Abstract
Small G-proteins belonging to the Arf (ADP-ribosylation factor) family serve as regulatory proteins for numerous cellular processes through GTP-dependent recruitment of effector molecules. In the present study we demonstrate that proteins in this family regulate, and are regulated by, membrane curvature. Arf1 and Arf6 were shown to load GTP in a membrane-curvature-dependent manner and stabilize, or further facilitate, changes in membrane curvature through the insertion of an amphipathic helix.
Keywords: ADP-ribosylation factor family (Arf family), amphipathic helix, clathrin-mediated endocytosis, coatamer protein (COP), membrane curvature sensing and generation, small G-protein
Abbreviations: AP, adaptor protein; Arf, ADP-ribosylation factor; Arl, Arf-like; BAR, Bin/Amphiphysin/Rvs; COP, coatamer protein; GAP, GTPase-activating protein; GEF, guanine-nucleotide-exchange factor; PIP5K, phosphatidylinositol-4-phosphate 5-kinase; Sf9, Spodoptera frugiperda 9
INTRODUCTION
Small G-proteins cycle between a GTP-bound active state and a GDP-bound inactive state [1,2]. In the active state, these proteins are able to recruit a wide variety of effector molecules that participate in various cellular processes. The GTP-loaded status is regulated positively by GEFs (guanine-nucleotide-exchange factors) and negatively by GAPs (GTPase-activating proteins). A common theme for small G-proteins is that they localize to membranes, and it is here that they execute their functions. Most members of the Ras superfamily of small G-proteins acquire a C-terminal lipid modification (geranylgeranylation or farnesylation) that anchors these proteins to the membrane. Arf (ADP-ribosylation factor) and Arl (Arf-like) family proteins have a myristoylated (C14:0) N-terminal helix that is thought to facilitate membrane binding. The myristoyl moiety is attached to the second residue (a glycine) following removal of the first residue (a methionine).
The six mammalian Arfs can be categorized into three subclasses based on sequence similarities. Class I Arfs (Arf1–Arf3) are involved in the recruitment of membrane-budding machineries, whereas the roles of Class II Arfs (Arf4 and Arf5) remain unclear. Arf6 alone constitutes the third class and this protein has been shown to regulate trafficking in the endocytic system, actin polymerization and cell migration. Structural details of Arf1 and Arf6 have been described through crystallographic interrogation of the GDP- and GTP-bound proteins [3–5]. Surprisingly, the rearrangements induced by GTP loading are relatively minor, but must be sufficient to provide information for the specific recruitment of effectors. In addition, the precise localization of the Arfs has been suggested to allow for specific effector recruitment [6]. The exposure of the N-terminal helix is thought to be closely linked to the GDP/GTP cycle of Arfs. Here binding of GTP causes a shift in the intermolecular switch region, which suggestively displaces the helix that is normally buried in a hydrophobic pocket in the GDP-bound state.
Arf1 and Arf6 are each vital for particular membrane-trafficking events, although it seems that they act via slightly different mechanisms. Arf1 is absolutely required for the recruitment of COPI (coatamer protein I), AP-1 (adaptor protein-1) and GGA (Golgi-localizing γ-adaptin ear homology domain Arf-binding proteins) coat proteins to the Golgi. Furthermore, Arf1 has been suggested to recruit AP-3 and AP-4 [7]. However, the precise mechanism of action of Arf1 at the budding vesicle remains elusive. It has been shown that, in addition to recruiting coat proteins, Arf1 couples the generation of vesicles to the incorporation of cargo molecules [8,9]. It has previously been demonstrated that, when cargo is present, the association between Arf1 and AP-1 is stabilized and ArfGAP1 activity is inhibited [10].
