Abstract
The rate of meiotic recombination in the yeast Saccharomyces cerevisiae varies widely in different regions of the genome with some genes having very high levels of recombination (hotspots). A variety of experiments done in yeast suggest that hotspots are a feature of chromatin structure rather than a feature of primary DNA sequence. We examined the effects of mutating a variety of enzymes that affect chromatin structure on the recombination activity of the well-characterized HIS4 hotspot including the Set2p and Dot1p histone methylases, the Hda1p and Rpd3p histone deacetylases, the Sin4p global transcription regulator, and a deletion of one of the two copies of the genes encoding histone H3–H4. Loss of Set2p or Rpd3p substantially elevated HIS4 hotspot activity, and loss of Hda1p had a smaller stimulatory effect; none of the other alterations had a significant effect. The increase of HIS4 hotspot activity in set2 and rpd3 strains is likely to be related to the recent finding that histone H3 methylation by Set2p directs deacetylation of histones by Rpd3p.
2. Introduction
The rate of meiotic recombination varies considerably at different positions in the yeast genome (1,2). This variation reflects differences in the frequency of local meiosis-specific double-strand DNA breaks (DSBs), the recombination-initiating lesion (3,4) catalyzed by Spo11p and associated proteins (5). There are several lines of evidence that the frequency of DSBs is regulated by elements of chromatin structure rather than primary DNA sequence (2). First, recombination hotspots exhibit hypersensitivity to nucleases (6–10). Some loci (8,9) undergo meiosis-specific alterations in nuclease sensitivity prior to DSB formation, although no such changes are observed at other loci (6). Since all nuclease-hypersensitive regions are not meiotic recombination hotspots (6,10), “open” chromatin appears necessary, but not sufficient, for meiotic recombination hotspot activity. Second, insertion of a nucleosome-excluding sequence into the yeast genome creates a recombination hotspot (11). Third, the activity of some hotspots requires the binding of transcription factors, but not high levels of transcription (2). This result has been interpreted as indicating that chromatin modifications associated with transcription factor binding stimulate both transcription and recombination. Finally, some mutants that affect chromatin modifications reduce the frequency of meiosis-specific DSBs. In the yeast S. pombe, mutations in a histone acetylase or in a chromatin-remodeling activity reduce the activity of the ade6-M26 recombination hotspot (12). In S. cerevisiae, loss of Set1p (a histone methyltransferase) or Rad6p (a ubiquitin-conjugating enzyme) reduce DSB formation (13,14).
In our genetic background, the HIS4 gene is one of the strongest recombination hotspots in the S. cerevisiae genome (15) and is associated with a high-frequency DSB located upstream of HIS4 (16,17). In cells sporulated at 25°, this DSB is responsible for initiating about half of HIS4 conversion events (16). The binding of four transcription factors (Bas1p, Bas2p, Rap1p, and Gcn4p) is required for optimal levels of this DSB (16,18–20). One role of these transcription factors is presumably to recruit chromatin-modifying complexes to the HIS4 promoter. The transcription factor Rap1p directly alters the positioning of nucleosomes in the HIS4 upstream region (21,22). Based on the evidence that HIS4 hotspot activity is likely related to a particular chromatin structure, in this study, we analyzed the effects of mutations affecting chromatin modifications on HIS4 hotspot activity.
Two broad classes of protein complexes maintain or alter local chromatin structure: proteins that mediate post-translational modification of histones and other chromatin-related proteins, and ATP-dependent nucleosome remodeling complexes (23). The amino-terminal ‘tails’ of histones are subject to various covalent modifications including acetylation, methylation, phosphorylation, ubiquitination, and ADP-ribosylation (24). These modifications affect chromatin structure by altering histone-DNA or histone-histone contacts (25), and/or by collectively establishing a code that is recognized by downstream effector proteins and complexes (24). The chromatin-modifying activities are recruited to certain loci by sequence-specific transcription factors (26,27), or by association with RNA polymerase II (27,28).
Since there are many proteins involved in modifying chromatin, our analysis emphasized mutants that were reported to affect the expression of HIS4 or to alter its chromatin structure. Mutations in RPD3, encoding an H4 histone deacetylase (26), were reported to reduce HIS4 expression about three-fold in one study (29), although no significant change was observed in another study (30). Deletion of the HDA1-encoded histone H3 and H2B deacetylase (31), resulted in a three-fold increase of HIS4 gene expression in one study (30), but had no significant effect on expression in another study (29). The deletion of SIN4, a transcription factor that is part of the Mediator complex, reduced HIS4 transcription about 15-fold and altered HIS4 chromatin structure (32). In addition, Wyrick et al. (33) found that depletion of histone H4 decreased HIS4 expression two-fold.
In addition to examining these mutants previously demonstrated to affect HIS4 expression, we analyzed the effects of several other related mutants. We examined mutants of SET2 (encoding an H3K36 histone methylase, 34), since it has been recently reported (35–37) that Set2p-dependent methylation recruits an Rpd3p-containing repressive complex (Rpd3C[S]). We also examined the effects of eliminating Dot1p, an H3K79 histone methyltransferase (38,39). H3K79 methylation is at low levels at yeast telomeres (40), but occurs at high levels at actively-transcribed genes (41). As described below, we found that deletion of SET2 or RPD3 strongly stimulated HIS4 hotspot activity, whereas deletion of HDA1 had a more subtle stimulatory effect. None of the other mutants examined had any significant effect. We also showed that strains that lacked either SET2 or RPD3 had elevated levels of H3K27 acetylation near the HIS4 recombination hotspot in meiosis, and elevated meiotic HIS4 gene expression.
2. Materials and methods
2.1. Yeast strains
All strains used were derived by transformation from the haploid strains AS4 (∝trp1 arg4 tyr7 ade6 ura3) and AS13 (a leu2 ura3 ade6 rme1) (42). Derivatives of AS4 and AS13 are listed in Table 1; diploids constructed by crossing AS4 and AS13 derivatives are described in Table 2. Deletions were constructed by replacing the relevant gene with a kanMX4 cassette (43) or hphMX4 cassette (44), which contain a gene that confers geneticin or hygromycin resistance, respectively. The PCR synthesis of the cassettes and subsequent transformation were performed as described by Wach et al. (43), using the plasmid pFA6-kanMX4 (kanMX4 cassette; 43) or pAG32 (hphMX4 cassette; 44) and the primer pairs listed in Table 3. One pair of set2::hphMX4 strains, ER4 and ER5, had complete deletions of SET2 and was constructed with primers set2Δ-F and set2Δ-R. A second pair of strains, DCB4 and DCB6, had a substitution that deleted the last 100 amino acids of the Set2p (set2::kanMX4). In all the assays in our study, these two types of set2 mutations had the same phenotypes. All constructions, unless noted otherwise, were checked by PCR. The rad50S derivatives were constructed as described by Alani et al. (45).
Table 1.
