Abstract
The tobacco hornworm Manduca sexta is widely used as a model organism to investigate the biochemical basis of insect physiological processes but little transcriptome information is available. To get a broad view of the larval hemolymph proteins, particularly those related to immunity, we synthesized and sequenced cDNA fragments from a mixture of eight total RNA samples: fat body and hemocytes from larvae injected with killed bacteria, fat body, hemocytes, integument and trachea from naïve larvae, and fat body and hemocytes from wandering larvae. Using massively parallel pyrosequencing, we obtained 95,458 M. sexta expressed sequence tags (ESTs) at an average size of 185 bp per read. A majority of the sequences (69,429 reads) could be assembled into 7,231 contigs with an average size of 300 bp, 1178 of which had significant similarity with Drosophila genes from various functional groups. Only ~8% (606) of the contigs matched known M. sexta cDNA sequences, representing 186 of the 375 unique NCBI entries. The remaining 6,625 contigs represented newly discovered cDNA segments from this well studied biochemical model insect. A search of the 7,231 contigs using Tribolium castaneum, Drosophila melanogaster, and Bombyx mori immunity-related sequences revealed 424 cDNA contigs with significant similarity (E-value < 1×10−5). These included 218 previously unknown M. sexta sequences coding for putative defense molecules such as pattern recognition receptors, serine proteinases, serpins, Spätzle, Toll-like receptors, intracellular signaling molecules, and antimicrobial peptides.
Keywords: insect immunity; hemolymph proteins; gene discovery; transcript profiling; 454, sequencing
1. Introduction
Having a large body size and hemolymph volume, the tobacco hornworm Manduca sexta has been extensively used as a model organism to investigate the biochemical basis of insect physiological processes including cuticle formation, neural transmission, hormonal regulation, intermediary metabolism, nutrient transport, environmental perception and immune responses (Hopkins et al., 2000; Shield and Hildebrand, 2001; Riddiford et al., 2003; Kanost et al., 1990 and 2004; Jiang, 2008). While M. sexta has significantly contributed to our understandings of insect biochemistry and molecular biology, there is no genome project available for this species. A small EST project on odorant-binding proteins (Robertson et al., 1999) and a differential expression study on defense molecules (Zhu et al., 2003) generated sequences from 375 and 238 cDNA clones, respectively. In the era of systems biology, this situation has largely limited the future development of M. sexta as a major contributor for insect biochemistry and molecular biology.
Over the past three years, massively parallel pyrosequencing has emerged as an alternative approach for high-throughput sequence determination (Margulies et al., 2005), now that instruments based on this technology are available from 454/Roche. While this new technology has been applied to genotyping and genome re-sequencing (Isler et al., 2007), there are only a few reports describing EST-based transciptome studies (Gowda, et al., 2006; Bainbridge, et al. 2006; Emrich et al, 2006; Cheung et al, 2006; Weber et al., 2007). Since cDNA does not contain A/T-rich introns, intergenic regions or repetitive elements which cause problems in sequencing and data interpretation (Wicker et al., 2006) and a large portion codes for polypeptides, determination of expressed sequence tags (ESTs) is an effective approach to study the transcriptome and for gene discovery. By applying 454-based pyrosequencing to an organism, the large number of randomly selected cDNA fragments that are partially sequenced, often leads to the identification of heretofore undescribed proteins encoded by this organism and expressed only at very low levels, as well as those moderately and highly expressed enzymes for biosynthesis of a broad spectrum of metabolites that give the organism its unique phenotype. Previous EST projects for species spanning diverse phylogenetic groups have yielded rich datasets essential for structural, functional and comparative genomic analyses, and variations in protein sequences obtained by back translation of the ESTs have been used to identify new conserved motifs, active site residues, and substrate-binding sites (Mayer et al., 2005).
In order to expand our knowledge on M. sexta larval hemolymph proteins, especially those participating in antimicrobial responses, we isolated total RNA from hemocytes, fat body and other tissues which may constitutively synthesize and secrete defense molecules. Because several defense-related genes are thought to be only expressed during an immune response or in naïve wandering larvae of M. sexta (Kanost et al., 2004; Jiang et al., 2008), we also prepared fat body and hemocyte total RNA from these insects. To take full advantage of the large capacity of pyrosequencing, we combined these RNA samples at certain ratios for mRNA isolation, cDNA synthesis, and sequence determination. In this paper, we report our analysis of over 95,000 ESTs determined by 454-based pyrosequencing and their assembly into 7,231 contigs. A similarity search with Drosophila melanogaster (Diptera), Tribolium castaneum (Coleoptera) and Bombyx mori (Lepidoptera) sequences provides an overview of cellular and plasma proteins in the larval hemolymph, particularly those involved in immune responses. We also discuss the advantages and limitations of pyrosequencing as well as potential applications of this approach to rapidly obtain sequence information for non-model organisms.