Arf6 localizes to the plasma membrane and endosomal system, where it is known to regulate both clathrin-dependent and -independent endocytic pathways, as well as endomembrane recycling mechanisms. This is not thought to be facilitated via direct recruitment of coat proteins, but rather through the generation of distinct membrane domains [11]. Arf6 activates phospholipase D and PIP5K (phosphatidylinositol-4-phosphate 5-kinase). Arf6 and PIP5K act synergistically to generate membrane domains enriched in PtdIns(4,5)P2, thereby helping to recruit the protein-budding machineries that rely on this phospholipid.
Generation of membrane curvature is a prerequisite for the budding of vesicles from a flat membrane. Recent evidence suggests that Arf effectors and Arfs themselves might be directly involved in membrane remodelling. The recruitment of the COPII (coatamer protein II) budding machinery to the endoplasmic reticulum is dependent on Sar1, a small G-protein that is closely related to the Arfs. Sar1 was fairly recently shown to induce membrane curvature [12]. In addition, overexpression of regulatory GAPs (centaurins) or GEFs [EFA6 (exchange factor for Arf6)] for Arfs have been found to result in the tubulation of cellular membranes [13,14]. This, coupled with our previous observations of amphipathic helix-mediated membrane curvature generation [15], led us to study the role of Arfs as membrane curvature regulators. In the present study we have used purified proteins to study the role of Arfs in generating membrane curvature and how this ability is influenced by, and influences, GTP loading.
EXPERIMENTAL
cDNA construct preparation
cDNA constructs encoding human Arf1 and Arf6 were obtained from the cDNA Resource Center, Guthrie Research Institute, Sayre, PA, U.S.A. Full-length Arf1 (residues 1–181), Arf1 Δ N-terminus (residues 18–181) and full-length Arf6 (residues 1–175) were subsequently cloned into the pBac4×1 vector for baculovirus expression or the pET15b vector for bacterial expression. The Arf6 K3E mutation was generated by site-directed mutagenesis. The plasmid encoding N-myristoyltransferase was kindly provided by Dr Sean Munro of this laboratory.
Protein expression and protein purification
Histidine-tagged myristoylated Arf1, Arf6 and the Arf6 K3E mutant were purified from baculovirus-infected Sf9 (Spodoptera frugiperda 9) insect cells using nickel–agarose beads and gel filtration. Arf1 was expressed in Escherichia coli BL21 strain with or without the N-myristoyltransferase-encoding plasmid mentioned above and purified as described in [16].
Liposome binding and tubulation
Generation of liposomes from total brain lipids (Folch fraction I) (Sigma–Aldrich) or synthetic lipids (Avanti Polar Lipids), and specification of liposome diameter (made by sequential filtration) were performed as previously described [13]. The sequential filtration results in slightly lower concentrations of smaller-diameter versus larger-diameter liposomes being present in assays. Liposome-binding assays were performed as previously described [13]. Briefly, proteins were incubated together with 1 mg/ml liposomes, followed by centrifugation and analysis of the pellet and supernatant by SDS/PAGE and Coomassie Blue staining. In vitro liposome-tubulation assays were performed and the results analysed as previously described [13].
Real-time fluorescence GTP-loading assay
GTP loading of Arfs was measured as described in [17]. Briefly, purified GDP-loaded Arfs (0.5 μM) were incubated at 37 °C in the presence or absence of liposomes (final concn. 0.1 mg/ml Folch lipids) and HKM buffer (50 mM Hepes, pH 7.2, 120 mM potassium acetate and 1 mM MgCl2) and loading was induced by sequential addition of 1 mM GTP and 2 mM EDTA. Tryptophan fluorescence was measured at 340 nm after excitation at 297.5 nm in a FluoroMax-2 fluorimeter (Horiba Jobin Yvon Ltd.). Data was normalized by setting the fluorescence value before induction of GTP loading to zero and the value obtained after 1000 s for the 50 nm liposomes and Arf1/Arf6 respectively as the maximum value in each experimental set. All data were processed identically and are presented as a percentage of the maximum value to demonstrate differences in GTP loading.