Genotypes of haploid yeast strains
| Strain | Relevant genotype; reference | Name of rad50S derivative; reference |
|---|---|---|
| AS4-derived haploids | ||
| AS4 | WT; 43 | DNY107; 16 |
| ER4 | set2::hphMX4 | JDM205 |
| DCB4 | set2::kanMX4 | DCB14 |
| MD136 | rpd3::kanMX4 | JDM210 |
| ER2 | dot1::hphMX4 | |
| JDM166 | hda1::kanMX4 | |
| MD131 | hht2-hhf2::kanMX4 | |
| MD133 | sin4::kanMX4 | |
| JDM214 | set2::hphMX4 rpd3::kanMX4 rad50S | JDM214 |
| AS13-derived haploids | ||
| AS13 | WT; 42 | |
| DNY9 | his4-lop; 16 | HF1; 16 |
| DNY25 | his4-lopc; 16 | HF4; 16 |
| ER5 | his4-lop set2::hphMX4 | JDM204 |
| DCB6 | his4-lopc set2::kanMX4 | DCB15 |
| MD135 | his4-lop rpd3::kanMX4 | JDM131 |
| ER1 | his4-lop dot1::hphMX4 | |
| MD141 | his4-lop hda1::kanMX4 | |
| MD132 | his4-lop hht2-hhf2::kanMX4 | |
| MD134 | his4-lop sin4::kanMX4 | |
| JDM213 | his4-lop set2::hphMX4 rpd3::kanMX4 rad50S | JDM213 |
All haploid strains were derived from AS4 (α arg4-17 trp1 tyr7-1 ade6 ura3) and AS13 (a leu2 ade6 ura3 rme1) by transformation or by crosses with isogenic strains. The mutant his4-lop and his4-lopc are 26 bp palindromic insertions that result in inefficiently-repaired mismatches (61). Only those markers that are different from the haploid progenitor strains are shown. If no reference is given, the strains were constructed in this study.
Table 2.
Diploid yeast strains used in this study
| Strain | Relevant homozygous mutations | Cross; referencea |
|---|---|---|
|
Strains heterozygous for his4-lop
| ||
| DNY11 | WT | AS4 X DNY9; 16 |
| FX1 | rad50S::URA3 | DNY107 X HF1; 16 |
| ER6 | set2::hphMX4 | ER4 X ER5 |
| JDM1093 | set2::hphMX4 rad50S::URA3 | JDM205 X JDM204 |
| JDM1094 | rpd3::kanMX4 | MD136 X MD135 |
| JDM1095 | rpd3::kanMX4 rad50S::URA3 | JDM210 X JDM131 |
| ER3 | dot1::hphMX4 | ER2 X ER1 |
| JDM1076 | hda1::kanMX4 | JDM166 X MD141 |
| MD137 | hht2-hhf2::kanMX4 | MD131 X MD132 |
| MD138 | sin4::kanMX4 | MD133 X MD134 |
| JDM1096 | set2::hphMX4 rpd3::kanMX4 rad50S::URA3 | JDM214 X JDM213 |
|
| ||
|
Strains heterozygous for his4-lopc
| ||
| DNY26 | WT | AS4 X DNY25; 16 |
| FX3 | rad50S::URA3 | DNY107 X HF4; 16 |
| DCB9 | set2::kanMX | DCB4 X DCB6 |
| DCB16 | set2::kanMX rad50S::URA3 | DCB14 X DCB15 |
Genotypes of the haploids used in the constructions are given in Table 1. In crosses, AS4- and AS13-derived strains are shown on the left and the right of the X, respectively.
Table 3.
Primers used to make deletions with kanMX4 or hphMX4 cassettesa and for other types of analysis.
| Primer name | Sequence (5′-3′) |
|---|---|
| set2Δ-F | AGTCGTGCTGTCAAACCTTTCTCCTTTCCTGGTTGTTGTTTTACGTGATCCGTACGCTGCAGGTCGAC |
| set2Δ-R | CTTTGGGACAGAAAACGTGAAACAAGCCCCAAATATGCATGTCTGGTTAAATCGATGAATTCGAGCTCG |
| Set2 del UP | TCACATTACCTATCATTACAGTGCTACTGATAGCCCACTAATGCGGATATCGTACGCTGCAGGTCGAC |
| Set2 del DN | TCTCCCAGTCCCAAAGACTAGAGCACAACTGGAATAAATTCTTTGCATCTATCGATGAATTCGAGCTCG |
| rpd3Δ-F | TGCGCCATACAAAACATTCGTGGCTACAACTCGATATCCGTGCAGCGTACGCTGCAGGTCGAC |
| rpd3Δ-R | TTGTTTCACATTATTTATATTCGTATATACTTCCAACTCTTTTTTATCGATGAATTCGAGCTC |
| dot1Δ-F | AAGGAGGTCACCAGTAATTGTGCGCTTTGGTTACATTTTGTTGTACAGTACGTACGCTGCAGGTCGAC |
| dot1Δ-R | TATTTCTACTTAGTTATTCATACTCATCGTTAAAAGCCGTTCAAAGTGCCATCGATGAATTCGAGCTCG |
| hda1Δ-F | GAGAAAGGGAAAGTTGAGCACTGTAATACGCCGAACAGATTAAGCCGTACGCTGCAGGTCGAC |
| hda1Δ-R | GGCATGAAGGTTGCCGAAAAAAAATTATTAATGGCCAGTTTTTCCATCGATGAATTCGAGCTC |
| hht2-hhf2Δ-F | CCCCCAGTCTAAATGCATAGAAAAAAAAAAATTCCCGCTTTATATCGTACGCTGCAGGTCGAC |
| hht2-hhf2Δ-Rb | GGCATGAAAATAATTTCAACACCGATTGTTTAACCACCGATTGTTATCGATGAATTCGAGCTCG |
| sin4Δ-F | AGAAAAGAAACTAGCAGACCTGACCTTCTGTTGGTAAATATTAGTCGTACGCTGCAGGTCGAC |
| sin4Δ-R | ATGTTTAAAACAATTCTATACAAAACTATGCTATAGTACTAATAAATCGATGAATTCGAGCTCG |
| BIK1codpre350F | GGATAGATATCAAAGAAAGATAGGATGTTTC |
| BIK1codpre350R | GGGATTTAGGATCATCCATGGG |
| BIK1codpost300F | GGTGCCAACGAAGATGTCG |
| BIK1codpost300R | GAAAATTGGCAACGATTCCAC |
| HIS4pro242F | GGCAGTCGAACTGACTCTAATAGTG |
| HIS4pro242R | CTATTGTATTACTATTACACAGCGCAG |
| HIS4codpost180F | CGTGTGGATATTGTTCGTAAATG |
| HIS4codpost180R | GTAAGCACCCACAAATACGGAC |
| IntergenicV-1 | GGCTGTCAGAATATGGGGCCGTAGTA |
| IntergenicV-2 | CACCCCGAAGCTGCTTTCACAATAC |
| ChrIII-15830 | CTGGAAATATGAAATTGTCAGCGAC |
| ChrIII-16773 | TGAATGCCTTCAACCAAGTGGCTCCTTC |
| his4_185_U | CGTTGCTGCTATGGCTTACG |
| his4_185_L | CCACATCGGCATCTTCATCG |
| BIK1-388-F | CACAGCGGTAATCAACAGTC |
| BIK1-531-R | GCTGGTGGTGTTGTGATTAG |
| RNA1_128_U | CTGCTGGTTCAGATGAAGTC |
| RNA1_128_L | TTCCATAGCCGGTAAGAAGG |
Gene deletions were made using PCR fragments containing a selectable marker flanked by sequences from the gene to be deleted (43, 44). For most of these constructions, the sequences of the oligonucleotides match those located immediately upstream and downstream of the gene to be deleted. The only exceptions are the Set2 del UP and Set2 del DN oligonucleotides. Set2 DN UP is located within SET2, about 300 bp from the end of the gene, and Set2p UP is located about 150 bp downstream of SET2. Sequences homologous to the region flanking the target gene are shown in bold-face type. Sequences homologous to the selectable cassette used to make the disruption are shown in regular type. The primers labeled “BIK1”, “HIS4”, and “Intergenic” were used in chromatin immunoprecipitation experiments.
