Abstract
A gene encoding an α-l-arabinofuranosidase, designated SaAraf43A, was cloned from Streptomyces avermitilis. The deduced amino acid sequence implies a modular structure consisting of an N-terminal glycoside hydrolase family 43 module and a C-terminal family 42 carbohydrate-binding module (CBM42). The recombinant enzyme showed optimal activity at pH 6.0 and 45°C and was stable over the pH range of 5.0–6.5 at 30°C. The enzyme hydrolyzed p-nitrophenol (PNP)-α-l-arabinofuranoside but did not hydrolyze PNP-α-l-arabinopyranoside, PNP-β-d-xylopyranoside, or PNP-β-d-galactopyranoside. Debranched 1,5-arabinan was hydrolyzed by the enzyme but arabinoxylan, arabinogalactan, gum arabic, and arabinan were not. Among the synthetic regioisomers of arabinofuranobiosides, only methyl 5-O-α-l-arabinofuranosyl-α-l-arabinofuranoside was hydrolyzed by the enzyme, while methyl 2-O-α-l-arabinofuranosyl-α-l-arabinofuranoside and methyl 3-O-α-l-arabinofuranosyl-α-l-arabinofuranoside were not. These data suggested that the enzyme only cleaves α-1,5-linked arabinofuranosyl linkages. The analysis of the hydrolysis product of arabinofuranopentaose suggested that the enzyme releases arabinose in exo-acting manner. These results indicate that the enzyme is definitely an exo-1,5-α-l-arabinofuranosidase. The C-terminal CBM42 did not show any affinity for arabinogalactan and debranched arabinan, although it bound arabinan and arabinoxylan, suggesting that the CBM42 bound to branched arabinofuranosyl residues. Removal of the module decreased the activity of the enzyme with regard to debranched arabinan. The CBM42 plays a role in enhancing the debranched arabinan hydrolytic action of the catalytic module in spite of its preference for binding arabinofuranosyl side chains.
Keywords: α-l-Arabinofuranosidase, Glycoside hydrolase family 43, Carbohydrate binding module family 42, Arabinan binding module, Streptomyces avermitilis
Introduction
l-Arabinose residues are widely distributed in plant cell walls, where they are present in polymers such as arabinans, arabinoxylans, arabinogalactans, and arabinogalactan proteins (Carpita and Gibeaut 1993). Plant cell walls are becoming important because worldwide attention has now focused on bioethanol production to combat global warming and to safeguard global energy. Because of competition between food and fuel production, lignocellulose is expected to be utilized for fuel ethanol production in the future. Generally, lignocellulose is composed of almost 40% cellulose together with about 20% hemicelluloses which mainly consist of pentoses such as xylose and arabinose (Saha 2003). Hemicelluloses are often detrimental to the production of bio-ethanol because pentoses are less efficiently converted to ethanol than are hexoses (Aristidou and Penttilä 2000; Skoog and Hahn-Hägerdal 1988).
In contrast, l-arabinose has potential as a useful sugar in the food industry. The sugar has a sweet taste but is not readily absorbed by the body (Seri et al. 1996). l-Arabinose selectively inhibits intestinal sucrase in a noncompetitive manner and reduces the glycemic response after sucrose ingestion in animals (Seri et al. 1996; Sanai et al. 1997; Osaki et al. 2001). Thus, l-arabinose may be useful in preventing excess sucrose utilization.
Because the structure of l-arabinose-containing polysaccharides are highly variable and complex, a wide variety of α-l-arabinofuranosidases (EC 3.2.1.55) which have various substrate specificities are necessary for the hydrolysis of such polysaccharides and the production of l-arabinose. We have previously purified some α-l-arabinofuranosidases and elucidated their substrate specificities towards structurally defined substrates (Kaneko et al. 1993; Kaneko et al. 1994; Kaneko et al. 1995; Kaneko et al. 1998a; Kaneko et al. 1998b; Kaneko et al. 1998c; Kaneko et al. 1998d; Kaneko and Kusakabe 1995; Kawabata et al. 1995; Kusakabe et al. 1975; Matsuo et al. 2000; Yang et al. 2006; Yoshida et al. 1994). The substrate specificities of the α-l-arabinofuranosidases studied were very broad; however, one of the enzymes (α-l-arabinofuranosidase II) from Streptomyces chartreusis (Matsuo et al. 2000) has strict substrate specificity. This enzyme hydrolyzed only the α-1,5-linkages of linear arabinan and arabinooligosaccharides in an exo-acting manner (Matsuo et al. 2000). The amino acid sequence of the enzyme indicated that this enzyme belongs to family 43 (GH43) of the glycoside hydrolase family (http://www.cazy.org/CAZY/). Because the substrate specificity and the amino acid sequence were quite distinct from known α-l-arabinofuranosidases, it represents a novel type of enzyme as an exo-1,5-α-l-arabinofuranosidase (Matsuo et al. 2000).
The GH43 family incorporates a wide variety of enzyme activities including β-xylosidase (EC 3.2.1.37), α-l-arabinofuranosidase (exo-1,5-α-l-arabinofuanosidase), bifunctional β-xylosidase/α-l-arabinofuranosidase, endo-arabinanase (EC 3.2.1.99), β-xylanase (EC 3.2.1.8), and exo-β-1,3-galactanase (EC 3.2.1.145). Therefore, a detailed functional characterization of α-l-arabinofuranosidase II using the recombinant enzyme with its mutants would be interesting because all the enzymes belonging to GH43 have the same holding and catalysis in the same inverting mechanism even though the enzyme activities are different. We have attempted heterologous expression of α-l-arabinofuranosidase II from S. chartreusis, but unfortunately, the effort was unsuccessful. When the amino acid sequence of exo-1,5-α-l-arabinofuranosidase from S. chartreusis was subjected to a BLAST search, a similar gene from Streptomyces avermitilis (SAV1043) was identified. The gene designated SaAraf43A gene encodes not only a GH43 catalytic domain similar to α-l-arabinofuranosidase II from S. chartreusis but also the family 42 carbohydrate-binding module (CBM42) at the C-terminus. In the present study, the gene was cloned from S. avermitilis, and the recombinant protein expressed in Escherichia coli was characterized.
Materials and methods
Substrates
p-Nitrophenyl α-l-arabinofuranoside (PNP-α-l-Araf), p-nitrophenyl α-l-arabinopyranoside (PNP-α-l-Arap), p-nitrophenyl β-d-galactopyranoside (PNP-β-d-Galp), p-nitrophenyl β-d-xylopyranoside (PNP-β-d-Xylp) and larch wood arabinogalactan, oat spelts xylan, and birchwood xylan were purchased from Sigma Chemical Company (St. Louis, MO, USA). Debranched arabinan, wheat arabinoxylan, and arabinopentaose were obtained from Megazyme International (Wicklow, Ireland). Gum arabic was obtained from Nacali Tesque (Kyoto, Japan). Nihon Syokuhin Kakoh (Fuji, Japan) supplied corn hull arabinoxylan. Sugar-beet arabinan was prepared according to methods described elsewhere (Kusakabe et al. 1975). Methyl 2-O-, methyl 3-O-, and methyl 5-O-α-l-arabinofuranosyl-α-l-arabinofuranosides (arabinofuranobiosides) were synthesized as described previously (Kawabata et al. 1995). β-1,3-Galactan was prepared as previously reported (Ichinose et al. 2005).