2. Methods and materials
2.1. Insect rearing, bacterial challenge and RNA isolation
M. sexta eggs, purchased from Carolina Biological Supply, were hatched and reared on an artificial diet (Dunn and Drake, 1983). Each of day 2, 5th instar larvae (20) was injected with a mixture of formaldehyde-killed Escherichia coli (2×107 cells), Micrococcus luteus (20 µg) and curdlan (20 µg)(insoluble β-1,3-glucan from Alcaligenes faecallis) in 30 µl H2O. Total RNA samples were isolated from the hemocytes and fat body 24 h later using TRIZOL Reagent (Invitrogen Life Technology). Hemocyte and fat body total RNA were also prepared from day 3, 5th instar naïve (40) and bar-stage wandering (20) larvae. Similarly, integuments and trachea of the 5th instar naïve larvae were dissected for total RNA isolation. The RNA samples (A260/A280 > 1.8) were combined at the following percentages: hemocytes (10%), fat body (15%), integument (5%) and trachea (5%) from naïve larvae, hemocytes (20%) and fat body (35%) from injected larvae, hemocytes (5%) and fat body (5%) from wandering larvae. mRNA was purified from the pooled total RNA (500 µg) by binding to oligo(dT) cellulose twice (Poly(A)Purist, Ambion).
2.2. cDNA synthesis
For first strand synthesis, the purified mRNA (2 µl, 2.6 µg/µl), random pentadecamers (2 µl, 1 µg/µl)(Stangegaard et al., 2006), and H2O (4 µl) were denatured at 70°C for 10 min, rapidly chilled on ice, mixed with 250 mM Tris-HCl, pH 8.3, 375 mM KCl and 15 mM MgCl2 (4 µl), 0.1 M DTT (2 µl), dNTPs (1 µl, 10 mM each), and SuperScript™ II Reverse Transcriptase (5 µl, 200 U/µl)(Invitrogen Life Technology). Following incubation at 25°C for 10 min and 42°C for 50 min, cDNA synthesis was stopped by placing the tube on ice. For second strand cDNA synthesis, H2O (91 µl), 100 M Tris-HCl, pH 6.9, 450 mM KCl, 23 mM MgCl2, 0.75 mM β-NAD+ and 50 mM (NH4)2SO4 (30 µl), dNTPs (3 µl, 10 mM each), E. coli DNA ligase (1 µl, 10 U/µl), E. coli DNA polymerase I (4 µl, 10 U/µl), and E. coli RNase H (1 µl, 2 U/µl) were mixed with the first strand synthesis reaction and incubated at 16°C for 120 min. For end blunting, T4 DNA polymerase (4 µl, 5 U/µl) was incubated with the second strand synthesis reaction at 16°C for 5 min. The cDNA (10 µg) was purified using MinElute PCR Purification Kit (Qiagen) and phosphorylated by T4 polynucleotide kinase (5 µl, 10 U/µl)(New England Biolabs) at 37°C for 30 min. After purification, the DNA was eluted from the MinElute spin column in 30 µl of elution buffer (EB)(10 mM Tris-HCl, pH 8.5) and stored at −20°C.
2.3. Adaptor attachment, bead binding, PCR amplification, and pyrosequencing
About 3–5 µg of cDNA (15 µl) was ligated to double-stranded, 5′ overhung adaptors A (CCATCTCATCCCT GCGTGTCCCATCTGTTCCCTCCCTGTCTCAG) and B (5′-biotinylated CCTATCCCCTGTG TGCCTTGCCTATCCCCTGTTGCGTGTCTCAG)(1 µl, 200 pmol/µl for each) in the presence of 20 µl 2×ligase buffer, and 4 µl of DNA ligase (2000 U/µl). After incubation at 25°C for 15 min, the DNA was recovered in 25 µl EB using MinElute PCR Purification Kit.
After 100 µl M-280 streptavidin-coated beads (Dynal) were equilibrated with B&W Buffer (5 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 1 M NaCl)(twice, 200 µl each) and resuspended in 100 µl of 2×B&W Buffer and 75 µl H2O, the DNA sample (25 µl) was thoroughly mixed with the magnetic beads at room temperature for 20 min. Upon buffer removal on a magnetic particle collector, the immobilized cDNA fragments were end-repaired by incubating at 37°C for 20 min with 40 µl H2O, 5 µl 10×polymerase buffer, 2 µl dNTPs (10 mM each) and 3 µl T4 DNA polymerase (5 U/µl). With the reaction mixture removed, the beads were washed twice with B&W Buffer (100 µl each) and then suspended in 50 µl of melt solution (125 µg NaOH in 9.875 ml H2O) to denature the immobilized DNA. After thorough mixing at room temperature for 3 min, the solution containing the single-stranded DNA was separated from the beads and added to the neutralization solution (500 µl of Qiagen PB buffer mixed with 3.8 µl of 20% acetic acid. The single-stranded DNA library was bound to a MinElute column and eluted in 15 µl EB.
The single-stranded DNA (15 µl, 500 ng/µl) was mixed with 1.5 million capture beads and annealed at a temperature gradient of 70, 60, and 50°C (Margulies et al., 2005). The captured, single-stranded DNA was then added to 400 µl emulsion oil containing 181.62 µl amplification reaction mixture, 10 µl of 2 mM MgSO4, 2.08 µl primer mixture (CCATCTCATCCCTGCGTGTC and CCTATCCCCTGTGTGCCTTG, 200 µM each), 0.3 µl thermal stable pyrophosphatase (2 U/µl), and 6 µl Platinum Hi-Fi Taq Polymerase (5 U/µl). After shaking for 5 min at 15 rps on a TissueLyser MM300 (Retsch GmbH), the emulsified amplification mixture was thermocycled as follows: 94°C for 4 min, 40 cycles of 94°C for 30 s, 58°C for 60 s and 68°C for 90 s, and 13 cycles of 94°C for 30 s and 58°C for 360 s. Then, the emulsion was broken with isopropanol and the beads were recovered for second strand removal by alkaline denaturation and washing. Upon elimination of null beads, the sequencing primer (CCATCTGTTCCCTCCCTGTC) was annealed to single stranded DNA associating with the beads. Deposition of the DNA and enzyme beads to fiber-optic wells was followed by eight-four cycles of delivery of the pyrosequencing reagents, incubation and washes, achieved by pre-programmed operation of the fluidics system (Margulies et al., 2005).