RESULTS
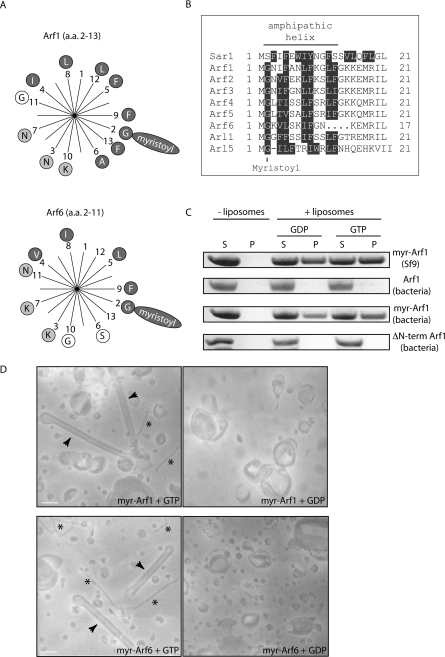
In the Arf–GDP crystal structure, amino acids 2–10 form a helical structure buried in a hydrophobic pocket in the core of the protein (PDB code: 1HUR). However, upon the structural rearrangements induced by GTP binding, this helix is thought to be displaced from its groove. It was previously shown that myristoylation of the N-terminal helix is absolutely required for membrane binding in the GDP-bound state and that point mutations in this helix decreased binding to membranes [17,18]. Helical-wheel presentations of the N-terminal amino acids of Arf1 and Arf6 show that, when the myristoylation of the first glycine residue is taken into account, the helices have an amphipathic nature (Figure 1A). Sequence comparison of the N-termini of Arfs and Arls shows that the amphipathic properties of the first 13 amino acids are conserved in many family members (Figure 1B). We analysed the binding of purified Arf1 to artificially generated liposomes in the presence of GTP or GDP. When Arf1 was purified from insect cells or bacteria co-expressing N-myristoyltransferase, which enables myristoylation, robust membrane binding was observed. This was true for both GTP- and GDP-loaded Arf1, although more efficient binding was observed when GTP was included in the assay. However, if Arf1 was purified without myristoylation or without the N-terminus, no significant binding could be detected (Figure 1C). These results are in agreement with the findings of previous studies and demonstrate that our proteins are functional and behave as would be expected [16,17]. Similar results were obtained for Arf6, but Arf6 bound even more strongly to membranes in the absence of GTP, probably due to binding sites on the protein core ([19,20] and results not shown).
Figure 1. Arfs induce membrane curvature in a GTP-dependent manner.
(A) Helical-wheel representation of amino acids (a.a.) 2–13 in Arf1 (top wheel) and 2–11 in Arf6 (bottom wheel). Hydrophobic and charged amino acids are highlighted by dark grey circles or light grey circles respectively. Myristoylation of the second residue (glycine) is depicted by the long oval containing the word ‘myristoyl’. (B) Sequence alignment of the N-terminal amino acids of Arf/Arl family members. Hydrophobic amino acids are highlighted in dark grey. (C) Liposome co-sedimentation assays. Liposomes generated from brain-derived lipids (Folch fraction I) were incubated with the indicated proteins and GTP or GDP before centrifugation and analysis of supernatants (S) and pellets (P) by SDS/PAGE and Coomassie Blue staining. A control experiment where protein was incubated in the absence of liposomes is shown. The proteins used were myristoylated Arf1 purified from Sf9 insect cells [myr-Arf1 (Sf9)], Arf1 purified from bacteria [Arf1 (bacteria)], myristoylated Arf1 purified from bacteria co-expressing N-myristoyltransferase [myr-Arf1 (bacteria)] and N-terminally truncated Arf1 purified from bacteria [ΔN-term Arf1 (bacteria)]. (D) Electron micrographs of negatively stained 800 nm liposomes incubated in the presence of equal concentrations of proteins (10 μM) and nucleotides (100 μM) as indicated. Arrowheads indicate tubules that are approx. 45 nm in diameter and asterisks indicate tubules that are 13–17 nm wide. Scale bars represent 100 nm. Abbreviation: myr, myristoylated.
