine ring
Residues Asp197 and Ser97 in the active site of S. pneumoniae MTAN (described below) form a network of hydrogen bonds with each other and with N6 and N7 of MT-ImmA. Changes in the conjugation pattern of the 9-deazaadenine ring or the substitution of N6 and N7 may influence binding affinity of Immucillins and DADMe-Immucillins. Replacing the 9-deazaadenine ring of p-Cl-PhT-DADMe-ImmA (360 pM inhibitor) [21] with an indole ring to give p-Cl-PhT-DADMe-Indole [39] reduced the binding affinity to 4.0 μM, 11,000-fold weaker binding than p-Cl-PhT-DADMe-ImmA [21]. The p-Cl-phenylthio group is not sufficient for tight binding and requires cooperative interactions from the 9-deazaadenine group. MT-ImmH [16] alters only the 6-amino substituent relative to MT-ImmA and does not inhibit at 50 μM. A N3 to C3 substitution in MT-DADMe-3-deaza-ImmA [40] causes a 63-fold reduction in binding affinity compared to MT-DADMe-ImmA [34]. There are no enzymatic contacts to N3 in the crystal structure (see below) thus reduction in binding affinity may be attributed to altered pKa values and decreased H-bond interactions at N1, N6 and N7.
8-aza Immucillins and DADMe-Immucillins
The crystal structure of S. pneumoniae MTAN shows Ser97 near C8 of MT-ImmA, similar to Ser196 of E. coli MTAN in its interaction with N8 of formycin-A (51). Bn-DADMe-Pz-ImmA [24] binds 2.4-fold better than Bn-DADMe-ImmA [28], supporting a similar interaction in S. pneumoniae MTAN. This difference is significant in design of specificity for bacterial MTAN since inhibitors of MTAN also inhibit human MTAP (31). Human MTAP lacks a hydrogen bonding partner at the 8-position in the leaving group and 8-aza substituted Immucillins and DADMe-Immucillins are poor inhibitors of human MTAP. The tighter binding of pyrazolo-Immucillins and DADMe-Immucillins can be exploited for the design of bacteria-specific inhibitors.
Thioether substitutions
The methylthio binding pocket of S. pneumoniae MTAN is hydrophobic (Met9, Ile50, Phe207, Phe105, Ala113, Val102, and Tyr107; see below) and sulfur ethers interact more favorably with hydrophobic regions than carbon or oxygen substituents. BnT-DADMe-ImmA [28] with a Ki* of 2.4 nM binds 20-fold more tightly than 5′-BnO-DADMe-ImmA [36] and approximately 10,000 times more tightly than 5′-d-Bn-DADMe-ImmA [38], binding with a Ki of 23 μM (Figure 7). Similar patterns were observed for MT-DADMe-ImmA [34], 5′-d-Et-DADMe-ImmA [35] and MeO-DADMe-ImmA [36] with Ki values of 24 nM, 38 nM and 50 nM respectively. Likewise, MT-ImmA [11] binds more favorably than 5′-MeO-ImmA [14] or 5′-d-ImmA [15].
Figure 7.
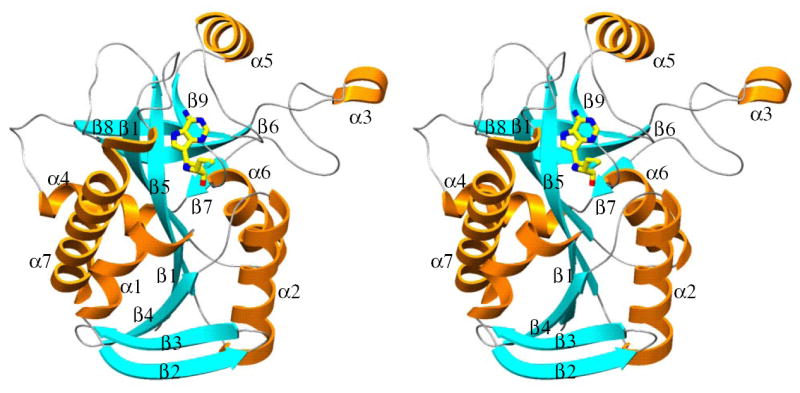
Stereo diagram of monomer of S. pneumoniae MTAN·MT-ImmA [11] is shown to indicate the position of active site and the position of α- and β- segments of the enzyme. Figures 6-9 are generated using Setor (53).
Overall structure of MTAN•MT-ImmA
The overall fold of S. pneumoniae MTAN subunits is similar to that reported earlier for E. coli MTAN as expected from 40% amino acid sequence identity (Figure 6; 49, 50)4. The monomer of S. pneumoniae MTAN is folded into a single domain structure containing ten β strands and six α helices (Figure 7). The core consists of a mixed 10-stranded β sheet (β1, 2-7; β2, 20-28; β3, 30-38; β4, 40-47; β5, 70-80; β6, 89-97; β7, 119-121; β8, 140-147; β9, 168-172; and β10, 189-197). The β sheet core is surrounded by seven α helices (α1, 9-18; α2, 52-66; α3, 103-106; α4, 123-132; α5, 155-164; α6, 175-184; and α7, 203-229). The dimer interface of S. pneumoniae MTAN is also similar to that of the E. coli MTAN, and buries 1700 Å2 surface area from each monomer. Contacts between the adjacent subunits involve the α3 and α2 from one monomer, with the loop connecting β8 to α5 and α2 from the adjacent monomer and contain a mixture of hydrogen bonds and hydrophobic interactions.