2.4. Sequence assembling and functional categorization based on Drosophila gene ontology
After image recording and signal processing, flows from the 454 sequencer were first trimmed to three sets of reads (60, 80, and 100 bp long) and then assembled with Newbler, a de novo sequence assembly software using flow signals (Margulies et al., 2005) to reduce the number of artificial contigs produced when the sequence reads have poor quality at the end of contigs. The results from the three Newbler assemblies were then assembled into the final contig set using Phrap (Ewing et al., 1998a and b). The contigs were analyzed using BLASTX against Drosophila proteins. The output was employed to reconstruct M. sexta metabolic profile using Kyoto Encyclopedia for Genes and Genomes (KEGG)(Kanehisa et al., 2004).
2.5. Comparison with M. sexta and other insect sequences
Complete or partial M. sexta cDNA sequences were downloaded from NCBI (http://www.ncbi.nlm.nih.gov/). After manual removal of redundant sequences, the remaining ones were classified into two groups: immunity-related and -unrelated. The comparison with the 7,231 contigs was performed using TBLASTX at an E-value cutoff of 1×10−20. TBLASTX is a part of BLAST 2.2.14 downloaded from the NCBI site. The EST contigs were searched by TBLASTX at E-value ≤ 1×10−5, using the coding sequences of T. castaneum, D. melanogaster and B. mori immunity-related genes as queries (Zou et al., 2007; Sackton et al., 2007; Chen et al., 2008). The silkworm dataset was established using the sequences retrieved from NCBI based on a PubMed search of the literature on B. mori immunity. The M. sexta EST contigs were also compared with B. mori, Spodoptera frugiperda, D. melanogaster, T. castaneum and Apis mellifera ESTs downloaded from NCBI EST database (2007.12).
3. Results
To get an overview of M. sexta hemolymph proteins including those induced upon injection of bacteria (Gram-positive and -negative) and β-1,3-glucan (a fungal cell wall component), we isolated total RNA samples from fat body and hemocytes of the 5th instar naïve and immune challenged larvae. We also prepared fat body and hemocyte RNA from wandering stage larvae as well as integument and tracheal total RNA from the 5th instar insects. These eight samples were combined at a ratio of 3:2:7:4:1:1:1:1 for mRNA purification, cDNA synthesis, and sequence determination. We purified 39 µg mRNA from 2.0 mg total RNA, synthesized 19.3 µg cDNA using 5.2 µg mRNA, and obtained 95,358 high-quality reads using 3–5 µg cDNA (Table 1). At an average size of 185 bp per read, we acquired over 17.6 million bases of cDNA at a cost of ~$10,000. A majority of these ESTs (69,427 or 72.8%) were assembled into 7,231 contigs ranging from 85–3909 bp. The total number of bases covered by these contigs is 2.17 million with an average length of 300 bp.
Table 1.
Summary statistics for pyrosequencing M. sexta ESTs
| Number of instrument runs | 1 |
| Size of fiber-optic slide | 6 × 6 cm2 |
| Run time/number of cycles | 453 min/84 |
| High-quality reads | 95,358 |
| Average size of reads | 185 bp |
| Total number of contigs | 7,231 |
| Contigs size (average and range) | 300 and 85~3909 bp |
| Total reads within contigs | 69,429 |
| Singlet reads | 25,929 |
| Contigs with functional assignment | 1,178 |
| Contigs matching known M. sexta sequences | 606 |
| Contigs for immunity-related proteins | 424 |
Using BLASTN, we compared the 7,231 contigs with a total of 902,165 ESTs from B. mori, S. frugiperda, T. castaneum, D. melanogaster, and A. mellifera (Table 2) The total sequence matches were more with the two lepidopteran species (B. mori: 2,427; S. frugiperda: 1,739) than with the coleopteran, dipteran and hymenopteran insects (734, 930, and 488). The silkworm B. mori and armyworm S. frugiperda had similar numbers of matches with the M. sexta contigs in the first three E-value categories (0 to 10−150, 10−150 to 10−100, and 10−100 to 10−50), even though there is a major difference in their EST repository sizes (184,509 for B. mori and 32,217 for S. frugiperda). In the next two categories (10−50 to 10−20 and 10−20 to 10−5), the silkworm had 789 and 1,085 matches with the M. sexta sequences. Nonetheless, 4,804 (66.4%) and 5,492 (76.0%) of the contigs did not match the ESTs of B. mori and S. frugiperda, respectively. This high percentage of no match, as previously reported between the silkworm and armyworm (Deng et al., 2006), further confirmed that Lepidoptera is a highly diverse order of insects.
Table 2.