The association of Arfs with membranes even in the GDP-bound state suggested that membrane binding of the myristoylated protein does not necessarily involve the exposure of the amphipathic helix. Thus the helix itself might not be absolutely required for binding, but may instead have a more elaborate role. We reasoned that, following GTP-dependent release of the amphipathic helix, it might insert into the lipid bilayer and subsequently impose positive membrane curvature by acting as a wedge [21]. To study this, liposomes were incubated together with myristoylated Arf1 or Arf6 and GTP or GDP and analysed using electron microscopy (Figure 1D). We found that the normal spherical morphology of the liposomes always persisted after incubation with either GDP-loaded Arf1 or Arf6. However, after incubation with either GTP-loaded Arf1 or Arf6, we consistently observed a drastic change in the shape of many of the liposomes. Frequently, straight tubular structures with an apparent diameter of 45±5 nm were observed (Figure 1D; arrowheads). In addition, smaller 13–17-nm-diameter tubular structures were detected (Figure 1D; asterisks). We have observed similar structures on rare occasions with the N-BAR domain of amphiphysin (results not shown). [N-BAR domains are BAR (Bin/Amphiphysin/Rvs) domains with an N-terminal amphipathic helix.] The nature of such small structures is uncertain, but might result from single layers of lipids being forced into tubular micelles. These results suggest that Arfs generate high membrane curvature in a GTP-dependent manner through the insertion of a helix in the lipid bilayer.
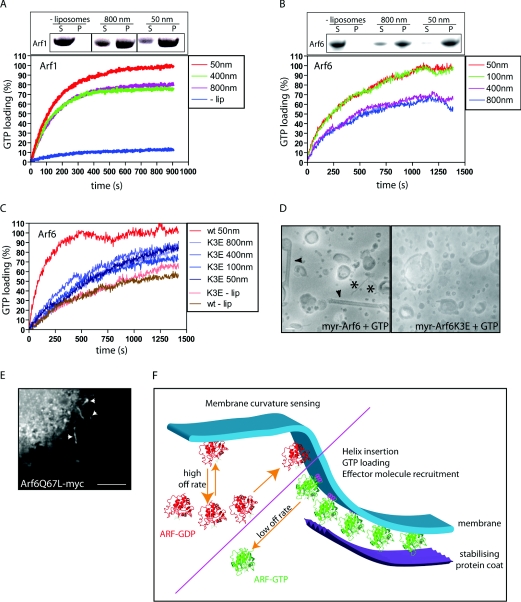
In theory, the insertion of a helix into the membrane would be favoured by positive membrane curvature, since the lipid headgroups apposing the helix would be more separate from one another. This would be reflected by increased membrane binding as the liposome diameter decreases. To test this, we generated liposomes with low curvature (800 nm in diameter) and high curvature (50 nm in diameter) and assayed binding of Arf1–GTP and Arf6–GTP to these liposomes (see the insets to Figures 2A and 2B and see the Discussion). The results indicate that both Arf1 and Arf6 show a small increase in binding in the presence of higher-curvature liposomes (insets to Figures 2A and 2B). Curvature-sensitivity in this assay may reflect an increased amount of GTP loading, since amphipathic helix insertion occurs in response to GTP binding (Figure 1D), which would decrease protein off-rates from membranes. We therefore studied GTP loading in the presence of liposomes of different diameters. The structural rearrangements in Arfs upon GTP binding can be detected by an increase in tryptophan fluorescence [22]. This can be used to analyse GTP loading in real time [17]. No increase in tryptophan fluorescence was observed on the binding of GDP-loaded Arf1/Arf6 to membranes (results not shown). This strongly suggests that the amphipathic helix does not change conformation upon lipid binding per se, a finding consistent with the observation that this helix is buried in the core of the GDP-bound Arf [15]. By incubating Arf1 with liposomes of different diameters in the presence of GTP, we were able to observe that Arf1 was loaded more quickly with GTP, and to a greater extent, in the context of higher liposome curvature (Figure 2A). As was previously observed, GTP loading was very slow in the absence of membranes [17,23]. Similar results were obtained for the loading of Arf6 (Figure 2B). Thus curvature-sensitive GTP loading may be a general property of Arf-family proteins. Liposomes of different diameters did not on their own yield any significant change in fluorescence (Figure 2C and results not shown). The increase in the extent of GTP loading in the context of higher membrane curvature probably reflects a change in the steady-state distribution of Arfs, with more being found bound to the more-highly-curved membranes.