Figure 6.
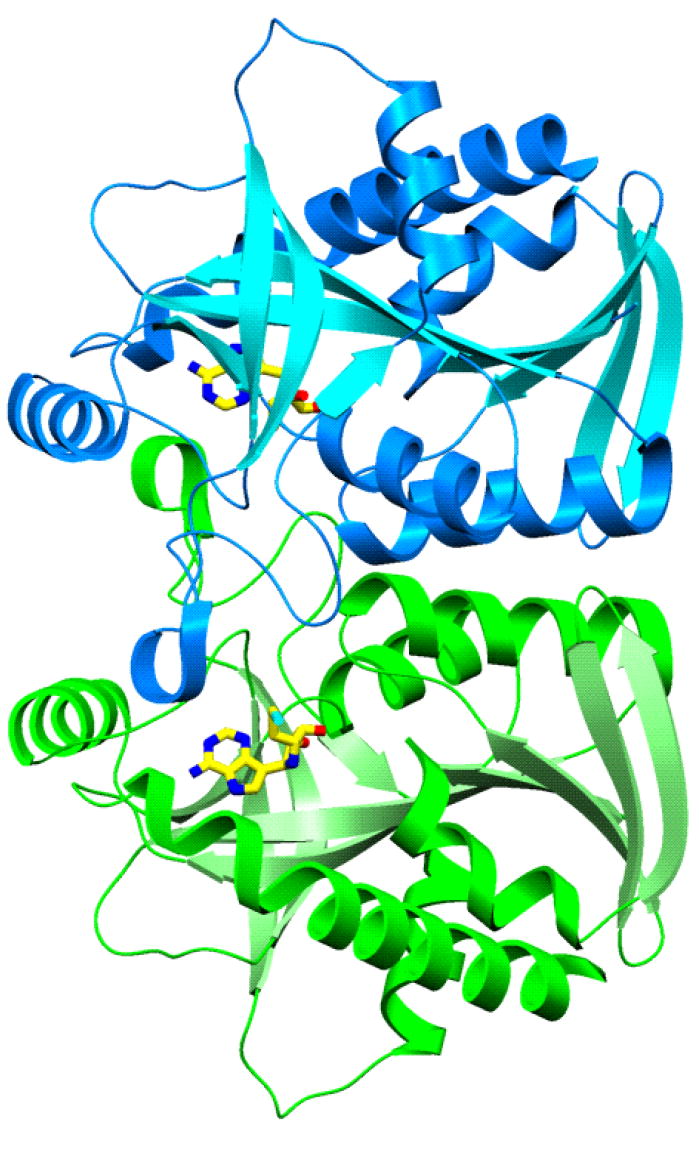
S. pneumoniae MTAN dimer.
Interactions with the transition state analogue MT-ImmA
The catalytic sites of S. pneumoniae MTAN are located near the subunit interfaces. The base and the sugar binding sites are composed of residues contributed entirely from a single monomer. The 5′-methylthio binding site, however, is located at the dimer interface and is formed by residues contributed from both monomers. The 9-deazaadenine base of MT-ImmA [11] is wedged between the backbones of β5 and β9, and the side chain of Phe151 (Figure 8). The side chain of Asp197 forms favorable hydrogen bonds with N6 and N7 of 9-deazaadenine ring (2.6 Å and 2.8 Å, respectively). The iminoribitol ring is anchored through favorable hydrogen bonds between the O2′ and O3′ hydroxyls and the side chain of Glu174 (both are 2.6 Å), and by the hydrogen bond between O2′ and the side chain of Arg193 (3.0 Å). Four ordered solvent molecules are located in the active sites of S. pneumoniae MTAN and two of these water molecules are in direct contact with MT-ImmA [11]. The N4′ of the iminoribitol ring is the mimic of the oxycarbenium transition state and forms a weak hydrogen bond with the side chain of Ser76 (3.2 Å). The hydrophobic pocket that accommodates the 5′-methylthio group of MT-ImmA [11] includes Met9, Ile50, Phe207 from the main monomer, and Phe105, Ala113, Val102, and Tyr107 from the adjacent monomer.
Figure 8.