Comparative analysis of M. sexta contigs with ESTs from five insect species
| Similarity | S. frugiperda | B. mori | T. castaneum | D. melanogaster | A. mellifera | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | n | % | |
| E ≤ 10−150 | 119 | 1.6 | 112 | 1.5 | 17 | 0.2 | 51 | 0.7 | 20 | 0.3 |
| E ≤ 10−100 | 104 | 1.4 | 104 | 1.4 | 35 | 0.5 | 48 | 0.7 | 34 | 0.5 |
| E ≤ 10−50 | 330 | 4.6 | 337 | 4.7 | 84 | 1.2 | 149 | 2.1 | 53 | 0.7 |
| E ≤ 10−20 | 560 | 7.7 | 789 | 10.9 | 205 | 2.8 | 229 | 3.2 | 97 | 1.3 |
| E ≤ 10−5 | 626 | 8.7 | 1085 | 15.0 | 393 | 5.4 | 453 | 6.3 | 284 | 3.9 |
| Total matched | 1739 | 24.0 | 2427 | 33.6 | 734 | 10.2 | 930 | 12.9 | 488 | 6.7 |
| No match | 5492 | 76.0 | 4804 | 66.4 | 6497 | 89.8 | 6301 | 87.1 | 6743 | 93.3 |
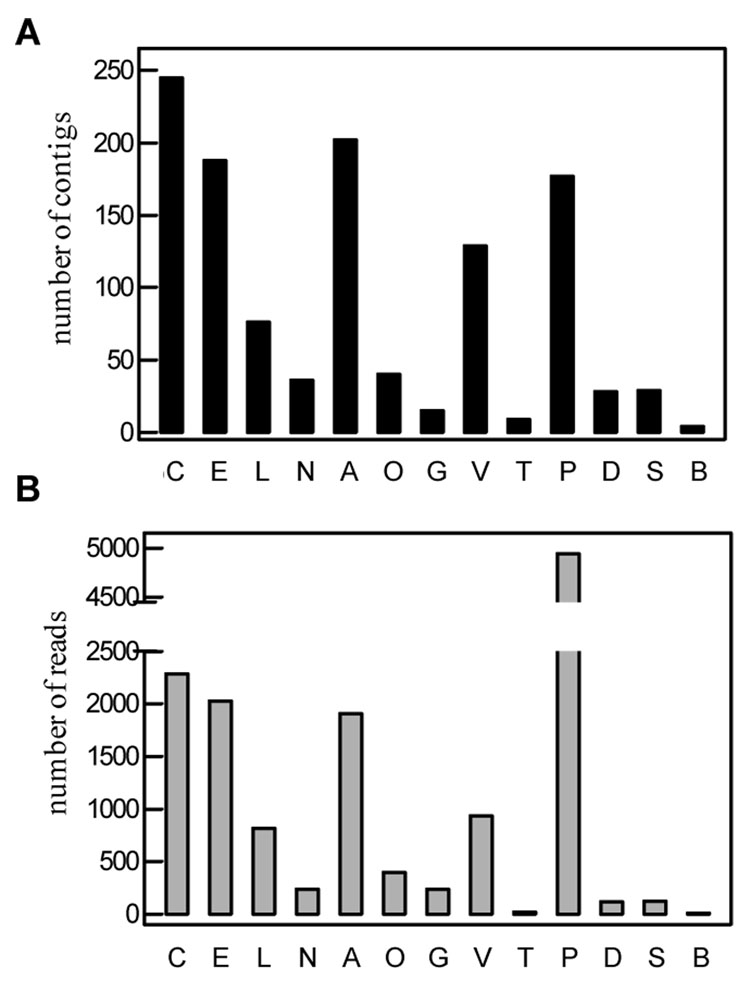
A Drosophila-based gene ontology search indicated that 1,178 of the 7,231 contigs can be categorized into 13 functional groups (Table S1 and Fig. 1A). Enzymes involved in metabolism of carbohydrates (245, 21%), energy (188, 16%), amino acids (202, 17%) and vitamins (129, 11%) represent the largest group in the 1,178 contigs with putative function. While metabolism-related contigs (891) account for 79% of the total, transcription- and translation-associated ones constitute the second largest group (16%): most of its members encode ribosomal proteins (177, 15%). Poor representation of other functional groups (e.g. environmental information processing and other cellular processes) is probably caused by their high sequence divergence.
Fig. 1. Distribution of the M. sexta cDNA contigs (A) and reads (B) coding for proteins in different functional groups.
C, carbohydrate metabolism; E, energy metabolism; L, lipid metabolism; N, nucleotide metabolism; A, amino acid metabolism; O, other amino acid metabolism; G, glycan biosynthesis and metabolism; V, vitamin and cofactor metabolism; T, transcription; P, protein synthesis; D, protein sorting and degradation; S, signal transduction; B behaviors and development. Black and gray bars represent the numbers of sequence contigs (A) and reads (B), respectively.
Frequencies of sequence reads partly reflect their relative mRNA abundance (Fig. 1B). The ratio of ribosomal protein reads to contigs (28) is the highest, and the ratio for non-ribosomal proteins is 9. When we examined the other major groups with >30 contigs, the ratios ranged from 7 to 11. Significant deviations were found in the following minor groups: glycan biosynthesis and metabolism (16), transcription (3), behavior and development (2).