Figure 2. Arf-family proteins are activated by high positive membrane curvature.
(A)–(C) Indicated proteins were incubated together with or without liposomes (−lip) of the indicated diameters and GTP. GTP loading was analysed in real time by measuring tryptophan fluorescence (see the Experimental section). Fluorescence is presented as a percentage of the value at 1000 s for 50 nm liposomes and Arf1 and Arf6 respectively. Insets in (A) and (B) shows liposome binding assays of Arf1 (A) and Arf6 (B) in the absence or presence of liposomes of different diameters. S, supernatant; P, pellet. Results shown in (A)–(C) are representative of three independent experiments with the same set of proteins and lipids. (D) Electron micrographs of negatively stained liposomes incubated together with GTP-loaded Arf6 (left) and Arf6 K3E (right). The scale bar represents 100 nm. The asterisks and arrowheads have the same meaning as in Figure 1. myr, myristoylated. (E) Epifluorescent micrograph of a HeLa cell expressing a myc-tagged constitutively active Arf6 mutant (Arf6 Q67L-myc). Arrowheads indicate tubular localization. The scale bar represents 5 μm. (F) Schematic model of the GTP-dependent role of Arfs as curvature sensors at the rim of a budding vesicle. Arfs loaded with GDP (ARF–GDP) display a transient membrane binding while Arfs bound to GTP (ARF–GTP) stay membrane-associated within the diffusion barrier generated by positive membrane curvature.
Our present results suggest that the N-terminus of Arfs is responsible for both curvature sensing and generation. We therefore created a point mutant (Arf6 K3E) in the helix to disrupt a positively charged residue that would be expected to align with the negatively charged lipid headgroups once the helix is inserted in the membrane. We used Arf6 as a model, since this protein showed greater curvature-sensitivity than did Arf1. We chose to mutate this particular residue since, according to the structure, this mutation should not affect myristoylation or localization of the helix in the GDP-bound state (having no interaction with other residues in the crystal structure), while being sufficiently N-terminal to be readily accessible for membrane insertion. When the Arf6 K3E mutant was assayed for its ability to load GTP, we found that this protein loads GTP at the same rate as wild-type Arf6 in the absence of liposomes (Figure 2C). However, when liposomes of differing diameters were added to the reaction mixture, we found that Arf6 K3E showed a slight increase in the GTP-loading rate, but this increase was independent of liposome diameter. This strongly suggests that the introduced mutation renders the helix unable to properly insert in the membrane and therefore Arf6 K3E cannot respond to membrane curvature. We next examined whether the K3E mutation affected membrane curvature generation. Liposomes that had been incubated together with wild-type Arf6 or Arf6 K3E were analysed by electron microscopy. Indeed we found that, in contrast with wild-type Arf6, Arf6 K3E was unable to tubulate liposomes (Figure 2D). To resolve whether GTP loading might result in a tubular localization of Arf6 in vivo, we expressed constitutively GTP-associated Arf6 (Arf6 Q67L-myc). This protein was indeed found on tubular structures in cells, which is consistent with curvature-sensitive membrane binding of this protein.