The primer hht2-hhf2Δ-R is missing a single base relative to the region downstream of HHF2, which creates a single base pair deletion when the PCR fragment is inserted in the locus. The correct sequence is GGCATGAAAATAATTTCAAACACCGATTGTTTAACCACCGATTGTT, with the deleted base underlined.
2.2. Genetic analysis
Standard materials and methods were used (46) except where noted. Diploids were sporulated on plates at 25°C. Although the HIS4 hotspot is stronger at 18°C (16), not all of the mutants sporulated at this temperature. Following tetrad dissection on plates with rich growth medium (YPD), the spore colonies were replica-plated to various omission media; spore colonies on medium lacking histidine were examined microscopically in order to detect small sectors.
2.3. Southern analysis
Cells were harvested from rad50S diploid strains just prior to being placed on a sporulation plate (0 hr) or after 24 hr at 25° C. Cells were washed with 0.5 ml 10 mM Tris (pH 8.0)-1 mM EDTA, and stored at −80° C. DNA isolation and Southern blot procedures were performed as described by Nag and Petes (17). The HIS4 DSB was examined using a BglII digest, and an XhoI-BglII fragment of HIS4 derived from pDN42 as a hybridization probe (17). The ARG4 DSB was examined using a BglII digest, and an EcoRV-BglII fragment derived from pAK1 as a hybridization probe (16). Hybridization was quantitated using a PhosphorImager (Molecular Dynamics), and the percent of molecules with a DSB was calculated as described by Kirkpatrick et al. (47). Since the percentage of molecules that receive a DSB is affected by the sporulation efficiency, the percentage of DSBs at HIS4 was normalized to the percentage of DSBs at ARG4.
We also did Southern analysis of intact chromosomal DNA molecules. For these experiments, DNA molecules were separated using CHEF (contour-clamped homogeneous electric field) gel electrophoresis as described previously (48). DNA was isolated from cells immediately after transfer to solid sporulation medium or after 24 hours in solid sporulation medium (room temperature, 23°–25° C). Following electrophoresis, DNA was transferred to a Nylon membrane and hybridized to a probe derived from left end of chromosome III, within the CHA1 gene. The probe was generated by PCR amplification of yeast genomic DNA with the primers ChrIII-15830 and ChrIII-16773 (Table 3).
2.4. Analysis of histone modifications using chromatin immunoprecipitation (ChIP)
Cell extracts were prepared from the diploid strains used for ChIP analysis (FX1, JDM1093, JDM1095, and JDM1096) as described by Xiao et al. (49). Prior to extraction, the cells were incubated on solid sporulation medium for six hours at room temperature (23°–25° C.). Cells at this time point are beginning to form DSBs and binucleate cells appear at about 10 hours (17). Immunoprecipitation of formaldehyde-fixed chromatin was done with the following antibodies: anti-H3K36me3 (AB9050, Abcam), anti-H3K27ac (07–360, Upstate), and anti-H3 (AB1791, Abcam). Following the immunoprecipitation, we reversed the crosslinks by treatment of the samples at 65° C and performed multiplex PCR to amplify sequences in the HIS4 and BIK1 region.
Four sets of primers were used (sequences in Table 3). A PCR reaction with primers BIK1codpre350F and BIK1codpre350R results in a 350 bp fragment (termed “A”) derived from the 5′ end of BIK1. The 300 bp “B” fragment (located at the 3′ end of BIK1) and the 242 bp “C” fragment (located in the intergenic region upstream of HIS4) flank the site of the DSB associated with the HIS4 hotspot. The primers used to produce the “B” and “C” fragments are BIK1codpost300F and BIK1codpost300R, and HIS4pro242F and HIS4pro242R, respectively. The fourth primer pair, which generates the 180 bp “D” fragment are HIS4codpost180F and HIS4codpost180R. The “D” sequences are derived from the 3′ end of HIS4. We also utilized primers (IntergenicV-1 and IntergenicV-2) to amplify a control locus on chromosome V that was used in previous studies (49); this region has a low intrinsic level of histone methylation. The multiplex procedure involved using all five pairs of primers (four from the HIS4-BIK1 region and the control pair) in one reaction. The products of the PCR reaction were analyzed on 12% polyacrylamide gels that were stained with ethidium bromide. The number of pixils in each band was measured using the Kodak 100 Imaging System.
The levels of histone modifications for the four fragments near the HIS4 recombination hotspot were calculated as a ratio (a/b) as described previously (49). The a value is also a ratio: (w/x)/(y/z), where w is the amount of the PCR fragment from the HIS4 region in the ChIP sample, x is the amount of control PCR fragment in the ChIP sample, y is the amount of PCR fragment from the HIS4 region in the input DNA, and z is the amount of PCR fragment from the control locus in the input DNA. The b values were calculated similarly using the data obtained with antibodies directed against H3 histone; the b values are used to control for histone occupancy at each site. At least three independent experiments were done for each strain. For each experiment, we calculated separately the (a/b) ratio and the ratios were averaged. In summary, we calculate the level of histone modifications in the HIS4 region relative to a locus on chromosome V and corrected for the level of histone occupancy at each position.
2.5. Analysis of meiotic expression of HIS4 and BIK1
We examined the meiotic mRNA levels of HIS4 and BIK1 in cells harvested after six hours incubation in sporulation medium at 23° C. RNA was isolated using the Yeast RNA Kit (Omega Biotech) and these RNA samples were reverse-transcribed into DNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the instructions of the manufacturer. The real-time PCR was performed with the Power SYBR Green PCR Master Mix (Applied Biosystems) using the Applied Biosystems Step One Plus PCR machine. After a 10 minute 95°C “start-up” incubation, we used 40 cycles of PCR with the following temperatures and step times: 95° C, 15 seconds; 60° C, 1 minute.
Three sets of primers were used for this analysis. The pairs of primers used to monitor the HIS4 and BIK1 cDNAs were his4_185_U and his4_185_L, and BIK1-388-F and BIK1-531-R, respectively (Table 3). We measured the cDNA levels of the control RNA1 gene with primers RNA1_128_U and RNA1_128_L. The RNA1 gene was chosen as a control because its mRNA level is not affected by sporulation (50) or by the rpd3 mutation (29,30). In our analysis, we first normalized the levels of BIK1 and HIS4 cDNAs to the level of RNA1 cDNA. We subsequently normalized the levels of BIK1 and HIS4 cDNAs to the level of HIS4 cDNA in the wild-type strain.