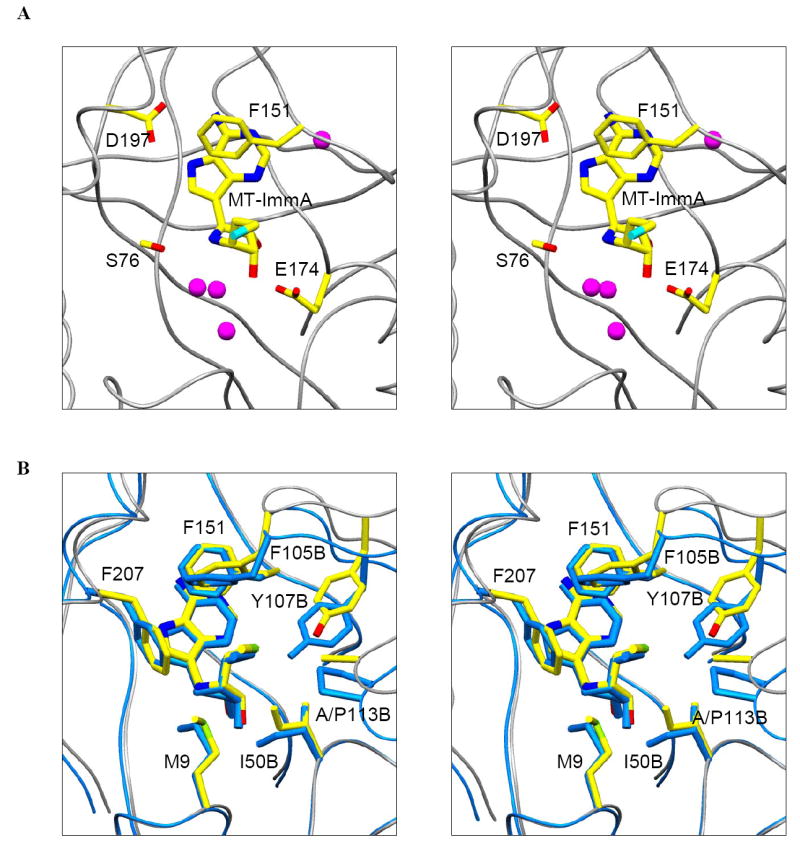
Stereo diagram of the active site of S. pneumoniae MTAN (A), and superposition of the methylthio binding sites of S. pneumoniae MTAN and E. coli MTAN (B).
Comparison of S. pneumoniae•MT-ImmA and E. coli MTAN•MT-ImmA complexes
The overall structures of S. pneumoniae MTAN•MT-ImmA and E. coli MTAN•MT-ImmA are similar with an RMS deviation of 1.647 Å for 229 Cα atoms. S. pneumoniae MTAN and E. coli MTAN share 41% sequence identity and the active site residues are almost completely conserved. Even though the active site structures are almost identical these two enzymes exhibit very different inhibition constants with Immucillins. For instance, MT-ImmA and phenylthio-ImmA bind to E. coli MTAN 104-105 times tighter than to S. pneumonia MTAN under the same assay condition. Although the active site residues which interacts with 9-deazaadenine and 4′-iminoribotol of MT-ImmA are largely conserved, the part of the catalytic site that interacts with 5′-methylthio group of MT-ImmA exhibits some significant differences. Residues Phe105 and Tyr107 from the adjacent monomer form part of the 5′-methylthio binding site. The side chains of the two aromatic residues are in position to interact with the phenyl group in phenylthio-ImmA. In the S. pneumonia MTAN•MT-ImmA structure, the loop which includes these two aromatic residues is approximately 2 Å further away from the methylthio group of MT-ImmA [11] compared to the loop position in the E. coli MTAN•MT-ImmA structure. In addition Ala113 and Val152 in S. pneumonia MTAN are Pro113 and Ile152 in E. coli MTAN, and these are the only residues in the active site that are not conserved in the two enzymes.
Comparison of binding affinities of Immucillins and DADMe-Immucillins for E. coli MTAN and S. pneumoniae MTAN
Wolfenden et. al. (27, 28) proposed that binding affinity of transition state analogues is related to catalytic potential of enzymes and it is proportional to catalytic acceleration imposed by enzyme relative to uncatalyzed reaction. A comparison of catalytic potential indicates that the E. coli MTAN (kcat = 4 s-1) is catalytically more efficient than S. pneumoniae MTAN (kcat = 0.25 s-1), and the catalytic efficiency (kcat/Km) differs by a factor of 845 for MTA as substrate. Therefore, MTA mimics of the transition states are expected to bind better to E. coli MTAN than to S. pneumoniae MTAN. MT-DADMe-Immucilln-A [34], binds with a Ki* of 2 pM to E. coli MTAN, a factor of 12,000 tighter, compared to the Ki* of 24 nM for S. pneumoniae MTAN. Also, p-Cl-PhT-DADMe-ImmA [21], the tightest binding inhibitor of both E. coli MTAN and S. pneumoniae MTAN, binds ~7,660 fold tighter to E. coli MTAN. Likewise, DADMe-Immucillins such as BuT-DADMe-ImmA [26], PhT-DADMe-ImmA [27] and PrT-DADMe-ImmA [31] bind tighter to E. coli MTAN by a factor of, 6,600, 1,000 and 6,900, respectively. Similar tighter binding for E. coli MTAN was observed with most DADMe-Immucillins and Immucillins suggesting an extra 3-4 kcals/mol stabilization of the transition state by E. coli MTAN. Although kcat/Km comparison only predicts an approximately 1000 fold increase in binding affinity for transition state analogue inhibitors to E. coli MTAN, an extra 5 to 12 fold tighter binding observed for E. coli MTAN is may be due to differences in the transition state structures of E. coli and S. pneumoniae MTAN or in kinetic factors. An alternative explanation is that the kcat for these two enzymes does not faithfully represent the chemical step. Thus, a non-chemical, rate-limiting step that differs by a factor of 5 to 12 between the enzymes can also account for this observation. Rate-limiting product release is one such example. The transition state of S. pneumoniae MTAN is more dissociative than E. coli MTAN. At the transition state the leaving group is 3.0 Å (23) from the oxacarbenium ion in the E. coli MTAN transition state and is > 3.5 Å in S. pneumoniae MTAN. DADMe-Immucillins with a N1-C9 bond distance of 2.5 Å are a good match to the observed C1′-N9 distance of the E. coli MTAN transition state, but this distance is shorter than the C1′-N9 distance observed in the transition state of S. pneumoniae MTAN. Therefore, DADMe-Immucillins are closer mimics of the E. coli MTAN transition state than to the transition state of S. pneumoniae MTAN, easily accounting for the 5 to 12 fold extra binding affinity than predicted for E. coli MTAN. In future inhibitor design programs, it may be possible to increase the binding affinity of DADMe-Immucillins for S. pneumoniae MTAN by increasing the distance between N1′ and C9.