Taken together, our data are consistent with a model in which Arf family proteins function as membrane curvature sensors and further facilitate membrane budding through direct GTP-dependent membrane remodelling (Figure 2E). In the GDP-bound state, myristoyl-enabled membrane binding of Arfs is insufficient, owing to a relatively high off-rate. This can be overcome by the local high positive membrane curvature seen, for example, at a budding vesicle. In addition, GEFs function to induce local GTP loading of Arfs that subsequently generate curvature via insertion of the helix, thereby creating a positive-feedback loop. Once the helix is inserted, a diffusion barrier is probably created, since Arfs are probably unable move into the negative curvature surrounding the positive curvature of the budding membrane (tubule or vesicle). A high local concentration of Arf–GTP with lowered off-rate is obtained that can recruit effector molecules to this precise location where budding is in progress. Subsequently, coat proteins are recruited that help to stabilize the budding vesicle.
DISCUSSION
Cellular processes such as morphogenesis and signalling are complex, and regulatory proteins are required to orchestrate and drive the membrane remodelling, cargo sorting and cytoskeletal rearrangements that must occur during such events. Small G-proteins of the Arf and Arl families function as such regulatory units, owing to their ability to define processes in a spatial and temporal manner and transmit this information to effector molecules.
In the present study we have shown that Arf family proteins generate and sense membrane curvature through an amphipathic helix that can be inserted into the lipid bilayer in a GTP-driven manner. Given that amphipathic helix insertion leads to the induction of local positive membrane curvature, this can result in curvature-sensitivity in the rate of Arf insertion. Furthermore, once a helix has inserted, this will help to drive the curvature preferred by the next Arf molecules. Through this mechanism, GTP-loaded Arfs are organized in high concentrations at local sites of positive membrane curvature. In this context, membrane curvature functions as a base for protein recruitment. This ability makes Arfs unique among small G-proteins and might explain why they are used in membrane budding. Previous studies on Arf1 and Sar1p have shown that mutations in the helix that renders it less hydrophobic results in an increased off-rate from the membrane [12,17]. These findings are consistent with our results and show that mutations in the helix affect both membrane binding and curvature-sensitivity. Also, the binding of Arf–GTP to a membrane feeds back to generate or stabilize even higher curvature, which will drive the progression of membrane budding both directly and indirectly through increased recruitment of curvature effector proteins. Since insertion of the helix is dependent upon the GDP/GTP status of Arf, the procedure can be arrested or promoted by GAPs and GEFs respectively. Interestingly several Arf-specific GAPs and GEFs are predicted to contain BAR domains that are known to sense, stabilize and generate membrane curvature [13]. This is in agreement with our model for the function and regulation of Arfs. Our results could also suggest that the need for GEFs in membrane budding might be to initially recruit Arfs and that, once the curvature is generated, Arfs can drive the process via their curvature-sensitivity. It has been proposed that ArfGAP1, which is curvature-sensitive owing to helix insertion, promotes hydrolysis of Arf–GTP at the positive curvature on the vesicle, but that Arf–GTP at the vesicular neck, with negative curvature, is unaffected [24]. However, from our results it is difficult to envisage how Arf-GTP can be located at sites of negative curvature while ArfGAP cannot, since they both localize better to increasing positive membrane curvature.
Arf6 has been ascribed several roles, which include the regulation of membrane trafficking, actin rearrangements and membrane–cytoskeleton interactions [2,25]. We found that the amphipathic helix of Arf6 is shorter than the helix in other Arf-family members. In addition, Arf6 is thought to be continuously associated with membranes, perhaps since core residues in Arf6 bind more efficiently to membranes than those of others such as Arf1. This suggests that the role of Arf6 may have evolved to function more as a sensor than as a generator of membrane curvature. Arf6 has been shown to function in clathrin-independent endocytic and recycling pathways for MHC class I proteins and CD59 [26,27]. Morphologically, these pathways are characterized by tubular rather than vesicular structures. Our results suggest that Arf6 is adapted to localize and function at such membrane tubules. It is possible that the shorter helix in Arf6 is more suitable in this system. The role of Arf6 has been suggested to be largely dependent upon effects on PtdIns(4,5)P2 levels [14]. Our in vitro data, however, show that Arf6 also has a direct effect on lipid packing, resulting in, and depending upon, membrane shape modulation.