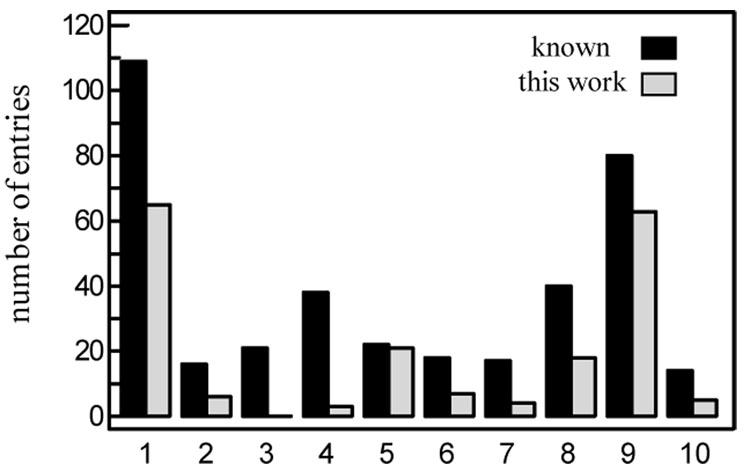
We retrieved from GenBank all the M. sexta sequence entries, compared them with our EST dataset, and identified contigs cloned previously. After removing genomic sequences and redundant cDNAs, we organized the remaining 375 sequences into ten functional groups (Fig. 2). These sequences largely reflect our current understandings of this insect at the molecular level, which account for only 8.4% of the EST contigs we determined in this project (Fig. 3).
Fig. 2. Distribution of known M. sexta cDNA sequences encoding proteins involved in various physiological processes or system.
1, cellular processes; 2, cuticle formation; 3, neural transmission; 4, hormonal regulation; 5, circulatory system; 6, digestive system; 7, development; photo- and chemoreception; 9, immune responses; 10, others. Black and gray bars represent numbers of sequence entries in GenBank and EST contigs from this study, respectively.
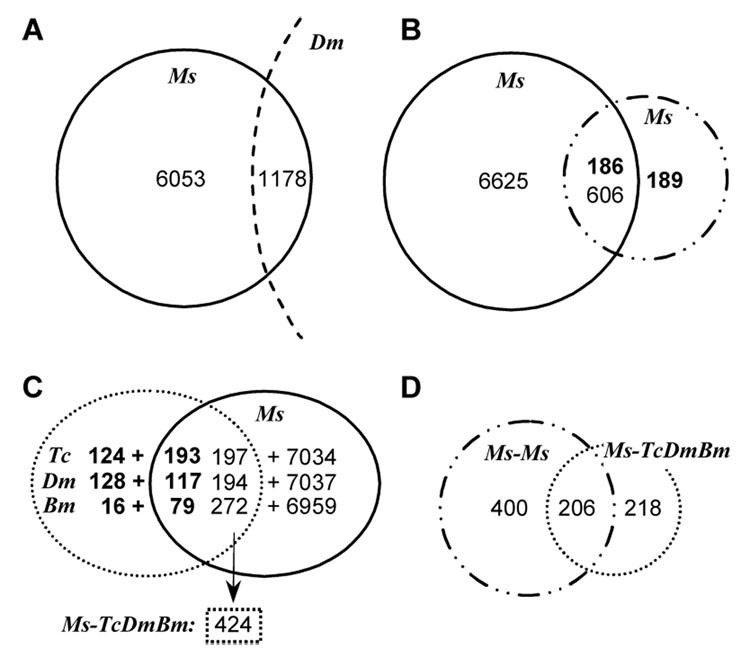
Fig. 3. Venn diagrams of M. sexta ESTs compared with D. melanogaster genes (A), known M. sexta cDNAs (B), and immunity-related genes from T. castaneum, D. melanogaster, and B. mori (C).
EST contig numbers are in regular font whereas numbers of known cDNA/gene sequences are in bold. A comparison of the overlapping regions from Panel B (known M. sexta EST contigs) and Panel C (M. sexta immunity-related EST contigs, non-redundant) results in Panel D, which shows the number of EST contigs encoding unknown, putative defense proteins.
Proteins associating with various cell processes represent the largest group of known sequences (109). These processes include intermediary metabolism (of carbohydrates and lipids for instance), drug resistance (e.g. cytochrome P450s), ion/metabolite transport (e.g. channel proteins), cell structure (e.g. integrins) and others (Table S2). Sixty percent of these entries have at least one matching contig. While cuticle formation (16), neurotransmission (21), hormonal regulation (38), digestion (18), development (17), photo- and chemoreception (40) have been quite well studied in M. sexta, their percentages of matching range from 0 to 45% (Fig. 2). This is probably because the combined RNA sample is mainly from fat body and hemocytes. For the same reason, 80% of the immunity-related sequences and 95% of the hemolymph protein sequences are present in our EST collection.
Although 80 (or 21%) of the 375 M. sexta proteins in the NCBI database participate in immune responses, these molecules appear to represent only a small portion of the M. sexta immune system. From the recently annotated T. castaneum genome, we selected 317 proteins which may take part in the antimicrobial responses (Zou et al., 2007). A search of the EST data collection with these genes indicated that 193 of the beetle sequences are homologous to 197 of the 7,231 M. sexta contigs (Fig. 3). Similar comparisons with the D. melanogaster and B. mori immunity-related genes showed that 117 and 79 of the fly and silkworm sequences are homologous to 194 and 272 of M. sexta EST contigs, respectively. After removing the redundant ones from the combined list, we found that 206 of the 424 contigs had already been identified in M. sexta whereas the other 218 may encode defense proteins previously unknown.