Expression of recombinant SaAraf43A
The oligonucleotide primers were designed from the terminal of position 1318577–1319938 on the S. avermitilis genome sequence (on the website at http://avermitilis.ls.kitasato-u.ac.jp) for ligation into the NdeI-HindIII site of the expression vector pET30 (Novagen, Madison, WI, USA). The 5′-primer (5′-AACATATGACCGCCCCGGCCTCGCCCTC-3′) contained two adenine residues to prevent removal of the terminal nucleotide by the exonuclease activity of high-fidelity DNA polymerase, the NdeI site (shown in italics) and a stretch of 20 nucleotide residues encoding the N-terminal amino acids of mature SaAraf43A. The 3′-primer (5′-AAAAGCTTTTCTGCGTAGAACGTGGCGTCC-3′) contained two adenine residues, the HindIII site (shown in italics) and 22 nucleotide residues encoding C-terminal amino acids of the mature SaAraf43A. For amplification of the catalytic domain of SaAraf43A, a pair of primers was designed as follows. The 5′-primer (5′-AACATATGACCGCCCCGGCCTCGCCCTC-3′) contained the NdeI site (shown in italics) and a stretch of 20 nucleotide residues encoding the N-terminal amino acids of mature SaAraf43A. The 3′-primer (5′-AAAAGCTTATCAGCCACTGGAATACCGA -3′) contained two adenine residues, the HindIII site (shown in italics) and 20 nucleotide residues encoding C-terminal amino acids of the catalytic domain of SaAraf43A. Polymerase chain reactions (PCRs) were performed with the pair of primers described above using KOD-plus-DNA polymerase (TOYOBO, Osaka, Japan). In the reactions, genomic DNA from S. avermitilis was used as a template. The amplified products were subcloned into pGEM-T Easy vector (Promega, Madison, WI, USA) followed by sequencing analysis. The target genes were digested with NdeI and HindIII and ligated into the corresponding restriction site of the pET30 vector.
E. coli strain BL21 Gold (DE3) (Stratagene, La Jolla, CA, USA) was transformed by the resultant plasmids. The transformants were grown in 100 ml of Luria–Bertani medium at 37°C until they had reached an optimal optical density of 0.2 at 600 nm; expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 24 h at 25°C. The cells were harvested and then resuspended in 5 ml of 50 mM phosphate buffer (pH 7.2) followed by sonication for 5 min. After centrifugation to remove insoluble material, the supernatant was used as the crude enzyme which was purified on a Ni-nitrilotriacetic acid (NTA) agarose (QIAGEN GmbH, Hilden, Germany) column (5 × 50 mm). The eluted enzyme was applied to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and the relevant fractions were pooled and dialyzed against deionized water. The final preparation thus obtained was used as the purified enzyme.
Enzyme assay and measurement of protein
The enzyme assay mixture contained 25 μl 2 mM PNP-α-l-Araf, 20 μl McIlvaine buffer, pH 6.0 (0.2 M Na2HPO4/0.1 M citric acid) and 5 μl of the enzyme solution. The reactions were carried out at 45°C for 10 min and were terminated by the addition of 50 μl of 0.2 M Na2CO3. The amount of PNP released was determined at 400 nm with an extinction coefficient of 19,608 M−1 cm−1. One unit of enzyme activity is defined as the amount of enzyme that released 1 μmol of PNP per minute from PNP-α-l-Araf under these conditions. The protein concentration was determined using the BCA Protein Assay Kit (PIERCE, Rockford, IL, USA) with bovine serum albumin as the standard.
Enzymatic properties
The effects of pH on the activity and stability of SaAraf43A were examined in a series of McIlvaine buffers ranging in pH from 3.0 to 8.0 and in Atkins-Pantin buffers (0.2 M boric acid/0.2 M KCl/0.2 M Na2CO3) ranging from pH 8.0 to 10.0. The activities were assayed under the conditions described for the standard method. In order to determine the effect of pH on enzyme stability, the enzymes were preincubated at various pH values in the absence of substrate at 30°C for 60 min, and the residual activity was then assayed using the standard method. The effects of temperature on the activity of α-l-arabinofuranosidase were determined from 4 to 50°C. With the exception of temperature, the assay conditions were the same as described for the standard method. For the temperature-stability measurements, the enzymes were pre-incubated at various temperatures at pH 6.0 for 60 min, and the residual activity was determined using the standard method.
Substrate specificities
To determine the substrate specificities, various PNP-glycosides were used as substrates. In these cases, the assay method was identical to that described for PNP-α-l-Araf. For polysaccharide and arabinofuranobioside substrates, a solution was prepared containing 5 μl of enzyme solution (0.0015 unit), 20 μl McIlvaine buffer (pH 6.0) and 25 μl 0.5% (w/v) substrate (β-1,3-galactan, gum arabic, larch arabinogalactan, debranched arabinan, arabinan, oat spelt xylan, birchwood xylan, wheat arabinoxylan, corn hull arabinoxylan, or arabinofuranobiosides). After incubation for the appropriate reaction time at 30°C, the reaction was stopped by boiling for 5 min. Analysis of the hydrolysis products of arabinofuranobiosides was carried out by determining the concentration of l-arabinose released by the enzyme using the Somogyi–Nelson method (Somogyi 1952). For the hydrolysis products analysis of debranched arabinan, the enzyme (75 micro units) was incubated with 0.25% (w/v) the substrate at 30°C. The reaction mixtures were subjected to high-performance anion-exchange chromatography and identified using a pulsed amperometric detection (HPAEC-PAD) system. The samples were analyzed using a CarboPac™ PA1 column (4 × 250 mm, Dionex, Sunnyvale, CA, USA) and elution with 0.1 M NaOH (0–5 min), followed by a linear gradient (5–35 min) of sodium acetate (0–0.2 M) at a flow rate of 1 ml/min. For the hydrolysis of arabinopentaose, 25 μl of 0.5% (w/v) arabinopentaose was incubated with 5 μl of the enzyme solution (75 micro units) in the reaction mixture. After incubation for the appropriate reaction time at 30°C, the reaction was stopped by boiling for 5 min. The hydrolysis products were analyzed by HPAEC-PAD analysis as described above.
The catalytic activities of the enzymes with and without CBM42 towards PNP-α-l-Araf and debranched arabinan were compared as follows. The enzyme assay mixture contained 25 μl 2 mM PNP-α-l-Araf, 20 μl McIlvaine buffer, pH 6.0, and 5 μl of 1 μM enzyme solution. The reactions were carried out at 20°C and for periods of up to 20 min. The amount of PNP released was determined at 400 nm with an extinction coefficient of 19,608 M−1 cm−1. For debranched arabinan, a solution was prepared containing 5 μl of 1 μM enzyme solution, 20 μl McIlvaine buffer (pH 6.0), and 25 μl of 0.5% (w/v) substrate. After incubation for the appropriate reaction time at 20°C, the reaction was stopped by boiling for 5 min. For the hydrolysis products of debranched arabinan, the concentration of l-arabinose released by hydrolysis from debranched arabinan was determined using the Somogyi–Nelson method (Somogyi 1952).