In conclusion, we propose that proteins in the Arf and Arl families of small G-proteins serve as GTP-dependent sensors and drivers of membrane shape. This ability will help to define local areas of activity that are fine-tuned by the characteristics of effector molecules. The unresolved functions of the less-studied Arfs will probably also require the ability to sense membrane curvature. Whether this includes direct roles in vesicular budding, or indirect regulation of membrane remodelling, remains to be seen.
Acknowledgments
We thank Dr Sven Carlsson (Department of Medical Biochemistry and Biophysics, Umeå University, Umeå, Sweden) for advice. R. L. was supported by the Swedish Research Council, the Medical Faculty of Umeå University and the Magn. Bergvalls Foundation. G. J. D. was supported by a Trinity College Cambridge Internal Graduate Studentship. We declare no competing interests.
References
- 1.Gillingham A. K., Munro S. The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- 2.D'souza-Schorey C., Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 3.Pasqualato S., Menetrey J., Franco M., Cherfils J. The structural GDP/GTP cycle of human Arf6. EMBO Rep. 2001;2:234–238. doi: 10.1093/embo-reports/kve043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amor J. C., Harrison D. H., Kahn R. A., Ringe D. Structure of the human ADP-ribosylation factor 1 complexed with GDP. Nature. 1994;372:704–708. doi: 10.1038/372704a0. [DOI] [PubMed] [Google Scholar]
- 5.Menetrey J., Macia E., Pasqualato S., Franco M., Cherfils J. Structure of Arf6–GDP suggests a basis for guanine nucleotide exchange factors specificity. Nat. Struct. Biol. 2000;7:466–469. doi: 10.1038/75863. [DOI] [PubMed] [Google Scholar]
- 6.Peters P. J., Hsu V. W., Ooi C. E., Finazzi D., Teal S. B., Oorschot V., Donaldson J. G., Klausner R. D. Overexpression of wild-type and mutant ARF1 and ARF6: distinct perturbations of nonoverlapping membrane compartments. J. Cell Biol. 1995;128:1003–1017. doi: 10.1083/jcb.128.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson M. S. Adaptable adaptors for coated vesicles. Trends Cell Biol. 2004;14:167–174. doi: 10.1016/j.tcb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Bremser M., Nickel W., Schweikert M., Ravazzola M., Amherdt M., Hughes C. A., Sollner T. H., Rothman J. E., Wieland F. T. Coupling of coat assembly and vesicle budding to packaging of putative cargo receptors. Cell. 1999;96:495–506. doi: 10.1016/s0092-8674(00)80654-6. [DOI] [PubMed] [Google Scholar]
- 9.Lanoix J., Ouwendijk J., Lin C. C., Stark A., Love H. D., Ostermann J., Nilsson T. GTP hydrolysis by Arf-1 mediates sorting and concentration of Golgi resident enzymes into functional COPI vesicles. EMBO J. 1999;18:4935–4948. doi: 10.1093/emboj/18.18.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer D. M., Crottet P., Maco B., Degtyar E., Cassel D., Spiess M. Oligomerization and dissociation of AP-1 adaptors are regulated by cargo signals and by ArfGAP1-induced GTP hydrolysis. Mol. Biol. Cell. 2005;16:4745–4754. doi: 10.1091/mbc.E05-06-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donaldson J. G. Arfs, phosphoinositides and membrane traffic. Biochem. Soc. Trans. 2005;33:1276–1278. doi: 10.1042/BST0331276. [DOI] [PubMed] [Google Scholar]