The case with SAH as a substrate differs from that of MTA in terms of catalytic efficiency for the S. pneumoniae and E. coli MTANs with the E. coli MTAN being 57-fold more efficient in terms of kcat/Km. This relationship predicts that transition state analogues based on the SAH scaffold would prefer the E. coli enzyme by a smaller difference that those based on the MTA structure. Homocys-DADMe-ImmA [22] is a 610 pM inhibitor of S. pneumoniae MTAN and a 6 pM inhibitor of E. coli MTAN (29), in good agreement with the kcat/Km values for these enzymes and substrates. We are not aware of other examples that demonstrate this close relationship between kcat/Km and transition state analogue binding with closely related isozymes and different substrates. This result supports the conversion of catalytic potential into transition state analogue binding energy.
Structural rationale for tighter binding of DADMe-Immucillins
A comparison of crystal structures of E. coli and S. pneumoniae MTANs with MT-ImmA [11] revealed little structural difference (Figure 9). In addition, the residues in contact with bound MT-ImmA at the catalytic sites of E. coli and S. pneumoniae MTANs are completely conserved with the exception of the conservative Ile to Val substitution at amino acid 152 (Figure 9, 10). Therefore, transition state features are responsible for tighter binding of MT-ImmA [11] and MT-DADMe-ImmA [34] to E. coli MTAN than to S. pneumoniae MTAN. Two transition state features, namely the cationic character of the transition state and increased pKa of the adenine leaving group are responsible for tight binding of MT-ImmA [11] and MT-DADMe-ImmA [34] to E. coli MTAN (50). The 1′-pyrrolidine nitrogen of DADMe-Immucillins has a pKa of > 9.2, and is fully protonated at physiological pH whereas the 4′-iminoribitol group (pKa ~ 6.9) has been shown bind to nucleoside hydrolase as the neutral species (51) but to be protonated in the active sites of human PNP (47). In E. coli MTAN the tight binding of MT-ImmA is due to an ion-pair electrostatic interaction between the cationic 4′-iminoribitol group and the nucleophilic water molecule (50). It was proposed that in E. coli MTAN, the water nucleophile is activated by Glu12. An ion-pair interaction is proposed for the nucleophilic water hydroxyl ion and the protonated 4′-iminoribitol group to cause tight binding of MT-ImmA [11] (50). The crystal structure of S. pneumoniae MTAN also has a nucleophilic water molecule that interacts with MT-ImmA [11] suggesting a similar mechanism. However, energetic stabilization by ion pair interactions are tens of kcals/mol whereas MT-ImmA [11] only binds 23 fold tighter than MTA [19]. Therefore, the water nucleophile found between the 4′-iminoribitol of MT-ImmA [19] and S. pneumoniae Glu12 is not likely to be ionized to the hydroxyl. DADMe-Immucillins are more powerful inhibitors of MTANs than are the Immucillins. Transition state parameters more optimized in DADMe-Immucillins include the increased pKa of the 1′-pyrrolidine nitrogen and greater separation between the oxacarbenium ion and the leaving group. MT-DADMe-ImmA [34] is a 2 nM inhibitor of S. pneumoniae MTAN and binds 11,500 fold tighter than substrate. Although crystallographic information is not yet available for S. pneumoniae MTAN in complex with DADMe-Immucillin, the structure of MT-DADMe-ImmA [34] with E. coli MTAN suggests enhanced ion pair interaction between the hydroxyl nucleophile and the N1′ cation as well as favorable interactions with the leaving group (50). Considering the near identity of the active sites of E. coli and S. pneumoniae MTANs, a similar mechanism explains the tight binding of DADMe-Immucillins to S. pneumoniae MTAN, but the affinity is reduced because of the reduced catalytic efficiency of S. pneumoniae MTAN and the greater distance between C1’ and the leaving group in the S. pneumoniae MTAN transition state.
Figure 9.