These newly discovered sequences include proteins with putative functions in immunity, including recognition of pathogen-associated molecular patterns (e.g., peptidoglycans, β-1,3-glucan, galactose and other sugar moieties) and mediation or modulation of extracellular signals stimulated by pathogen invasion (e.g. serine proteinases, serpins and serine proteinase homologs) (Table 3). We discovered one contig encoding Spätzle and five encoding Toll-like receptors. We found six putative components of the intracellular signal transduction pathways. These proteins are similar in sequence to Drosophila pelle, pellino, Traf2, basket, HOP, and IKKb (Wang and Ligoxygakis, 2006). In addition, we identified fifteen EST contigs that may encode transcription factors (e.g. Dif, Relish, Jra, and Domino). Similar to Drosophila Dif and Relish, some of these proteins may dissociate from their partners, translocate into the nucleus and regulate expression of immunity-related genes. There are several mechanisms that kill invading microorganisms: phagocytosis, antimicrobial peptides, reactive oxygen/nitrogen species, and melanization. We found ~50 contigs for proteins which may participate in these processes (Table 3).
Table 3.
M. sexta cDNA contigs encoding putative immune proteins
| Family Name | Contig No. (E-value < 1 × 10−5) |
|---|---|
| Recognition | |
| PGRP | 3683 |
| GNBP | 1635, 3092, 4565, 6004, 6134, 6422 |
| C-type lectin | 510, 1242, 3822, 5488 |
| SR/CTL | 2397, 4003, 4647, 5031, 5660, 5932, 6716, 7151 |
| lectin | 147, 221, 537, 597, 2784, 3594, 3917, 4417, 4695, 4975, 5213, 5366, 5548, 5686, 5827, 5833, 5967, 5968, 6410, 6656, 6749, 6196, 6493, 6578, 6615, 6819, 6826, 6845, 6962, 7018, 7064, 7089, 7093, 7114, 7175, 7198 |
| LPS-binding protein | 25, 4662, 5054, 5486 |
| multi-binding protein | 778, 1853, 4547, 5528 |
| Nimrod | 3001, 4465, 5381, 6380, 7023, 7206 |
| Galectin | 1485, 1486 |
| SR-C | 294, 2630, 3141, 4888, 5106, 6096, 6157, 6745 |
| SR-B14 | 2693 |
| Signaling | |
| SP/SPH | 1128, 1958, 2469, 2495, 5177, 5587, 6919 |
| serpin | 477, 574, 736, 1483, 2972, 3437, 3611, 4324, 4857, 5142, 5627 |
| Kazal-type inhibitor | 5044, 6104, 6145, 6341, 6427, 6740, 6844 |
| Serrate | 2065 |
| Spätzle | 4514 |
| Toll-like Receptor | 1038, 1295, 1752, 6106, 6618 |
| Notch | 4202 |
| MD2-like protein | 6412, 6724 |
| Pelle/HOP | 418 |
| pellino | 4423 |
| Lesswright | 2497, 3500, 4596 |
| Mask | 3823,4772, 4989 |
| SAE2 or Uba2 | 1351 |
| Smt3 (SUMO) | 3605 |
| Stam | 1792 |
| Rac1 | 3154, 3498, 3808, 4771, 5452, 5569, 6011, 6238 |
| Ras85D | 838, 5168, 5651 |
| Uev1A | 5088 |
| aPKC | 2789, 3068 |
| Mekk1 | 2744 |
| bendless | 295, 2401, 5848 |
| Traf2 | 1069 |
| IKKb | 777 |
| Dif | 3981 |
| Rel | 415, 2427 |
| brahma (brm) | 3017 |
| lozenge | 2837 |
| serpent | 5422 |
| Domino | 381, 5943 |
| Pointed | 5814, 5949, 6002 |
| Helicase 89B | 717 |
| Jra | 368, 3362, 4395 |
| basket | 2164 |
| Thor | 5050 |
| Execution | |
| proPO | 212, 5511, 5588 |
| hexamerin | 7, 45, 360, 2850, 3633, 5212, 5544, 5621 |
| catalase | 219, 6229, 6645, 6824 |
| heme peroxidase | 3517 |
| Pale | 2749, 4691, 5331 |
| peroxiredoxin | 987, 2951, 5419, 6551 |
| superoxide dismutase | 746, 2012, 3655, 4799, 5246, 5532, 5790, 6463 |
| transferrin | 241, 6923, 6975, 7065, 7152 |
| I-type lysozyme | 6834 |
| WAP | 168, 1367, 5863 |
| cecropin | 2488, 4701, 4933, 5774, 6013, 6560 |
| lebocin | 5813, 6639, 6760, 6851 |
| neucin | 854, 5178 |
| 6tox | 3760, 6308, 6646 |
4. Discussion
As a biochemical model insect, M. sexta has contributed a wealth of knowledge to insect biochemistry and molecular biology (Kanost et al., 1990). Abundant hemolymph proteins (those with concentrations greater than 5% of the total plasma protein concentration) were isolated from larvae and characterized biochemically twenty years ago. Since then, efforts have been made to expand our knowledge of plasma factors, particularly those involved in defense responses (Jiang, 2008). We managed to clone hundreds of cDNAs from the larval fat body and hemocytes (Zhu et al., 2003; Jiang et al., 2005) and purify several additional proteins, including active proteinases from the plasma (Jiang et al., 2003). Even so, our understanding of the physiological processes in this insect is still rudimentary. As shown in this study (Table 1, Fig. 1 and Fig. 3), over 90% of the unique EST contigs were previously unknown in M. sexta. While 87–93% of the M. sexta (Lepidoptera, Bombycoidea, Sphingidiae) sequences fall into the group of no match (E ≥ 10−5) in the cross-order EST comparisons, 76% and 66% of the 7,231 contigs have no significant match with ESTs of S. frugiperda (Lepidoptera, Noctuoidae, Noctuidae) and B. mori (Lepidoptera, Bombycoidea, Bombycidae) in the cross-superfamily and cross-family analyses (Table 2), respectively. In other words, the silkworm genome project could be insufficient to cover Lepidoptera, a highly diverse order of insects.