Affinity gel electrophoresis
The affinity of SaAraf43A for a range of soluble polysaccharides was determined by affinity gel electrophoresis. The method was essentially as described previously (Takeo 1984), with the exception of the buffer system, and used arabinoxylan from wheat, arabinogalactan from larch, arabinan from sugar beet pulp, and debranched arabinan as the polysaccharides. The separating gels were made up of 7% (w/v) acrylamide in 0.375 M Tris (pH 9.4), and the stacking gels consisted of 2.2% (w/v) acrylamide in 0.125 M Tris (pH 6.8). For the separating gels containing ligands, polysaccharide was added to gel mixtures at concentrations ranging from 0.0005% to 0.2% prior to polymerization. The proteins (5 μg) were electrophoresed at 10 mA/gel for 3 h at 4°C. The cathode buffer (pH 8.9) contained 0.042 M Tris and 0.045 M glycine, and the anode buffer (pH 8.0) was prepared with 0.12 M Tris. SDS-PAGE Standard Low Range (Bio-Rad, Hercules, CA) was used as a negative, non-interacting control. Proteins were stained with Coomassie Brilliant Blue R-250. Quantitative assessment of binding was carried out as described in the previous report (Bolam et al. 2004).
Results
Nucleotide and amino acid sequences of SaAraf43A and expression of recombinant protein
Using the amino acid sequence of α-l-arabinofuranosidase II from S. chartreusis, a gene from S. avermitilis (GenBank accession number BAC68753) encoding a putative protein (SAV1043, annotated as a putative secreted α-l-arabinofuranosidase II in the NCBI database) was found. The DNA sequence was 1,443 bp long and putatively encodes a 481-amino acid protein. The deduced amino acid sequence was compared with sequences in the protein database using BLAST (on a National Center for Biotechnology Information website, http://www.ncbi.nlm.nih.gov/BLAST/). The sequence (residues 28 to 481), SaAraf43A, resembled the following sequences, in order of decreasing similarity: putative α-arabinofuranosidase II from Stigmatella aurantiaca DW4/3-1 (68% identity and 82% similarity, GenBank accession number, EAU68983.1); Clostridium cellulosome enzyme, dockerin type I: glycoside hydrolase, family 43: α-l-arabinofuranosidase B from Clostridium thermocellum ATCC 27405 (62% identity and 79% similarity, GenBank accession number, EAM44536.1); α-arabinofuranosidase II from S. chartreusis [60% identity and 74% similarity, DNA Data Bank of Japan (DDBJ) accession number, BAA90772.1]; hypothetical protein BH1867 from Bacillus halodurans C-125 (59% identity and 74% similarity, DDBJ accession number, BAB05586.1); hypothetical protein from Neurospora crassa OR74A (58% identity and 74% similarity, GenBank accession number, EAA29844.1); and hypothetical protein CHGG_03607 from Chaetomium globosum CBS 148.51 (55% identity and 70% similarity, GenBank accession number, EAQ91672.1). The sequence at the N-terminus (amino acids 1 to 27) was predicted to be a signal sequence (SOSUI[http://bp.nuap.nagoya-u.ac.jp/sosui/]), and residues 28 to 315 of SaAraf43A showed sequence similarity to the GH43 enzymes, while residues 349 to 481 showed similarity with the CBM42 (Fig. 1a,b).
Fig. 1.
a Multiple alignment of the catalytic domains of SaAraf43A and several enzymes belonging to GH43. The alignment was achieved with ClustalW. Identical amino acid residues are enclosed in black boxes. The asterisks indicate the putative catalytic amino acids in CjArb43A and BsArb43A. Boxes L1 to L9 are insertion or deletion loops between exo-1,5-α-l-arabinofuranosidases and α-l-arabinanases. ScAraf43A, exo-1,5-α-l-arabinofuranosidase from S. chartreusis (accession number, BAA90772); CjArb43A, arabinanase from C. japonicus (CAA71485); BsArb43A, arabinanase from B. subtilis (CAA99586). b Multiple alignments of SaCBM42 with other known members of CBM42. The alignment was achieved using ClustalW. Conserved amino acids are shown in black boxes, and asterisks indicate the amino acids related to the substrate binding. AkCBM42, CBM42 of α-l-arabinofuranosidase from A. kawachii (BAB96816); HjCBM42, CBM42 of α-l-arabinofuranosidase from Hypocrea jecorina (CAA93243)
To confirm that SaAraf43A is an exo-1,5-α-l-arabinofuranosidase, the SaAraf43A gene was expressed and then characterized. The DNA fragments encoding the mature region of SaAraf43A and the catalytic domain of SaAraf43A (the CBM42-deleted mutant) were amplified by PCR and then cloned into the expression vector pET30, respectively. The recombinant SaAraf43A was successfully produced in E. coli BL21(DE3) at a level of approximately 280 mg/l after the induction for 24 h at 25°C. The recombinant protein was purified by a Ni-NTA column chromatography and gave a single band on SDS-PAGE when visualized by staining with CBB R-250 (Fig. 2, lane 1). The CBM42-deleted mutant also could be expressed in E. coli in its active form (Fig. 2, lane 2). The molecular mass of both recombinant SaAraf43A and the CBM42-deleted mutant as estimated from SDS-PAGE were found to be 53 and 35 kDa, respectively. These corresponded to their predicted molecular mass (52 and 33 kDa).
Fig. 2.
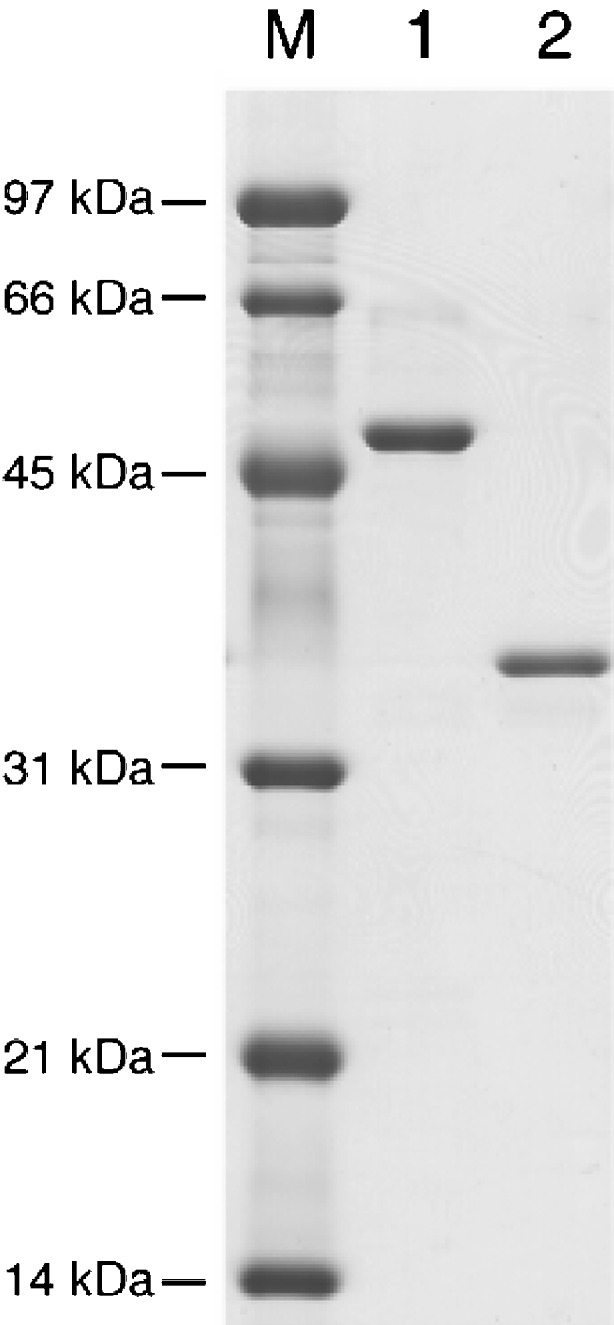
SDS-PAGE of purified SaAraf43A. M Molecular mass standard; lane 1 recombinant SaAraf43A; lane 2 CBM-deficient mutant of SaAraf43A. Approximately 1 μg of each sample was separated on 12% polyacrylamide gel
Properties and substrate specificities of SaAraf43A
When recombinant SaAraf43A was incubated with PNP-α-l-arabinofuranoside, α-l-arabinofuranosidase activity was detected, and the specific activity was 2.92 U/mg. This value is similar to that of α-l-arabinofuranosidase II from S. chartreusis (3.16 U/mg in Matsuo et al. 2000). The enzyme showed maximal activity at pH 6.0 and was stable between pH 5.0 and 6.5, under which conditions more than 80% of the activity was retained (data not shown). The enzyme achieved maximal activity at 45 °C, and more than 80% activity was retained after incubation at 30°C for 60 min (data not shown).