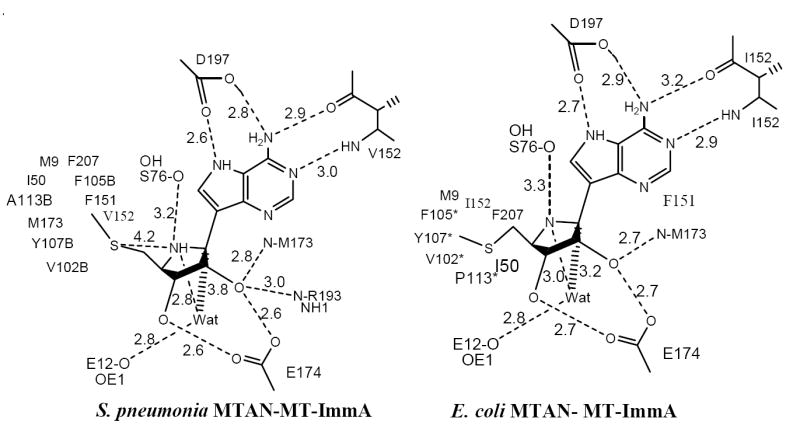
Catalytic site contacts for S. pneumoniae MTAN·MT-ImmA and E. coli MTAN ·MT-ImmA complexes. Distances are in angstroms. Data for E. coli MTAN·MT-ImmA is from (50).
Figure 10.
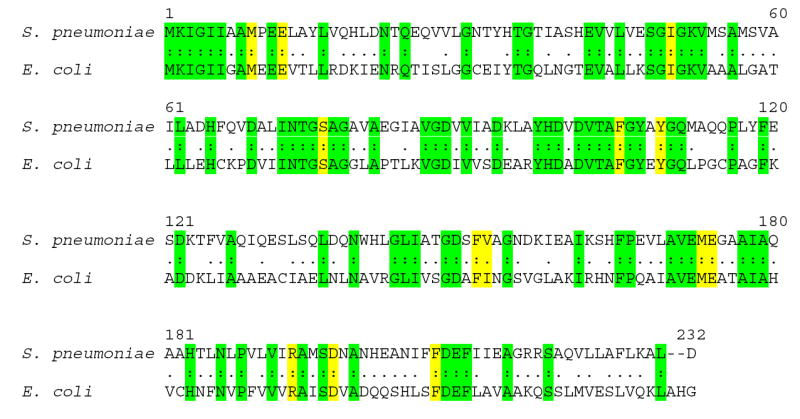
Alignment of the amino acid sequences of Streptococcus pneumoniae (AAK99698) and Escherichia coli (NP_414701) methylthioadenosine S-adenosylhomocysteine nucleosidases. Residues in contact with MT-ImmA at the catalytic site of S. pneumoniae MTAN are shown in yellow and other regions of amino acid identity are shown in green. There is 40.9% identity and 75.4% similarity between the S. pneumoniae and E. coli enzymes.
MTAP as a quorum sensing target
S. pneumoniae deleted in luxS cannot make AI2 molecules and shows diminished but not eliminated virulence in a mouse internasal infection model (9). Since MTAN is the only known reaction to generate ribosyl-homocysteine, the LuxS substrate, its inhibition is required to have the same result. In preliminary studies, we find that selected members of the DADMe-Immucillins block synthesis of quorum sensing molecules in cultured S. pneumoniae as detected by a Vibrio harveii bioassay.5
Summary and Conclusions
Transition state structures derived from the kinetic isotope effects and computational studies provide a molecular skeletal framework for the design of powerful transition state analogue inhibitors. Structural information from X-ray crystallography is useful to dissect the binding affinity interactions for transition state analogue inhibitors. Transition state analogues designed for MTANs based on the oxacarbenium ion transition state of other N-ribosyltransferases are also powerful inhibitors of S. pneumoniae MTAN. Binding affinity extends into the picomolar range for the DADMe-Immucillins, and suggests a highly dissociative transition state for S. pneumoniae MTAN, a conclusion also derived from kinetic isotope effect studies.2 p-Cl-PhT-DADMe-ImmA [21] with a Ki* of 360 pM is the tightest binding inhibitor of S. pneumoniae MTAN. Binding affinity of transition state inhibitors is related to the catalytic potential of the enzyme and this principle is clearly illustrated with the E. coli and S. pneumoniae MTANs. E. coli MTAN is 845 times more catalytically efficient than S. pneumoniae MTAN with MTA as substrate and both Immucillins and DADMe-Immucillins are generally 103 more powerful inhibitors of E. coli MTAN than of S. pneumoniae MTAN. With SAH as substrate, catalytic efficency is 57-fold greater for E. coli MTAN and the one transition state analogue of this group binds the E. coli enzyme 100-fold better than MTAN from S. pneumoniae.
The nearly-identical crystal structures of S. pneumoniae and E. coli MTANs with MT-ImmA [11] bound at the catalytic sites suggests that the catalytic efficiency of the MTAN catalytic sites is related to electrostatic or structural elements beyond the primary amino acid contacts to bound reactants. One possibility is that protein changes throughout the protein may contribute to the dynamic contributions to transition state formation (52).
Acknowledgments
This work was supported by NIH Research Grant GM41916. The authors recognize the chemical synthetic expertise provided by Drs. Gavin Painter, Dirk Lenz and Simon Mee of Industrial Research Ltd., New Zealand.