2.6. Data analysis
Statistical analysis was performed using Instat 1.12 (GraphPad Software) for the Macintosh. The Fisher’s exact test with a two-tailed P value or Chi-squared analysis (for comparisons that involve more than two experimental measures) were used for all comparisons, and P<0.05 was considered statistically significant. For the ChIP experiments, standard errors of the mean were calculated using Excel.
3. Results
We used two different assays to monitor the effects of mutations affecting chromatin structure on the recombination activity of the HIS4 hotspot. First, we monitored the rate of aberrant segregation of a heterozygous HIS4 marker in the wild-type strain and strains homozygous for various mutations. Second, for some strains, we examined the rate of meiosis-specific double-strand DNA breaks (DSBs) at the HIS4 and ARG4 loci. In our previous studies (16) and those of others (51), these two assays for recombination activity generally yield the same result.
3.1. Tetrad analysis of strains with mutations affecting chromatin structure
When a yeast strain that is heterozygous for alleles A and a undergoes meiosis, most tetrads segregate 2A:2a spores (Mendelian segregation). There are two classes of aberrant segregation events, gene conversion and postmeiotic segregation (PMS) events. In most models of recombination (52), both gene conversion and PMS events reflect heteroduplex formation involving the heterozygous site, followed by mismatch repair (gene conversion) or failure to repair (PMS events). Thus, the frequency of aberrant segregation tetrads is a measurement of the rate of local recombination events. Although the average rates of conversion for markers in most genomic regions are low (less than 5% of the tetrads), the rates of conversion for markers at the HIS4 and ARG4 loci in our genetic background are very high. At the HIS4 locus, more than 50% of the tetrads have aberrant segregation when the cells are sporulated at 18°C. (16). When the cells are sporulated at 25° C., the rate of aberrant segregation is approximately halved. In the experiments described below, we used the higher temperature of sporulation because the set2 strains failed to sporulate at 18° C.
Our analysis is summarized in Table 4. The set2 mutation in two closely-related genetic backgrounds resulted in a very significant (p < 0.0001) stimulation of the rate of aberrant segregation for the HIS4, but not the ARG4, marker. Since the rate of aberrant segregation is much higher for HIS4 than ARG4 (which affects the power of the statistical test), we cannot rule out the possibility that both loci are affected by the set2 mutation to similar extents; this possibility, however, is excluded by the physical analysis of DSBs described in Section 3.2. The wild-type strain DNY11 is isogenic (except for the set2 mutation) with ER6 and the wild-type strain DNY26 strain is isogenic with DCB9. We observed a significant increase in crossing over in the HIS4-LEU2 interval for the DNY26/DCB9 comparison, although the increase for the DNY11/ER6 comparison was not significant. We also found that a mutation of the Hda1p deacetylase significantly stimulated the rate of aberrant segregation of the HIS4 marker, although the magnitude of this stimulation was half of that observed for the set2 mutation. Loss of the Dot1p methylase, the Sin4p transcription factor, and loss of one copy of the H3-H4 histone pair had no effect on the hotspot activity associated with the HIS4 gene.
Table 4.
Rates of meiotic recombination (aberrant segregation and crossovers) in strains with mutations affecting chromatin structure
| Strain | Relevant genotype | # Tetrads | HIS4 Ab. Seg. (%) | ARG4 Ab. seg. (%) | HIS4-LEU2 map dist. (cM)c |
|---|---|---|---|---|---|
| DNY11a | wild-type | 475 | 25 | 6 | 24 |
| DNY26a | wild-type | 515 | 27 | 6 | 24 |
| ER6 | set2::hphMX4 | 647 | 38b | 8 | 26 |
| DCB9 | set2::kanMX4 | 556 | 40b | 7 | 30c |
| ER3 | dot1::hphMX4 | 250 | 26 | 4 | 19 |
| JDM1076 | hda1::kanMX4 | 537 | 33b | 9 | 25 |
| MD137 | hht2-hhf2::kanMX4 | 115 | 23 | 12 | 24 |
| MD138 | sin4::kanMX4 | 109 | 25 | 6 | 19 |
The diploids DNY11 and DNY26 are isogenic except that DNY11 is heterozygous for the his4-lop allele and DNY26 is heterozygous for the his4-lopc allele. DCB9 is isogenic with DNY26; all other diploids are isogenic with DNY11.
Chi-square tests were performed using the numbers of aberrant and non-aberrant tetrads for DNY11 versus ER6, ER3, JDM1076, MD137, and MD138, and DNY26 versus DCB9. The p values for these six comparisons were ranked and the Benjamini and Hochberg (62) procedure was applied with an initial α value of 0.05. p values of less than 0.012 were considered significant. The comparisons with such p values were: ER6 and DNY11 (p ≤ 0.0001), DCB9 and DNY26 (p ≤ 0.0001), and JDM1076 and DNY11 (p = 0.006).
Calculated using the standard equation (63). The DCB9 strain has a significantly (p = 0.001) elevated map distance compared to DNY26. All other alterations are statistically insignificant (p ≥ 0.05).
As described in the Introduction, Set2p-dependent methylation of H3K36 is involved in the recruitment of the histone deacetylase Rpd3p. Thus, one explanation of the effects of the set2 mutation is that loss of the Set2p-dependent recruitment of Rpd3p results in hyper-acetylated chromatin that allows more efficient entry of the Spo11p recombination machinery and, therefore, more DSBs in the HIS4 region. This model predicts that loss of Rpd3p would also result in a higher level of aberrant segregation. We could not check this prediction by tetrad analysis, since (as reported previously, 53) rpd3 mutants did not complete meiosis and failed to produce viable spores. An adjustment of the sporulation conditions that allowed rpd3 mutants to complete sporulation in some genetic backgrounds (54) did not allow sporulation in our genetic background. To confirm our conclusions about the effects of the set2 mutation on recombination and to examine the effect of rpd3 mutants on HIS4 recombination activity, we measured the rate of meiosis-specific DSBs at HIS4 in wild-type, set2, rpd3, and set2 rpd3 double mutant strains.
3.2. Measurement of meiosis-specific DSBs at the HIS4 and ARG4 loci in wild-type, set2, rpd3, and set2 rpd3 diploid strains
Since meiotic recombination is initiated by a DSB (3,4), the frequency of local DSBs measured by Southern analysis is a direct measurement of recombination activity. We constructed derivatives of the wild-type, set2, rpd3, and set2 rpd3 diploids (Table 2) that were homozygous for the rad50S mutation, which prevents subsequent processing of DSBs (45). These strains were sporulated at 25° C., and DSBs were examined at both the HIS4 and ARG4 loci. The Southern analysis for one set of experiments in which we examined DSBs at the HIS4 locus is shown in Fig. 1. Since the efficiency of cells undergoing meiotic DSB formation can vary from one experiment to another, we normalized the levels of DSBs measured at the HIS4 locus to the levels observed at the ARG4 locus (as described in Materials and Methods). Normalizing the ratio of HIS4 to ARG4 DSBs to 1 for the wild-type strains FX1 and FX3, we found this ratio averaged 3.3 for the set2 strains JDM1093 and DCB16, 2.7 for the rpd3 strain JDM1095, and 4.1 for the set2 rpd3 strain JDM1096. We also examined the % DSBs relative to the unbroken parental fragments for HIS4 and ARG4. These values are somewhat more variable than the normalized HIS4/ARG4 ratio, since the fraction of cells in sporulation medium that undergo DSB formation is somewhat variable from culture to culture. The % DSBs for HIS4 and ARG4, respectively, in the various strains were: 4.5, 2.6 (FX1, wild-type); 3.0, 0.6 (FX3, wild-type), 12.9, 2.7 (JDM1093, set2); 8.3, 0.4 (DCB16, set2); 19.1, 3.9 (JDM1095, rpd3); 18, 2.1 (JDM1096, set2 rpd3). These values represent the average of two independent experiments for each strain, except for FX3 and DCB16 which represent a single experiment. Although our analysis does not rule out the possibility that set2 and rpd3 mutations stimulate DSB formation at ARG4, the degree of stimulation is clearly stronger at HIS4.