The initial analysis of the EST dataset provides new candidates for functional tests (Table 3), including pathogen recognition, proteinase cascades and modulation, intracellular signaling pathways, and microbe killing. While predicted functions of these EST contigs obviously need confirmation, we have already used the sequence information to isolate corresponding full-length cDNA clones and are making further breakthroughs in understanding the molecular basis of insect immunity using pyrosequencing-generated data. We anticipate the entire genome of M. sexta will be determined by pyrosequencing probably in a few years at less than one-tenth of the current cost and time for a 4×108-nucleotide genome.
Implementation of a massively parallel pyrosequence-based approach provides rapid, cost-effective DNA sequence acquisition for organisms lacking detailed genomic sequence data. Since molecular cloning is not involved, technical difficulties, labor, reagents, and supplies associated with library construction, DNA normalization, colony picking, plasmid isolation, and Sanger sequencing are eliminated, this approach is particularly useful for cDNAs that are short, unstable, toxic or difficult to clone. Also, by bypassing biological cloning procedures the time from RNA isolation to completion of pyrosequencing is significantly shortened to approximately two weeks from mRNA isolation to EST sequence data analysis.
As a new technology, pyrosequencing also has several limitations including a measurable rate of deletions and insertions (~4%) and shorter sequence reads than those obtained by Sanger sequencing (~185 bp versus reads approaching 1 kb). Open-reading-frame shifts can cause difficulties in similarity-based searches for evolutionarily distant, i.e. less conserved genes. However, as the pyrosequencing technology rapidly evolves, this powerful method for sequence acquisition and function categorization will become extremely useful for expression studies in non-model organisms.
EST data resulting from pyrosequencing have a number of potential applications. We have observed that most assembled contigs fell within the known ontology groups including highly conserved, housekeeping genes (Table S1). The massive sequence information can be used, for instance, in microarray experiments for transcript profiling. This is quite appealing for species with unknown genomes but with major socioeconomic implications, such as most agricultural pests as well as human and domestic animal disease vectors. Since pyrosequencing can be used for comparative expression profiling, i.e. comparing cDNAs from control and treatment groups, the frequencies of individual reads grouped by sequence similarities relative to the frequencies of reads of a house-keeping gene can be directly compared to find changes occurring after a treatment. A second application involves the conversion of ESTs to a database of amino acid sequences, which facilitates protein identification in protein-sequencing-based proteomic research, especially for organisms lacking genomic sequence information.
Supplementary Material
Acknowledgments
We wish to dedicate this paper to Dr. Michael Wells, who devoted a major part of his life investigating the basic biochemical processes in insects including M. sexta. We also greatly appreciate insightful suggestions from Dr. Udaya Desilva in the Department of Animal Science at Oklahoma State University. We thank Drs. Michael Kanost, Jack Dillwith, Udaya Desilva, and Maureen Gorman for their critical comments on the manuscript. This work was supported by National Institutes of Health Grants GM58634 (to H. Jiang). This article was approved for publication by the Director of the Oklahoma Agricultural Experiment Station and supported in part under project OKLO2450.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bainbridge MN, Warren RL, Hirst M, Romanuik T, Zeng T, Go A, Delaney A, Griffith M, Hickenbotham M, Magrini V, Mardis ER, Sadar MD, Siddiqui AS, Marra MA, Jones SJ. Analysis of the prostate cancer cell line LNCaP transcriptome using a sequencing-by-synthesis approach. BMC Genomics. 2006;7:246. doi: 10.1186/1471-2164-7-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung F, Haas BJ, Goldberg SM, May GD, Xiao Y, Town CD. Sequencing Medicago truncatula expressed sequenced tags using 454 Life Sciences technology. BMC Genomics. 2006;7:272. doi: 10.1186/1471-2164-7-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng TC, Zhang YL, Liu C, Xu PZ, Gao ZH, Xia QY, Xiang ZH. Identification and analysis of Toll-related genes in the domesticated silkworm, Bombyx mori. Dev. Comp. Immunol. 2008;32:464–475. doi: 10.1016/j.dci.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Deng Y, Dong Y, Thodima V, Clem RJ, Passarelli AL. Analysis and functional annotation of expressed sequence tags from the fall armyworm Spodoptera frugiperda. BMC Genomics. 2006;7:264. doi: 10.1186/1471-2164-7-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn P, Drake D. Fate of bacteria injected into naïve and immunized larvae of the tobacco hornworm, Manduca sexta. J. Invertebr. Pathol. 1983;41:77–85. [Google Scholar]