Footnotes
Abbreviations and Footnotes: Abbreviations: MTA, 5′-methylthioadenosine; SAH, S-adenosylhomocysteine; MTR, methylthioribose; MTAN, 5’-methylthioadenosine/S-adenosylhomocysteine nucleosidase; ImmA, Immucillin-A, (1S)-1-(9-deazaadenin-9-yl)-1,4-dideoxy-1,4-imino-D-ribitol; MT-ImmA, MT-Immucillin-A, (1S)-1-(9-deazaadenin-9-yl)-1,4-dideoxy-1,4-imino-5-methylthio-d-ribitol; p-Cl-PhT-ImmA, (1S)-5-(4-chlorophenylthio)-1-(9-deazaadenin-9-yl)-1,4-dideoxy-1,4-imino-d-ribitol; p-TolT-ImmA, (1S)-1-(9-deazaadenin-9-yl)-1,4-dideoxy-1,4-imino-5-(4-methylphenylthio)-d-ribitol; m-TolT-ImmA, (1S)-1-(9-deazaadenin-9-yl)-1,4-dideoxy-1,4-imino-5-(3-methylphenylthio)-D-ribitol; MT-DADMe-ImmA, 5′-methylthio-DADMe-Immucillin-A, (3R,4S)-1-[(9-deazaadenin-9-yl)methyl]-3-hydroxy-4-(methylthiomethyl)pyrrolidine; BnT-DADMe-ImmA, (3R,4S)-4-(benzylthiomethyl)-1-[(9-deazaadenin-9-yl)methyl]-3-hydroxypyrrolidine; p-Cl-PhT-DADMe-ImmA, (3R,4S)-4-(4-chlorophenylthiomethyl)-1-[(9-deazaadenin-9-yl)methyl]-3-hydroxypyrrolidine; BnT-DADMe-8-azaImmA, (3R,4S)-1-[(8-aza-9-deazaadenin-9-yl)methyl]-4-(benzylthiomethyl)-3-hydroxy-pyrrolidine. The IUPAC names of other inhibitors are readily derived from the examples given above. Ki*, dissociation constant for the equilibrium complex of enzyme-inhibitor following slow-onset inhibition; Ki, dissociation constant for the equilibrium between enzyme and inhibitor before slow-onset inhibition.
Singh, V. and Schramm, V. L., unpublished observations.
Immucillin-H is (1S)-1-(9-deazahypoxanthin-9-yl)-1,4-dideoxy-1,4-imino-D-ribitol and has been shown to have a pKa > 10 at N7 (47). MT-ImmA is chemically similar in the 9-deazaadenine ring and is expected to have similar pKa.
The atomic coordinates and structure factors for S. pneumoniae MTAN-MT-ImmA (PDBID 1ZOS) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Unpublished observations, Crowder, T. and Schramm, V. L.
References
- 1.Ragione D, Porcelli FM, Carteni-Farina M, Zappia V, Pegg AE. Escherichia coli S-adenosylhomocysteine/5’-methylthioadenosine nucleosidase: purification, substrate specificity, and mechanism of action. Biochem J. 1985;232:335–341. doi: 10.1042/bj2320335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller CH, Duerre JA. S-Ribosylhomocysteine cleavage enzyme from Escherichia coli. J Biol Chem. 1968;243:92–97. [PubMed] [Google Scholar]
- 3.Tabor CW, Tabor H. Polyamines. Methods Enzymol. 1983;94:294–297. doi: 10.1016/s0076-6879(83)94019-3. [DOI] [PubMed] [Google Scholar]
- 4.Xavier KB, Bassler BL. LuxS quorum sensing: more than just a numbers game. Curr Opin Microbiol Rev. 2003;6:191–197. doi: 10.1016/s1369-5274(03)00028-6. [DOI] [PubMed] [Google Scholar]
- 5.Chen X, Schauder S, Potier N, Dorsselaer VA, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 6.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–99. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 7.Borchardt RT. S-adenosyl-L-methionine-dependent macromolecule methyltransferases: Potential targets for the design of chemotherapeutic agents. J Med Chem. 1980;23:347–357. doi: 10.1021/jm00178a001. [DOI] [PubMed] [Google Scholar]
- 8.Cadieux N, Bradbeer C, Reeger-Schneider E, Koster W, Mohanty AK, Wiener MC, Kadner RJ. Identification of the periplasmic cobalamin-binding protein BtuF of Escherichia coli. J Bacteriol. 2002;184:706–717. doi: 10.1128/JB.184.3.706-717.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stroeher UH, Paton AW, Ogunniyi AD, Paton JC. Mutation of luxS of Streptococcus pneumoniae affects virulence in a mouse model. Infect Immun. 2003;71:3206–3212. doi: 10.1128/IAI.71.6.3206-3212.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- 11.Winzer K, Hardie KR, Burgess N, Doherty N, Kirke D, Holden MT, Linforth R, Cornell KA, Taylor AJ, Hill PJ, Williams P. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl -3(2H)-furanone. Microbiology. 2002;148:909–922. doi: 10.1099/00221287-148-4-909. [DOI] [PubMed] [Google Scholar]
- 12.Zhao G, Wan W, Mansori S, Alfarno JF, Bassler BL, Cornell KA, Zhou ZS. Chemical synthesis of S-ribosyl-L-homocysteine and activity assay as a LuxS substrate. Bioorg Med Chem Lett. 2003;13:3897–3900. doi: 10.1016/j.bmcl.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Withers H, Swift HS, Williams P. Quorum sensing as an integral component of gene regulatory networks in Gram-negative bacteria. Curr Opin Microbiol. 2001;4:186–193. doi: 10.1016/s1369-5274(00)00187-9. [DOI] [PubMed] [Google Scholar]