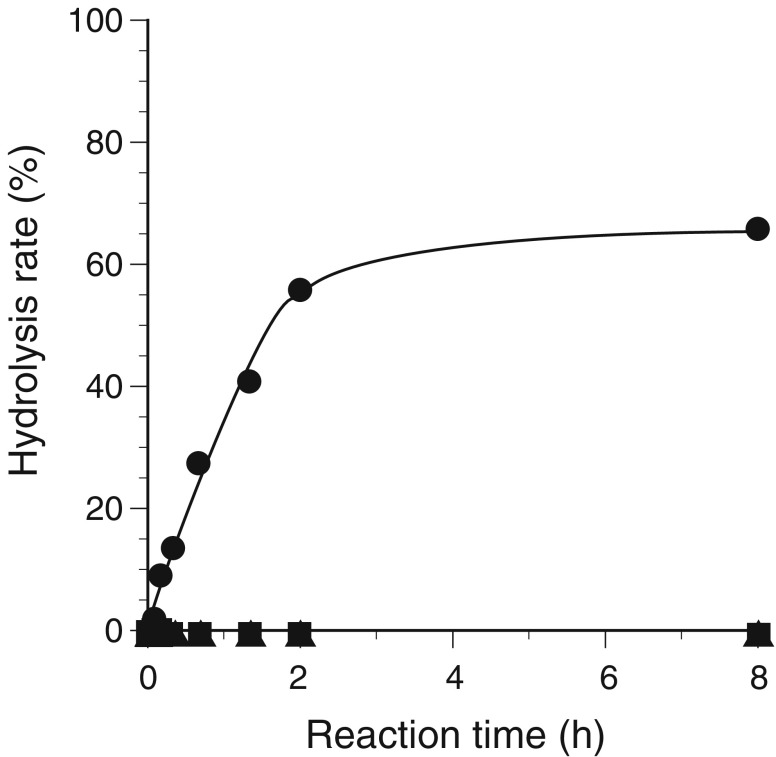
To determine the enzyme activities included in the GH43 family, SaAraf43A was incubated with appropriate substrates (β-1,3-galactan, gum arabic, larch arabinogalactan, debranched arabinan, arabinan, oat spelt xylan, birchwood xylan, wheat arabinoxylan, corn hull arabinoxylan, or PNP-glycosides). The enzyme only showed significant activity toward PNP-α-l-arabinofuranoside among tested PNP-glycosides. When the enzyme was incubated with polysaccharides, SaAraf43A only had significant activity on debranched arabinan which mainly consists of α-1,5-linked arabinofuranosyl residues among the polysaccharides tested. Therefore, the detailed substrate specificities of the enzyme were characterized by using various types of structurally defined arabinofuranooligosaccharides. The degrees of the hydrolysis of three kinds of regioisomer of arabinofuranobiosides by SaAraf43A was monitored over time (Fig. 3). The enzyme hydrolyzed methyl α-l-1,5-arabinofuranobioside to arabinose and methyl α-l-arabinofuranoside but did not hydrolyze the 1,2- and 1,3-linkages (Fig. 3). Those data suggest that SaAraf43A discriminates between the different types of linkage and that the enzyme is specific for 1,5-linked α-arabinofuranosyl residues.
Fig. 3.
Course of the hydrolysis of regioisomeric arabinofuranobiosides by SaAraf43A. Closed square Methyl 2-O-α-l-arabinofuranosyl-α-l-arabinofuranoside; closed triangle methyl 3-O-α-l-arabinofuranosyl-α-l-arabinofuranoside; closed circle methyl 5-O-α-l-arabinofuranosyl-α-l-arabinofuranoside. The enzyme was incubated with 0.5% (w/v) arabinofuranobioside at 30°C for the appropriate time. The hydrolysis rate of the substrate was estimated by the amount of released l-arabinose
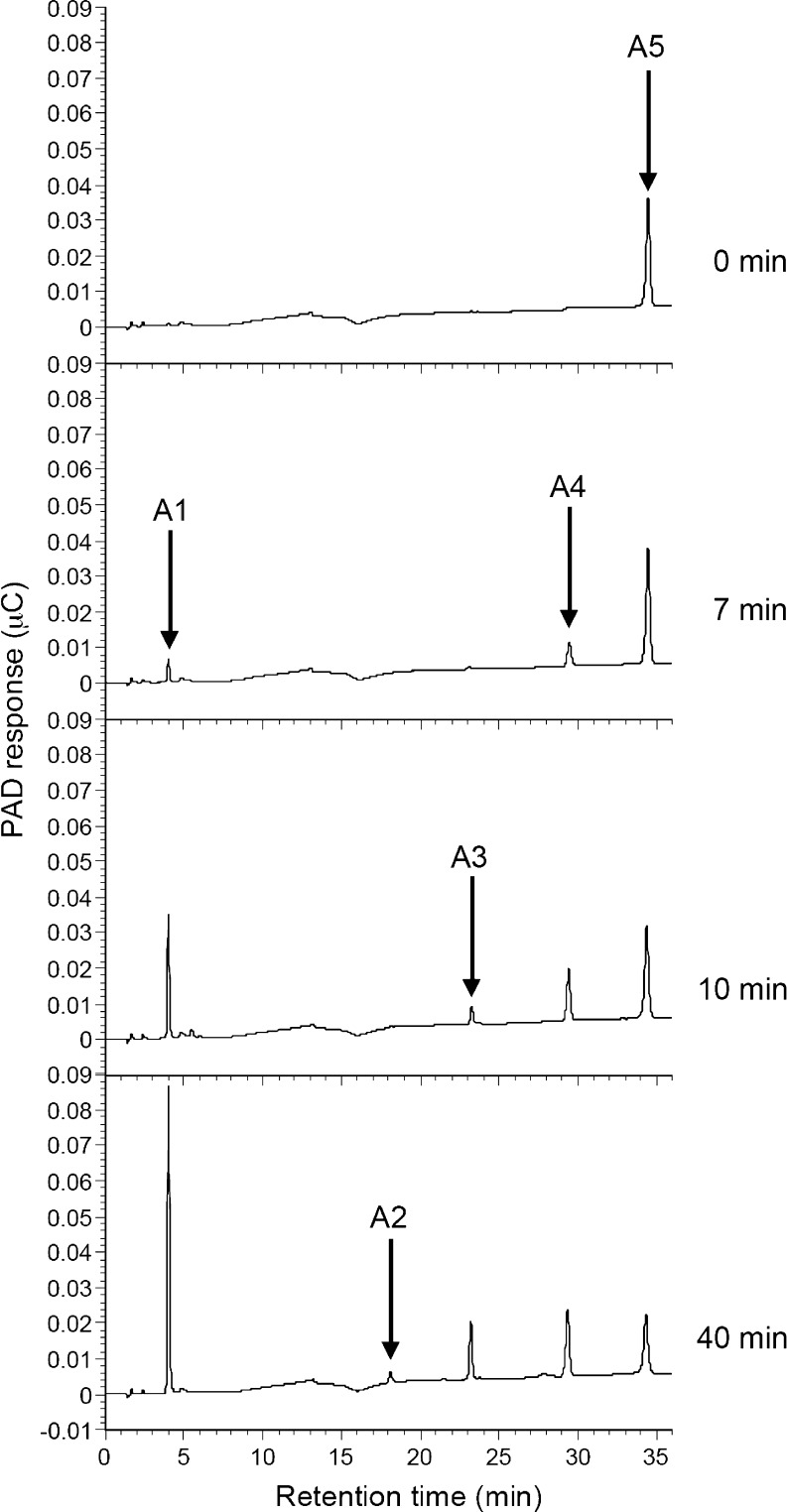
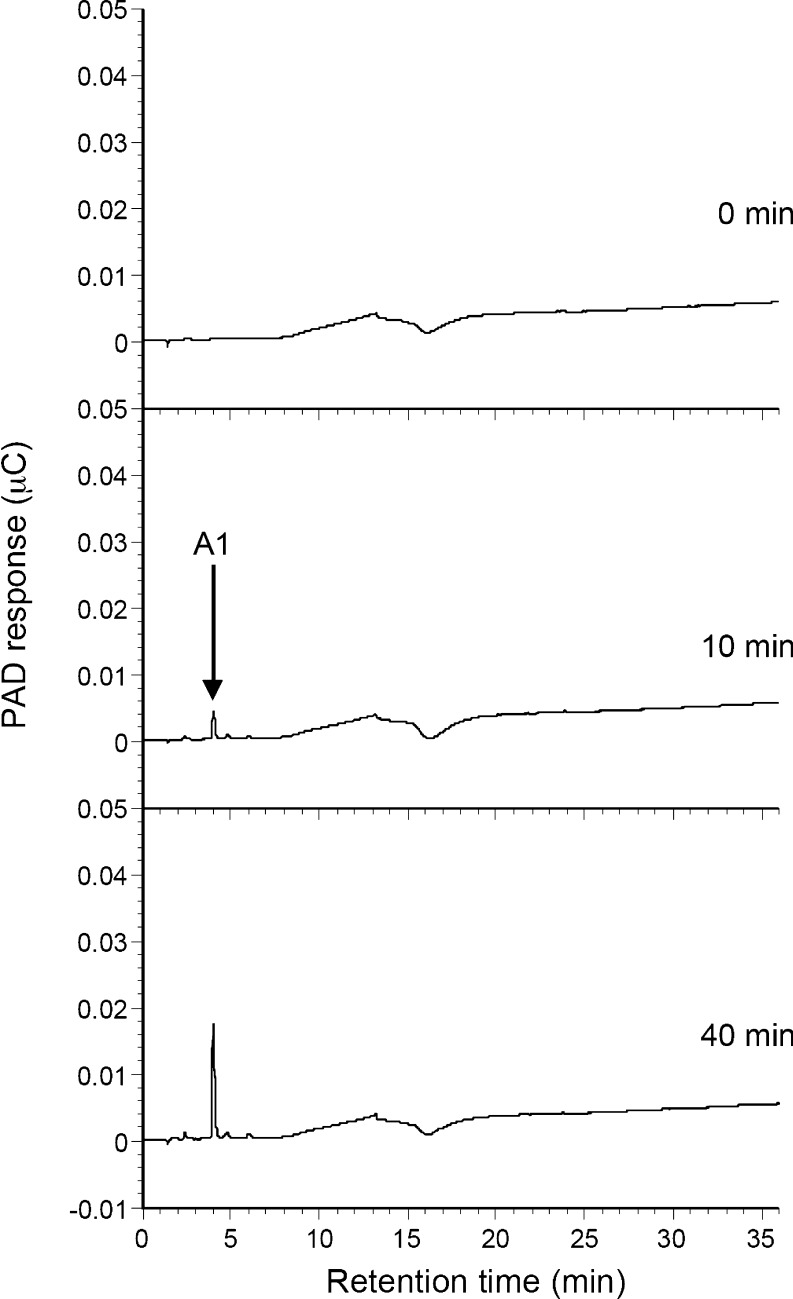