Figure 1. Southern analysis of HIS4 meiosis-specific DSBs in chromatin structure mutants.
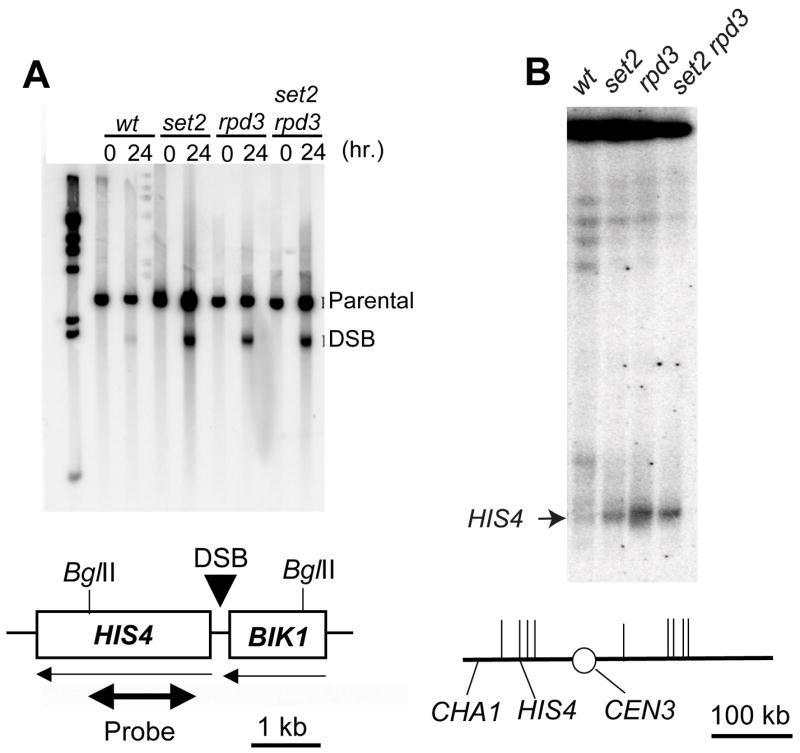
DNA was isolated from cells just prior to (0 hr) and after a 24 hr (Fig. 1A) or 48 h (Fig. 1B) incubation period on solid sporulation medium. Samples were examined by standard gel electrophoresis (Fig. 1A) or CHEF gel electrophoresis (Fig. 1B).
A. The DNA was digested with BglII, and examined by Southern analysis using a XhoI-BglII fragment of HIS4 as a hybridization probe (shown as a double-headed arrow in the lower part of the figure). The parental band (at roughly 3 kb) represents the unbroken BglII fragments, while the DSB band (at roughly 1.9 kb) represents fragments with a meiosis-specific DSB at HIS4. All strains are homozygous for the rad50S mutation. FX1 has no mutations in genes affecting chromatin structure. JDM1093, JDM1095, and JDM1096 are isogenic derivatives of FX1 homozygous for set2Δ, rpd3Δ, and set2Δ rpd3Δ mutations, respectively. In the schematic, horizontal arrows indicate the direction of transcription and the vertical arrow shows the DSB site associated with the HIS4 hotspot.
B. For CHEF gel electrophoresis, DNA was prepared and the resulting molecules separated by electrophoresis using the methods described by Lobachev et al. (48). Following electrophoresis, the chromosomal DNA molecules were transferred to a Nylon membrane and hybridized to CHA1 sequences, a gene located near the left end of chromosome III. The band of hybridization near the top of the gel represents chromosome III molecules without a DSB. The chromosomal DNA with a DSB at the HIS4 hotspot is shown with an arrow. Chromosome III is depicted in the lower part of the figure. The short vertical lines represent the positions of meiotic recombination hotspots determined by microarray analysis (15).
These results confirm the conclusion, based on tetrad analysis, that the set2 mutation elevates recombination rates at the HIS4 hotspot. This elevation does not reflect the presence of novel DSBs near HIS4, but the strengthening of the same DSB site observed in the wild-type strain (Fig. 1A). The set2 and rpd3 mutations had similar effects on the frequency of HIS4-associated DSBs, and the set2 rpd3 double mutant strain had an effect that was only slightly stronger than the single mutants. This result suggests that Set2p and Rpd3p may be acting in the same pathway to stimulate HIS4 hotspot activity. It should be noted that the increase in DSB formation observed in the mutant strains is substantially greater than the increase in gene conversion rates. In part, this difference is attributable to the observation that only half of the conversion events at HIS4 in cells sporulated at 25° are a consequence of the DSB site shown in Fig. 1A (16).
We also examined DSBs on chromosome III by Southern analysis of chromosomal DNA molecules separated by CHEF (contour-clamped homogeneous electric field) gel electrophoresis (Fig. 1B). As observed previously (55), DSBs on chromosome III occurred in two clusters, one on the left arm (which includes the HIS4 DSB) and one on the right arm. From the analysis shown in Fig. 1B, it is clear that mutations in set2 and rpd3 substantially elevated the DSB frequency of the HIS4 hotspot relative to other DSBs on chromosome III. In Fig. 1B, DSBs on the right arm of III appear to be repressed in strains with the set2 and rpd3 mutations. It is possible that strengthening of the HIS4 DSB reduces the frequency of DSBs on the same chromosome; we and others have previously observed local competition between DSB sites. Alternatively, it is possible that Rpd3p- and Set2p-mediated modifications repress meiotic recombination at some loci and stimulate recombination at others.
3.3. Histone modifications near the HIS4 hotspot in wild-type, set2, rpd3, and set2 rpd3 strains
Loss of the Rpd3p deacetylase results in increased acetylation of many sites in H3 and H4 histones (56,57), whereas loss of the Set2p methylase directly affects only methylation of H3K36 (34). The Set2p-mediated methylation, however, helps recruit a repressive Rpd3p-containing complex to chromatin, and elevated histone acetylation was observed for several loci in set2 mutant strains (35,37). Consequently, we examined the histone acetylation levels at H3K27 by chromatin immunoprecipitation (ChIP analysis) in the diploids FX1 (wild-type), JDM1093 (set2), JDM1095 (rpd3), and JDM1096 (set2 rpd3). For this analysis, chromatin was isolated from cells sporulated for six hours, about the time of DSB formation (17). We used four different PCR products (labeled A–D in Fig. 2A) to look at H3K27 acetylation levels at four sites in the HIS4 region. The A fragment (located near the 5′ end of BIK1) and the D fragment (located near the 3′ end of HIS4) are more than 1 kb from the site of the DSB. The B and C fragments are within about 200 bp of the DSB site. ChIP experiments were done with antibodies directed against acetylation of H3K27, and unmodified histone H3 (as a histone occupancy control). In addition, we used an antibody directed against tri-methylation of H3K36 in extracts derived from the wild-type, rpd3, and set2 strains.