- 14.Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- 15.Parsek MR, Val DL, Hanzelka BL, Cronan JE, Jr, Greenberg EP. Acyl homoserine-lactone quorum-sensing signal generation. Proc Natl Acad Sci. 1999;96:4360–4365. doi: 10.1073/pnas.96.8.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pajula RL, Raina A. Methylthioadenosine, a potent inhibitor of spermine synthase from bovine brain. FEBS Lett. 1979;99:153–156. doi: 10.1016/0014-5793(79)80988-6. [DOI] [PubMed] [Google Scholar]
- 17.Hibasami H, Borchardt RT, Chen SY, Coward JK, Pegg AE. Studies of inhibition of rat spermidine synthase and spermine synthase. Biochem J. 1980;187:419–428. doi: 10.1042/bj1870419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams-Ashman HG, Seidenfeld J, Galletti P. Trends in the biochemical pharmacology of 5’-deoxy-5’-methylthioadenosine. Biochem Pharmacol. 1982;31:277–288. doi: 10.1016/0006-2952(82)90171-x. [DOI] [PubMed] [Google Scholar]
- 19.Myers RW, Abeles RH. Conversion of 5-S-methyl-5-thio-D-ribose to methionine in Klebsiella pneumoniae. Stable isotope incorporation studies of the terminal enzymatic reactions in the pathway. J Biol Chem. 1990;265:16913–16921. [PubMed] [Google Scholar]
- 20.Horenstein BA, Parkin DW, Estupinan B, Schramm VL. Transition-state analysis of nucleoside hydrolase from Crithidia fasciculate. Biochemistry. 1991;30:10788–10795. doi: 10.1021/bi00108a026. [DOI] [PubMed] [Google Scholar]
- 21.Chen XY, Berti PJ, Schramm VL. Ricin A-Chain: Kinetic Isotope Effects and Transition State Structure with Stem-Loop RNA. J Am Chem Soc. 2000;122:1609–1617. [Google Scholar]
- 22.Mentch F, Parkin DW, Schramm VL. Transition-state structures for N-glycosidic hydrolysis of AMP by acid and by AMP nucleosidase in the presence and absence of allosteric activator. Biochemistry. 1987;26:921–930. doi: 10.1021/bi00377a037. [DOI] [PubMed] [Google Scholar]
- 23.Singh V, Lee JL, Nunez S, Howell PL, Schramm VL. Transition State Structure of 5′-Methylthioadenosine/S-Adenosylhomocysteine Nucleosidase from Escherichia coli and its similarity to transition state analogues. Biochemistry. 2005;44:11647–11649. doi: 10.1021/bi050863a. [DOI] [PubMed] [Google Scholar]
- 24.Lewandowicz A, Schramm VL. Transition state analysis for human and Plasmodium falciparum purine nucleoside phosphorylases. Biochemistry. 2004;43:1458–1468. doi: 10.1021/bi0359123. [DOI] [PubMed] [Google Scholar]
- 25.Kline PC, Schramm VL. Purine nucleosidase phosphorylases Catalytic mechanisms and transition state analysis of the arsenolysis reaction. Biochemistry. 1993;32:13212–13219. doi: 10.1021/bi00211a033. [DOI] [PubMed] [Google Scholar]
- 26.Schramm VL. Enzymatic transition state poise and transition state analogues. Acc Chem Res. 2003;36:588–596. doi: 10.1021/ar0200495. [DOI] [PubMed] [Google Scholar]
- 27.Wolfenden R. Transition state analogues for enzyme catalysis. Nature. 1969;223:704–705. doi: 10.1038/223704a0. [DOI] [PubMed] [Google Scholar]
- 28.Wolfenden R, Snider MJ. The depth of chemical time and the power of enzymes as catalysts. Acc Chem Res. 2001;34:938–45. doi: 10.1021/ar000058i. [DOI] [PubMed] [Google Scholar]
- 29.Singh V, Evans GB, Lenz DH, Mason JM, Clinch K, Mee S, Painter GF, Tyler PC, Furneaux RH, Lee JE, Howell PL, Schramm VL. Femtomolar transition state analogue inhibitors of 5’-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Escherichia coli. J Biol Chem. 2005;280:18265–18273. doi: 10.1074/jbc.M414472200. [DOI] [PubMed] [Google Scholar]
- 30.Cornell KA, Swarts WE, Barry RD, Riscoe MK. Characterization of recombinant Escherichia coli 5’-methylthioadenosine/S -adenosylhomocysteine nucleosidase: analysis of enzymatic activity and substrate specificity. Biochem Biophys Res Commun. 1996;21:228, 724–732. doi: 10.1006/bbrc.1996.1723. [DOI] [PubMed] [Google Scholar]
- 31.Singh V, Shi W, Evans GB, Tyler PC, Furneaux RH, Almo SC, Schramm VL. Picomolar transition state analogue inhibitors of human 5’ -methylthioadenosine phosphorylase and X-ray structure with MT-Immucillin-A. Biochemistry. 2004;43:9–18. doi: 10.1021/bi0358420. [DOI] [PubMed] [Google Scholar]
- 32.Evans GB, Furneaux RH, Schramm VL, Singh V, Tyler PC. Targeting the polyamine pathway with transition-state analogue inhibitors of 5’- methylthioadenosine phosphorylase. J Med Chem. 2004;47:3275–3281. doi: 10.1021/jm0306475. [DOI] [PubMed] [Google Scholar]