The hydrolysis products of α-1,5-arabinofuranopentaose and debranched arabinan were analyzed by HPAEC-PAD (Fig. 4, 5). When SaAraf43A was incubated with α-1,5-arabinofuranopentaose (Fig. 4), l-arabinose and arabinotetraose were detected as the initial hydrolysis products (after 7 min incubation), and the levels of shorter oligosaccharides such as arabinotriose and arabinobiose then gradually increased as the period of incubation progressed. In contrast, only arabinose was released from debranched arabinan (Fig. 5). These results indicate that the enzyme released arabinofuranose from the substrate in an exo-type mode of action. Thus, SaAraf43A is definitely an exo-1,5-α-l-arabinofuranosidase.
Fig. 4.
HPAEC-PAD analysis of the hydrolysis products of α-1,5-arabinopentaose by SaAraf43A. The enzyme was incubated with 0.5% (w/v) arabinopentaose at 30°C for the appropriate time; the samples were subjected to HPAEC-PAD analysis. A1l-arabinose; A2 α-1,5-l-arabinobiose; A3 α-1,5-l-arabinotriose; A4 α-1,5-l-arabinotetraose; A5, α-1,5-l-arabinopentaose
Fig. 5.
HPAEC-PAD analysis of the hydrolysis products of debranched arabinan by SaAraf43A. The enzyme was incubated with 0.25% (w/v) debranched arabinan at 30°C for the appropriate time; the samples were analyzed by HPAEC-PAD. A1 indicates l-arabinose
Sugar-binding properties of SaCBM42 and contribution to catalytic activity
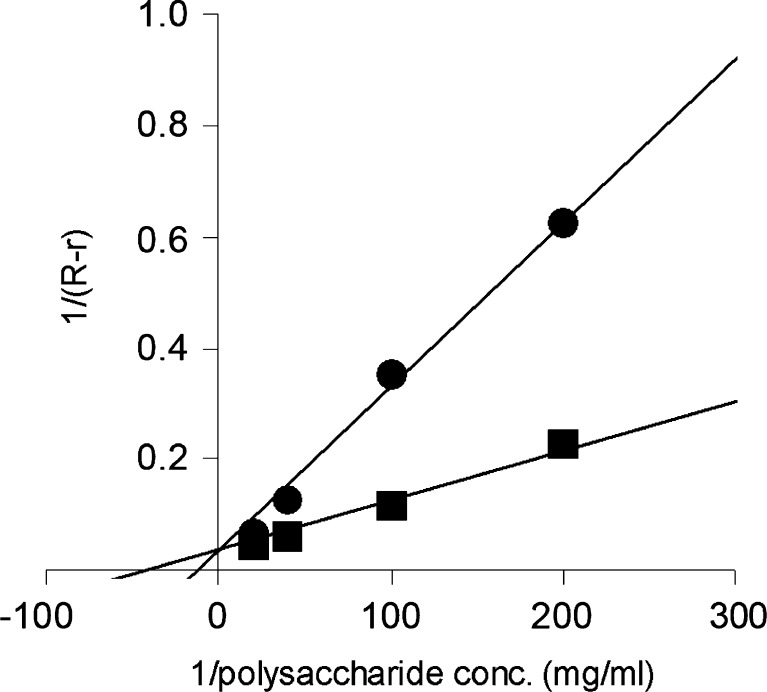
To determine SaCBM42 binding to polysaccharides, affinity gel electrophoresis was performed (Fig. 6). SaCBM42 showed affinity for arabinan and wheat arabinoxylan but did not bind to arabinogalactan. Interestingly, SaCBM42 did not bind to debranched arabinan even though it is a good substrate for SaAraf43A. The K d value for arabinan (0.023 mg/ml) was slightly smaller than that for arabinoxylan (0.075 mg/ml). In contrast, the CBM42-deleted mutant did not show any affinity for all substrates tested (data not shown).