Figure 2. Chromatin modifications near the HIS4 hotspot examined by chromatin immunoprecipitation.
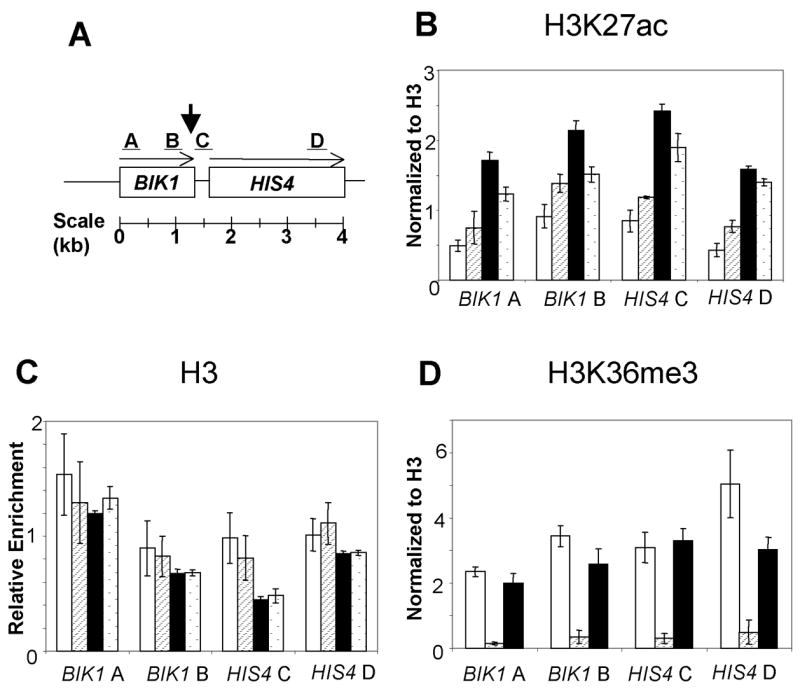
Cell extracts were prepared from the same diploid strains that were analyzed for DSBs: FX1 (wild-type, white bars), JDM1093 (set2, striped bars), JDM1095 (rpd3, black bars), and JDM1096 (set2 rpd3, stippled bars). The levels of histone modifications for four different fragments (A–D, Fig. 2A) were monitored by multiplex PCR. The arrow in Fig. 2A shows the approximate location of the recombination-associated DSB. Antibodies against H3K27 acetylation (panel B), histone H3 (panel C), or H3K36 tri-methylation (panel D) were used to precipitate chromatin harvested from meiotic cells incubated for six hours in sporulation medium. As described in the Materials and Methods, the levels of acetylation and methylation were normalized to a control region on chromosome V. In addition, the levels of acetylation and methylation were normalized for histone occupancy, using the data derived from the immunoprecipitations shown in Fig. 2C. Error bars represent the standard error of the mean.
The ChIP data were normalized in two ways: by comparison to a control DNA fragment (an intergenic region on chromosome V) and by comparison to the level of nucleosome occupancy (monitored using an antibody directed against histone H3); details of these normalizations are described in Materials and Methods. We found that the regions near the DSB sites were hyper-acetylated at the H3K27 sites in the set2, rpd3, and set2 rpd3 strains (Fig. 2B). The acetylation was elevated most strongly for the sites (B and C fragments) located near the site of the DSB. As expected, the region near the DSB site had lower nucleosome occupancy than the regions represented by the A and D fragments (Fig. 2C). Finally, we found that the tri-methylation of H3K36 was not substantially affected in rpd3 strains, but was, as expected, eliminated in the set2 strain (Fig. 2D). The interpretation of these results will be described below.
3.4. Meiotic expression of HIS4 and BIK1 is elevated in strains with set2 or rpd3 mutations
Although there is not a direct relationship between gene expression and the level of meiotic recombination (2), some mutations reduce both gene expression and meiotic recombination. For example, at HIS4, deletion of the Bas2p transcription factor binding site, which reduces the level of HIS4 expression, also substantially reduces the HIS4 recombination hotspot activity (19). Consequently, we used real-time PCR to measure the level of BIK1 and HIS4 mRNA in wild-type, set2, rpd3, and set2 rpd3 strains incubated for six hours in sporulation medium. As shown in Fig. 3, expression levels for both genes (BIK1 levels shown by white bars, HIS4 by stippled bars) were significantly elevated in the mutant strains relative to the wild-type strain. These results indicate that the chromatin alterations associated with the rpd3 and set2 mutations at the HIS4 locus elevate both meiotic recombination and meiotic gene expression.
Figure 3. Meiotic gene expression of HIS4 and BIK1 in wild-type, set2, rpd3, and set2 rpd3 strains.
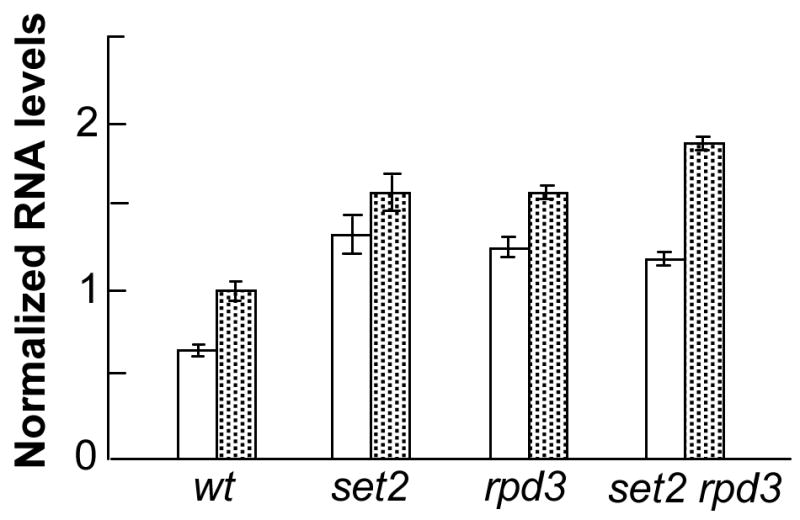
RNA was isolated from wild-type (FX1), set2 (JDM1093), rpd3 (JDM1095), and set2 rpd3 strains that had been incubated for six hours in sporulation medium. Following reverse transcription of the RNA, we measured the levels of the resulting BIK1 (shown by the white bars) and HIS4 (shown by the stippled bars) cDNAs by real-time PCR (details in Materials and Methods). Following normalization of the amounts to the level of RNA1 (a gene whose expression is unaffected by set2 or rpd3 mutations), the expression levels were normalized to the level of HIS4 expression in the wild-type strain. Bars represent 95% confidence limits.