- 33.Whatmore AM, Barcus VA, Dowson CG. Genetic diversity of the streptococcal competence (com) gene locus. J Bacteriol. 1999;181:3144–3154. doi: 10.1128/jb.181.10.3144-3154.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.Lee JE, Cornell KA, Riscoe MK, Howell PL. Structure of Escherichia coli 5’-methylthioadenosine/ S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis. Structure. 2001;9:941–953. doi: 10.1074/jbc.M210836200. [DOI] [PubMed] [Google Scholar]
- 37.Navaza J. AMoRe: an automated package for molecular replacement. Acta Cryst Sect A. 1994;50:157–163. [Google Scholar]
- 38.Kissinger CR, Gehlhaar DK, Fogel DB. Rapid automated molecular replacement by evolutionary search. Acta Cryst D Biol Crystallogr. 1999;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- 39.Brunger AT, Adam PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Cryst D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 40.Jones TA. Diffraction methods for biological molecules. Interactive computer graphics: FRODO. Meths Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- 41.Laskowski RA, MacArthur MW, Moss DS, Thorton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 42.Evans GB, Furneaux RH, Tyler PC, Schramm VL. Synthesis of a transition state analogue inhibitor of purine nucleoside phosphorylase via the Mannich reaction. Org Letters. 2003;5:3639–3640. doi: 10.1021/ol035293q. [DOI] [PubMed] [Google Scholar]
- 43.Evans GB, Furneaux RH, Hausler H, Larsen JS, Tyler PC. Imino-C-nucleoside synthesis: heteroaryl lithium carbanion additions to a carbohydrate cyclic imine and nitrone. J Org Chem. 2004;69:2217–2220. doi: 10.1021/jo035744k. [DOI] [PubMed] [Google Scholar]
- 44.Evans GB, Furneaux RH, Schramm VL, Singh V, Tyler PC. Targeting the polyamine pathway with transition-state analogue inhibitors of 5′-methylthioadenosine phosphorylase. J Med Chem. 2004;47:3275–3281. doi: 10.1021/jm0306475. [DOI] [PubMed] [Google Scholar]
- 45.Evans GB, Furneaux RH, Lenz DH, Painter GF, Schramm VL, Singh V, Tyler PC. Second generation transition-state analogue inhibitors of human 5′-methylthioadenosine phosphorylase. J Med Chem. 2005;48:4679–4689. doi: 10.1021/jm050269z. [DOI] [PubMed] [Google Scholar]
- 46.Miles RW, Tyler PC, Furneaux RH, Bagdassarian CK, Schramm VL. One-third-the-sites transition-state inhibitors for purine nucleoside phosphorylase. Biochemistry. 1998;37:8615–8621. doi: 10.1021/bi980658d. [DOI] [PubMed] [Google Scholar]
- 47.Sauve AA, Cahill SM, Zech SG, Basso LA, Lewandowicz A, Santos DS, Grubmeyer C, Evans GB, Furneaux RH, Tyler PC, McDermott A, Girvin ME, Schramm VL. Ionic states of substrates and transition state analogues at the catalytic sites of N-ribosyltransferases. Biochemistry. 2003;42:5694–5705. doi: 10.1021/bi034003a. [DOI] [PubMed] [Google Scholar]
- 48.Zhou GC, Parikh SL, Tyler PC, Evans GB, Furneaux RH, Zubkova OV, Benjes PA, Schramm VL. Inhibitors of ADP-ribosylating bacterial toxins based on oxacarbenium ion character at their transition states. J Am Chem Soc. 2004;126:5690–5698. doi: 10.1021/ja038159+. [DOI] [PubMed] [Google Scholar]
- 49.Lee JE, Cornell KA, Riscoe MK, Howell PL. Structure of Escherichia coli 5’-methylthioadenosine/ S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis. J Biol Chem. 2001;278:8761–8770. doi: 10.1074/jbc.M210836200. [DOI] [PubMed] [Google Scholar]
- 50.Lee JE, Singh V, Evans GB, Tyler PC, Furneaux RH, Cornell KA, Riscoe MK, Schramm VL, Howell PL. Structural rationale for the affinity of pico-and femtomolar transition state analogues of E. coli 5’-methylthioadenosine/s-adenosylhomocysteine nucleosidase. J Biol Chem. 2005;280:18274–18282. doi: 10.1074/jbc.M414471200. [DOI] [PubMed] [Google Scholar]
- 51.Parkin DW, Schramm VL. Binding modes for substrate and a proposed transition state analogue of protozoan nucleoside hydrolase. Biochemistry. 1995;34:13961–13966. doi: 10.1021/bi00042a030. [DOI] [PubMed] [Google Scholar]
- 52.Schramm VL. Enzymatic transition state and transition state analogues. Curr Opin Struct Biol. 2005;15:1–10. doi: 10.1016/j.sbi.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 53.Evan SV. SETOR: hardware lighted three dimensional solid model representation of Macromolecules. J Mol Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]