Fig. 6.
Plots of the affinity gel electrophoresis data used to determine the affinity of SaAraf43A for wheat arabinoxylan and arabinan. Closed circle wheat arabinoxylan; closed square arabinan; R relative mobility of SaAraf43A compared with the standard in the absence of the sugars; r relative mobility of SaAraf43A compared with the standard in the presence of the sugars
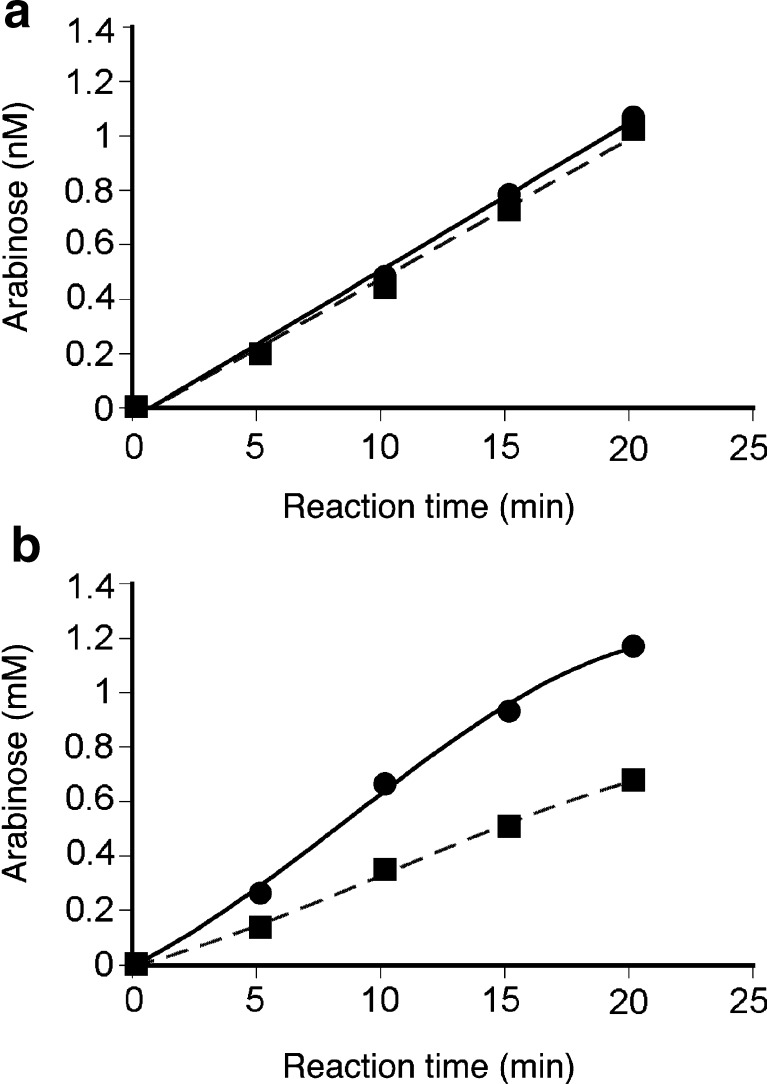
To understand the role of SaCBM42 in the activity of SaAraf43A, the CBM42-deleted mutant of SaAraf43A was constructed. The mutant showed almost the same activity as the wild-type with regard to PNP-α-l-Araf, but there was a significant reduction in the hydrolysis rate when the mutant was incubated with debranched arabinan (Fig. 7). The data suggest that SaCBM42 binds to small amount of arabinosyl side chains that were left on debranched arabinan and thus increases the enzyme concentration around the substrate.
Fig. 7.
Effect of SaCBM42 on hydrolysis rate of SaAraf43A. a Hydrolysis of PNP-α-l-arabinofuranoside; b hydrolysis of debranched arabinan. Closed circle SaAraf43A; closed square the catalytic domain of SaAraf43A. The enzyme (1 μM) was incubated with 1 mM PNP-α-l-Araf or 0.25% (w/v) debranched arabinan at 20°C for up to 20 min. The hydrolysis rate of the substrate was estimated by the amount of released PNP or l-arabinose
Discussion
GH43 includes several kinds of α-l-arabinanases. The crystal structure of α-l-arabinanases from Cellvibrio japonicus (CjArb43A) and Bacillus subtilis (BsArb43A) have been determined (Nurizzo et al. 2002; Proctor et al. 2005). BsArb43A is a typical endo-type of enzyme which randomly hydrolyzes linear 1,5-arabinan, while CjArb43A is exo-acting and releases arabinotriose from the 1,5-arabinan. Therefore, the amino acid sequences of SaAraf43A and α-l-arabinofuranosidase II from S. chartreusis were compared with those of CjArb43A and BsArb43A (Fig. 1a). The most obvious sequential difference in SaAraf43A (exo-1,5-α-l-arabinofuranosidase) with BsArb43A (endo-arabinanase) is the presence of three loop insertions (indicated as L4, L6, and L9 in Fig. 1a) and six loop deletions (indicated as L1, L2, L3, L5, L7, and L8 in Fig. 1a). The five-residue loop (L1) which is relating to the endo-mode of action in BsArb43A (Proctor et al. 2005) was not observed in SaAraf43A, while the large loop insertion (L6) that seals the putative aglycon end of the substrate binding groove in CjArb43A (Proctor et al. 2005) was also present in the exo-1,5-α-l-arabinofuranosidases. The important acidic residues for the catalysis such as E221 (catalytic acid), D38 (catalytic base), D158 (function unknown) in CjArb43A which were demonstrated by the mutagenesis study (Nurizzo et al. 2002) were completely conserved in SaAraf43A (Fig. 1a). In contrast, the amino acids which were shown to be important in substrate binding in CjArb43A (Nurizzo et al. 2002; Proctor et al. 2005) were not completely conserved in SaAraf43A. The W94 in CjArb43A was conserved, but phenylalanines such as F114 and F176 in CjArb43A were not conserved, suggesting that the manner in which exo-1,5-α-l-arabinofuranosidase binds the substrate is different from that of the arabinanases.