4. Discussion
The main conclusions from our study are: 1) loss of the Set2 and Rpd3 proteins strongly elevates HIS4 hotspot activity, whereas loss of the Hda1p has a weaker stimulating effect, and 2) loss of the Dot1 and Sin4 protein, or a reduction in the amount of histones H3 and H4 do not affect HIS4 hotspot activity. These results will be discussed further below.
Since mutations in SET2 and RPD3 stimulate meiosis-specific DSBs at the HIS4 hotspot to approximately the same extent and since the double mutant strain has a level of DSBs that is not substantially greater than the single mutants, we favor the possibility that Set2p and Rpd3p act in the same pathway to repress hotspot activity. Similarly, the BIK1 expression level is elevated to about the same extent in the double mutant compared to both single mutants, although HIS4 expression is somewhat greater in the double mutant than in either single mutant (Fig. 3). Based on the recent evidence that Set2p-mediated methylation of H3K36 is involved in the recruitment of a repressive Rpd3 complex (35–37), the simplest explanation of these observations is that this complex negatively regulates both meiotic recombination and transcription. This negative regulation is likely to be a consequence of a closed chromatin structure caused by Rpd3p-mediated deacetylation. Our chromatin immunoprecipitation experiments, although limited in scope, are also consistent with this possibility. We found that acetylation of H3K27 was elevated in the set2, rpd3, and set2 rpd3 strains near the site of the HIS4 DSB in meiotic cells. These results suggest that the elevated recombination at HIS4 reflects a more open chromatin structure resulting from increased acetylation of histones.
There are a number of caveats concerning this explanation. In our study, as in most studies of the effects of chromatin-modifying enzymes, it is difficult to distinguish direct from indirect effects. For example, microarray analysis indicates rpd3Δ affects the expression of over 400 genes (29) and one of these gene products could indirectly stimulate meiotic recombination. Although we cannot exclude indirect effects of the set2 and rpd3 mutations, a number of arguments suggest that the effects at the HIS4 hotspot may be direct, in addition to the ChIP data discussed above. First, Robyr et al. (58) found that the level of histone H4K12 acetylation (a modification regulated by Rpd3p; 57) in the region adjacent to the HIS4 DSB site is increased approximately three-fold in an rpd3Δ strain relative to wild type. Second, Kurdistani et al. (59) found that Rpd3p binds in the HIS4 intergenic region. Third, in our study, the set2 and rpd3 mutations differentially affect the rate of meiotic recombination at HIS4 and ARG4. The frequency of DSB formation in the rpd3 and set2 mutants is clearly more elevated at HIS4 than at ARG4. In addition, the frequency of DSBs at HIS4 is elevated relative to many other DSB sites on chromosome III (Fig. 1B). Thus, our results are unlikely to be explained by the argument that the elevated level of HIS4 recombination reflects increased expression of Spo11p and other components of the meiotic recombination machinery in rpd3 and set2 strains.
Our observation that meiotic expression of HIS4 is somewhat elevated in rpd3 and set2 strains contrasts with the observation that the level of HIS4 gene expression is reduced by the rpd3 mutation (29), although no such reduction was observed in another study (30). These differences may reflect differences in the pattern of mitotic and meiotic expression and/or strain differences. We also found that the hda1 mutation significantly elevated the activity of the HIS4 hotspot (Table 4), although this effect was smaller than that observed for the set2 and rpd3 mutations. Since the Hda1p histone deacetylase has somewhat different activities than the Rpd3p deacetylase (56), this result may argue that the general level of histone acetylation is more important in regulating HIS4 hotspot activity than a specific type of modification. Although we cannot rule out an indirect effect of hda1 on recombination, HIS4 gene expression is elevated about three-fold by the hda1 mutation in one study (30), although no effect on expression was found in another study (29).
As described in the Introduction, many mutations that reduce HIS4 transcription (deletion of transcription factors or their binding sites upstream of HIS4) also reduce HIS4 meiotic recombination (11, 18–20,47). White et al. (60) showed, however, that deletion of the TATAA sequence upstream of HIS4 significantly decreased HIS4 expression, but had no effect on meiotic recombination. In this study, we found that the sin4 deletion, which reduces HIS4 expression about 15-fold (32), had no effect on HIS4 hotspot activity. Similarly, a reduction in the level of histones H3 and H4 that stimulates HIS4 gene expression had no effect on HIS4 hotspot activity. These results support our earlier conclusion that, although some features of chromatin structure favor both gene expression and recombination, other features may specifically stimulate one process or the other.
Whatever the precise mechanism responsible for the elevated recombination in the set2 and rpd3 strains, our results demonstrate that the activity of one of the strongest hotspots in the genome can be further elevated. In an analysis of the ade6-M26 hotspot in S. pombe, Yamada et al. (12) reported that loss of the SpGcn5 histone acetylase or deletion of the Snf22 histone-remodeling proteins reduced the activity of the hotspot. Since the loss of a histone acetylase would be expected to decrease acetylation at the ade6-M26 hotspot, the mechanism by which loss of SpGcn5 reduces hotspot activity may be related to the mechanism by which loss of Set2p, Rpd3p, and Hda1p stimulate hotspot activity.
Acknowledgments
We thank J. Lieb for discussions concerning chromatin structure. This research was supported by National Institutes of Health grants GM24110 (TDP) and GM68088 (BDS). B. D. Strahl is a Pew Scholar in the Biomedical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
None.