- Emrich SJ, Barbazuk WB, Li L, Schnable PS. Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res. 2007;17:69–73. doi: 10.1101/gr.5145806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl M, Green P. Basecalling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998a;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Basecalling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998b;8:186–194. [PubMed] [Google Scholar]
- Gowda M, Li H, Alessi J, Chen F, Pratt R, Wang GL. Robust analysis of 5′- transcript ends (5′-RATE): a novel technique for transcriptome analysis and genome annotation. Nucleic Acids Res. 2006;34(19):e126. doi: 10.1093/nar/gkl522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins TL, Krchma LJ, Ahmad SA, Kramer KJ. Pupal cuticle proteins of Manduca sexta: characterization and profiles during sclerotization. Insect Biochem. Mol. Biol. 2000;30:19–27. doi: 10.1016/s0965-1748(99)00091-0. [DOI] [PubMed] [Google Scholar]
- Isler JA, Vesterqvist OE, Burczynski ME. Analytical validation of genotyping assays in the biomarker laboratory. Pharmacogenomics. 2007;8:353–368. doi: 10.2217/14622416.8.4.353. [DOI] [PubMed] [Google Scholar]
- Jiang H. The biochemical basis of antimicrobial responses in Manduca sexta. Insect Sci. 2008;15:53–66. [Google Scholar]
- Jiang H, Wang Y, Yu X-Q, Kanost MR. Prophenoloxidase-activating proteinase-2 (PAP-2) from hemolymph of Manduca sexta: a bacteria-inducible serine proteinase containing two clip domains. J. Biol. Chem. 2003;278:3552–3561. doi: 10.1074/jbc.M205743200. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Gu Y, Guo X, Zou Z, Scholz F, Trenczek TE, Kanost MR. Molecular identification of a bevy of serine proteinases in Manduca sexta hemolymph. Insect Biochem. Mol. Biol. 2005;35:931–943. doi: 10.1016/j.ibmb.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanost MR, Kawooya JK, Law JH, Ryan RO, Van Heusden MC, Ziegler R. Insect hemolymph proteins. Advances in Insect Physiol. 1990;22:299–396. [Google Scholar]
- Kanost MR, Jiang H, Yu X-Q. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol. Rev. 2004;198:97–105. doi: 10.1111/j.0105-2896.2004.0121.x. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Bader JS, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437(7057):376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KM, McCorkle SR, Shanklin J. Linking enzyme sequence to function using conserved property difference locator to identify and annotate positions likely to control specific functionality. BMC Bioinformatics. 2005;6:284. doi: 10.1186/1471-2105-6-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddiford LM, Hiruma K, Zhou X, Nelson CA. Insights into the molecular basis of the hormonal control of molting and metamorphosis from Manduca sexta and Drosophila melanogaster. Insect Biochem. Mol. Biol. 2003;33:1327–1338. doi: 10.1016/j.ibmb.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Martos R, Sears CR, Todres EZ, Walden KK, Nardi JB. Diversity of odorant binding proteins revealed by an expressed sequence tag project on male Manduca sexta moth antennae. Insect Mol. Biol. 1999;8:501–518. doi: 10.1046/j.1365-2583.1999.00146.x. [DOI] [PubMed] [Google Scholar]
- Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG. Dynamic evolution of the innate immune system in Drosophila. Nat. Genet. 2007;39:1461–1468. doi: 10.1038/ng.2007.60. [DOI] [PubMed] [Google Scholar]
- Shields VD, Hildebrand JG. Recent advances in insect olfaction, specifically regarding the morphology and sensory physiology of antennal sensilla of the female sphinx moth Manduca sexta. Microsc. Res. Tech. 2001;55:307–329. doi: 10.1002/jemt.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangegaard M, Dufva IH, Dufa M. Reverse transcription using random pentadecamer primers increases yield and quality of resulting cDNA. BioTechniques. 2006;40:649–657. doi: 10.2144/000112153. [DOI] [PubMed] [Google Scholar]
- Wang L, Ligoxygakis P. Pathogen recognition and signaling in the Drosophila innate immune response. Immunobiol. 2006;211:251–261. doi: 10.1016/j.imbio.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Weber AP, Weber KL, Carr K, Wilkerson C, Ohlrogge JB. Sampling the Arabidopsis transcriptome with massively-parallel pyrosequencing. Plant Physiol. 2007;144:32–42. doi: 10.1104/pp.107.096677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Schlagenhauf E, Graner A, Close TJ, Keller B, Stein N. 454 sequencing put to the test using the complex genome of barley. BMC Genomics. 2006;7:275. doi: 10.1186/1471-2164-7-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Evans J, Lu Z-Q, Zhao P-C, Williams M, Sumathipala N, Hetru C, Hultmark D, Jiang H. Comparative genomic analysis of the Tribolium immune system. Genome Biol. 2007;8:R177. doi: 10.1186/gb-2007-8-8-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Johnson TJ, Myers AA, Kanost MR. Identification by subtractive suppression hybridization of bacteria-induced genes expressed in Manduca sexta fat body. Insect Biochem. Mol. Biol. 2003;33:541–559. doi: 10.1016/s0965-1748(03)00028-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.