The substrate recognition mechanism of exo-1,5-α-l-arabinofuranosidase will be different from the other α-l-arabinofuranosidases. Crystal structures of α-l-arabinofuranosidases belonging to GH51 and GH54 suggest that these α-l-arabinofuranosidases have an active site pocket which is suitable to recognize monosaccharide (Hövel et al. 2003; Taylor et al. 2006; Miyanaga et al. 2004). The substrate specificities of these enzymes are rather broad so that these enzymes do not recognize aglycon side so strictly. In contrast, exo-1,5-α-l-arabinofuranosidase specifically cleaves the internal α-1,5-linkage of two arabinofuranosyl residues. Therefore, the enzyme would possess at least two subsites capable to bind α-1,5-linked arabinofuranosyl residues. In general, the arabinans consist of a linear chain of α-1,5-linked arabinofuranosyl residues that may be alternately substituted at the O-3 and/or O-2 with additional arabinofuranosyl residues. Therefore, exo-1,5-α-l-arabinofuranosidase could not hydrolyze sugar beet arabinan by the steric hindrance caused by 1,3- and/or 1,2-linked α-arabinofuranosyl side chains at the subsite + 1 or −1.
On the other hand, CBM42 was first found associated to GH54 α-l-arabinofuranosidase from Trichoderma reesei as a xylan-binding module (Nogawa et al. 1999). The crystal structure of Aspergillus kawachii GH54 α-l-arabinofuranosidase was resolved, and it was demonstrated that the CBM in the C-terminal of the GH54 enzyme binds to arabinose (Miyanaga et al. 2004). Miyanaga et al. (2006) characterized the CBM in detail and found that CBM42 binds to arabinose side chains in arabinoxylan and arabinan. CBM42 consists of three similar repeated peptides of about 40 residues, each referred to as subdomains α, β, and γ, and these three units assemble around the pseudo threefold axis to form a globular structure. This combination of three subdomains results in a fold similar to the “β-trefoil fold”. This fold is common to CBM13 and certain plant galactose-binding lectins such as ricin B-chain and abrin B-chain (Boraston et al. 2004). The residues involved in arabinose-binding in AkCBM42 which were demonstrated in the AkCBM42/arabinose complex (Miyanaga et al. 2004) showed that most of the residues are strictly conserved among all three subdomains of SaCBM42 (Fig. 1b). In subdomain β, aromatic amino acid in AkCBM42 (Y417) stacking with arabinose was replaced by Lys in SaCBM42 (K382). However, subdomain β of SaCBM42 would be able to bind to the substrate because the other residues involved in substrate binding were conserved. Actually, the properties of SaCBM42 was quite similar to AkCBM42. Generally, the α-l-arabinofuranosidases belonging to GH54 preferentially remove arabinosyl side chains from arabinan. Therefore, the properties of CBM42 which specifically bind the arabinofuranose side chains of hemicelluloses would be suitable for the substrate specificity of the catalytic module. In contrast, exo-1,5-α-l-arabinofuranosidase hydrolyzes debranched arabinan (linear 1,5-arabinan) and does not cleave the arabinosyl side chains of arabinan. Synergism in arabinan hydrolysis would be expected if GH54 α-l-arabinofuranosidase and SaAraf43A were present together. However, no GH54 enzymes were found in genome database of S. avermitilis. Therefore, it would be interesting to determine whether SaCBM42 enhances the activity of SaAraf43A. To elucidate this, the CBM42-deleted mutant of SaAraf43A was constructed, and the activity of the mutant for debranched arabinan was tested (Fig. 7). The activity of SaAraf43A was significantly higher than the CBM42-deleted mutant, suggesting that the binding ability of SaCBM42 for only small amounts of arabinosyl side chains remaining in debranched arabinan increases the enzyme concentration around the substrate. The debranched arabinan had been obtained by treating the arabinan with arabinofuranosidase, which removed the 1,3- and 1,2-linked α-arabinofuranosyl residues. The methylation analysis of arabinan and enzyme-digested arabinan revealed that 32–45% of side chains were left on debranched arabinan (Kaneko et al. 1998a, c, d), indicating the possibility of SaCBM42 to bind the debranched arabinan. In order to obtain more detailed information about this unique function, structure–function analysis of SaCBM42 would be required.
The open reading frame (ORF) SAV1043 lies adjacent to ORF SAV1044 and forms an operon in the S. avermitilis genome. The expressions of both genes were regulated by the same promoter. BLAST search revealed that the ORF SAV1044 might encode an α-l-arabinofuranosidase which would have different substrate specificity from SAV1043. In this study, we showed the ORF SAV1043 encode an exo-1,5-α-l-arabinofuranosidase. The proximity of the ORFs may suggest that the actinomycete has advantage for the hydrolysis of polysaccharides including arabinose.
In conclusion, we demonstrated in this work that SaAraf43A is definitely an exo-1,5-α-l-arabinofuranosidase. SaAraf43A is a modular enzyme possessing CBM42 at its C terminus. We also demonstrated that SaCBM42 binds to the arabinofuranosyl side chains of arabinan and arabinoxylan. Interestingly, it plays a role in enhancing the debranched arabinan hydrolytic action of the catalytic module in spite of its preference for binding arabinofuranosyl side chains.
SaAraf43A provides a good model for investigating the hydrolytic activity of the modular enzyme. In the future, we hope to provide additional information that should be of use in further determinations of the structure–function relationships of SaAraf43A.
Acknowledgment
This work was supported financially by a grant-in-aid (Development of biomass utilization technologies for revitalizing rural areas) from the Ministry of Agriculture, Forestry and Fisheries of Japan.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