- 12.Lee M. C., Orci L., Hamamoto S., Futai E., Ravazzola M., Schekman R. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 2005;122:605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 13.Peter B. J., Kent H. M., Mills I. G., Vallis Y., Butler P. J., Evans P. R., McMahon H. T. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 14.Brown F. D., Rozelle A. L., Yin H. L., Balla T., Donaldson J. G. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J. Cell Biol. 2001;154:1007–1017. doi: 10.1083/jcb.200103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ford M. G., Mills I. G., Peter B. J., Vallis Y., Praefcke G. J., Evans P. R., McMahon H. T. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 16.Franco M., Chardin P., Chabre M., Paris S. Myristoylation of ADP-ribosylation factor 1 facilitates nucleotide exchange at physiological Mg2+ levels. J. Biol. Chem. 1995;270:1337–1341. doi: 10.1074/jbc.270.3.1337. [DOI] [PubMed] [Google Scholar]
- 17.Antonny B., Beraud-Dufour S., Chardin P., Chabre M. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry. 1997;36:4675–4684. doi: 10.1021/bi962252b. [DOI] [PubMed] [Google Scholar]
- 18.Randazzo P. A., Terui T., Sturch S., Fales H. M., Ferrige A. G., Kahn R. A. The myristoylated amino terminus of ADP-ribosylation factor 1 is a phospholipid- and GTP-sensitive switch. J. Biol. Chem. 1995;270:14809–14815. doi: 10.1074/jbc.270.24.14809. [DOI] [PubMed] [Google Scholar]
- 19.Paleotti O., Macia E., Luton F., Klein S., Partisani M., Chardin P., Kirchhausen T., Franco M. The small G-protein Arf6GTP recruits the AP-2 adaptor complex to membranes. J. Biol. Chem. 2005;280:21661–21666. doi: 10.1074/jbc.M503099200. [DOI] [PubMed] [Google Scholar]
- 20.Amor J. C., Horton J. R., Zhu X., Wang Y., Sullards C., Ringe D., Cheng X., Kahn R. A. Structures of yeast ARF2 and ARL1: distinct roles for the N terminus in the structure and function of ARF family GTPases. J. Biol. Chem. 2001;276:42477–42484. doi: 10.1074/jbc.M106660200. [DOI] [PubMed] [Google Scholar]
- 21.Campelo F., McMahon H. T., Kozlov M. M. The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys. J. 2008. [DOI] [PMC free article] [PubMed]
- 22.Kahn R. A., Gilman A. G. The protein cofactor necessary for ADP-ribosylation of Gs by cholera toxin is itself a GTP binding protein. J. Biol. Chem. 1986;261:7906–7911. [PubMed] [Google Scholar]
- 23.Weiss O., Holden J., Rulka C., Kahn R. A. Nucleotide binding and cofactor activities of purified bovine brain and bacterially expressed ADP-ribosylation factor. J. Biol. Chem. 1989;264:21066–21072. [PubMed] [Google Scholar]
- 24.Antonny B., Bigay J., Casella J. F., Drin G., Mesmin B., Gounon P. Membrane curvature and the control of GTP hydrolysis in Arf1 during COPI vesicle formation. Biochem. Soc. Trans. 2005;33:619–622. doi: 10.1042/BST0330619. [DOI] [PubMed] [Google Scholar]
- 25.Doherty G. J., McMahon H. T. Mediation, modulation, and consequences of membrane-cytoskeleton interactions. Annu. Rev. Biophys. 2008;37:65–95. doi: 10.1146/annurev.biophys.37.032807.125912. [DOI] [PubMed] [Google Scholar]
- 26.Radhakrishna H., Donaldson J. G. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J. Cell Biol. 1997;139:49–61. doi: 10.1083/jcb.139.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naslavsky N., Weigert R., Donaldson J. G. Characterization of a nonclathrin endocytic pathway: membrane cargo and lipid requirements. Mol. Biol. Cell. 2004;15:3542–3552. doi: 10.1091/mbc.E04-02-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]