References
- 1.Lichten M, Goldman ASH. Meiotic recombination hotspots. Ann Rev Genet. 1995;29:423–444. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- 2.Petes TD. Meiotic recombination hot spots and cold spots. Nature. 2001;2:360–369. doi: 10.1038/35072078. [DOI] [PubMed] [Google Scholar]
- 3.Sun H, Treco D, Schultes NP, Szostak JW. Double strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 4.Szostak JW, Orr-Weaver TL, Rothstein R, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 5.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 6.Fan QQ, Petes TD. Relationship between nuclease-hypersensitive sites and meiotic recombination hot spot activity at the HIS4 locus of Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2037–2043. doi: 10.1128/mcb.16.5.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keeney S, Kleckner N. Communication between homologous chromosomes: genetic alterations at a nuclease-hypersensitivity site can alter meiotic chromatin structure at that site both in cis and in trans. Genes Cells. 1996;1:475–489. doi: 10.1046/j.1365-2443.1996.d01-257.x. [DOI] [PubMed] [Google Scholar]
- 8.Mizuno K, Emura Y, Baur M, Kohli J, Ohta K, et al. The meiotic recombination hotspot created by the single-base substitution ade6-M26 results in remodeling of chromatin structure in fission yeast. Genes Dev. 1997;11:876–886. doi: 10.1101/gad.11.7.876. [DOI] [PubMed] [Google Scholar]
- 9.Ohta K, Shibata T, Nicolas A. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J. 1994;13:5754–5763. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu TC, Lichten M. Meiosis-induced double-strand breaks determined by yeast chromatin structure. Science. 1994;263:515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- 11.Kirkpatrick DT, Wang YH, Dominska M, Griffith JD, Petes TD. Control of meiotic recombination and gene expression in yeast by a simple repetitive DNA sequence that excludes nucleosomes. Mol Cell Biol. 1999;19:7661–7671. doi: 10.1128/mcb.19.11.7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamada T, Mizuno K, Hirota K, Kon N, Wahls WP, et al. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 2004;23:1792–1803. doi: 10.1038/sj.emboj.7600138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sollier J, Lin W, Soustelle C, Suhre K, Nicolas A, et al. Set1 is required for meiotic S-phase onset, double-strand break formation and middle gene expression. EMBO J. 2004;23:1957–1967. doi: 10.1038/sj.emboj.7600204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamashita K, Shinohara M, Shinohara A. Rad6-Bre1-mediated histone H2B ubiquitylation modulates the formation of double-strand breaks during meiosis. Proc Natl Acad Sci USA. 2004;101:11380–11385. doi: 10.1073/pnas.0400078101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerton J, Derisi J, Shroff R, Lichten M, Brown PO, et al. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan QQ, Xu F, Petes TD. Meiosis-specific double-strand DNA breaks at the HIS4 recombination hot spot in the yeast Saccharomyces cerevisiae: control in cis and trans. Mol Cell Biol. 1995;15:1679–1688. doi: 10.1128/mcb.15.3.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nag DK, Petes TD. Physical detection of heteroduplexes during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2324–2331. doi: 10.1128/mcb.13.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdullah MFF, Borts RH. Meiotic recombination frequencies are affected by nutritional states in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2001;98:14524–14529. doi: 10.1073/pnas.201529598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White MA, Dominska M, Petes TD. Transcription factors are required for the meiotic recombination hotspot at the HIS4 locus in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:6621–6625. doi: 10.1073/pnas.90.14.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White MA, Wierdl M, Detloff P, Petes TD. DNA-binding protein RAP1 stimulates meiotic recombination at the HIS4 locus in yeast. Proc Natl Acad Sci USA. 1991;88:9755–9759. doi: 10.1073/pnas.88.21.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devlin C, Tice-Baldwin K, Shore D, Arndt K. RAP1 is required for BAS1/BAS2- and GCN4-dependent transcription for the yeast HIS4 gene. Mol Cell Biol. 1991;11:3642–3651. doi: 10.1128/mcb.11.7.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu L, Morse RH. Chromatin opening and transactivator potentiation by RAP1 in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:5279–5288. doi: 10.1128/mcb.19.8.5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional activation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 24.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 25.Hansen JC, Tse C, Wolffe AP. Structure and function of the core histone N-termini: more than meets the eye. Biochemistry. 1998;37:17637–17641. doi: 10.1021/bi982409v. [DOI] [PubMed] [Google Scholar]
- 26.Kadosh D, Struhl K. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, et al. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:1–10. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernstein BE, Tong JK, Schreiber SL. Genomewide studies of histone deacetylase function in yeast. Proc Natl Acad Sci USA. 2000;97:13708–13713. doi: 10.1073/pnas.250477697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 31.Wu J, Carmen AA, Kobayashi R, Suka N, Grunstein M. HDA2 and HDA3 are related proteins that interact with and are essential for the activity of the yeast histone deacetylase HDA1. Proc Natl Acad Sci USA. 2001;98:4391–4396. doi: 10.1073/pnas.081560698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang YW, Stillman DJ. Regulation of HIS4 expression by the Saccharomyces cerevisiae SIN4 transcriptional regulator. Genetics. 1995;140:103–114. doi: 10.1093/genetics/140.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyrick JJ, Holstege FCP, Jennings EG, Causton HC, Shore D, et al. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- 34.Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carrozza MJ, Li B, Florens L, Suganua T, Swanson SK, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 36.Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 37.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, et al. Cotranscriptional Set2 methylation of histone H3 Lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 38.Feng Q, Wang H, Ng HH, Erdjument H, Tempst P, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 39.Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, et al. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng HH, Ciccone DN, Morshead KB, Oettinger MA, Struhl K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc Natl Acad Sci USA. 2003;100:1820–1825. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 42.Stapleton A, Petes TD. The Tn3 beta-lactamase gene acts as a hotspot for meiotic recombination in yeast. Genetics. 1991;127:39–51. doi: 10.1093/genetics/127.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wach AA, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 44.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 45.Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- 46.Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1983. [Google Scholar]
- 47.Kirkpatrick DT, Fan QQ, Petes TD. Maximal stimulation of meiotic recombination by a yeast transcription factor requires the transcription activation domain and a DNA binding domain. Genetics. 1999;152:101–115. doi: 10.1093/genetics/152.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 49.Xiao T, Shibata Y, Rao B, Laribee RN, O’Rourke R, et al. The RNA polymerase II kinase Ctk1 regulates positioning of a 5′ histone methylation boundary along genes. Mol Cell Biol. 2007;27:721–731. doi: 10.1128/MCB.01628-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, et al. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 51.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petes TD, Malone RE, Symington LS. Recombination in Yeast. In: Broach J, Jones E, Pringle J, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Energetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1991. pp. 407–521. [Google Scholar]
- 53.Dora EG, Rudin N, Martell JR, Esposito MS, Ramirez RM. RPD3 (REC3) mutations affect mitotic recombination in Saccharomyces cerevisiae. Curr Genet. 1999;35:68–76. doi: 10.1007/s002940050434. [DOI] [PubMed] [Google Scholar]
- 54.Burgess SM, Ajimura M, Kleckner N. GCN5-dependent histone H3 acetylation and RPD3-dependent histone H4 deacetylation have distinct, opposing effects on IME2 transcription, during meiosis and during vegetative growth in budding yeast. Proc Natl Acad Sci USA. 1999;96:6835–6840. doi: 10.1073/pnas.96.12.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zenvirth D, Arbel T, Sherman A, Goldway M, Klein S, et al. Multiple sites for double-strand breaks in whole meiotic chromosomes of Saccharomyces cerevisiae. EMBO J. 1992;11:3441–3447. doi: 10.1002/j.1460-2075.1992.tb05423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, et al. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. New antibodies for sites of acetylation determine novel histone site usage in heterochromatin and euchromatin. Mol Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- 58.Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, et al. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell. 2002;109:437–446. doi: 10.1016/s0092-8674(02)00746-8. [DOI] [PubMed] [Google Scholar]
- 59.Kurdistani SK, Robyr D, Tavazoie S, Grunstein M. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat Genet. 2002;31:248–254. doi: 10.1038/ng907. [DOI] [PubMed] [Google Scholar]
- 60.White MA, Detloff P, Strand M, Petes TD. A promoter deletion reduces the rate of mitotic, but not meiotic, recombination at the HIS4 locus in yeast. Curr Genet. 1992;21:109–116. doi: 10.1007/BF00318468. [DOI] [PubMed] [Google Scholar]
- 61.Nag DK, White MA, Petes TD. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature. 1989;340:318–320. doi: 10.1038/340318a0. [DOI] [PubMed] [Google Scholar]
- 62.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 63.Perkins DD. Biochemical mutants in the smut fungus Ustilago maydis. Genetics. 1949;34:607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]