H. Ichinose and M. Yoshida contributed equally to this work.
References
- Aristidou A, Penttilä M. Metabolic engineering applications to renewable resource utilization. Curr Opin Biotechnol. 2000;11:187–198. doi: 10.1016/S0958-1669(00)00085-9. [DOI] [PubMed] [Google Scholar]
- Bolam DN, Xie H, Pell G, Hogg D, Galbraith G, Henrissat B, Gilbert HJ. X4 modules represent a new family of carbohydrate-binding modules that display novel properties. J Biol Chem. 2004;279:22953–22963. doi: 10.1074/jbc.M313317200. [DOI] [PubMed] [Google Scholar]
- Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 2004;382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313X.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Hövel K, Shallom D, Niefind K, Belakhov V, Shoham G, Baasov T, Shoham Y, Schomburg D. Crystal structure and snapshots along the reaction pathway of a family 51 α-l-arabinofuranosidase. EMBO J. 2003;22:4922–4932. doi: 10.1093/emboj/cdg494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose H, Yoshida M, Kotake T, Kuno A, Igarashi K, Tsumuraya Y, Samejima M, Hirabayashi J, Kobayashi H, Kaneko S. An exo-β-1,3-galactanase having a novel β-1,3-galactan binding module from Phanerochaete chrysosporium. J Biol Chem. 2005;280:25820–25829. doi: 10.1074/jbc.M501024200. [DOI] [PubMed] [Google Scholar]
- Kaneko S, Kusakabe I. Substrate specificity of α-l-arabinofuranosidase from Bacillus subtilis 3–6 toward arabinofurano-oligosaccharides. Biosci Biotechnol Biochem. 1995;59:2132–2133. doi: 10.1271/bbb.59.2132. [DOI] [PubMed] [Google Scholar]
- Kaneko S, Shimasaki T, Kusakabe I. Purification and some properties of intracellular α-l-arabinofuranosidase from Aspergillus niger 5–16. Biosci Biotechnol Biochem. 1993;57:1161–1165. doi: 10.1271/bbb.57.1161. [DOI] [PubMed] [Google Scholar]
- Kaneko S, Sano M, Kusakabe I. Purification and some properties of α-l-arabinofuranosidase from Bacillus subtilis 3–6. Appl Environ Microbiol. 1994;60:3425–3428. doi: 10.1128/AEM.60.9.3425-3428.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Kawabata Y, Ishii T, Gama Y, Kusakabe I. The core trisaccharide of α-l-arabinofuranan: synthesis of methyl 3,5-di-O-α-l-arabinofuranosyl-α-l-arabinofuranoside. Carbohydr Res. 1995;268:307–311. doi: 10.1016/0008-6215(94)00327-C. [DOI] [PubMed] [Google Scholar]
- Kaneko S, Kuno A, Matsuo N, Ishii T, Kobayashi H, Hayashi K, Kusakabe I. Substrate specificity of α-l-arabinofuranosidase from Trichoderma reesei. Biosci Biotechnol Biochem. 1998;62:2205–2210. doi: 10.1271/bbb.62.2205. [DOI] [PubMed] [Google Scholar]
- Kaneko S, Higashi K, Yasui T, Kusakabe I. Substrate specificity of α-l-arabinofuranosidase from Streptomyces diastatochromogenes 065 toward arabinose-containing oligosaccharides. J Ferment Bioengin. 1998;85:519–521. doi: 10.1016/S0922-338X(98)80073-1. [DOI] [Google Scholar]
- Kaneko S, Arimoto M, Ohba M, Kobayashi H, Ishii T, Kusakabe I. Purification and substrate specificities of two α-l-arabinofuranosidases from Aspergillus awamori IFO 4033. Appl Environ Microbiol. 1998;64:4021–4027. doi: 10.1128/AEM.64.10.4021-4027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Ishii T, Kobayashi H, Kusakabe I. Substrate specificities of α-l-arabinofuranosidases produced from two species of Aspergillus niger. Biosci Biotechnol Biochem. 1998;62:695–699. doi: 10.1271/bbb.62.695. [DOI] [PubMed] [Google Scholar]
- Kawabata Y, Kaneko S, Kusakabe I, Gama Y. Synthesis of regioisomeric methyl α-l-arabinofuranobiosides. Carbohydr Res. 1995;267:39–47. doi: 10.1016/0008-6215(94)00290-V. [DOI] [PubMed] [Google Scholar]
- Kusakabe I, Yasui T, Kobayashi T. Some properties of araban degrading enzymes produced by microorganisms and enzymatic preparation of arabinose from sugar beet pulp. Nippon Nogeikagaku Kaishi. 1975;49:295–305. doi: 10.1271/nogeikagaku1924.49.295. [DOI] [Google Scholar]
- Matsuo N, Kaneko S, Kuno A, Kobayashi H, Kusakabe I. Purification, characterization and gene cloning of two α-l-arabinofuranosidases from Streptomyces chartreusis GS901. Biochem J. 2000;346:9–15. doi: 10.1042/bj3460009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanaga A, Koseki T, Matsuzawa H, Wakagi T, Shoun H, Fushinobu S. Crystal structure of a family 54 α-l-arabinofuranosidase reveals a novel carbohydrate-binding module that can bind arabinose. J Biol Chem. 2004;279:44907–44914. doi: 10.1074/jbc.M405390200. [DOI] [PubMed] [Google Scholar]
- Miyanaga A, Koseki T, Miwa Y, Mese Y, Nakamura S, Kuno A, Hirabayashi J, Matsuzawa H, Wakagi T, Shoun H, Fushinobu S. The family 42 carbohydrate-binding module of family 54 α-l-arabinofuranosidase specifically binds the arabinofuranose side chain of hemicellulose. Biochem J. 2006;399:503–511. doi: 10.1042/BJ20060567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogawa M, Yatsui K, Tomioka A, Okada H, Morikawa Y. An alpha-l-arabinofuranosidase from Trichoderma reesei containing a noncatalytic xylan-binding domain. Appl Environ Microbiol. 1999;65:3964–3968. doi: 10.1128/AEM.65.9.3964-3968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurizzo D, Turkenburg JP, Charnock SJ, Roberts SM, Dodson EJ, Mckie VA, Taylor EJ, Gilbert HJ, Davies GJ. Cellvibrio japonicus α-l-arabinanase 43A has a novel five-blade β-propeller fold. Nat Struct Biol. 2002;9:665–668. doi: 10.1038/nsb835. [DOI] [PubMed] [Google Scholar]
- Osaki S, Kimura T, Sugimoto T, Hizukuri S, Iritani N. l-Arabinose feeding prevents increases due to dietary sucrose in lipogenic enzymes and triacylglycerol levels in rats. J Nutr. 2001;131:796–799. doi: 10.1093/jn/131.3.796. [DOI] [PubMed] [Google Scholar]
- Proctor MR, Taylor EJ, Nurizzo D, Turkenburg JP, Lloyd RM, Vardakou M, Davies GJ, Gilbert HJ. Tailored catalysts for plant cell-wall degradation: redesigning the exo/endo preference of Cellvibrio japonicus arabinanase 43A. Proc Natl Acad Sci USA. 2005;102:2697–2702. doi: 10.1073/pnas.0500051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha BC. Hemicellulose bioconversion. J Ind Microbiol Biotechnol. 2003;30:279–291. doi: 10.1007/s10295-003-0049-x. [DOI] [PubMed] [Google Scholar]
- Sanai K, Seri K, Inoue S. Inhibition of sucrose digestion and absorption by l-arabinose in rats. J Jpn Soc Nutr Food Sci. 1997;50:133–137. doi: 10.4327/jsnfs.50.133. [DOI] [Google Scholar]
- Seri K, Sanai K, Matsuo N, Kawakubo K, Xue C, Inoue S. l-Arabinose selectively inhibits intestinal sucrase in an uncompetitive manner and suppresses glycemic response after sucrose ingestion in animals. Metabolism. 1996;45:1368–1374. doi: 10.1016/S0026-0495(96)90117-1. [DOI] [PubMed] [Google Scholar]
- Skoog K, Hahn-Hägerdal B. Xylose fermentation. Enzyme Microb Technol. 1988;10:66–80. doi: 10.1016/0141-0229(88)90001-4. [DOI] [Google Scholar]
- Somogyi M. Notes on sugar determination. J Biol Chem. 1952;195:19–23. [PubMed] [Google Scholar]
- Takeo K. Affinity electrophoresis: Principles and applications. Electrophoresis. 1984;5:187–195. doi: 10.1002/elps.1150050402. [DOI] [Google Scholar]
- Taylor EJ, Smith NL, Turkenburg JP, D’Souza S, Gilbert HJ, Davies GJ. Structural insight into the ligand specificity of a thermostable family 51 arabinofuranosidase, Araf51, from Clostridium thermocellum. Biochem J. 2006;395:31–37. doi: 10.1042/BJ20051780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Ichinose H, Nakajima M, Kobayashi H, Kaneko S. Synergy between an α-l-arabinofuranosidase from Aspergillus oryzae and an endo-arabinanase from Streptomyces coelicolor for degradation of arabinan. Food Sci Technol Res. 2006;12:43–49. doi: 10.3136/fstr.12.43. [DOI] [Google Scholar]
- Yoshida S, Kawabata Y, Kaneko S, Nagamoto Y, Kusakabe I. Detection of α-l-arabinofuranosidase activity in isoelectric focused gels using 6-bromo-2-naphtyl α-l-arabinofuranoside. Biosci Biotechnol Biochem. 1994;58:580–581. doi: 10.1271/bbb.58.580. [DOI] [Google Scholar]